Abstract
Vaccinia virus (VACV) K1L and C7L function equivalently in many mammalian cells to support VACV replication and antagonize antiviral activities induced by type I interferons (IFNs). While K1L is limited to orthopoxviruses, genes that are homologous to C7L are found in diverse mammalian poxviruses. In this study, we showed that the C7L homologues from sheeppox virus and swinepox virus could rescue the replication defect of a VACV mutant deleted of both K1L and C7L (vK1L−C7L−). Interestingly, the sheeppox virus C7L homologue could rescue the replication of vK1L−C7L− in human HeLa cells but not in murine 3T3 and LA-4 cells, in contrast to all other C7L homologues. Replacing amino acids 134 and 135 of the sheeppox virus C7L homologue, however, made it functional in the two murine cell lines, suggesting that these two residues are critical for antagonizing a putative host restriction factor which has some subtle sequence variation in human and murine cells. Furthermore, the C7L family of host range genes from diverse mammalian poxviruses were all capable of antagonizing type I IFN-induced antiviral activities against VACV. Screening of a library of more than 350 IFN-stimulated genes (ISGs) identified interferon-regulated factor 1 (IRF1) as an inhibitor of vK1L−C7L− but not wild-type VACV. Expression of either K1L or C7L, however, rendered vK1L−C7L− resistant to IRF1-induced antiviral activities. Altogether, our data show that K1L and C7L antagonize IRF1-induced antiviral activities and that the host modulation function of C7L is evolutionally conserved in all poxviruses that can readily replicate in tissue-cultured mammalian cells.
INTRODUCTION
Poxviruses are a family of complex DNA viruses that replicate entirely in the cytoplasm and dedicate a substantial amount of their genome coding capacity for modulating host immune responses (16). Based on differences in genetics and host range, poxvirus family members that infect mammalian hosts are classified into seven genera: Orthopoxvirus, Parapoxvirus, Capripoxvirus, Leporipoxvirus, Suipoxvirus, Molluscipoxvirus, and Yatapoxvirus (16). Vaccinia virus (VACV), a member of the Orthopoxvirus genus, serves as the vaccine for smallpox and is the prototypical poxvirus. VACV has a broad host range in vivo and in cell lines, as it is capable of entering nearly all cell types examined to date and initiating the synthesis of the early set of viral proteins (11). However, whether viral replication proceeds to completion or not depends on the host cell type and often requires virus-carried host range genes, including K1L and C7L (11). VACV requires either K1L or C7L for productive replication in most mammalian cells (3, 17), but it requires neither for replication in avian cells and in a few specific mammalian cell lines, such as human hepatoma Huh7 cells (13). When Huh7 cells are pretreated with type I interferon (IFN), however, K1L or C7L is required for replication beyond early gene expression (13), suggesting that K1L and C7L support VACV replication by antagonizing some antiviral factors that are expressed constitutively in most mammalian cells but are induced by IFN only in some cell lines, such as Huh7 (13).
K1L and C7L function equivalently in supporting VACV replication in most mammalian cells, but they share no sequence similarity. The K1 protein has 284 amino acids and is comprised entirely of ankyrin repeats (8), a protein motif that is involved in protein-ligand interaction (19). In contrast, the C7 protein has 150 amino acids and no discernible sequence motif. K1L is present only in orthopoxviruses, but homologues that are 20 to 30% identical to C7L at the amino acid level are found in five of the seven genera of mammalian poxviruses (12). The only exceptions are molluscipoxviruses and parapoxviruses, which have no or very limited capacity for replicating in mammalian cells in tissue culture. Previously, we showed that both myxoma virus (MYXV) (a leporipoxvirus that infects rabbits) and Yaba-like disease virus (YLDV) (a yatapoxvirus that infects monkeys) carry C7L homologues that function equivalently to C7L in supporting VACV replication in human and murine cells (12). To complete the studies of C7L homologues from mammalian poxviruses, we characterized the C7L homologues from a suipoxvirus (infects swine) and a capripoxvirus (infects sheep and goats) in the current study. In addition, we examined the C7L homologues from all mammalian poxviruses for the ability to antagonize IFN. We also screened a large IFN-stimulated gene (ISG) library in an effort to identify the host factor(s) antagonized by K1L and C7L. Our results showed that K1L and C7L antagonize antiviral activities induced by interferon-regulated factor 1 (IRF1) and that the host modulation function of C7L is evolutionally conserved in all poxviruses that are capable of replicating in mammalian tissue-cultured cells.
MATERIALS AND METHODS
Cells and viruses.
Vero (ATCC CCL-81) cells were cultured in minimum essential medium with Earle's balanced salts (Invitrogen) supplemented with 10% fetal bovine serum (FBS). HeLa, Huh7, NIH 3T3, and LA-4 (ATCC CCL196; obtained from Peter Dube) cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) with 10% FBS. P815 cells (ATCC TIB-64; obtained from Ellen Kraig) were cultured in RPMI 1640 with 10% FBS. All vaccinia viruses were propagated on Vero cells.
Construction of recombinant vaccinia viruses.
vK1L−C7L−/GFP+, WT-GFP+, vK1L−C7L−, K1L-rev, C7L-rev, vVACV-C7L, vYLDV-67R, vMYXV-M62R, vMYXV-M63R, vMYXV-M64R, and vCPXV-020 were described previously (12–14).
vSPPV-063 and vSWPX-064 were derived from vK1L−C7L− through homologous recombination with specific transfer plasmids as described previously (12) (Fig. 1). Each transfer plasmid contained (i) 300 bp of the downstream flanking region of C7L, (ii) the green fluorescent protein (GFP) gene under the control of the VACV late promoter P11, (iii) sheeppox virus (SPPV) open reading frame (ORF) 063 or swinepox virus (SWPV) ORF 064 with a C-terminal V5 tag, and (iv) 300 bp of the upstream flanking region of C7L, including the C7L promoter. The plasmids were assembled together through recombinant PCR as described previously (12). Sheeppox virus ORF 063 or swinepox virus ORF 064 was PCR amplified from genomic DNA of a sheeppox virus isolated from Nigeria or swinepox virus strain Kasza by use of specific primers (sheeppox 063F, AATAAGTAAAATGGGTATCAGACACGAG; sheeppox 063R, GTTTATCCATAGATACTGCTTCCAT; swinepox 064F, GTCGTTCATGTTACTTAACCTGAAA; and swinepox 064R, GGTTTATCTATTGACAATGGTTCCA). The ORF was placed immediately downstream of the C7L promoter.
