Abstract
We recently reported that human immunodeficiency virus type 1 (HIV-1) carrying PTAP and LYPXnL L domains ceased budding when the nucleocapsid (NC) domain was mutated, suggesting a role for NC in HIV-1 release. Here we investigated whether NC involvement in virus release is a property specific to HIV-1 or a general requirement of retroviruses. Specifically, we examined a possible role for NC in the budding of retroviruses relying on divergent L domains and structurally homologous NC domains that harbor diverse protein sequences. We found that NC is critical for the release of viruses utilizing the PTAP motif whether it functions within its native Gag in simian immunodeficiency virus cpzGAB2 (SIVcpzGAB2) or SIVsmmE543 or when it is transplanted into the heterologous Gag protein of equine infectious anemia virus (EIAV). In both cases, virus release was severely diminished even though NC mutant Gag proteins retained the ability to assemble spherical particles. Moreover, budding-defective NC mutants, which displayed particles tethered to the plasma membrane, were triggered to release virus when access to the cell endocytic sorting complex required for transport pathway was restored (i.e., in trans expression of Nedd4.2s). We also examined the role of NC in the budding of EIAV, a retrovirus relying exclusively on the (L)YPXnL-type L domain. We found that EIAV late budding defects were rescued by overexpression of the isolated Alix Bro1 domain (Bro1). Bro1-mediated rescue of EIAV release required the wild-type NC. EIAV NC mutants lost interactions with Bro1 and failed to produce viruses despite retaining the ability to self-assemble. Together, our studies establish a role for NC in the budding of retroviruses harboring divergent L domains and evolutionarily diverse NC sequences, suggesting the utilization of a common conserved mechanism and/or cellular factor rather than a specific motif.
INTRODUCTION
In human immunodeficiency virus type 1 (HIV-1) and most retroviruses, the Gag precursor polyprotein is the major structural component that orchestrates the assembly and release of virus particles from the plasma membrane. It carries three main domains, namely, matrix (MA), capsid (CA), and nucleocapsid (NC). The MA domain directs the interaction of Gag with the plasma membrane (11, 34, 59, 79), whereas the CA domain carries regions responsible for Gag-Gag multimerization during assembly (28, 31, 35, 37, 46, 64). The NC domain (also called NC here) interacts with the viral genomic RNA, as well as cellular RNAs that can serve as a scaffold for Gag-Gag assembly (1, 8, 12–14, 23, 37, 46). In addition to the three main domains, Gag proteins carry short peptides, called late (L) domains, involved in virus budding and separation from infected cells (10, 15, 21, 33, 39, 42, 57).
A total of three different core sequences have been identified as essential elements for L-domain function: P(T/S)AP, PPXY, and LYPXnL (32, 49, 52, 68, 73, 74). They serve as docking sites for cellular proteins that associate or function in the endocytic sorting complex required for transport (ESCRT) pathway (51, 68, 74). The C-terminal p6 domain of HIV-1 Gag carries two L domains, PTAP and LYPXnL, that recruit Tsg101 and Alix, respectively, and both cellular factors are involved in virus budding (22, 32, 39, 52, 68, 73). Tsg101 functions in HIV-1 budding as part of “early”-acting ESCRT-I, a cellular complex believed to recruit downstream members of the “late”-acting ESCRT-III pathway named the charged multivesicular body proteins (CHMPs) (4, 32, 51, 56, 68, 74). CHMP isoforms catalyze membrane modeling events that promote virus separation from the cell. Alix binds the ESCRT-III member CHMP4B through its Bro1 domain, thus serving as a direct cellular bridge between the viral LYPXnL motif and the ESCRT pathway (30, 41, 43, 53, 68, 72). Both Tsg101- and Alix-mediated budding pathways require the activity of the AAA ATPase VPS4 (5, 6, 51, 70, 74).
In addition to HIV-1, other retroviruses utilize Tsg101- and/or Alix-binding L domains for virus release. These include simian immunodeficiency virus (SIV), equine infectious anemia virus (EIAV), and Rous sarcoma virus (24, 65, 68, 77, 78). For example, SIVcpzGAB2 harbors a PTAP motif in the p6 domain located adjacent to a canonical LYPXnL motif (9, 78) whereas HIV-2 and SIVsmm/mac display a different distribution of L domains. They recruit Alix through p6-located L domains that were recently designated type 3 (7, 68, 78) and contribute to virus budding events. EIAV harbors only one L domain, YPDL (within the NC-adjacent p9 domain of Gag), that binds Alix directly and mediates the recruitment of ESCRT-III members necessary for virus release (65, 68, 77).
While it has been established that L domains play a key role in retroviral budding and separation from the host cell membrane, recent studies have suggested the involvement of the NC domain in these steps. Indeed, in the presence of an intact p6 domain, mutation or deletion of the NC domain interferes with PTAP-mediated virus budding in model cells and the physiologically relevant T cells while preserving the ability to assemble particle (27, 37, 60, 75). These observations indicated a specific interplay between NC and L domains due to their adjacent positions and suggested a mutual functional interdependence in the process of virus budding and release. Moreover, we and others have shown that NC interacts with the Bro1 domain of Alix (26, 62), which mediates the recruitment of the cellular fission machinery components CHMP4 and VPS4 (26). Interestingly, this interaction is critical for the HIV-1 LYPXnL-driven budding pathway (26, 62, 66, 67), emphasizing the role of NC in Alix-mediated HIV-1 budding.
Several roles have been attributed to NC in the HIV-1 life cycle; they include reverse transcription, integration, trafficking, virus assembly, viral genome encapsidation, and cell-to-cell virus transmission (19, 38, 44, 47, 54, 58, 71). We recently obtained evidence supporting a role for NC in virus budding. However, it is not known whether NC involvement in budding is specific to HIV-1 or a general requirement of other retroviruses utilizing divergent L domains and evolutionarily diverse NC regions. In this study, we investigated the function of NC in lentiviruses whose release is stimulated by Tsg101- and/or Alix-binding motifs PTAP and (L)YPXnL in various contexts. We examined the role of NC in the release of SIVcpzGAB2 and SIVsmmE543, two lentiviruses that contain PTAP and (L)YPXnL motifs (in contexts similar to that found in HIV-1) and NC domains that are homologous but not identical to HIV-1 NC. A role for NC in virus budding mediated exclusively through the PTAP motif was also assessed using a chimeric EIAV whose YPDL L domain motif was functionally replaced with the HIV-1 PTAP motif positioned next to the heterologous EIAV NC, which shares only little sequence homology with HIV-1 NC. We previously reported that a WT NC is also required for HIV-1 release mediated via the Alix-binding LYPXnL motif. These studies were conducted using Alix overexpression in virus rescue assays. To examine the role of NC in Alix-binding L domain-mediated virus release in a natural setting, we used EIAV, which relies solely on a (L)YPXnL motif and cellular Alix for virus production. We found that a functional WT NC domain is required for (L)YPXnL-mediated virus release. Together, the data invoke the importance of a common conserved feature of NC (i.e., its three-dimensional structure) for the release of viruses utilizing divergent L domain motifs and containing evolutionarily diverse NC protein sequences.
MATERIALS AND METHODS
Cells and transfection.
293T cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 1% l-glutamine. Transfection was carried out using Lipofectamine 2000 (Invitrogen) and following the manufacturer's protocol.
Proviral and protein expression constructs.
