Abstract
After initiation of antiretroviral therapy (ART), HIV loads and frequencies of HIV epitope-specific immune responses decrease. A diverse virus-specific T cell receptor (TCR) repertoire allows the host to respond to viral epitope diversity, but the effect of antigen reduction as a result of ART on the TCR repertoire of epitope-specific CD8+ T cell populations has not been well defined. We determined the TCR repertoires of 14 HIV-specific CD8+ T cell responses from 8 HIV-positive individuals before and after initiation of ART. We used multiparameter flow cytometry to measure the distribution of memory T cell subsets and the surface expression of PD-1 on T cell populations and T cell clonotypes within epitope-specific responses from these individuals. Post-ART, we noted decreases in the frequency of circulating epitope-specific T cells (P = 0.02), decreases in the number of T-cell clonotypes found within epitope-specific T cell receptor repertoires (P = 0.024), and an overall reduction in the amino acid diversity within these responses (P < 0.0001). Despite this narrowing of the T cell response to HIV, the overall hierarchy of dominant T cell receptor clonotypes remained stable compared to that pre-ART. CD8+ T cells underwent redistributions in memory phenotypes and a reduction in CD38 and PD-1 expression post-ART. Despite extensive remodeling at the structural and phenotypic levels, PD-1 was expressed at higher levels on dominant clonotypes within epitope-specific responses before and after initiation of ART. These data suggest that the antigen burden may maintain TCR diversity and that dominant clonotypes are sensitive to antigen even after dramatic reductions after initiation of ART.
INTRODUCTION
Successful antiretroviral therapy (ART) reduces viral loads and decreases the level of T cell activation, but the effect of HIV antigen reduction on the T cell receptor (TCR) repertoire of epitope-specific T cell responses remains poorly defined (36). The level of generalized T cell activation as measured by expression of CD38 is a strong, independent predictor of disease progression (16, 28). More recent work has shown a positive correlation between increased expression of PD-1 on T cells and the level of viremia (14, 37). In addition to being an independent risk factor for disease progression in the absence of ART, sustained high levels of immune activation in the presence of ART are associated with poorer levels of CD4+ T cell recovery (18). Other phenotypic markers, such as CD45R0, CCR7, CD27, and CD28, define T cell memory subsets which are altered as a result of HIV infection (1, 5, 12). After the initiation of ART, the distribution of memory T cell subsets improves, indicating broad remodeling of T cell populations with successful treatment and antigen reduction (44). Few studies have evaluated the effect of ART on HIV-specific T cell populations in detail, and even these have not analyzed the epitope-specific TCR repertoire in detail (5, 11, 15, 21, 36, 38, 57).
Virus-specific CD8+ T cell responses are a critical component of the natural immune response to HIV (20, 25, 48). However, quantitative features of T cell responses, such as the frequency or magnitude of HIV-specific T cell responses, do not correlate well with control of viral replication or disease progression (2, 7). On the other hand, qualitative features of T cell responses, such as epitope-specific proliferation (34) and breadth of T cell effector function, have been shown to correlate well with control of viremia (8) and may represent important determinants of disease outcome. Indeed, qualitatively superior, polyfunctional HIV-specific T cells have been shown to emerge after suppression of viremia with ART (42); however, after initiation of ART, the magnitude of HIV epitope-specific immune responses also contracts (11, 21, 35). Clonotypic and amino acid diversity within the epitope-specific TCR repertoire is a qualitative feature of T cell responses that may be associated with control of viremia and selection of immune escape variants (32, 41), but there are limited data to inform our understanding of how the TCR repertoire may be affected by ART.
We hypothesized that the structural composition and clonotypic phenotype of epitope-specific responses may also be altered in individuals undergoing ART. In this study, we compared amino acid and clonotypic diversity within the HIV epitope-specific TCR repertoire before and after initiation of ART. Furthermore, we analyzed memory phenotypes and expression of CD38 and PD-1 on dominant and subdominant clonotypes from these epitope-specific responses. After initiation of ART, we noted changes in memory subset distributions and decreased activation of CD4+ and CD8+ T cell populations. The magnitude of HIV epitope-specific T cell responses decreased after ART, and this was accompanied by a reduction in TCR repertoire diversity as measured by the number of discrete clonotypes found within epitope-specific responses, as well as by measurement of T cell receptor beta variable region (TRBV) CDR3 amino acid diversity. However, dominant T cell clonotypes typically remained dominant even post-ART, and these T cell populations retained a PD-1high phenotype compared to subdominant clonotypes. These findings provide insights into the forces which likely drive the evolution of the TCR repertoire, namely, high levels of persistent antigen exposure may drive TCR repertoire diversity, and even in the setting of sustained reduction in circulating antigens, dominant T cell clonotypes maintain a phenotype consistent with exhaustion.
