Abstract
Herpesviruses have an icosahedral nucleocapsid surrounded by an amorphous tegument and a lipoprotein envelope. The tegument comprises at least 20 proteins destined for delivery into the host cell. As the tegument does not have a regular structure, the question arises of how its proteins are recruited. The herpes simplex virus 1 (HSV-1) tegument is known to contact the capsid at its vertices, and two proteins, UL36 and UL37, have been identified as candidates for this interaction. We show that the interaction is mediated exclusively by UL36. HSV-1 nucleocapsids extracted from virions shed their UL37 upon incubation at 37°C. Cryo-electron microscopy (cryo-EM) analysis of capsids with and without UL37 reveals the same penton-capping density in both cases. As no other tegument proteins are retained in significant amounts, it follows that this density feature (∼100 kDa) represents the ordered portion of UL36 (336 kDa). It binds between neighboring UL19 protrusions and to an adjacent UL17 molecule. These observations support the hypothesis that UL36 plays a major role in the tegumentation of the virion, providing a flexible scaffold to which other tegument proteins, including UL37, bind. They also indicate how sequential conformational changes in the maturing nucleocapsid control the ordered binding, first of UL25/UL17 and then of UL36.
INTRODUCTION
Adistinctive feature of herpesviruses is their tegument, a proteinaceous layer enclosed between the nucleocapsid and viral envelope (reviewed in references 1, 3, 14, and 22). Teguments vary somewhat in composition among members of the herpesvirus family but in the well-studied herpes simplex virus 1 (HSV-1) system, it contains about 20 different viral proteins, accounting for some 40% of total virion protein (35). Upon cell entry, most of the tegument is released into the cytoplasm of the host cell (21, 43) where at least some of these proteins perform functions needed early in the infection process. Released molecules are designated “outer tegument” proteins. A few components—primarily, the UL36 (also called VP1/2), UL37, and US3 proteins—remain associated with the nucleocapsid and participate in its transport to the nucleus by the microtubule/dynein motor system (28, 32, 34). These three molecules are referred to as “inner tegument proteins.”
Unlike the icosahedrally symmetric capsid, the tegument is not a highly ordered structure. As visualized by cryo-electron tomography (cryo-ET), it is ∼35 nm thick on one side of extracellular HSV-1 virions but no more than 5 nm thick on the opposite side (13). Cell-associated virions have a more symmetrical distribution of tegument material (24), but no regular substructure has been reported. Several observations have implicated the vertices as the capsid attachment points for the HSV-1 tegument (the triplexes are directly involved in the case of cytomegalovirus [40]). An icosahedrally symmetrized cryo-electron microscopy (cryo-EM) reconstruction of intraviral capsids detected extended density features associated with the vertices (44). Averaging intraviral capsids depicted in cryo-electron tomograms revealed that compared with reconstructions of isolated capsids (13), they have additional density features capping their pentons. In both studies, UL36 was suggested to contribute to the density features that had been visualized, but no data were presented to identify the protein(s) involved. Subsequently, a study combining cryo-EM with biochemical analysis revealed a vertex-adjacent density feature, ∼11 nm long by ∼4 nm in diameter, on C-capsids (DNA-filled nuclear capsids) and identified it as a heterodimer of UL25 (63 kDa) and UL17 (75 kDa) (41). This feature was named the CCSC (C-capsid-specific component). Identification of its components has subsequently been confirmed by cryo-EM mapping studies with a UL25 construct mutated to have an additional domain (8, 10) and a UL17 construct with green fluorescent protein fused to its C terminus (37). The CCSC appears to represent the vertex-distal portion of the density features seen in the reconstruction from whole virions (44).
Previously, we developed a procedure based on extracting purified virions with the Triton X-100 detergent, which yields DNA-filled capsids that retain only two tegument proteins in significant amounts—UL36 and UL37 (23). These particles were called T36 capsids. In the present study, we analyzed their structure by cryo-EM and three-dimensional image reconstruction. In addition, we identified conditions for the removal of UL37, and these capsids were also reconstructed. Comparison of the two reconstructions allows us to assign density features visualized on the outer surfaces of pentons to UL36. The observed interactions indicate how the buildup of proteins on and around the nonportal vertices is orchestrated by sequential conformational changes in the maturing capsid.