The wild-type (WT) full-length EIAV molecular clone EIAVUK and its derivative EIAVUKPTAP (designated eWT and ePTAP, respectively, here) were previously described (18, 45). The p9 L domain mutant eYP− contained the YPDL-to-SRGA substitution (45). eRKI and ePTAP-RKI mutants were generated by changing NC residues K10, K15, K20, and K30 to alanines in the eWT and ePTAP proviruses, respectively. Residues K30, R39, K42, K45, K47, and K54 were replaced with alanines to generate eRKII and ePTAP-RKII mutants. The full-length sequence of the SIVcpzGab2 strain has been reported previously (9). To construct the molecular clone, strain-specific primers were designed for nested-PCR amplification of the 5′ and 3′ halves from uncultured peripheral blood mononuclear cell genomic DNA. Unique restriction enzyme sites were introduced into the primers used for the second round of PCR amplification to facilitate cloning. Amplification products were digested with the appropriate restriction enzymes, agarose gel purified, and ligated into the pCR-XL vector (Invitrogen). Plasmids were purified from 1.5-ml volumes of bacterial cultures (STBL2) in LB-kanamycin (100 μg/ml) grown for 18 h at 30°C. Plasmids containing full-length inserts were selected by restriction enzyme digestion and tested for the production of infectious viral particles by transfecting 293T cells. Supernatants were collected 48 h posttransfection and assayed for the presence of infectious virus particles using the TZM-BL assay. A total of 104 full-length molecular clones were screened, and 6 clones with the highest infectious titers were further tested for replication competence in activated human CD4+ lymphocytes. Among them, clone 62, used in this study, was selected for exhibiting growth characteristic comparable to those of control HIV-1 strains (GenBank accession no. AF382828). The p6 L domain sPTAP− and sYP− mutants contained the PTAP-to-LIAP and YPSL-to-SLSL substitutions, which were introduced using the QuikChange XL site-directed mutagenesis kit (Stratagene) according to the manufacturer's protocol. The SIVcpzGAB2 sRKI and sRKII NC mutants carry alanine substitutions of residues R2, R6, R10, R11, K13, K19, and R25 and residues R25, K28, R31, R32, K33, K37, K40, K46, and R53, respectively. In SIVsmmE543-3 (36), N-terminal basic residues K2, R5, R6, K9, K15, and R21 and C-terminal residues R24, R27, R28, K36, K43, and R47 were mutated to alanines to generate the smRKI and smRKII NC mutants, respectively, in the context of the previously described SIVsm YL mutant (7). Glutathione S-transferase (GST) fusion constructs encoding EIAV NC-p9, NCRKI-p9, and NCRKII-p9 were obtained by subcloning DNA fragments corresponding to EIAVUK Gag amino acid residues 359 to 486 into the BamHI-EcoRI sites of pGEX-5-X2 (GE Healthcare). The hemagglutinin (HA)-tagged Nedd4.2s and Bro1 expression vectors were previously described (26).
Virus release assay.
293T cells (3 × 106) were transfected with either 500 ng (EIAV) or 1 μg (SIV) of proviral DNA. At 24 h posttransfection, culture supernatants were filtered and virions were pelleted at 151,000 × g for 1 h on a 20% (wt/vol in phosphate-buffered saline [PBS]) sucrose cushion. Cells were washed once with cold PBS and lysed in lysis buffer (1% [vol/vol] Igepal, 50 mM Tris [pH 8.0], 150 mM NaCl, protease inhibitor cocktail [Complete; Roche]). Viral and cellular proteins were analyzed by SDS-PAGE and immunoblotting. Virus release efficiency was calculated as the ratio of virion-associated Gag to total cellular Gag as determined by densitometry analysis of Western blot films using ImageJ software (W. S. Rasband, NIH, Bethesda, MD; http://rsb.info.nih.gov/ij). The antibodies used were monoclonal anti-HA (HA-7; Sigma), anti-tubulin (DM1A; Sigma), and anti-Tsg101 (BD Biosciences). Cellular Alix was detected by using rabbit anti-Alix serum. EIAV was detected using horse anti-EIAV serum (55), and SIV was detected using anti-SIV antiserum.
RNAi knockdown.
Knockdown of cellular Alix and Tsg101 by RNA interference (RNAi) was performed as previously described (7, 66). Briefly, 293T cells (2.25 × 106) were seeded into T25 flasks and transfected with 0.2 nmol of Tsg101 or Alix RNAi oligonucleotides (Invitrogen) using Lipofectamine 2000. After 24 h, cells were cotransfected with the same amount of RNAi oligonucleotides and 500 ng of EIAVUK-PTAP proviral DNA. Cells and virus were harvested and processed as outlined above.
GST pulldown assays.
The DNA of pGEX-NC-p9, RKI-p9, RKII-p9, and GST–empty-vector constructs was transformed into Escherichia coli BL21(DE3)/pLysS (Stratagene). GST pull-down assays were performed as previously described (67). Briefly, cultures in the exponential phase of growth were induced with isopropyl-β-d-thiogalactopyranoside. Bacterial pellets were resuspended in bacterial lysis buffer (BLB) containing lysozyme. Samples were sonicated and clarified by centrifugation. Supernatants containing GST fusion proteins were incubated with glutathione-Sepharose 4B beads (GE Healthcare). The beads were washed in BLB and incubated with mammalian cell lysates containing the proteins of interest. The complexes were washed in mammalian lysis buffer and eluted with reduced Glutathione (Sigma). Eluates and cell lysates (input fractions) were analyzed by SDS-PAGE and Western blot analysis with the appropriate antibodies.
Electron microscopy (EM).
293T cells were transfected with either 500 ng (eWT, eRKI, and eRKII) or 1 μg (ePTAP, ePTAP-RKI, and ePTAP-RKII) of proviral DNA. Thirty hours posttransfection, 293T cells were fixed for 2 h at room temperature in 2% (vol/vol) glutaraldehyde in 0.1 M cacodylate buffer (pH 7.4). The cells were then rinsed in cacodylate buffer and postfixed in 1% (vol/vol) osmium tetroxide in the same buffer. The samples were subsequently rinsed again in 0.1 N sodium acetate buffer (pH 4.2), stained with 0.5% uranyl acetate (vol/vol) in the same buffer, dehydrated in graded ethanol, and then infiltrated with pure epoxy resin overnight. The wells were embedded in fresh resin the next day and cured at 55°C. Blocks were cut from the cured samples and mounted appropriately for ultramicrotomy. Thin sections were stained with uranyl acetate and lead citrate and stabilized by carbon evaporation. Images were obtained with a Hitachi H7600 electron microscope equipped with an AMT XL41M digital camera. Each EM experiment was performed at least twice independently, and multiple grids were scanned to determine the predominant phenotype before images were selected for figures.
RESULTS
NC is required for PTAP-mediated budding of SIVcpzGAB2 and SIVsmmE543.