Fig 1.
Schematic diagram of the experimental design for testing the functionalities of C7L homologues from different mammalian poxviruses. vK1L−C7L−, a previously described VACV mutant with deletions in both K1L and C7L (14), was used as the parental virus for constructing VACV with K1L deleted and C7L replaced with a C7L homologue. Homologous recombination of vK1L−C7L− with a plasmid containing the C7L homologue and the GFP gene resulted in recombinant viruses that expressed C7L homologues under the control of the C7L promoter. GFP expression, under the control of the VACV late promoter P11, served as an indicator for viral late protein synthesis. The functionality of the C7L homologues was determined by testing the replication capacity of the viruses in representative human and murine cell lines and by testing the IFN sensitivity of the viruses in specific cell lines that were otherwise permissive for the replication of the viruses.
To make viruses expressing chimeras of SPPV 063 and SWPV 064 or mutants of SPPV 063 or VACV C7L, the relevant chimeras or mutants were constructed by recombinant PCR and subcloned into the above-mentioned transfer plasmids after restriction enzyme digestions. The viruses were then derived from vK1L−C7L− through homologous recombination with the transfer plasmids.
Growth curve analysis.
Cells in 12-well plates were incubated with 0.5 or 5 PFU of virus per cell for 2 h at room temperature. Following adsorption, the cells were washed twice with phosphate-buffered saline (PBS) and moved to a 37°C incubator to initiate viral entry and replication. The cells were harvested at 0, 24, and 48 h postinfection (hpi). The viral titers in the cell lysates were determined by duplicate plaque assays on Vero cells.
Western blot analysis.
Cells were infected with VACV at a multiplicity of infection (MOI) of 5 for 8 h. The cells were then lysed for Western blot analysis as described previously (14). The detection antibodies were mouse monoclonal antibodies (MAbs) against V5 (Sigma-Aldrich), Hsp70 (Santa Cruz), and WR148 (15) and rabbit polyclonal antibodies against VACV D12 (1).
Interferon sensitivity assays.
The sensitivity of the viruses to human IFN-β was tested with Huh7 cells as described previously (13). Briefly, Huh7 cells were treated with 200 U/ml of human IFN-β (PBL Biomedical Laboratories) for 24 h and then infected with wild-type (WT) or mutant VACV at an MOI of 5. At 1 hpi, the medium was removed and fresh medium containing no IFN was added. At 24 hpi, the cells were harvested and the viral titers were determined by duplicate plaque assays on Vero cells. The sensitivity of the viruses to murine IFN-β was tested with P815 cells as described above, except that the cells were treated with 200 U/ml of murine IFN-β (PBL Biomedical Laboratories) and the expression of GFP under the control of the VACV late promoter was measured as follows. At 24 hpi, the cells were washed with PBS, resuspended in 1 ml of PBS, pelleted, and resuspended in 600 μl of PBS. Two hundred microliters of cells was added to a well of a black opaque 96-well assay plate (Costar). Fluorescence intensity from GFP was measured with a Synergy 2 plate reader (Biotek). The measurement was made from the top of the wells with an excitation filter of 485/20 nm, an emission filter of 528/20 nm, and a 510-nm dichroic mirror. The background value obtained with uninfected cells was subtracted from the readouts.
Screening of ISGs that inhibit VACV replication.
The generation of vesicular stomatitis virus glycoprotein (VSV-G)-pseudotyped lentiviral pseudoparticles coexpressing an ISG and TagRFP and lentiviral transduction of Huh7 cells were performed as described previously (18). Briefly, cells were transduced with pseudoparticles in 24-well plates and split 1:2 at 48 h posttransduction. At 72 h, transduced cells were infected with either vK1L−C7L−/GFP+ or WT-GFP+ at a dose that would yield approximately 50% infected cells in control populations expressing firefly luciferase (Fluc) instead of an ISG. At 8 hpi, the infected cells were collected into 96-well plates and processed for fluorescence-activated cell sorter (FACS) analysis as described previously (18). Briefly, the cells were fixed with 1% paraformaldehyde fixation solution for 1 h, pelleted, resuspended in cold 1× PBS containing 3% FBS, and stored at 4°C until FACS analysis was carried out. Samples were analyzed with an LSRII-HTS flow cytometer (BD Biosciences) equipped with a 561-nm laser for detection of TagRFP. Data were analyzed using FlowJo software (Treestar) with a 0.1% compensation matrix. Virus replication was determined by the percentage of GFP-positive cells in the red fluorescent protein (RFP)-positive population. Replication levels in Fluc-expressing cells were used as a non-ISG control to normalize the data.
Testing the effects of IRFs on VACV replication.
The expression plasmid for IRF7 was originally from Luwen Zhang (22). Expression plasmids for all other IRFs were constructed by PCR amplifying the IRFs from cDNAs (obtained from Openbiosystem or from Ashok Aiyar) and cloning the PCR fragments between NheI and HindIII sites of pcDNA3.1. The IRF ORF in the plasmid was confirmed by sequencing. Huh7 cells grown in 12-well culture plates were transfected with 1 μg of pYW31 (14), which contains GFP under the control of the VACV late promoter P11, and 1 μg of either pcDNA3.1 or a mammalian expression plasmid for an IRF. Forty-eight hours after transfection, one set of cells were harvested for Western blot analysis using antibodies against IRF1 (SC-7453; Santa Cruz), IRF2 (SC-101069; Santa Cruz), IRF3 (2324-1; Epitomics), IRF7 (3384-1; Epitomics), and IRF9 (SC-101070; Santa Cruz). Another set of cells were infected with WT or mutant VACV at an MOI of 0.1 or 1 PFU/cell. At 24 hpi, the infected cells expressing GFP were visualized with an inverted fluorescence microscope and photographed. The cells were then harvested, and fluorescence intensity from GFP was measured with a Synergy 2 plate reader (Biotek) as described above.