MATERIALS AND METHODS
Sample preparation.
T36 capsids were prepared as described previously (23). Typically, 10 μl of purified virus at a 1-mg/ml protein concentration was diluted into 80 μl of TNE buffer (10 mM Tris-HCl, 0.5 M NaCl, 1 mM EDTA [pH 7.4]) and 10 μl of 10% Triton X-100 was added, along with dithiothreitol (DTT), giving final concentrations of 1% Triton X-100 and 10 mM DTT. Two tubes were prepared: one was incubated on ice, and the other was incubated at 37°C for 1 h. The contents of each tube were loaded onto a gradient of 20 to 50% sucrose and TNE buffer made in a 650-μl ultraclear tube. These tubes were spun for 40 min at 23,000 rpm in an SW50.1 rotor (Beckmann) equipped with nylon tube adapters and then fractionated by bottom puncture and drop collection. The fractions containing the major light-scattering band (containing the capsids) from each tube were diluted 1:3 with TNE buffer, and the capsids were pelleted at 23,000 rpm in a TLA100 rotor. For SDS-polyacrylamide gel electrophoresis (PAGE), pellets were resuspended in 25 μl TNE buffer, boiled with dissociation buffer, run on an 8.5% polyacrylamide gel, and stained with Coomassie blue as described previously (19).
Cryo-electron microscopy.
Cryo-EM was performed as described previously (6, 7). Micrographs were acquired on a CM200-FEG microscope (FEI, Beaverton, OR) operated at 120 kV. Images were recorded under low-dose conditions on film. The nominal defocus range was between 0.5 μm and 1.5 μm.
Image analysis.
Micrographs of T36 capsids were digitized on an SCAI scanner (Z/I Imaging, Huntsville, AL), giving a pixel size of 1.4 Å, and subsequently binned to 4.2 Å/pixel. Micrographs of other specimens were digitized with a Nikon Super CoolScan 9000 ED scanner, giving a pixel size of 1.67 Å at the specimen, and binned to a final size of 3.34 Å. Particles were picked semiautomatically with the e2boxer program in EMAN2 (36). The remaining preprocessing steps, consisting of particle extraction, normalization, and contrast transfer function (CTF) estimation and correction (by phase flipping), were performed using Bsoft (15). Orientation parameters were determined by iterative projection matching as implemented in PFT2 (4). For an initial model, we used a preexisting reconstruction of nuclear C-capsids (41) after scaling it to match the current data. Density maps were calculated with EM3DR2 (http://people.chem.byu.edu/belnap/). Iterations continued until the resolution could not be improved further. Resolution (Table 1) was assessed in terms of Fourier shell correlation (29). Before calculating the resolution, the two half-data-set maps were filtered with a soft-edge mask to keep only the density features in the radial zone between 418 Å and 728 Å.
Table 1.
Reconstruction parameters and occupancy levels of UL36 and the CCSC
| Type of capsid | No. of particles selected/total no. of particles | Resolution (Å) (0.3/0.5)a | CCSC occupancy (%) | UL36 occupancy (%) |
|---|---|---|---|---|
| T36 capsids isolated at 37°C | 1,236/2,551 | 19.0/21.3 | ∼100 | ∼40 |
| T36 capsids | 952/1,696 | 21.3/22.2 | ∼100 | ∼50 |
| T36 capsids with low occupancy of UL36 | 320/1,696 | 22.7/24.5 | ∼80 | ∼25 |
| T36 capsids with high occupancy of UL36 | 319/1,696 | 22.5/24.2 | ∼100 | ∼75 |
Resolution is specified by the Fourier shell correlation coefficient at thresholds of 0.3 and 0.5, respectively.
Occupancy and classification analyses.