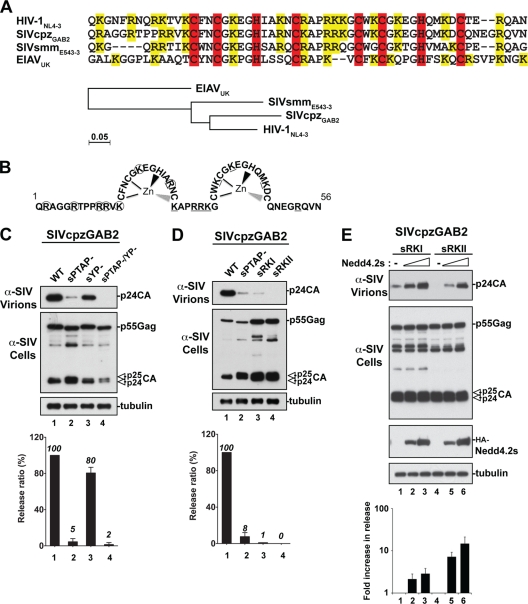
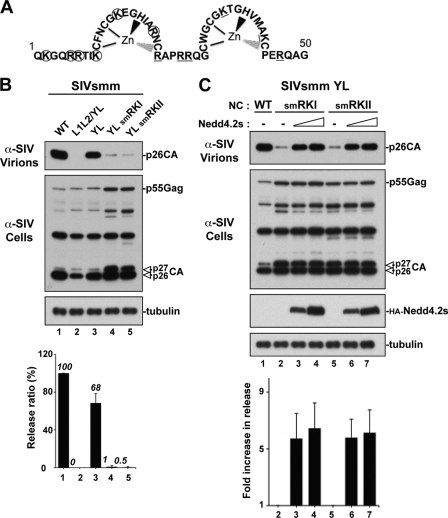
We recently showed that NC is involved in the PTAP-mediated viral release of HIV-1 (27). To address whether this is a property specific to HIV-1 NC or a general functional requirement of other lentiviruses, we investigated the role of NC in the release of other lentiviruses with NC domains that are homologous (i.e., conservation of zinc fingers and basic residues) but bear evolutionarily diverse protein sequences (Fig. 1A). SIVcpzGAB2 and SIVsmmE543, whose NC domains share 81 and 76% sequence homology with HIV-1 NC, respectively, were also selected because they contain a p6 domain with the consensus PTAP L domain located adjacent to NC domains in contexts similar to that found in HIV-1 (9, 36). Indeed, SIVcpzGAB2 relies mainly on the PTAP L domain for efficient viral release from 293T cells, as disruption of the PTAP motif (sPTAP−) caused a near elimination of SIVcpzGAB2 release (Fig. 1C, lane 2), in contrast to the LYPXnL motif mutant (sYP−), which had only 20% viral release reduction (lane 3). Similarly, about 70% of SIVsmmE543 release is mediated by the p6-bound PTAP motif (7) (Fig. 2B, YL mutant). We concluded that SIVcpzGAB2 and SIVsmmE543 exhibite functional release determinants similar to those of HIV-1 and constitute good models to examine the role of NC domains in PTAP-mediated virus release. To conduct this analysis, we mutated basic residues in the N- and the C-terminal regions of the SIVcpzGAB2 (named sRKI and sRKII; Fig. 1B) and SIVsmmE543 (named smRKI and smRKII; Fig. 2A) NC domains and examined their effects on virus release. Both the sRKI and sRKII NC mutants exhibited a severe reduction in virus release that is comparable to that observed with the sPTAP− mutant (Fig. 1D). Similarly, the smRKI and smRKII mutations of SIVsmmE543 lacking the Alix-binding YL L domain (YL mutant [7])—and therefore relying exclusively on the PTAP motif for L domain function—eliminated virus release (Fig. 2B, lanes 4 and 5). These results indicated that the NC domains of SIVcpzGAB2 and SIVsmmE543 play an important role in PTAP-mediated virus release, a property they share with HIV-1 (27), suggesting that NC involvement in virus release is not limited to HIV-1.
Fig 1.
The NC domain is required for PTAP-mediated budding of SIVcpzGAB2. (A) Protein sequence alignment of the HIV-1 NL4-3, SIVcpzGAB2, SIVsmmE543-3, and EIAV core NC regions. The conserved zinc finger CCHC motifs and basic residues are highlighted in red and yellow, respectively. The phylogenetic analysis of the four NC protein core sequences is shown at the bottom. (B) Schematic representation of the SIVcpzGAB2 NC domain (amino acids 1 to 56). Lysine (K) and arginine (R) residues replaced with alanine in the sRKI and sRKII mutants are circled and underlined, respectively. (C and D) Comparison of virus release among WT SIVcpzGAB2, single L domain PTAP (sPTAP−) and LYPXnL (sYP−) mutants, a double L domain mutant (sPTAP/YP−), and NC mutants. 293T cells were transfected with either WT SIVcpz or the indicated mutant. Cell lysates and viral fractions were analyzed by Western blotting using the indicated antibodies. Relative virus release efficiencies calculated as described in Materials and Methods are shown at the bottom of panels C to E (± the standard deviations; n = 3). (E) Release defects of SIVcpzGAB2 NC mutant viruses are rescued by ectopic expression of Nedd4.2s. 293T cells were transfected with the indicated NC mutants alone (lanes 1 and 4) or in the presence of increasing amounts of Nedd4.2s (lanes 2, 3, 5, and 6). The n-fold increase in viral release upon cotransfection with Nedd4.2s for each mutant was calculated after determining the individual relative virus release efficiencies as in panels C and D and setting viral release in the absence of Nedd4.2s arbitrarily to 1 (± the standard deviations; n = 3).
Fig 2.
The NC domain is required for PTAP-mediated budding of SIVsmmE543. (A) Schematic representation of the SIVsmmE543 NC domain (amino acids 1 to 50). Lysine (K) and arginine (R) residues replaced with alanine in the smRKI and smRKII mutants are circled and underlined, respectively. (B) Comparison of virus release among WT SIVsmmE543, the L domain mutant L1L2/YL (lacking all known L domains), and the Alix binding site mutant YL (relying only on PTAP for release) in the context of a WT or mutated NC. 293T cells were transfected with either WT SIVsmmE543 or the indicated mutant. Cell lysates and viral fractions were analyzed by Western blotting using the indicated antibodies. Relative virus release efficiencies are shown at the bottom (± the standard deviations; n = 3). (C) Release defects of SIVsmmE543/YL NC mutant viruses are rescued by ectopic expression of Nedd4.2s. 293T cells were transfected with the indicated NC mutant alone (lanes 1 and 2) or in the presence of increasing amounts of Nedd4.2s (lanes 3, 4, 6, and 7). Pelleted virion and cell lysate fractions were analyzed by Western blotting using the indicated antibodies. Quantification of the stimulatory effect of Nedd4.2s on viral release is shown at the bottom. The n-fold increase in viral release upon cotransfection with Nedd4.2s for each mutant was calculated as described in the legend to Fig. 1.
To ascertain that mutations in NC impeded release and not viral assembly, we ectopically expressed Nedd4.2s under conditions analogous to those used for HIV-1 RKI and RKII mutants (27). Importantly and similarly to HIV-1 NC mutants, expression of Nedd4.2s rescued the release defects of the SIVcpzGAB2 sPTAPYP−/ mutant (data not shown); the SIVsmmE543 L1L2/YL mutant, lacking all known L domains (the two PTAP motifs and the type 3 Alix-binding motif YL/LXXLF [7]); and the sRKI/sRKII NC and smRKI/smRKII NC mutants (Fig. 1E and 2B, respectively). Virus rescue in three separate experiments was quantified as shown in the graphs below the Western blot analyses. Together, these results indicate that NC plays a critical role in the release of SIVcpzGAB2 and SIVsmmE543 and suggest the key involvement of a functional NC in the release of retroviruses utilizing the PTAP motif near NC domains that are homologous but not identical to those of HIV-1 NC.
NC is required for virus release when PTAP is transplanted into a heterologous Gag protein.