Generation of MEFs.
IRF1−/− and IRF1+/+ mouse embryonic fibroblasts (MEFs) were generated from embryos of IRF1+/− × IRF1+/− (B6.129S2-Irf1tm1Mak/J) (10) mice according to standard protocols. MEFs were propagated in high-glucose DMEM (Gibco) and supplemented with 10% heat-inactivated FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin (Gibco). To generate immortalized IRF1−/− and IRF1+/+ MEFs, cell were lentivirally transduced with the simian virus 40 (SV40) large T antigen. All mice were bred and maintained under specific-pathogen-free conditions at the Laboratory Animal Research Center of the Rockefeller University; all mouse protocols were approved by the Rockefeller University IACUC.
RESULTS
The C7L homologue from swinepox virus is functional in both human and murine cell lines, while the C7L homologue from sheeppox virus is functional in human but not murine cell lines.
Previously, we found that MYXV M062 and YLDV 067 could functionally replace C7L as host range genes for VACV in both human and murine cell lines, while MYXV M063, MYXV M064, and cowpox virus (CPXV) 020 failed to do the same (12). This left the C7L homologues from the Suipoxvirus and Capripoxvirus genera as the only two that were not characterized regarding their function as host range genes. We thus examined whether the C7L homologues from swinepox virus (SWPV 064) and sheeppox virus (SPPV 063) could replace C7L as host range genes for VACV. Similar to what we described previously (12), and as diagramed in Fig. 1, we inserted the ORF for SWPV 064 or SPPV 063, along with a C-terminal V5 epitope tag, into the C7L locus of vK1L−C7L− (14), a VACV mutant deleted of the host range genes K1L and C7L. The recombinant viruses, referred to as vSWPV-064 and vSPPV-063, were isolated on Vero cells, on which neither K1L nor C7L is required for VACV replication. The native C7L promoter controlled the expression of SWPV 064 or SPPV 063, and the expression was confirmed by Western blotting with an antibody against the epitope tag (Fig. 2B).
Fig 2.
Swinepox virus 064 can functionally replace C7L in both human and murine cell lines, while sheeppox virus 063 can functionally replace C7L only in human cells, not in murine cells. (A) Growth curves of recombinant VACVs on the indicated cell lines at an MOI of 0.5 (HeLa) or 5 (3T3 and LA-4) PFU/cell. The recombinant VACV is referred to by the name of the ORF present at the C7L locus. Virus yields at 0, 24, and 48 hpi were determined by plaque assay on permissive Vero cells. (B) vSPPV-063 is defective in viral protein synthesis in nonpermissive murine cells. HeLa or murine LA-4 cells were infected with the indicated viruses at an MOI of 5 PFU/cell. At 8 hpi, the levels of various viral or cellular proteins in the cell lysates were determined by Western blotting with the indicated antibodies. C7 and its homologues were tagged at the C terminus with a V5 epitope. The predicted molecular sizes of the tagged C7 homologues are listed on the right. C7 and D12 are early proteins, while WR148 (6) is a late protein. The host cell protein Hsp70 served as a control for gel loading. The sizes of the molecular markers (in kilodaltons) are shown on the left.
The host range of recombinant viruses in various cell lines was then examined by plaque assay (not shown) and growth curve analysis (Fig. 2A). In human HeLa and mouse 3T3 cells, the parental vK1L−C7L− virus failed to replicate, while a recombinant virus expressing VACV C7L, vVACV-C7L (12), replicated productively (Fig. 2A). Similar to vVACV-C7L, vSWPV-064 replicated productively in both HeLa and 3T3 cells, indicating that SWPV 064 could function as a host range gene in these cells. However, vSPPV-063 replicated productively only in HeLa cells and failed to replicate in 3T3 cells (Fig. 2A). This was surprising, as HeLa cells and 3T3 cells do not display differences in permissiveness for any other recombinant viruses expressing a C7L homologue (12). To test whether this difference was due to their derivation from different mammalian species, we then tested the replication of the recombinant viruses on additional murine cell lines. A murine lung epithelial cell line, LA-4, was found to be similar to 3T3 in that it restricted the replication of vK1L−C7L− but permitted the replication of vVACV-C7L. Also similar to 3T3 cells, LA-4 cells allowed the replication of vSWPV-064 but restricted the replication of vSPPV-063 (Fig. 2A), suggesting that the defect of vSPPV-063 is specific to murine cells.
In the nonpermissive murine cells, vSPPV-063 did not express GFP from the late P11 promoter (data not shown), suggesting that its replication was blocked before late protein synthesis. Indeed, WR148, a late protein of VACV, was not detected in LA-4 cells that had been infected with vSPPV-063 for 8 h, while it was expressed in cells that had been infected with either vVACV-C7L or vSWPV-064 (Fig. 2B). Furthermore, reduced amounts of D12, an early protein, and of SPPV 063 were detected in vSPPV-063-infected LA-4 cells, indicating that early protein synthesis was also reduced. This is consistent with recent reports that VACV lacking K1L and C7L failed to maintain the synthesis of early proteins such as E3 in nonpermissive cells (2, 12, 21).
Mapping of SPPV 063 residues responsible for its specific defect in murine cells.
SPPV 063 is unique among all C7L homologues in that it shows a specific defect in murine cells. This is probably due to some subtle difference between SPPV 063 and other C7L homologues at residues that interact with the putative host restriction factor. The human and murine versions of the host restriction factor, in turn, may have some subtle difference at residues that interact with the C7 family of viral proteins. Thus, in an effort to gain some insight into the host factor interacting site in the C7 family of viral proteins, we went on to map SPPV 063 residues that are responsible for its specific defect in murine cells. We took the approach of creating chimeras between SPPV 063 and SWPV 064, the latter of which is the closest homologue of SPPV 063 (47% amino acid identity) but is functional in both human and murine cells.