The levels of occupancy of the CCSC and the penton-associated density feature (UL36 [see below]) in each reconstruction were estimated by comparing their densities with the densities of constitutive capsid components after subtracting the background density. Variations in occupancy were assessed by classifying one-particle reconstructions (30). In brief, a density map was obtained for each particle included in the final reconstruction. These density maps were generated with a Fourier space reconstruction method based on the direct inversion algorithm, as implemented in Bsoft, and then binned by a factor of 2 and filtered to 40 Å−1. Binary masks, obtained by segmenting the regions of interest in the final reconstructions (from the full data set), were used to integrate the corresponding density in each one-particle reconstruction in a modified version of the method of Scheres et al. (30). These averages were compensated for bias from particle orientation by dividing them by the average density expected for the assigned orientation. Finally, the particles were sorted according to these compensated values, and the top third and bottom third were used to generate the high-occupancy and low-occupancy reconstructions, respectively.
Visualization and segmentation.
Surface rendering was performed using Chimera (12). All reconstructions were visualized at the isodensity contour corresponding to a half standard deviation. We devised a semiautomatic procedure, mostly within Chimera, to segment out pentons, hexons, triplexes, the CCSC, and UL36, allowing us to apply a molecule-specific color coding. An initial segmentation was obtained with Segger (27), and the interfaces were checked visually and refined manually for one asymmetric unit. The regions occupied by the CCSC and UL36 were assigned manually, based on comparison between T36 capsids with high occupancy of UL36 and those with low occupancy (see above). This result was propagated over the rest of the capsid by imposing icosahedral symmetry with the program bsym from Bsoft. The segmented maps were transformed into sets of markers (meshmol) and used to color surface contours (Color Zone). The segmentation was obtained, starting from the reconstruction of the T36 capsid treated at 37°C, and was used to color segment all of the reconstructions.
RESULTS
Protein composition of DNA-filled capsids obtained by detergent extraction of virions (virion-derived C-capsids).
When purified extracellular HSV-1 virions are treated with the Triton X-100 detergent by the protocol of Newcomb and Brown (23), the envelope and almost all of the tegument are stripped off. The residual particles, called T36 capsids, bear a close resemblance to C-capsids isolated from the nuclei of infected cells (Fig. 1A; compare to Fig. 5 of reference 40). When their protein composition is assessed by SDS-PAGE (Fig. 2), T36 capsids are seen to contain, in addition to the major capsid protein UL19 and the two triplex proteins, UL38 and UL18, two additional proteins, UL36 (336 kDa) and UL37 (121 kDa). The latter proteins are present in at most vestigial amounts on C-capsids (40). For T36 capsids, the intensity of the UL36 band relative to UL19 is substantially reduced compared with its intensity in virions (Fig. 2, compare the leftmost lane with the rightmost lane). Upon quantitating these bands, we found that 70% of UL36 was extracted by the detergent treatment. Taking its copy number in virions to be 134 (23), it follows that an average of ∼40 copies of UL36 per capsid are retained after the detergent extraction.
Fig 1.
Cryo-electron micrographs of HSV-1 T36 capsids isolated at 4°C (A) and at 37°C (B). Bar = 100 nm.
Fig 5.
T36 capsids were analyzed by content of UL36. (A and C) T36 capsid subset with the highest level of occupancy; (B and D) T36 capsid subset with the lowest level of occupancy. The UL36-related density is indicated by black arrows in panel A and gray arrows in panel B. The surface features in panels C and D are color coded as in Fig. 3. Bar = 20 nm.
Fig 2.

SDS-polyacrylamide gel electrophoresis comparing the protein compositions of purified HSV-1 virus (left lane) with T36 capsids prepared at 4°C (right) and at 37°C (middle). The molecular masses of the proteins in the bands labeled to the right of the gel are as follows: 336 kDa for UL36, 149 kDa for UL19, 121 kDa for UL37, 75 kDa for UL17, 63 kDa for UL25, and 50 kDa for UL38. UL36 is the virion protein with the highest mass. The UL37 band was identified by Western blotting and mass spectrometry (W. W. Newcomb, L. M. Jones, A. Dee, and J. C. Brown, unpublished results).