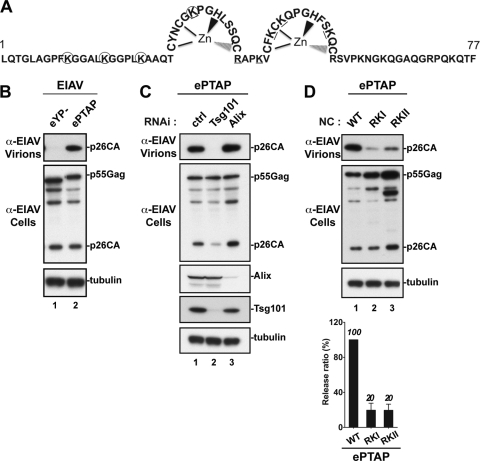
We next asked whether NC is required for virus release when the PTAP motif is transplanted into a heterologous Gag protein and near an NC domain that shares little sequence homology with HIV-1 NC. To answer this question, we made use of an EIAV chimera harboring a PTAP motif (named ePTAP here). In the latter construct, the EIAV YPDL motif was replaced with the HIV-1 PTAP motif to mediate PTAP-dependent viral release and test for its sensitivity to NC mutations in this context (Fig. 3A). Additionally, in the ePTAP chimera virus, the PTAP motif is located next to the EIAV NC domain, which shares only 32% sequence identity with HIV-1 NC. As previously reported (45), YPDL motif mutation (eYP) in WT EIAV induced a complete inhibition of viral production (Fig. 3B, lane 1). Conversely, YPDL replacement with the HIV-1-derived L domain PTAP (ePTAP) rescued viral release (Fig. 3B, lane 2). As expected and in contrast to the RNAi knockdown of cellular Alix, depletion of endogenous Tsg101 caused a dramatic inhibition of ePTAP release (Fig. 3C, lanes 2 and 3), demonstrating true dependence on the PTAP/Tsg101 pathway for virus release. Mutations in the N (ePTAP-RKI) or C (ePTAP-RKII) terminus of the ePTAP NC region (Fig. 3A) dramatically reduced viral release (Fig. 3D, lanes 2 and 3). Quantitative analysis of virus release efficiency indicated that mutations in NC reduced ePTAP virus release by 80% (bottom of panel D).
Fig 3.
NC is required for virus release when PTAP is inserted into a heterologous Gag protein. (A) Schematic representation of the EIAV NC domain (amino acids 1 to 77). Lysine (K) and arginine (R) residues replaced with alanine in the ePTAP-RKI and ePTAP-RKII mutants are circled and underlined, respectively. (B) 293T cells were transfected with L domain mutant EIAV (eYP−) or EIAV carrying PTAP as the L domain (ePTAP). Pelleted virion and cell lysate were analyzed by Western blotting using the indicated antibodies. (C) Depletion of Tsg101 or Alix in 293T cells and its effect on viral release. 293T cells were cotransfected as described in Materials and Methods, with ePTAP and control RNAi (ctrl, lane 1) or Tsg101 (lane 2)- or Alix (lane 3)-specific RNAi. Pelleted virion and cell lysate fractions were analyzed by Western blotting using the indicated antibodies. (D) Effects of NC mutations on EIAV release mediated exclusively via the PTAP L domain. 293T cells were transfected with ePTAP carrying a WT or mutated NC. Pelleted virion and cell lysate fractions were analyzed by Western blotting using the indicated antibodies. Relative virus release efficiencies are shown at the bottom (± the standard deviations; n = 3).
To ascertain that NC mutations affected a postassembly step, we analyzed the ability of ePTAP-RKI or ePTAP-RKII to coassemble with eYP−, which is release defective but provides a functional NC domain in trans. Coexpression of 20 to 40% of an eYP− Gag protein that contains a WT NC domain but lacks a functional L domain rescued the release of NC mutants (Fig. 4A and B), indicating that ePTAP NC mutants are assembly competent. In support of these findings, EM examination showed that spherical particles formed by ePTAP-RKI or ePTAP-RKII (Fig. 4C, parts c to f) remained tethered to the cell membrane, confirming a clear budding defect (Fig. 4C, parts d to f). Altogether, these data indicate that an intact NC is necessary for budding events of an EIAV chimera relying on a PTAP L domain for release and demonstrate that the requirement for WT NC is maintained when the PTAP L domain functions with an NC domain with little sequence homology with HIV-1 NC.
Fig 4.
Mutations in the EIAV-PTAP (ePTAP) NC domain interfere with virus budding. Coexpression of EIAV L domain mutant eYP−, which harbors a functional NC, rescues the release of ePTAP NC mutants ePTAP-RKI (A) and ePTAP-RKII (B). 293T cells were transfected with ePTAP (lane 1), eYP− (lane 2), or ePTAP-RKI or -RKII (lane 3) alone or cotransfected with the indicated NC mutant and eYP− plasmids (lanes 4 and 5). DNA ratios of the indicated NC mutant to the YP mutant used were 4:1 and 1.5:1, while the total amount of DNA remained constant. Viral fractions and cell lysates were analyzed by Western blotting using the indicated antibodies. Relative virus release efficiencies are shown at the bottom (± the standard deviations; n = 3). (C) ePTAP NC mutants displayed typical budding defects in 293T cells. Shown are transmission EM images of thin-sectioned 293T cells transfected with ePTAP-WT (a), ePTAP-RKI (b and d), or ePTAP-RKII (c and e). Arrows indicate either WT mature virions (a) or tethered arrested particles (b to e). Scale bars, 200 nm.
NC is involved in virus budding mediated by Alix-binding L domain.
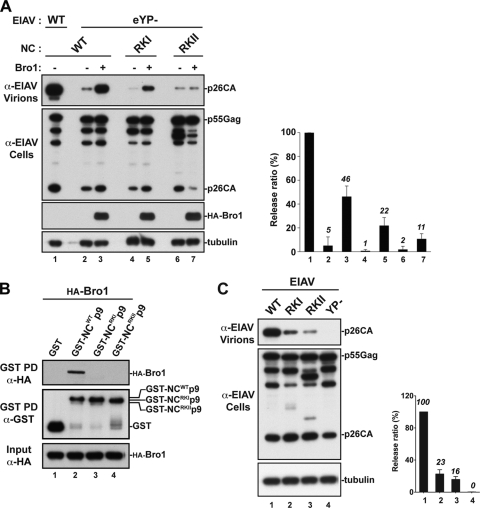
We previously presented evidence supporting a role for HIV-1 NC in the Alix-binding LYPXnL L domain budding pathway. We found that overexpression of the Alix Bro1 domain rescued virus release defects of an HIV-1 mutant lacking all L domains (26, 67). Similar results were obtained with the SIVcpzGAB2 mutant lacking all L domains (data not shown). In addition, Alix-mediated release of both human and simian viruses required an intact NC (26; data not shown), a region in Gag also involved in interactions with the Alix Bro1 domain (26, 62). In order to directly address the role of NC in the YPDL-driven pathway, we examined its function in EIAV budding. We tested whether the isolated Bro1 domain is sufficient to induce virus release and found that overexpression of Bro1 rescued release defects of an EIAV mutant lacking its YPDL motif (Fig. 5A, lanes 2 and 3). The results indicated that EIAV release can be stimulated independently of its sole L domain, the YPDL motif located in the p9 region of Gag.
Fig 5.