MATERIALS AND METHODS
Individual cohort and HLA typing.
The study cohort was organized within the Vanderbilt-Meharry CFAR and was comprised of patients recruited through the Comprehensive Care Center (Nashville, TN) who underwent initiation of ART during the course of follow-up. All individuals were typed for HLA class I by the DCI Tissue Typing Laboratory (Nashville, TN). This study was approved by the Institutional Review Board at Vanderbilt University, and all participating individuals provided written informed consent.
T cell sorting and TCR sequencing.
Epitope-specific T cells were labeled with appropriate major histocompatibility complex class I (MHC-I) tetramers and sorted by fluorescence-activated cell sorting (FACS) to >95% purity on a FACSAria cell sorter (BD). TCR repertoire sequencing was carried out as described previously (30). Briefly, total RNA was isolated from sorted class I tetramer-specific cells with RNA-STAT-60 (Teltest Inc., TX). Anchored reverse transcription-PCR (RT-PCR) was performed using a modified version of the SMART (switching mechanism at 5′ end of RNA transcript) procedure (Clontech, Mountain View, CA) and a TCRB constant region 3′ primer (5′-ATT CCT TTC TCT TGA CCA TG-3′). cDNA amplification was performed using a TCR constant region-based primer (5′-TTC ACC CAC CAG CTC AGC TC-3′) and 10× universal primer A mix (Clontech, Mountainview, CA). PCR products of 600 to 700 bp were gel purified (Qiagen, Valencia, CA), ligated into the TOPO TA vector (Invitrogen, Carlsbad, CA), and used to transform chemically competent Escherichia coli Top 10 cells (Invitrogen, Carlsbad, CA). Bacterial colonies were selected and screened for the presence of the insert by PCR with M13 primers. Selected colonies were sequenced with a Taq DyeDeoxy Terminator cycle sequencing kit (Applied Biosystems, Foster City, CA) and by capillary electrophoresis on a Prism automated sequencer (Applied Biosystems, Foster City, CA). In-frame TCR sequences were edited and aligned using Sequencher (Gene Codes Corp., Ann Arbor, MI) and were compared to the human TRBV gene database (http://imgt.org). The TRBV classification system of the international ImMunoGeneTics database (26) was used unless otherwise noted.
Measurement of T cell receptor diversity.
The diversity of the TCR CDR3 region was determined using the Shannon entropy index (H) (31, 49) calculation for protein sites as described previously (53), using the formula H = −Σpilog2 pi, where pi is the fraction of residues at a site that is amino acid type i. For amino acids in the CDR3 region, H can range from 0 (site contains only one amino acid in all sequences) to 4.32 (all amino acids are represented equally at this site). Positions that contained >50% gaps were excluded from analysis. Statistical values are expressed as means or medians ± standard errors of the means (SEM). The diversity of TCR clonotypes was also calculated using Simpson's diversity index (52, 55), using the formula Ds = 1 − Σ[ni(ni − 1)]/[N(N − 1)], where ni is the TCR clone size of the ith clonotype and N is the total number of TCR sequences sampled. Comparisons between groups were performed using the Mann-Whitney test or two-tailed Student t test, where appropriate. Correlations were calculated using the Spearman rank test. GraphPad Prism v5.03 software was used for statistical analysis.
Flow cytometric evaluation of lymphocyte surface molecules.
Lymphocyte subsets were evaluated using anti-CD3–Alexa Fluor 700 (BD), anti-CD4–phycoerythrin (PE)–Texas Red (Caltag), anti-CD8–Pacific Orange (Caltag), anti-CD14–peridinin chlorophyll protein (PerCP) (BD), anti-CD19–PerCP (BD), anti-CD38–PE–Cy7 (eBiosciences), anti-CD45RO–PE–Texas Red (eBiosciences), anti-CD56–PE–Cy5 (BD), anti-CCR7-pure (mouse IgG2 clone 150503; R&D Systems), Viaprobe (BD), anti-PD-1-pure (mouse IgG1 clone EH12:2H7; BioLegend), goat anti-mouse IgG–Pacific Blue (Molecular Probes), and anti-TRBV–PE–fluorescein isothiocyanate (FITC) (Beckman Coulter). Epitope-specific T cell populations were stained with MHC-I tetramers HLA-B*08-EI8 (EIYKRWII), HLA-B*08-FL8 (FLKEKGGL), HLA-B*15-GY9 (GLNKIVRMY), and HLA-B*15-TY11 (TQGYFPDWQNY), from the NIH Tetramer Core Facility (Atlanta, GA), and HLA-B*57-KF11 (KAFSPEVIPMF), HLA-B*57-IW9 (ISPRTLNAW), and HLA-B*57-QW9 (QASQEVKNW), from Beckman Coulter.