We subjected T36 capsids to a number of treatments, seeking to identify conditions that would selectively detach UL36 or UL37. Thus, we found that the relatively mild procedure of incubating for 30 min at 37°C removed UL37 quantitatively, while leaving the content of UL36 unaffected (compare the middle lane and the rightmost lane in Fig. 2).
Cryo-EM reconstructions of T36 capsids with and without UL37.
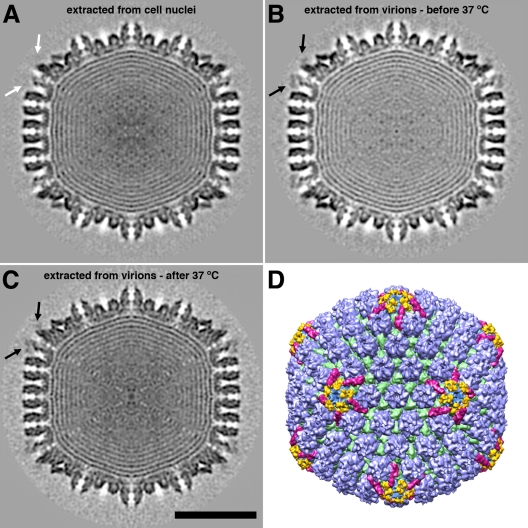
In unprocessed cryo-electron micrographs, the two capsids are indistinguishable (compare Fig. 1A and B). To examine them more closely, we reconstructed density maps to resolutions of about 20 Å (Table 1). In Fig. 3, central sections of the two capsids are compared with each other (Fig. 3B and C) and with the central section of C-capsids in Fig. 3A. The two T36 capsids have an additional density feature decorating the outer surfaces of pentons (black arrows in Fig. 3B and C) which has no counterpart on C-capsids (white arrows in Fig. 3A). Its shape and position on the capsid surface are shown by surface rendering in Fig. 3D and at higher magnification in Fig. 4 (density features shown in yellow). This density feature is indistinguishable in the two reconstructions: it is approximately bipartite and overlies the gap between the protrusion domains of two neighboring UL19 subunits in the penton.
Fig 3.
Three-dimensional reconstructions of DNA-containing HSV-1 capsids. (A to C) Central sections through capsids viewed along a 2-fold axis of icosahedral symmetry. (A) Nuclear C-capsid; (B) T36 capsid isolated at 4°C; (C) T36 capsid isolated at 37°C. The black arrows in panels B and C point to additional density features overlying pentons. There is no such density feature in panel A (white arrow). Bar = 50 nm. (D) Color-coded surface rendering of the reconstruction shown in panel C. The surface features are shown in color as follows: capsomer protrusions are blue, triplexes are green, CCSCs are magenta, and the additional penton-associated density features (part of UL36) found on the T36 capsid are yellow.
Fig 4.

Higher-magnification view of the region surrounding a vertex on the reconstruction of the T36 capsid isolated at 4°C, both before (A) and after (B) incubation at 37°C. The surface features were color coded as in Fig. 3. Bar = 10 nm.
The penton-associated density is substoichiometric, i.e., although well above background, it is lower than the peak densities in the triplexes and the UL19 protrusions (Fig. 3B and C). As the latter features can be assumed to have 100% occupancy, their density in the reconstruction can be used to calibrate the occupancy of the penton-associated densities. In this way, we find it to be 40 to 50%, corresponding to an average of 24 to 30 copies per capsid. Estimated in the same way, the occupancy of the CCSC was found to be close to 100% (Table 1), compared with an average of ∼50% in nuclear C-capsids (41).
All T36 capsids do not necessarily have the same content of the penton-associated protein. To assess its variability, we calculated an individual reconstruction for each particle in the data set, taking advantage of the property that icosahedral symmetry generates enough views (n = 60) to allow a reconstruction to be calculated (albeit to limited resolution) from a single particle. We could then classify the capsids according to their content of penton-associated density by measuring the density in the corresponding region of each one-particle reconstruction. The T36 capsids were ranked in this way, and reconstructions were calculated from the 33% of particles with the highest content and from the 33% with the lowest content (Fig. 5A and B). The former has an average occupancy of 75%, and the latter has an average occupancy of less than 25% (Table 1). The high-occupancy reconstruction, although diminished in resolution by the reduced number of particles used, gives a robust rendering of the penton-associated density, including its bipartite nature (Fig. 5C). As the CCSC content of T36 capsids does not vary much from particle to particle (∼100% occupancy [see above]), we could identify the boundary between the CCSC and the penton-capping density from a visual comparison of these two reconstructions.