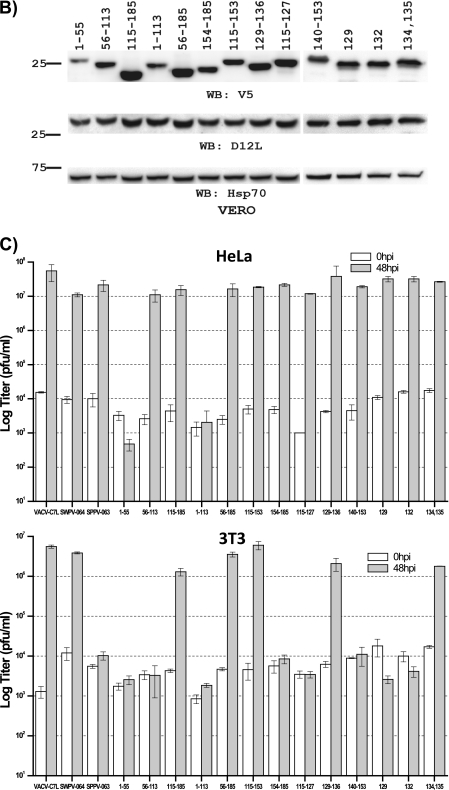
SPPV 063 was initially divided into three segments, each of which was individually replaced with the corresponding region of SWPV 064 (Fig. 3A). Recombinant viruses expressing the chimeras were constructed on Vero cells with vK1L−C7L− as the parental virus, and the replication capabilities of the viruses were assessed with HeLa and 3T3 cells (Fig. 3B and C). The replacement of the C-terminal one-third of SPPV 063 was sufficient to make it functional in both human and murine cells, as viruses expressing chimeras containing either the C-terminal one-third or C-terminal two-thirds of SPPV 063 were able to replicate in both HeLa and 3T3 cells (constructs 115-185 and 56-185 in Fig. 3). In contrast, replacement of the central one-third of SPPV 063 did not alter its functionality in human or murine cells (construct 56-113 in Fig. 3), while replacement of the N-terminal one-third or two-thirds of SPPV 063 disrupted its function in both human and murine cells (constructs 1-55 and 1-113 in Fig. 3). Since the latter two substitutions resulted in proteins that were defective in both human and murine cells, we then assessed whether the substitutions might affect protein folding, and consequently make the protein unstable, by comparing the levels of the chimeric proteins in Vero cells. In contrast to HeLa and 3T3 cells, Vero cells were permissive for all of the mutants. As a result, there was no difference in the replication capacities of the mutants in Vero cells that would have affected overall early protein expression levels. Therefore, any differences in chimeric protein levels would reflect a difference in the stability of the proteins. Similar levels of the viral early protein D12 were detected in Vero cells infected by all of the mutants, but reduced levels of chimeric protein were detected in Vero cells infected by viruses encoding SPPV 063 with substitutions at residues 1 to 55 or 1 to 113 (Fig. 3B), suggesting that these two substitutions affected protein folding and made the protein unstable.
Fig 3.
Mapping of SPPV 063 residues responsible for its specific defect in murine cells. (A) (Top) Schematic diagrams of SWPV 064 (white boxes) and SPPV 063 (shaded boxes) chimeras. (Bottom) Substitution mutations in SPPV 063 shown in the context of an amino acid sequence alignment of SWPV 064 and SPPV 063. The mutations are referred to by the positions of the SPPV 063 amino acids that are replaced. The phenotypes of the mutant viruses expressing these SPPV 063 mutations are summarized on the right. (B) Levels of mutated SPPV 063 proteins in Vero cells infected with mutant viruses for 8 h. The mutant viruses are referred to by the positions of the SPPV 063 mutations. Infections and Western blotting were performed as described in the legend to Fig. 2B. Expression levels of the VACV early protein D12L and the host protein Hsp70 were determined as controls for equal infection and gel loading, respectively. (C) Growth analysis of mutant viruses in HeLa cells and 3T3 cells. Cells were infected by the indicated mutant viruses at an MOI of 5 PFU/cell. Virus titers at 0 and 48 hpi were determined by plaque assay on permissive Vero cells.
Since replacement of the C-terminal one-third (residues 115 to 185) of SPPV 063 was sufficient to make it functional in both human and murine cells, we then further divided this section of SPPV 063 into two segments for substitutions. The replacement of residues 115 to 153 was sufficient to make the protein functional in both human and murine cells, while the replacement of residues 154 to 185 did not alter the functionality of the protein (Fig. 3). Further dividing the region of residues 115 to 153 into three segments for substitution revealed that the replacement of eight amino acids (residues 129 to 136) was sufficient to make the protein functional as a host range factor in both human and murine cells. Among the eight amino acids, only four are different between SPPV 063 and SWPV 064. Replacing Asn134 and Phe135 together was sufficient to make the protein functional in human and murine cells, while replacing either Arg129 or Cys132 had no effect on the function of the protein.
Altogether, the data showed that residues 134 and 135 were largely responsible for the specific defect of SPPV 063 in murine cells, suggesting that these two residues are critical for the function of the C7 family of host range proteins. VACV C7 has identical residues (Tyr135 and Ile136) to those of SWPV 064 at positions corresponding to residues 134 and 135 of SPPV 063. To assess whether these two residues are also critical for the function of C7, we generated a mutant of vVACV-C7L in which these two residues were replaced with alanines. The mutant replicated productively in Vero cells, but it failed to replicate in HeLa or 3T3 cells (see Fig. S1 in the supplemental material), indicating that Tyr135 and Ile136 are critical for the function of C7. A lower level of C7 protein, however, was detected in the permissive Vero cells, suggesting that Tyr135 and Ile136 may also play a structural role in C7.
The ability of C7L homologues to antagonize type I IFN correlates with their host range function.