The NC domain of EIAV Gag binds the Bro1 domain of Alix and is involved in virus release. (A) Release of the EIAV L domain mutant (eYP−) is rescued by Bro1 in an NC basic residue-dependent manner. 293T cells were transfected with WT EIAV (lane 1) or the eYP− mutant harboring either a WT NC (lanes 2 and 3) or a mutated NC (lanes 4 to 7) in the absence (lanes 1, 2, 4, and 6) or presence (lanes 3, 5, and 7) of HA-Bro1. Cell lysates and viral fractions were analyzed by Western blotting using the indicated antibodies. Relative virus release efficiencies are shown (± the standard deviation; n = 2). (B) Mutations of NC basic residues abrogate the interaction between Bro1 and NC. GST fusion proteins (NCWT-p9, NCRKI-p9, and NCRKII-p9) expressed in E. coli and captured on glutathione beads were incubated with 293T cell lysates expressing HA-Bro1. Interaction between Bro1 and the GST fusion proteins shown was analyzed by Western blotting using the indicated antibodies. (C) Mutations in NC interfere with the release of EIAV harboring a functional YPDL motif. 293T cells were transfected with WT EIAV or the indicated NC or L domain mutant. Viral fractions and cell lysates were analyzed by Western blotting. Relative virus release efficiencies are shown (± the standard deviations; n = 4).
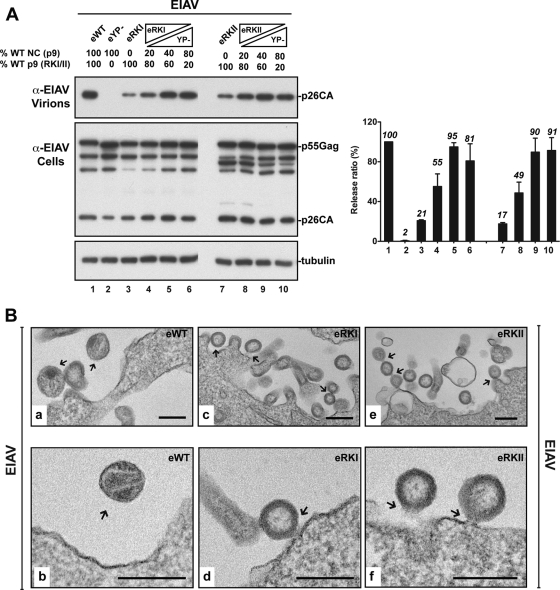
We, and others, previously obtained evidence suggesting a role for the NC in Alix-LYPXnL-mediated HIV-1 release, as Alix ceased to ectopically stimulate HIV-1 release in a virus rescue assay when mutations disrupted NC basic residues or its zinc fingers (26, 62). To address a role for NC in (L)YPDL-type-driven pathways directly and in a natural setting, we opted to examine this role in the release of EIAV, a virus that relies solely on an Alix-binding YPDL motif and utilizes endogenous Alix for virus budding. Mutations of basic residues to alanines were introduced into EIAV NC (eYP− RKI and RKII mutants), and their effect on Bro1-mediated virus rescue was assessed. In contrast to eYP−, which carries a WT NC, the eYP− RKI and RKII mutants were either partially rescued (Fig. 5A, lanes 5, RKI) or not rescued (lane 7, RKII) by the ectopic expression of Bro1. RKII mutation in NC (replacement of 6 basic residues with alanines) eliminated Bro1-induced virus rescue, whereas RKI mutation (4 basic residues substituted) displayed only a modest decrease. Virus rescue was examined in several independent experiments and quantified as shown in the right half of Fig. 5A. These data suggest that Bro1-mediated virus rescue involves NC basic residues directly and strongly suggest a requirement for a functional WT NC domain in EIAV budding.
Viral release rescue events due to overexpression of Bro1 had been attributed to its ability to bind NC (26, 67). To determine if NC is the actual domain required for Bro1 recruitment and rescue of EIAV release, we examined the interaction between Bro1 and the EIAV NC. We employed GST pulldown assays to examine the interaction between the WT or RKI or RKII mutant EIAV NC and Alix Bro1 (Fig. 5B). GST-tagged EIAV NC-p9 fusion protein was able to pull down HA-tagged Bro1 protein (Fig. 5B, lane 2). However, mutations introduced into the N or C terminus of NC eliminated binding to Bro1 (lanes 3 and 4). Specific interactions of GST-tagged and HA-tagged proteins were detected, since no discernible bands were observed in the control lane (Fig. 5B, lane 1). These data indicate that mutations in the EIAV NC prevented binding to Bro1 and therefore suggested that Alix interaction with NC might be a prerequisite for efficient EIAV release. To examine this hypothesis, we introduced mutations into the EIAV NC in the context of the whole virus carrying functional YPDL and examined their effect on virus release. EIAV NC mutants displayed a drastic reduction in virus release (Fig. 5C, lanes 2 and 3). The viral release efficiency of EIAV NC mutants was reduced by up to 6-fold compared to that of WT EIAV, as shown by the quantitative analysis of viral release (Fig. 5C, right side).
EIAV NC mutants display typical budding defects.
To verify that NC mutations affected viral release and not assembly, we performed complementation assays similar to those described in Fig. 3. Coexpression of 40 to 80% of a Gag protein that contains a WT NC domain but lacks a functional L domain efficiently rescued the release of NC mutants (Fig. 6A) compared to the controls (Fig. 6A, lanes 3 and 7). We concluded that EIAV NC mutants (RKI and RKII) coassembled with eYP−, indicating they are assembly competent and therefore their inability to release virus is due to a postassembly defect. To further analyze such a phenotype(s), we examined 293T cells expressing WT EIAV or the eRKI and eRKII mutants by EM (Fig. 6B). We frequently observed fully assembled particles at the plasma membrane in all cases. However, arrested budding virus particles that remained tethered to the plasma membrane (Fig. 6B, parts c, d, and f) or to each other (part e) accumulated at the surface of cells expressing either the eRKI or the eRKII NC mutant. These results indicate that mutation in NC caused typical budding defects and that NC is also involved in EIAV budding through the Alix/YPDL-mediated pathway.
Fig 6.
EIAV NC mutants are budding defective. Coexpression of EIAV L domain mutant eYP−, which harbors a functional NC, rescues the release of the eRKI and eRKII NC mutants. 293T cells were transfected with eWT (lane 1), eYP− (lane 2), eRKI (lane 3), or eRKII (lane 7) alone or cotransfected with the indicated NC mutant and eYP− plasmids (lanes 4, 5, 6, 8, 9, and 10). The transfected DNA ratios of the indicated NC mutant and the YP mutant were 4:1, 1.5:1, and 1:4 while the total amount of DNA remained constant. Viral fractions and cell lysates were analyzed by Western blotting using the indicated antibodies. Relative virus release efficiencies are shown (± the standard deviations; n = 3). (B) EIAV NC mutants displayed typical budding defects in 293T cells. Shown are transmission EM images of thin-sectioned 293T cells transfected with WT EIAV (a and b) or the eRKI (c and d) or eRKII (e and f) mutant. Arrows indicate either WT mature virions (a and b) or tethered arrested particles (c to f). Scale bars, 200 nm.
DISCUSSION
We recently obtained data supporting a key role for the NC domain of HIV-1 Gag in virus budding mediated through the Tsg101-binding PTAP and Alix-binding LYPXnL L domains (27). In the present study, examination of the role of NC in the release of retroviruses whose budding is controlled by the PTAP, the (L)YPXnL, or both L domains and utilizing evolutionarily diverse NC regions (Fig. 1A) confirmed the strict requirement for a functional WT NC domain in virus budding.