Correlating the SDS-PAGE results with the cryo-EM results, we conclude that the penton-associated density is contributed by UL36 and that no density feature can be assigned to UL37. The estimated volume of the penton-associated density would accommodate a protein mass of about 100 kDa. It follows that the major part of UL36 is not seen, presumably as a result of disorder. Similarly, the invisibility of UL37 also reflects disorder, perhaps arising from UL37 binding to the disordered part of UL36.
DISCUSSION
Is UL36 monomeric or dimeric?
According to quantitative SDS-PAGE, T36 capsids retain about a third of the UL36 present in virions, i.e., about 40 subunits per capsid, on average. Our cryo-EM reconstructions put the occupancy of the penton capping sites at about 40%. This would correspond to 24 to 30 subunits if the density visualized represents (part of) a monomer and 48 to 60 subunits if it represents (part of) a dimer. The two scenarios are equally consistent with the SDS-PAGE data.
Does UL36 also bind around the portal vertex, which is occupied by a dodecamer of UL6 instead of a pentamer of UL19 (25, 39)? In view of the dependence of UL36 binding on its interaction with UL19 (Fig. 4 and 5), this appears not to be the case, so the maximum number of vertex-associated UL36 molecules (monomers or dimers [see above]) should be 55, not 60. However, this consideration does not affect the copy numbers given in the preceding paragraph, as all 12 vertices were treated equally in calculating the icosahedrally symmetrized reconstructions.
Thus, the full virion complement of UL36, estimated at an average of 134 subunits from quantitative SDS-PAGE, can be explained in terms of about 55 dimers, allowing a reasonable margin of error (23). However, it can also be interpreted as 55 vertex-associated monomers plus about 80 more monomers bound to some other tegument component. Further, more-precise data are needed to settle the question of UL36 stoichiometry.
Ordered and disordered parts of UL36.
The volume of the density feature associated with UL36 corresponds to a protein mass of ∼100 kDa when the partial specific volume of protein is taken into account. This is only a minor fraction—28% in the monomer scenario and 14% in the dimer scenario—of its total mass. The remainder is unseen, presumably averaging out due to disorder. Indeed, thin flexible filaments have been observed emanating from the surfaces of negatively stained T36 capsids and tentatively assigned to UL36 (23). Functionally, this is an attractive hypothesis because an extended structure would potentially present many binding sites for other tegument components. It is also consistent with the fact that proteins as large as UL36 (3,164 amino acids) tend to consist of many concatenated domains, with potentially different functions (18, 19). However, the number of filaments seen per virion is much smaller than the number of UL36 molecules, and it remains possible that T36 capsids could retain a few copies of some other filamentous component, for example, actin (13, 17). Further characterization of these filaments and of isolated UL36 molecules is needed to clarify their status.
Interaction of UL36 with the CCSC.
Ultimately, a complex network of interactions builds up around the vertices of the late-stage HSV-1 nucleocapsid. The portion of UL36 that we have visualized binds to a bipartitite site between two penton protrusions and also to the vertex-proximal portion of the CCSC (Fig. 4). This is the portion occupied by UL17 (37). UL25 forms the vertex-distal portion of the CCSC, and our structural data show no sign of an interaction between this protein (UL25) and UL36 unless it were to involve disordered portions of one or both proteins that are not seen in the reconstructions.
In this context, evidence has been presented of an interaction between UL25 and the C-terminal 62 residues of UL36 (9), although a subsequent study has reported that a mutant lacking the C-terminal 167 residues of UL36 still bound to cytosolic, i.e., nucleus-exited, capsids (31). However, it remains possible that the fraction of virion UL36 that is extracted when T36 capsids are prepared is connected differently to the capsid surface in a way that might involve an interaction between its C-terminal region and UL25.