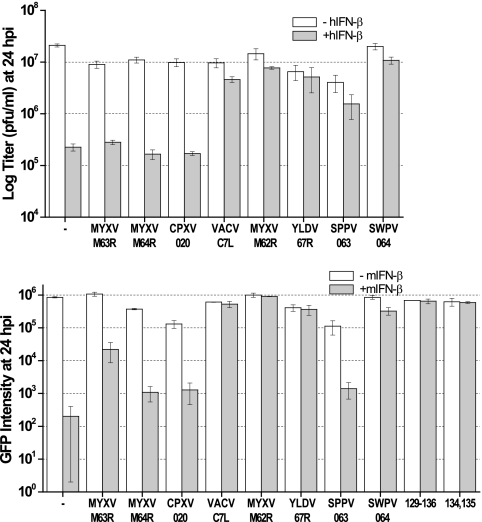
We previously showed that both K1L and C7L antagonize antiviral activities induced by type I IFN in Huh7 cells (13). However, it has not been determined whether C7L homologues from other mammalian poxviruses can also antagonize type I IFN activities. We thus tested the IFN sensitivity of a panel of recombinant VACVs that expressed different C7L homologues. All of the recombinant VACVs replicated productively in Huh7 cells (data not shown). As we showed previously, pretreating Huh7 cells with human IFN-β reduced the 24-h yield of vK1L−C7L− >10-fold, while only slightly reducing the yield of VACV-C7L (Fig. 4). Similar to vK1L−C7L−, the yields of recombinant viruses expressing MYXV M063, MYXV M064, or CPXV 020 were also reduced >10-fold with IFN-β pretreatment (Fig. 4). Similar to vVACV-C7L, the yields of viruses expressing MYXV M062, YLDV 67R, SPPV 063, or SWPV 064 were reduced only slightly by IFN-β. These viruses are also the ones that replicate productively in untreated human HeLa cells.
Fig 4.
Ability of C7L homologues to antagonize type I IFN correlates with their host range function. (Top) Huh7 cells were left untreated (−hIFN-β) or treated (+hIFN-β) with 200 U/ml of human IFN-β for 24 h and then infected with VACVs expressing different C7L homologues or the parental vK1L−C7L− virus (indicated by a dash) at an MOI of 5. Viral yields at 24 hpi were determined by plaque assay. (Bottom) Murine P815 cells were left untreated (−mIFN-β) or treated (+mIFN-β) with 200 U/ml of murine IFN-β for 24 h and then infected as described above. The level of GFP (arbitrary units) expressed by the virus at 24 hpi was measured with a fluorescence plate reader. GFP expression, which was under the control of the VACV late promoter P11, indicates viral late protein synthesis.
To test whether the recombinant viruses were sensitive to murine type I IFN, we first identified a murine cell line that is permissive for the replication of vK1L−C7L−. P815, a murine lymphoblast-like mastocytoma cell line, was found to support a >10-fold amplification in titer of vK1L−C7L− over 24 h of infection (data not shown). Pretreating P815 cells with murine IFN-β, however, blocked the replication of vK1L−C7L− before late protein synthesis, as indicated by a 2- to 3-log reduction in the intensity of GFP, which was expressed by the virus under the control of the P11 late promoter (Fig. 4). The replication of vMYXV-M63R, vMYXV-M64R, and vCPXV-020 was sensitive to murine IFN-β, as the GFP intensity in infected cells was reduced by approximately 2 log by murine IFN-β. The replication of vSPPV-063 was also sensitive to murine IFN-β, even though its replication was not sensitive to human IFN-β. This correlates with the specific defect of SPPV 063 as a host range factor in murine cells but not in human cells. All recombinant viruses that were able to replicate productively in 3T3 cells, including some substitution mutants of vSPPV-063, were insensitive to murine IFN-β (Fig. 4 and data not shown).
Altogether, the studies with human and murine IFN-β show a very consistent correlation between the replication capacities of the viruses in human and murine cell lines and their sensitivities to IFN-β in permissive cells.
Screening of ISGs that inhibit the replication of vK1L−C7L−.
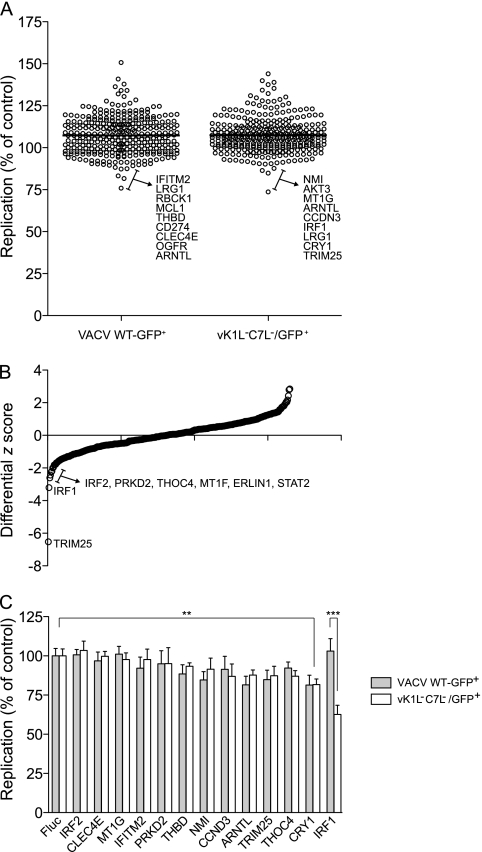
Our previous work suggested that K1L and C7L function by antagonizing products of ISGs (13). To find out which ISG product is the target of K1L and C7L, we screened an expression library consisting of over 350 ISGs for inhibitors of vK1L−C7L−. Similar to what was described recently (18), Huh7 cells were first transduced with a bicistronic lentiviral vector coexpressing an ISG and the red fluorescent protein TagRFP. Seventy-two hours later, the cells were infected with either VACV WT-GFP+ or vK1L−C7L−/GFP+, both of which encode GFP under the control of a VACV late promoter. Eight hours after VACV infection, viral replication (indicated by GFP expression) in lentivirus-transduced cells (indicated by TagRFP expression) was quantified by fluorescence-activated cell sorting. Viral replication in cells overexpressing an ISG (% GFP-positive cells in the RFP-positive population) was normalized to that of a control population overexpressing Fluc, and the results are presented as dot plots (Fig. 5A). Compared to previously reported ISG screens against a panel of positive-strand RNA viruses (18), both WT-GFP+ and vK1L−C7L−/GFP+ VACVs were remarkably resistant to ISG-mediated inhibition. Viral replication in the presence of most ISGs was similar to or higher than replication in control cells expressing Fluc (Fig. 5A; see Table S1 in the supplemental material). To determine if any ISGs preferentially inhibit vK1L−C7L−/GFP+, differential z scores from both screens were calculated, and several ISGs appeared to inhibit vK1L−C7L−/GFP+ more than WT-GFP+ (Fig. 5B). Select ISGs that targeted either virus or showed differential inhibition between the two viruses were rescreened by measuring the inhibition in four replicates, using fresh lentiviral stocks (Fig. 5C). IRF1 was the only ISG that was confirmed to significantly inhibit the replication of vK1L−C7L−/GFP+ but not WT-GFP+. Replication of both viruses was modestly inhibited by cryptochrome 1 (CRY1) (P < 0.01). We did not detect any ISGs that preferentially inhibited WT-GFP+ VACV over vK1L−C7L−/GFP+ VACV.