The role of NC in (L)YPXnL-mediated virus release.
A role for NC in HIV-1 budding through the Alix-binding LYPXnL L domain came to light with the discovery of interactions between the Bro1 domain of Alix and the NC domain of HIV-1 Gag (26, 62). The requirement of NC for efficient LYPXnL-mediated virus release is not limited to HIV-1, since mutations of NC domains belonging to viruses utilizing (L)YPXnL L domains of type 1 (and type 3 [7, 78; data not shown]) led to the accumulation of fully assembled budding particles at the cell surface (Fig. 6), indicating that Alix interaction with a functional NC is a general prerequisite for viruses utilizing Alix-binding motifs for budding.
The studies with Alix-mediated HIV-1 release were performed using overexpression and virus rescue assays posing questions regarding the importance of NC-Bro1 interactions in a natural setting. The requirement of a functional NC in EIAV budding suggested a critical role for NC-Bro1 interactions in the function of cellular Alix and provided the first hint to the clear relevance of Alix dual interactions with NC and p6 domains in vivo. Additionally, retaining the need for both interactions in the cell for EIAV release—despite the strong YPDL binding to the EIAV p9 domain (the highest-affinity binding among Alix-bindings motifs [77, 78])—demonstrated that NC is not an Alix secondary redundant binding site in Gag and is rather a key functional determinant of (L)YPXnL-type L domains.
The EIAV and HIV-1 NC domains share little sequence identity and display local fold differences. EIAV NC has the shortest linker among structurally determined retroviral NC proteins and fewer basic residues than HIV-1 NC (3) and SIVsmmE543 (Fig. 1A). Despite these differences, both NC proteins capture the Bro1 domain of Alix, suggesting that a feature common to HIV-1 and EIAV NC proteins (i.e., the conformational features of the CCHC motifs) is recognized for interactions with Bro1. It is interesting that in the cases of both HIV-1 and EIAV, Bro1-NC binding is sensitive to mutations of basic residues (which, by virtue of binding to RNA, are key determinants of NC folding), thus further suggesting a conserved mode of recognition that involves NC structure. How NC participates temporally and spatially in Alix-mediated virus release is not clear. However, a model involving a requirement of contacts between NC and the Bro1 domain of Alix during virus release is supported by the restoration of EIAV YPDL (lacking the YPDL motif) release following ectopic expression of the Bro1 domain and the latter's inability to rescue release when its binding to NC is compromised or eliminated (Fig. 5) (26, 67), thus drawing a direct correlation between the efficiency of Bro1-NC interactions and Alix activity in virus release. Interestingly, Alix access to NC via intermolecular interaction(s) with coassembling Gag proteins—rather than intramolecular binding within the same Gag protein—appears to be sufficient for function. Indeed, virus release was recovered when a budding-defective Gag protein carrying a mutated NC domain was coexpressed with a coassembling molecule bearing the WT NC (Fig. 6). Strikingly, this finding is similar to that previously obtained with L domain sequences (48).
Owing to its position in Alix, structural flexibility vis-à-vis other domains of Alix (30, 61), its boomerang fold, and the ability to link Gag to ESCRT-III members (30, 72), as well as other yet-to-be identified cellular proteins (67), the Bro1 domain of Alix might be binding NC to coordinate with p6 dynamic membrane modeling events necessary for virus budding. The onset of these events appears to take place during assembly, when Alix is recruited by EIAV Gag in the cytoplasm (40) to act later in the budding neck, possibly after subsequent recruitment(s) and local changes (i.e., CHMP polymerization and PR processing) that could position Alix and partners (67) at the budding neck for membrane fission.
The role of NC in PTAP-mediated virus release.
HIV-1 NC mutant viruses accumulated budding particles at the 293T cell surface (27), a context in which HIV-1 relies primarily on the PTAP motif to release virus, implying a role for NC in PTAP-mediated HIV-1 release. HIV-1 shares NC involvement in budding with SIVcpzGAB2 and SIVsmmE543, suggesting that the location of NC in Gag and its proximity to PTAP, as well as conserved structural features (zinc fingers and basic residues), are more important for function than is protein sequence identity, which varies among the NC domains studied (Fig. 1A).
Whereas the functional cross talk between the PTAP L domain and NC is supported by several lines of evidence, the mechanisms involved remain unclear. Conclusions have been confounded mainly by the overlapping roles the NC domain of Gag plays in both assembly and budding. Data in the present study, however, indicate that mutation of the NC domains of several retroviruses led to interference with virus separation from the cell despite the accumulation of assembled spherical virus particles at the cell membrane, further supporting interference with budding rather than assembly. Moreover, recognition of a specific sequence in NC is not required for function in virus release since NC domains belonging to evolutionarily divergent retroviruses sharing little sequence identity with HIV-1 NC (as much as 69% for SIVcpzGAB2 and as little as 32% for EIAV) are equally required for virus budding, suggesting the utilization of a conserved mechanism and/or cellular factor. Notably, basic residues are involved in RNA binding and NC structure “shaping” (2, 19, 20, 23). The role of these interactions in NC function during budding is being carefully evaluated.
Indeed, a PTAP L domain functional link with NC was found in human and simian lentiviruses and was maintained when the PTAP motif was transplanted into heterologous equine Gag protein (EIAV) and mutation of basic residues inhibited PTAP L domain activity in all contexts, further supporting the recognition of a structural entity rather than a specific sequence. It is noteworthy that in all cases the functional dependence of PTAP on NC varied with its localization in Gag, since optimal PTAP function requires positioning near NC (50, 69, 76), suggesting that both domains are likely involved in simultaneous protein-protein interactions to facilitate virus budding.
Consistent with such a model, NC mutants of viruses relying on the PTAP L domain in various contexts (reference 27 and this study) displayed clear virus budding defects, as they accumulated spherical budding particles that remained tethered to the cell membrane with remarkably elongated membranous stalks (Fig. 4) reminiscent of those seen following PTAP motif disruption (32) and demonstrated halted membrane fission. These observations indicate that NC and PTAP cooperate in the recruitment of cellular factors—belonging to the ESCRT pathway—necessary for virus release, a notion supported by findings that NC mutants' budding defects were alleviated by providing parallel access to the cell ESCRT via ectopic expression of Nedd4.2s. Accordingly, NC-p1 domains impose ESCRT requirement for PTAP-mediated HIV-1 release from 293T cells (63), thus arguing in favor of a role for NC (and possibly neighboring sequences) in PTAP-mediated ESCRT recruitment and/or function.
An NC feature shared by diverse retroviruses functions in assembly and budding.
In summary, we obtained evidence supporting the recognition of a feature common to diverse retroviral NC domains that is necessary for NC function with both the PTAP and (L)YPXnL L domains. This property shared among retroviral Gag proteins involves basic residues in NC that also mediate RNA incorporation, Gag-Gag multimerization, and distinct NC folding (2, 17, 19, 20, 23). The latter structure is shared among all of the retroviruses studied and might be involved in the recruitment of new cellular factors important for budding and/or known NC interacting partners such as ABCE1 (25, 80), Staufen1 (16), or Lyric (29), which notably are part of the ribonucleoprotein complex formed during Gag assembly. We propose a model in which a feature(s) of NC—in cooperation with sequences in p6—participates in dynamic and well-orchestrated recruitments and movements of proteins that begin during assembly and continue through budding, when host proteins are believed to ring the inside of the viral neck to promote membrane fission. Elucidating the nature of NC involvement and identifying cellular partners participating in such steps should further our understanding of its role in HIV-1 budding.