Cells were labeled with MHC-I tetramers at 21°C for 10 min. Anti-PD-1 or anti-CCR7 antibody was added to the suspension and incubated for a further 20 min. Cells were washed and labeled with Pacific Blue-conjugated goat anti-mouse antibody, washed, blocked with mouse normal immunoglobulin antibodies, and labeled with the remaining directly conjugated surface antibodies listed above. Cells were then washed a final time before acquisition on a BD FACSAria instrument. Data were analyzed using BD Diva v6.1 software. Fluorochrome panels were developed using fluorescence minus one (FMO) controls to adjust the compensation for each color. Positive and negative gates for phenotypic markers were set using bimodal expression patterns (29) evident on parent CD3+ T cell populations for each phenotypic marker of interest (PD-1, CD127, and CD38). These gates were then copied onto appropriate subpopulations for eventual analysis of surface expression.
RESULTS
Epitope-specific immune responses contract after ART.
ART dramatically reduces viral loads in HIV-positive individuals, and many individuals experience increases in CD4 T cell count or reductions in the phenotypic activation status of T cells. We determined the composition of the epitope-specific clonotypic TCR repertoire as well as the clonotypic cell surface phenotype for 14 epitope-specific CD8+ T cell responses from 8 HIV-positive individuals before and after the initiation of highly active ART (HAART) (Table 1). Six of eight individuals had increases in CD4+ T cell count, and the cohort had a mean reduction in viral load of 2.3 log (P = 0.01; paired t test). One of the two individuals who failed to reconstitute CD4+ T cells was patient 10004. Despite an undetectable viral load in the absence of ART, this individual had CD4+ T cell counts below 400 since inclusion in our cohort in 2003. This individual started ART in 2006 because of CD4+ T cell numbers that further declined to <200. Despite low CD4+ T cell numbers, patient 10004 has never had an AIDS-related opportunistic infection. The mean number of days before suppression of viremia to below 50 copies/ml was 170. All individuals were eventually suppressed to below 50 copies/ml, and 5 of 8 individuals were fully suppressed to undetectable levels at the time of repertoire and phenotypic analysis (Table 1).
Table 1.
HIV infection parameters for patients in this study
| Patient ID | TCR epitope(s) | Pretherapy value |
Posttherapy CD4 count | No. of days to reach <50 copies/ml after HAART | Day of TCR sequence sample after HAART | |
|---|---|---|---|---|---|---|
| CD4 count | Viral load (log10 copies/ml) | |||||
| 10001 | B*1501-GY9 and -TY11 | 378 | 4.5 | 598 | 288 | 288 |
| 10004 | B*5701-KF11 and -QW9 | 360 | 2.4 | 167 | 11 | 1,109 |
| 10027 | B*5701-KF11 and -QW9 and B*08-FL8 and -EI8 | 775 | 3.27 | 637 | 35 | 428 |
| 10086 | B*08-EI8 | 132 | 4.88 | 500 | 219 | 1,125 |
| 10105 | B*08-FL8 and -EI8 | 304 | 4.46 | 430 | 384 | 406 |
| 10138 | B*1501-GY9 | 291 | 3.2 | 305 | 190 | 137 |
| 10141 | B*1501-GY9 | 464 | 5.46 | 572 | 170 | 7 |
| 20023 | B*1501-GY9 | 538 | 3.54 | 413 | 60 | 46 |
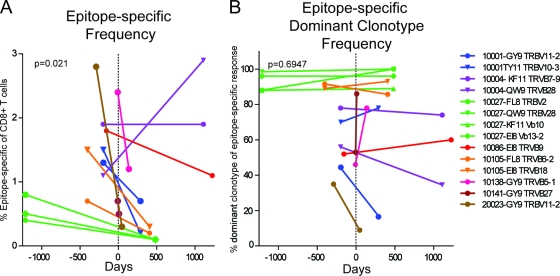
Using MHC-I tetramer reagents, we identified and tracked 14 distinct HIV epitope-specific CD8+ T cell populations before and after initiation of ART. Overall, epitope-specific T cell populations measured post-ART contracted as a percentage of the parent CD8+ T cell population compared to pre-ART measurements (73% median decrease; P = 0.02) (Fig. 1A). However, this effect was not universal. For example, the B*57-QW9 response in patient 10004 increased as a percentage of the parent CD8+ T cell population after initiation of ART, while the B*57-KF11 response in the same individual did not increase to a similar degree, indicating that epitope-specific responses within a single individual can respond independently after initiation of ART.
Fig 1.