Fig 5.
ISG screens against WT-GFP+ and vK1L−C7L−/GFP+ VACVs. (A) Huh7 cells were transduced with lentiviral vectors coexpressing an ISG and TagRFP. ISG-expressing cells were infected with the indicated GFP-expressing virus, and viral replication in the RFP+ population was monitored by two-color FACS. Data are presented as dot plots, with replication levels normalized to the Fluc control. A horizontal black line indicates the population mean. (B) The z scores from the screening data in panel A were determined, and differential z scores were plotted to show genes that preferentially inhibited vK1L−C7L−/GFP+. (C) A subset of ISGs were tested in confirmation assays using independent lentiviral preparations. Replication levels were normalized to the Fluc control (n = 4). Statistical significance was determined by one-way analysis of variance (ANOVA) (***, P < 0.001; **, P < 0.01).
Both K1L and C7L antagonize IRF1-mediated antiviral effects.
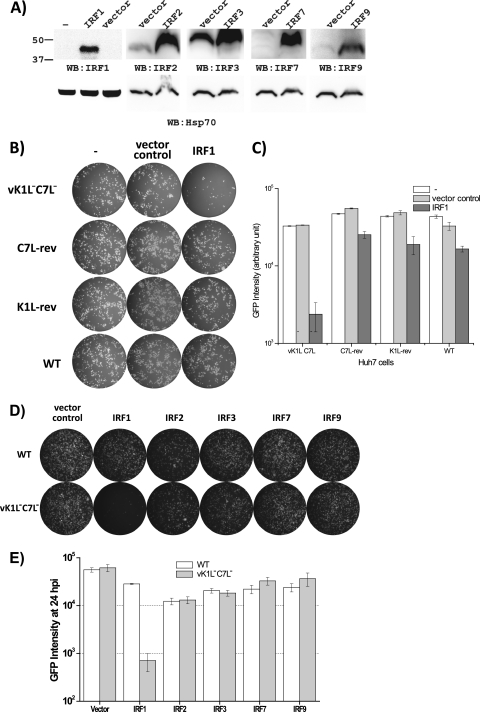
To find out whether K1L or C7L antagonizes IRF1-mediated antiviral activities, we examined the effects of IRF1 overexpression on the replication of two vK1L−C7L− revertants (K1L-rev and C7L-rev [13]) which had either the C7L or K1L gene added back to the mutant. Huh7 cells were transiently transfected with a mammalian expression vector for IRF1 together with a reporter plasmid encoding GFP under the control of a VACV late promoter. While endogenous IRF1 proteins in Huh7 cells were below the level of detection by an anti-IRF1 antibody in Western blots, transfection of the IRF1 plasmid resulted in a detectable level of IRF1 protein 48 h after transfection (Fig. 6A), when the cells were infected with WT or mutant VACV. At 24 h postinfection, GFP expression, which could occur only in cells that were both transfected and infected, was assessed qualitatively by fluorescence microscopy (Fig. 6B) and quantitatively by a fluorescence plate reader (Fig. 6C). While IRF1 greatly reduced GFP expression in vK1L−C7L−-infected cells, it only slightly reduced GFP expression in cells infected with WT VACV, K1L-rev, or C7L-rev, indicating that both K1L and C7L could antagonize IRF1-mediated antiviral activities.
Fig 6.
Both K1L and C7L antagonize IRF1-mediated antiviral effects. Huh7 cells were transfected with a reporter plasmid containing GFP under the control of the VACV P11 promoter by itself (−) or with the pcDNA3.1 vector (vector control) or an IRF expression plasmid based on pcDNA3.1 (IRF). (A) Forty-eight hours after transfection, Western blotting was performed on one set of cells, using the indicated antibodies. Another set of cells were infected with the indicated viruses at an MOI of 0.1. WT, wild-type VACV WR; K1L-rev and C7L-rev, revertant vK1L−C7L− viruses with restored K1L and C7L, respectively (12, 14). (B and D) At 24 hpi, GFP-expressing cells were photographed under a fluorescence microscope and are shown as white cells. (C and E) The intensity of GFP was also measured with a fluorescence plate reader and is shown in arbitrary units.
In addition to IRF1, the IRF2, IRF7, and IRF9 factors were also included in the initial large-scale ISG screen, but they did not show any specific inhibition of vK1L−C7L−. IRF3, a transcription factor associated with the induction of IFN, was not part of the initial screen, as it is not an ISG. To confirm the specificity of IRF1 in the inhibition of vK1L−C7L−, the effects of these IRFs on the replication of vK1L−C7L− were also examined. In contrast to IRF1, all other tested IRFs (IRF2, IRF3, IRF7, and IRF9) did not significantly reduce GFP expression in either WT VACV- or vK1L−C7L−-infected Huh7 cells (Fig. 6D and E), demonstrating the specificity of IRF1.