ACKNOWLEDGMENTS
We are grateful to Adam Harned (SAIC, NCI-Frederick) for assistance with EM processing and imaging, Ilnour Ourmanov and Vanessa Hirsch for reagents, Bernard Lafont for assistance with phylogenetic analysis, Alicia Buckler-White and her team for DNA sequencing, and Gisselle Medina for help in the preparation of the manuscript.
This work was supported in part by R37 AI050529 and the Intramural Research Program of the NIAID, NIH.
Footnotes
Published ahead of print 15 February 2012
REFERENCES
- 1. Aldovini A, Young RA. 1990. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J. Virol. 64:1920–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Amarasinghe GK, et al. 2000. NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the psi-RNA packaging signal. Implications for genome recognition. J. Mol. Biol. 301:491–511 [DOI] [PubMed] [Google Scholar]
- 3. Amodeo P, Castiglione Morelli MA, Ostuni A, Battistuzzi G, Bavoso A. 2006. Structural features in EIAV NCp11: a lentivirus nucleocapsid protein with a short linker. Biochemistry 45:5517–5526 [DOI] [PubMed] [Google Scholar]
- 4. Babst M, Katzmann DJ, Estepa-Sabal EJ, Meerloo T, Emr SD. 2002. Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev. Cell 3:271–282 [DOI] [PubMed] [Google Scholar]
- 5. Babst M, Sato TK, Banta LM, Emr SD. 1997. Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J. 16:1820–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Babst M, Wendland B, Estepa EJ, Emr SD. 1998. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 17:2982–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bello NF, et al. 2011. Distal leucines are key functional determinants of Alix-binding SIVsmE543 and SIVmac239 type 3 L domains. J. Virol. 85:11532–11537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Berkowitz RD, Luban J, Goff SP. 1993. Specific binding of human immunodeficiency virus type 1 gag polyprotein and nucleocapsid protein to viral RNAs detected by RNA mobility shift assays. J. Virol. 67:7190–7200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bibollet-Ruche F, et al. 2004. Complete genome analysis of one of the earliest SIVcpzPtt strains from Gabon (SIVcpzGAB2). AIDS Res. Hum. Retroviruses 20:1377–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bieniasz PD. 2009. The cell biology of HIV-1 virion genesis. Cell Host Microbe 5:550–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bryant M, Ratner L. 1990. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc. Natl. Acad. Sci. U. S. A. 87:523–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Burniston MT, Cimarelli A, Colgan J, Curtis SP, Luban J. 1999. Human immunodeficiency virus type 1 Gag polyprotein multimerization requires the nucleocapsid domain and RNA and is promoted by the capsid-dimer interface and the basic region of matrix protein. J. Virol. 73:8527–8540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Campbell S, Rein A. 1999. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. J. Virol. 73:2270–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Campbell S, Vogt VM. 1995. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J. Virol. 69:6487–6497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Carter CA. 2002. Tsg101: HIV-1's ticket to ride. Trends Microbiol. 10:203–205 [DOI] [PubMed] [Google Scholar]
- 16. Chatel-Chaix L, Boulay K, Mouland AJ, Desgroseillers L. 2008. The host protein Staufen1 interacts with the Pr55Gag zinc fingers and regulates HIV-1 assembly via its N-terminus. Retrovirology 5:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cimarelli A, Sandin S, Hoglund S, Luban J. 2000. Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA. J. Virol. 74:3046–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cook RF, et al. 1998. Development and characterization of an in vivo pathogenic molecular clone of equine infectious anemia virus. J. Virol. 72:1383–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Darlix JL, et al. 2011. Flexible nature and specific functions of the HIV-1 nucleocapsid protein. J. Mol. Biol. 410:565–581 [DOI] [PubMed] [Google Scholar]
- 20. De Guzman RN, et al. 1998. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science 279:384–388 [DOI] [PubMed] [Google Scholar]
- 21. Demirov DG, Freed EO. 2004. Retrovirus budding. Virus Res. 106:87–102 [DOI] [PubMed] [Google Scholar]
- 22. Demirov DG, Ono A, Orenstein JM, Freed EO. 2002. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc. Natl. Acad. Sci. U. S. A. 99:955–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. De Rocquigny H, et al. 1992. Viral RNA annealing activities of human immunodeficiency virus type 1 nucleocapsid protein require only peptide domains outside the zinc fingers. Proc. Natl. Acad. Sci. U. S. A. 89:6472–6476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dilley KA, Gregory D, Johnson MC, Vogt VM. 2010. An LYPSL late domain in the gag protein contributes to the efficient release and replication of Rous sarcoma virus. J. Virol. 84:6276–6287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dooher JE, Schneider BL, Reed JC, Lingappa JR. 2007. Host ABCE1 is at plasma membrane HIV assembly sites and its dissociation from Gag is linked to subsequent events of virus production. Traffic 8:195–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dussupt V, et al. 2009. The nucleocapsid region of HIV-1 Gag cooperates with the PTAP and LYPXnL late domains to recruit the cellular machinery necessary for viral budding. PLoS Pathog. 5:e1000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dussupt V, et al. 2011. Basic residues in the nucleocapsid domain of Gag are critical for late events of HIV-1 budding. J. Virol. 85:2304–2315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ehrlich LS, Agresta BE, Carter CA. 1992. Assembly of recombinant human immunodeficiency virus type 1 capsid protein in vitro. J. Virol. 66:4874–4883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Engeland CE, et al. 2011. The cellular protein Lyric interacts with HIV-1 Gag. J. Virol. 85:13322–13332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fisher RD, et al. 2007. Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell 128:841–852 [DOI] [PubMed] [Google Scholar]
- 31. Gamble TR, et al. 1997. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science 278:849–853 [DOI] [PubMed] [Google Scholar]
- 32. Garrus JE, et al. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55–65 [DOI] [PubMed] [Google Scholar]
- 33. Göttlinger HG, Dorfman T, Sodroski JG, Haseltine WA. 1991. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc. Natl. Acad. Sci. U. S. A. 88:3195–3199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Göttlinger HG, Sodroski JG, Haseltine WA. 1989. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. U. S. A. 86:5781–5785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gross I, Hohenberg H, Huckhagel C, Kräusslich HG. 1998. N-terminal extension of human immunodeficiency virus capsid protein converts the in vitro assembly phenotype from tubular to spherical particles. J. Virol. 72:4798–4810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hirsch V, et al. 1997. A molecularly cloned, pathogenic, neutralization-resistant simian immunodeficiency virus, SIVsmE543-3. J. Virol. 71:1608–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hogue IB, Hoppe A, Ono A. 2009. Quantitative fluorescence resonance energy transfer microscopy analysis of the human immunodeficiency virus type 1 Gag-Gag interaction: relative contributions of the CA and NC domains and membrane binding. J. Virol. 83:7322–7336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Houzet L, et al. 2008. Nucleocapsid mutations turn HIV-1 into a DNA-containing virus. Nucleic Acids Res. 36:2311–2319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Huang M, Orenstein JM, Martin MA, Freed EO. 1995. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J. Virol. 69:6810–6818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jouvenet N, Zhadina M, Bieniasz PD, Simon SM. 2011. Dynamics of ESCRT protein recruitment during retroviral assembly. Nat. Cell Biol. 13:394–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Katoh K, et al. 2003. The ALG-2-interacting protein Alix associates with CHMP4b, a human homologue of yeast Snf7 that is involved in multivesicular body sorting. J. Biol. Chem. 278:39104–39113 [DOI] [PubMed] [Google Scholar]