Regulation of capsid decoration by conformational changes of UL19.
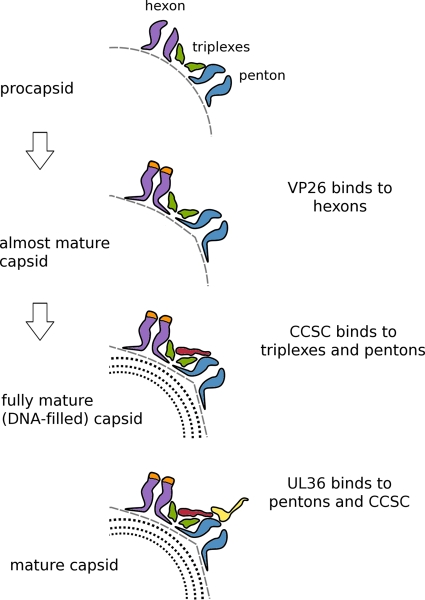
In the mature HSV-1 capsid, six monomers of the small accessory protein VP26 (12 kDa) bind around the outer rim of each hexamer of UL19, the major capsid protein (2, 42, 45). VP26 does not bind to UL19 in its procapsid conformation; it binds only after the hexon protrusions have reorganized into regular hollow hexameric cylinders (26). The same protein, UL19, also forms pentons at the capsid vertices, but VP26 does not bind to penton protrusions in either state (procapsid or mature capsid). Unlike the hexon protrusions, penton protrusions change little as the capsid matures, with adjacent subunits splayed apart compared to their mature hexon conformation (38). This conformational distinction leaves free access for UL36 to bind to its sites around the penton rim at a later stage of maturation. As the penton protrusions change little during maturation (16), it is noteworthy that UL36 does not bind to the procapsid (33). The reason appears to be that UL36 binding also needs the CCSC, and the CCSC does not bind to its vertex-adjacent sites in quantity until the capsid has undergone the additional conformational change induced by DNA packaging (5). Thus, the ordered addition of first CCSC and then UL36 is regulated by sequential conformational changes in the maturing capsid (Fig. 6).
Fig 6.
Cartoon summarizing regulation of the sequential addition of proteins to the HSV-1 capsid. In the first step, as the procapsid undergoes initial maturation, its shape alters from round to polyhedral, and the hexon protrusions close up and become 6-fold symmetric, creating binding sites for VP26. Late in DNA packaging, another less extreme transition takes place that affects the peripentonal triplexes and/or the penton so as to enhance the affinity of this composite binding site for the CCSC, whose binding promotes nuclear exit. Subsequently, (11, 20), UL36 binds to another composite site involving both the CCSC and UL19 penton tips. In this way, UL25/UL17 plays a functional role in the cytoplasm similar to that which it plays in the nucleus, allowing only DNA-filled capsids to progress efficiently along the exit pathway.
ACKNOWLEDGMENTS
This study was supported by the Intramural Research Program of NIAMS and by NIH grant R01AI041644 to J.C.B.