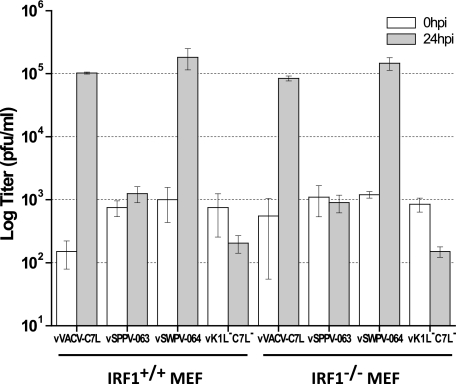
To determine whether IRF1 inhibition could be the mechanism by which the C7L family supports viral replication, we tested whether a functional C7L gene was required for VACV replication in IRF1−/− MEFs. While vVACV-C7L and vSWPV-064 replicated productively in MEFs, as measured by viral yields at 24 hpi, vK1L−C7L− and vSPPV-063 failed to replicate in IRF1+/+ or IRF1−/− MEFs (Fig. 7), indicating that a functional C7L gene is also required for VACV replication in cells deficient of IRF1. Similar results were also observed in IRF1+/+ and IRF1−/− MEFs that were immortalized with SV40 T antigen (data not shown). These data suggest that the C7L family functions not by directly inhibiting IRF1 but by inhibiting some IRF1-inducible factors that are nevertheless present at sufficient levels in MEFs even in the absence of IRF1.
Fig 7.
Functional C7L is required for VACV replication in cells deficient of IRF1. IRF1−/− and IRF1+/+ MEFs were infected with the indicated mutant viruses at an MOI of 0.5 PFU/cell. Virus titers at 0 and 24 hpi were determined by plaque assay on permissive Vero cells.
DISCUSSION
Among the poxvirus host range genes, C7L is one of the most conserved, with homologues present in five of the seven mammalian poxvirus genera (12). Previously, we found that the Yatapoxvirus and Leporipoxvirus genera both carry C7L homologues that function equivalently to C7L in supporting VACV replication in mammalian cells, suggesting that the C7L family of host range genes is essential for poxviruses to replicate in mammalian hosts (12). In the current study, we extended this idea further by demonstrating that the C7L homologues from two additional poxvirus genera, Capripoxvirus and Suipoxvirus, can also replace C7L to support VACV replication in mammalian cells. Thus, a host range gene that functions similarly to VACV C7L has been identified in all mammalian poxviruses except for molluscum contagiosum virus (MCV) and parapoxviruses. Both MCV and parapoxviruses have limited host ranges in nature and in cultured cells. MCV does not replicate in any tissue-cultured cells, while parapoxviruses replicate in only a very limited number of mammalian cell lines, such as Vero cells (4). Since the C7L family of host range genes are not required for mammalian poxviruses to replicate in avian cells and are absent in avian poxviruses, we postulate that acquiring a C7L-like host range gene is an important adaptation of mammalian poxviruses for replication in mammalian hosts. A recent study of MYXV M062 also illustrated the importance of the C7L family for poxvirus replication. The deletion of M062 from MYXV greatly reduced the host range of MYXV, resulting in a mutant that replicates abortively in its natural rabbit host as well as in many human cancer cell lines (9).
In this study, all C7L family members were studied in the context of VACV infection of human and murine cells. In this context, MYXV M062, YLDV 67R, and SWPX 064 functioned equivalently to VACV C7L, suggesting that these viral proteins target a host factor that is evolutionally conserved not only in natural hosts of these poxviruses but also in humans and mice. We were thus surprised to find that SPPV 063 failed to function in murine cells, even though it functioned in human cells and presumably in sheep cells, the natural host for SPPV. Recombinant VACV expressing SPPV 063 replicated productively in human HeLa cells and human IFN-treated Huh7 cells, but it failed to replicate in murine 3T3 or LA-4 cells, mouse embryo fibroblasts, or murine IFN-treated P815 cells. This suggests that a subtle sequence difference exists between the human and murine versions of the putative host restriction factor. The difference is substantial enough that the murine factor cannot be antagonized effectively by SPPV 063, but the difference overall is subtle enough that both the human and murine factors can be antagonized effectively by most other C7L family members. Indeed, simply replacing two residues (residues 134 and 135) of SPPV 063 is sufficient to make the protein functional in both human and murine cells. We postulate that the native sequence of SPPV 063 at residues 134 and 135 affects the binding interface with the putative host restriction factor and makes the binding interface ineffective or incompatible for binding to the murine factor. Replacing these two residues with their counterparts in SWPV 064 does not appear to affect protein structure, as the substitution did not affect the stability of the protein in Vero cells or the function of the protein in HeLa cells. Thus, the gain of function in murine cells resulting from the substitution is probably due to a subtle change in the binding interface that also makes it competent for binding to the murine factor. It is currently unclear whether these two residues directly participate in binding or somewhat indirectly affect the binding interface. Alanine substitutions of the corresponding residues in VACV C7 disrupted the function of C7 in both human and murine cells, suggesting that they are also critical for the function of C7. However, since the substitutions resulted in a lower C7 protein level in Vero cells, we cannot exclude the possibility that the alanine substitutions disrupted C7 function at least partly by affecting protein structure.