- 42. Kikonyogo A, et al. 2001. Proteins related to the Nedd4 family of ubiquitin protein ligases interact with the L domain of Rous sarcoma virus and are required for gag budding from cells. Proc. Natl. Acad. Sci. U. S. A. 98:11199–11204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kim J, et al. 2005. Structural basis for endosomal targeting by the Bro1 domain. Dev. Cell 8:937–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Levin JG, Mitra M, Mascarenhas A, Musier-Forsyth K. 2010. Role of HIV-1 nucleocapsid protein in HIV-1 reverse transcription. RNA Biol. 7:754–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li F, Chen C, Puffer BA, Montelaro RC. 2002. Functional replacement and positional dependence of homologous and heterologous L domains in equine infectious anemia virus replication. J. Virol. 76:1569–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li H, Dou J, Ding L, Spearman P. 2007. Myristoylation is required for human immunodeficiency virus type 1 Gag-Gag multimerization in mammalian cells. J. Virol. 81:12899–12910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Llewellyn GN, Hogue IB, Grover JR, Ono A. 2010. Nucleocapsid promotes localization of HIV-1 gag to uropods that participate in virological synapses between T cells. PLoS Pathog. 6:e1001167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Martin-Serrano J, Bieniasz PD. 2003. A bipartite late-budding domain in human immunodeficiency virus type 1. J. Virol. 77:12373–12377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Martin-Serrano J, Eastman SW, Chung W, Bieniasz PD. 2005. HECT ubiquitin ligases link viral and cellular PPXY motifs to the vacuolar protein-sorting pathway. J. Cell Biol. 168:89–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Martin-Serrano J, Perez-Caballero D, Bieniasz PD. 2004. Context-dependent effects of L domains and ubiquitination on viral budding. J. Virol. 78:5554–5563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Martin-Serrano J, Yarovoy A, Perez-Caballero D, Bieniasz PD. 2003. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc. Natl. Acad. Sci. U. S. A. 100:12414–12419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Martin-Serrano J, Zang T, Bieniasz PD. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7:1313–1319 [DOI] [PubMed] [Google Scholar]
- 53. McCullough J, Fisher RD, Whitby FG, Sundquist WI, Hill CP. 2008. ALIX-CHMP4 interactions in the human ESCRT pathway. Proc. Natl. Acad. Sci. U. S. A. 105:7687–7691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mirambeau G, Lyonnais S, Gorelick RJ. 2010. Features, processing states, and heterologous protein interactions in the modulation of the retroviral nucleocapsid protein function. RNA Biol. 7:724–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Montelaro RC, Parekh B, Orrego A, Issel CJ. 1984. Antigenic variation during persistent infection by equine infectious anemia virus, a retrovirus. J. Biol. Chem. 259:10539–10544 [PubMed] [Google Scholar]
- 56. Morita E, et al. 2011. ESCRT-III protein requirements for HIV-1 budding. Cell Host Microbe 9:235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Morita E, Sundquist WI. 2004. Retrovirus budding. Annu. Rev. Cell Dev. Biol. 20:395–425 [DOI] [PubMed] [Google Scholar]
- 58. Muriaux D, Darlix JL. 2010. Properties and functions of the nucleocapsid protein in virus assembly. RNA Biol. 7:744–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ono A, Ablan SD, Lockett SJ, Nagashima K, Freed EO. 2004. Phosphatidylinositol(4,5)bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. Proc. Natl. Acad. Sci. U. S. A. 101:14889–14894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ott DE, et al. 2003. Elimination of protease activity restores efficient virion production to a human immunodeficiency virus type 1 nucleocapsid deletion mutant. J. Virol. 77:5547–5556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Pires R, et al. 2009. A crescent-shaped ALIX dimer targets ESCRT-III CHMP4 filaments. Structure 17:843–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Popov S, Popova E, Inoue M, Göttlinger HG. 2008. Human immunodeficiency virus type 1 Gag engages the Bro1 domain of ALIX/AIP1 through the nucleocapsid. J. Virol. 82:1389–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Popova E, Popov S, Göttlinger HG. 2010. Human immunodeficiency virus type 1 nucleocapsid-p1 confers ESCRT pathway dependence. J. Virol. 84:6590–6597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Pornillos O, et al. 2009. X-ray structures of the hexameric building block of the HIV capsid. Cell 137:1282–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Puffer BA, Parent LJ, Wills JW, Montelaro RC. 1997. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J. Virol. 71:6541–6546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sette P, Jadwin JA, Dussupt V, Bello NF, Bouamr F. 2010. The ESCRT-associated protein Alix recruits the ubiquitin ligase Nedd4-1 to facilitate HIV-1 release through the LYPXnL L domain motif. J. Virol. 84:8181–8192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sette P, et al. 2011. The Phe105 loop of Alix Bro1 domain plays a key role in HIV-1 release. Structure 19:1485–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Strack B, Calistri A, Craig S, Popova E, Göttlinger HG. 2003. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114:689–699 [DOI] [PubMed] [Google Scholar]
- 69. Strack B, Calistri A, Göttlinger HG. 2002. Late assembly domain function can exhibit context dependence and involves ubiquitin residues implicated in endocytosis. J. Virol. 76:5472–5479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Stuchell-Brereton MD, et al. 2007. ESCRT-III recognition by VPS4 ATPases. Nature 449:740–744 [DOI] [PubMed] [Google Scholar]
- 71. Thomas JA, Bosche WJ, Shatzer TL, Johnson DG, Gorelick RJ. 2008. Mutations in human immunodeficiency virus type 1 nucleocapsid protein zinc fingers cause premature reverse transcription. J. Virol. 82:9318–9328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Usami Y, Popov S, Göttlinger HG. 2007. Potent rescue of human immunodeficiency virus type 1 late domain mutants by ALIX/AIP1 depends on its CHMP4 binding site. J. Virol. 81:6614–6622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. VerPlank L, et al. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag). Proc. Natl. Acad. Sci. U. S. A. 98:7724–7729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. von Schwedler UK, et al. 2003. The protein network of HIV budding. Cell 114:701–713 [DOI] [PubMed] [Google Scholar]
- 75. Wang SW, Aldovini A. 2002. RNA incorporation is critical for retroviral particle integrity after cell membrane assembly of Gag complexes. J. Virol. 76:11853–11865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Yuan B, Campbell S, Bacharach E, Rein A, Goff SP. 2000. Infectivity of Moloney murine leukemia virus defective in late assembly events is restored by late assembly domains of other retroviruses. J. Virol. 74:7250–7260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Zhai Q, et al. 2008. Structural and functional studies of ALIX interactions with YPX(n)L late domains of HIV-1 and EIAV. Nat. Struct. Mol. Biol. 15:43–49 [DOI] [PubMed] [Google Scholar]
- 78. Zhai Q, Landesman MB, Robinson H, Sundquist WI, Hill CP. 2011. Identification and structural characterization of the ALIX-binding late domains of simian immunodeficiency virus SIVmac239 and SIVagmTan-1. J. Virol. 85:632–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Zhou W, Parent LJ, Wills JW, Resh MD. 1994. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J. Virol. 68:2556–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Zimmerman C, et al. 2002. Identification of a host protein essential for assembly of immature HIV-1 capsids. Nature 415:88–92 [DOI] [PubMed] [Google Scholar]