Footnotes
Published ahead of print 15 February 2012
REFERENCES
- 1. Baines JD. 2007. Envelopment of herpes simplex virus nucleocapsids at the inner membrane, p 144–150 In Arvin A, et al. (ed), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 2. Booy FP, et al. 1994. Finding a needle in a haystack: detection of a small protein (the 12-kDa VP26) in a large complex (the 200-MDa capsid of herpes simplex virus). Proc. Natl. Acad. Sci. U. S. A. 91:5652–5656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Britt B. 2007. CMV maturation and egress, p 311–323 In Arvin A, et al. (ed), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom: [PubMed] [Google Scholar]
- 4. Bubeck D, et al. 2005. Structure of the poliovirus 135S cell entry intermediate at 10-angstrom resolution reveals the location of an externalized polypeptide that binds to membranes. J. Virol. 79:7745–7755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cardone G, Heymann JB, Cheng N, Trus BL, Steven AC. 2012. Procapsid assembly, maturation, nuclear exit: dynamic steps in the production of infectious herpesvirions, p 423–440 In Rossmann MG, Rao VB. (ed), Viral molecular machines. Springer, New York, NY: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cheng N, et al. 1999. Tetrairidium, a four-atom cluster, is readily visible as a density label in three-dimensional cryo-EM maps of proteins at 10-25 Å resolution. J. Struct. Biol. 127:169–176 [DOI] [PubMed] [Google Scholar]
- 7. Cheng N, et al. 2002. Handedness of the herpes simplex virus capsid and procapsid. J. Virol. 76:7855–7859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cockrell SK, Huffman JB, Toropova K, Conway JF, Homa FL. 2011. Residues of the UL25 protein of herpes simplex virus that are required for its stable interaction with capsids. J. Virol. 85:4875–4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Coller KE, Lee JI, Ueda A, Smith GA. 2007. The capsid and tegument of the alphaherpesviruses are linked by an interaction between the UL25 and VP1/2 proteins. J. Virol. 81:11790–11797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Conway JF, et al. 2010. Labeling and localization of the herpes simplex virus capsid protein UL25 and its interaction with the two triplexes closest to the penton. J. Mol. Biol. 397:575–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Desai PJ. 2000. A null mutation in the UL36 gene of herpes simplex virus type 1 results in accumulation of unenveloped DNA-filled capsids in the cytoplasm of infected cells. J. Virol. 74:11608–11618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Goddard TD, Huang CC, Ferrin TE. 2005. Software extensions to UCSF chimera for interactive visualization of large molecular assemblies. Structure 13:473–482 [DOI] [PubMed] [Google Scholar]
- 13. Grünewald K, et al. 2003. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science 302:1396–1398 [DOI] [PubMed] [Google Scholar]
- 14. Guo H, Shen S, Wang L, Deng H. 2010. Role of tegument proteins in herpesvirus assembly and egress. Protein Cell 1:987–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Heymann JB, Belnap DM. 2007. Bsoft: image processing and molecular modeling for electron microscopy. J. Struct. Biol. 157:3–18 [DOI] [PubMed] [Google Scholar]
- 16. Heymann JB, et al. 2003. Dynamics of herpes simplex virus capsid maturation visualized by time-lapse cryo-electron microscopy. Nat. Struct. Biol. 10:334–341 [DOI] [PubMed] [Google Scholar]
- 17. Johannsen E, et al. 2004. Proteins of purified Epstein-Barr virus. Proc. Natl. Acad. Sci. U. S. A. 101:16286–16291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kattenhorn LM, Korbel GA, Kessler BM, Spooner E, Ploegh HL. 2005. A deubiquitinating enzyme encoded by HSV-1 belongs to a family of cysteine proteases that is conserved across the family Herpesviridae. Mol. Cell 19:547–557 [DOI] [PubMed] [Google Scholar]
- 19. Kelly BJ, Fraefel C, Cunningham AL, Diefenbach RJ. 2009. Functional roles of the tegument proteins of herpes simplex virus type 1. Virus Res. 145:173–186 [DOI] [PubMed] [Google Scholar]
- 20. Klupp BG, Fuchs W, Granzow H, Nixdorf R, Mettenleiter TC. 2002. Pseudorabies virus UL36 tegument protein physically interacts with the UL37 protein. J. Virol. 76:3065–3071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Maurer UE, Sodeik B, Grünewald K. 2008. Native 3D intermediates of membrane fusion in herpes simplex virus 1 entry. Proc. Natl. Acad. Sci. U. S. A. 105:10559–10564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mettenleiter TC, Klupp BG, Granzow H. 2009. Herpesvirus assembly: an update. Virus Res. 143:222–234 [DOI] [PubMed] [Google Scholar]