While it has become clear that the C7L family is critical for poxvirus replication in mammalian cells, very little is known about the underlying mechanism for this critical function of C7L. Presumably, the C7L family antagonizes a mammalian cell factor that otherwise restricts poxvirus replication. Previously, we showed that both K1L and C7L antagonize antiviral activities induced by type I IFN and suggested that the putative host restriction factor is the product of an ISG (13). In the current study, we found that C7L homologues from four additional mammalian poxvirus genera all antagonize IFN-induced antiviral activities. There is a perfect correlation between the ability of a C7L homologue to antagonize IFN activities in Huh7 cells and its ability to support VACV replication in HeLa cells, lending further credence to the idea that the putative host restriction factor in HeLa cells is the same as the IFN-induced antiviral factor in Huh7 cells. We thus undertook a screen of a large ISG library and identified IRF1, a transcription factor in the IFN pathway, as an ISG that selectively inhibits the replication of vK1L−C7L−. We further showed that both K1L and C7L could antagonize IRF1-induced antiviral activities. However, IRF1 itself does not appear to be the target of C7L, as VACV replication in IRF1−/− mouse embryo fibroblasts also requires a functional C7L gene. In a similar screen of ISGs against RNA viruses, IRF1 was identified as one of the most broadly acting IFN effectors (18). It inhibited all viruses against which it was tested, even in STAT1−/− cells. Microarray analyses of Huh7 and STAT1−/− cells showed that IRF1 induces the expression of many ISGs but no type I IFN genes (18), suggesting that the effector mechanisms of IRF1 do not require IFN activity but partly overlap with those of IFN. The fact that IRF1 inhibits the replication of vK1L−C7L− but not WT VACV supports the hypothesis that K1L and C7L support VACV replication in mammalian cells by antagonizing an antiviral factor in the IFN pathway. Interestingly, MYXV M062 was recently found to interact with human sterile motif domain containing 9 (SAMD9), and knocking down SAMD9 in some human cancer cells partially rescued the replication of the M062 deletion mutant (9). SAMD9 is a protein that is expressed ubiquitously in various tissues, and its expression can be upregulated further by IFNs (5, 20). However, SAMD9 is not expressed in cells of mouse origin (7), and neither VACV C7L nor YLDV 67R interacts with SAMD9 (9). Therefore, although most C7L family members function equivalently in the context of VACV infection of human and murine cells, the mechanism of action might still be different for different C7L family members, and at least some C7L family members might target very different host factors in cells of different species. To identify the host factor that is targeted directly by VACV C7L, we will have to screen additional ISGs, especially the ones induced by both IFN and IRF1, for factors that can restrict the replication of vK1L−C7L− in Huh7 cells.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants AI079217 (Y.X.) and AI057158 (C.M.R.; Northeast Biodefense Center-Lipkin). Additional funding was provided by the Greenberg Medical Research Institute, the Starr Foundation, and the Ronald A. Shellow, M.D., Memorial Fund (C.M.R.). J.S. was supported by National Research Service award DK082155 from the National Institute of Diabetes and Digestive and Kidney Diseases.
Footnotes
Published ahead of print 15 February 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Arsenio J, Deschambault Y, Cao J. 2008. Antagonizing activity of vaccinia virus E3L against human interferons in Huh7 cells. Virology 377:124–132 [DOI] [PubMed] [Google Scholar]
- 2. Backes S, et al. 2010. Viral host-range factor C7 or K1 is essential for modified vaccinia virus Ankara late gene expression in human and murine cells, irrespective of their capacity to inhibit protein kinase R-mediated phosphorylation of eukaryotic translation initiation factor 2alpha. J. Gen. Virol. 91:470–482 [DOI] [PubMed] [Google Scholar]
- 3. Beattie E, et al. 1996. Host-range restriction of vaccinia virus E3L-specific deletion mutants. Virus Genes 12:89–94 [DOI] [PubMed] [Google Scholar]
- 4. Fischer T, Planz O, Stitz L, Rziha H-J. 2003. Novel recombinant parapoxvirus vectors induce protective humoral and cellular immunity against lethal herpesvirus challenge infection in mice. J. Virol. 77:9312–9323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hershkovitz D, et al. 2011. Functional characterization of SAMD9, a protein deficient in normophosphatemic familial tumoral calcinosis. J. Invest. Dermatol. 131:662–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jones-Trower A, et al. 2005. Identification and preliminary characterization of vaccinia virus (Dryvax) antigens recognized by vaccinia immune globulin. Virology 343:128–140 [DOI] [PubMed] [Google Scholar]
- 7. Li CF, et al. 2007. Human sterile alpha motif domain 9, a novel gene identified as down-regulated in aggressive fibromatosis, is absent in the mouse. BMC Genomics 8:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li Y, Meng X, Xiang Y, Deng J. 2010. Structure-function studies of vaccinia virus host range protein k1 reveal a novel functional surface for ankyrin repeat proteins. J. Virol. 84:3331–3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu J, Wennier S, Zhang L, McFadden G. 2011. M062 is a host range factor essential for myxoma virus pathogenesis and functions as an antagonist of host SAMD9 in human cells. J. Virol. 85:3270–3282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Matsuyama T, et al. 1993. Targeted disruption of IRF-1 or IRF-2 results in abnormal type I IFN gene induction and aberrant lymphocyte development. Cell 75:83–97 [PubMed] [Google Scholar]
- 11. McFadden G. 2005. Poxvirus tropism. Nat. Rev. Microbiol. 3:201–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Meng X, Chao J, Xiang Y. 2008. Identification from diverse mammalian poxviruses of host-range regulatory genes functioning equivalently to vaccinia virus C7L. Virology 372:372–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Meng X, et al. 2009. Vaccinia virus K1L and C7L inhibit antiviral activities induced by type I interferons. J. Virol. 83:10627–10636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Meng X, Xiang Y. 2006. Vaccinia virus K1L protein supports viral replication in human and rabbit cells through a cell-type-specific set of its ankyrin repeat residues that are distinct from its binding site for ACAP2. Virology 353:220–233 [DOI] [PubMed] [Google Scholar]
- 15. Meng X, et al. 2011. Generation and characterization of a large panel of murine monoclonal antibodies against vaccinia virus. Virology 409:271–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Moss B. 2007. Poxviridae: the viruses and their replication, p 2905–2946 In Knipe DM, et al. (ed), Fields virology, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 17. Perkus ME, et al. 1990. Vaccinia virus host range genes. Virology 179:276–286 [DOI] [PubMed] [Google Scholar]
- 18. Schoggins JW, et al. 2011. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472:481–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sedgwick SG, Smerdon SJ. 1999. The ankyrin repeat: a diversity of interactions on a common structural framework. Trends Biochem. Sci. 24:311–316 [DOI] [PubMed] [Google Scholar]
- 20. Tanaka M, Shimbo T, Kikuchi Y, Matsuda M, Kaneda Y. 2010. Sterile alpha motif containing domain 9 is involved in death signaling of malignant glioma treated with inactivated Sendai virus particle (HVJ-E) or type I interferon. Int. J. Cancer 126:1982–1991 [DOI] [PubMed] [Google Scholar]
- 21. Willis KL, Patel S, Xiang Y, Shisler JL. 2009. The effect of the vaccinia K1 protein on the PKR-eIF2alpha pathway in RK13 and HeLa cells. Virology 394:73–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang L, Pagano JS. 1997. IRF-7, a new interferon regulatory factor associated with Epstein-Barr virus latency. Mol. Cell. Biol. 17:5748–5757 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.