- 23. Newcomb WW, Brown JC. 2010. Structure and capsid association of the herpesvirus large tegument protein UL36. J. Virol. 84:9408–9414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Newcomb WW, Brown JC. 2009. Time-dependent transformation of the herpesvirus tegument. J. Virol. 83:8082–8089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Newcomb WW, Thomsen DR, Homa FL, Brown JC. 2003. Assembly of the herpes simplex virus capsid: identification of soluble scaffold-portal complexes and their role in formation of portal-containing capsids. J. Virol. 77:9862–9871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Newcomb WW, et al. 2000. Isolation of herpes simplex virus procapsids from cells infected with a protease-deficient mutant virus. J. Virol. 74:1663–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pintilie GD, Zhang J, Goddard TD, Chiu W, Gossard DC. 2010. Quantitative analysis of cryo-EM density map segmentation by watershed and scale-space filtering, and fitting of structures by alignment to regions. J. Struct. Biol. 170:427–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Roberts AP, et al. 2009. Differing roles of inner tegument proteins pUL36 and pUL37 during entry of herpes simplex virus type 1. J. Virol. 83:105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Saxton WO, Baumeister W. 1982. The correlation averaging of a regularly arranged bacterial cell envelope protein. J. Microsc. 127(Part 2):127–138 [DOI] [PubMed] [Google Scholar]
- 30. Scheres SH, et al. 2005. Classification of single-projection reconstructions for cryo-electron microscopy data of icosahedral viruses. J. Struct. Biol. 151:79–91 [DOI] [PubMed] [Google Scholar]
- 31. Schipke J, et al. 2012. The C terminus of the large tegument protein pUL36 contains multiple capsid binding sites that function differently during assembly and cell entry of herpes simplex virus. J. Virol. 86:3682–3700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shanda SK, Wilson DW. 2008. UL36p is required for efficient transport of membrane-associated herpes simplex virus type 1 along microtubules. J. Virol. 82:7388–7394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sheaffer AK, et al. 2000. Evidence for controlled incorporation of herpes simplex virus type 1 UL26 protease into capsids. J. Virol. 74:6838–6848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sodeik B, Ebersold MW, Helenius A. 1997. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J. Cell Biol. 136:1007–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Steven AC, Spear PG. 1997. Herpesvirus capsid assembly and envelopment, p 312–351 In Chiu W, Burnett RM, Garcea RL. (ed), Structural biology of viruses. Oxford University Press, New York, NY [Google Scholar]
- 36. Tang G, et al. 2007. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157:38–46 [DOI] [PubMed] [Google Scholar]
- 37. Toropova K, Huffman JB, Homa FL, Conway JF. 2011. The herpes simplex virus 1 UL17 protein is the second constituent of the capsid vertex-specific component required for DNA packaging and retention. J. Virol. 85:7513–7522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Trus BL, et al. 1996. The herpes simplex virus procapsid: structure, conformational changes upon maturation, and roles of the triplex proteins VP19c and VP23 in assembly. J. Mol. Biol. 263:447–462 [DOI] [PubMed] [Google Scholar]
- 39. Trus BL, et al. 2004. Structure and polymorphism of the UL6 portal protein of herpes simplex virus type 1. J. Virol. 78:12668–12671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Trus BL, Gibson W, Cheng N, Steven AC. 1999. Capsid structure of simian cytomegalovirus from cryoelectron microscopy: evidence for tegument attachment sites. J. Virol. 73:2181–2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Trus BL, et al. 2007. Allosteric signaling and a nuclear exit strategy: binding of UL25/UL17 heterodimers to DNA-filled HSV-1 capsids. Mol. Cell 26:479–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wingfield PT, et al. 1997. Hexon-only binding of VP26 reflects differences between the hexon and penton conformations of VP5, the major capsid protein of herpes simplex virus. J. Virol. 71:8955–8961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wolfstein A, et al. 2006. The inner tegument promotes herpes simplex virus capsid motility along microtubules in vitro. Traffic 7:227–237 [DOI] [PubMed] [Google Scholar]
- 44. Zhou ZH, Chen DH, Jakana J, Rixon FJ, Chiu W. 1999. Visualization of tegument-capsid interactions and DNA in intact herpes simplex virus type 1 virions. J. Virol. 73:3210–3218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhou ZH, et al. 1995. Assembly of VP26 in herpes simplex virus-1 inferred from structures of wild-type and recombinant capsids. Nat. Struct. Biol. 2:1026–1030 [DOI] [PubMed] [Google Scholar]