Epitope-specific responses contract while clonotypic dominance remains intact. (A) Epitope-specific T cell populations and constituent clonotypes were identified by colabeling with tetramer and anti-TRBV antibodies. Frequencies of epitope-specific populations as part of the CD8+ T cell population are noted at pre-ART and post-ART time points. (B) Dominant clonotypes were identified by sequencing and labeled with anti-TRBV antibodies, and the frequencies of dominant clonotypes as part of the epitope-specific population are noted at corresponding pre-ART and post-ART time points. The dotted vertical line at day 0 indicates initiation of ART. Subject, epitope identifier, and dominant TRBV family are indicated for each response studied.
T cell clonal dominance is maintained after ART.
Epitope-specific TCR repertoires were determined by labeling epitope-specific populations with MHC-I tetramers, isolating these cells using FACS, and directly sequencing the T cell receptor beta chains. Clonotypic dominance within the repertoire was determined by sequencing and confirmed by colabeling tetramer-positive populations with anti-TRBV antibodies. As our group has shown previously (13, 30, 51), TCR repertoire data generated independently using these two methods correspond well, with a high degree of significance. Our data reported here showed a similarly high correspondence (Pearson r = 0.85; P < 0.0001).
Several clonotypes varied in relative frequency within the epitope-specific repertoire after initiation of ART, but dominant clonotypes remained dominant after initiation of ART in the majority of cases (Fig. 1B). No statistically significant patterns emerged from our analysis of changes within the TCR repertoire after initiation of ART. Dominant clonotypes in patient 10001 B*15-TY11 and patient 20023 B*15-GY9 responses contracted significantly after initiation of ART, to the point that they made up only minor clonotypic populations in the TCR repertoire. We were unable to relate the contraction of these epitope-specific clonotypes after initiation of ART to changes in patterns of surface markers or to TCR tetramer binding levels. We next evaluated the TCR repertoire of these epitope-specific immune responses in more detail.
The epitope-specific T cell receptor repertoire narrows after ART.
We were interested in determining the effect of ART on the clonotypic composition and structural diversity of the T cell receptor repertoire within epitope-specific responses. Diversity within the TCR repertoire can be measured using several methods. Shannon entropy (H) provides a useful metric for assessing the degree of overall amino acid diversity within the TRBV CDR3 region (32, 49, 51, 53). Diversity analysis using Simpson's diversity index can account for both the abundance of a clonotype and its proportion within the repertoire (52, 55). We used both methods to evaluate T cell repertoire diversity before and after ART.
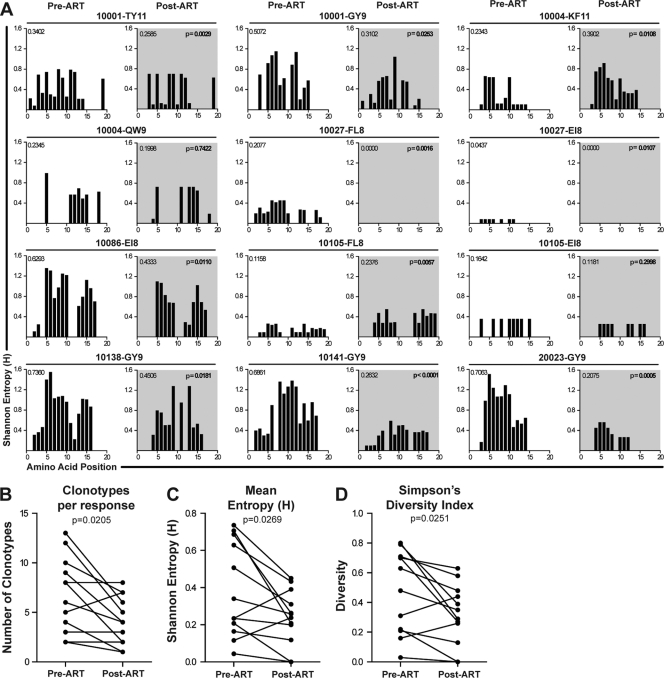
We calculated entropy (H) values for each amino acid position within the TRBV CDR3 region for 12 epitope-specific responses in 8 individuals before and after initiation of ART (Fig. 2A). The CDR3 entropy decreased significantly in 8 of 12 responses, and the mean entropy values across each CDR3 region (i.e., the average of the individual CDR3 amino acid entropy values) for epitope-specific populations post-ART decreased compared to pre-ART values (Fig. 2C) (P = 0.0269). The total number of clonotypes measured post-ART was reduced from a mean of 6.5 clonotypes/response to 3.9 clonotypes/response (Fig. 2B) (P = 0.0205). By measuring the number of clonotypes and their relative abundance in the epitope-specific TCR repertoire, we also showed that diversity was reduced after initiation of ART (Simpson's diversity index; P = 0.0251) (Fig. 2D). Additionally, it is important that despite the smaller magnitudes of epitope-specific responses after initiation of ART, there was no correlation between the number of cells sorted and the number of clonotypes determined either pretherapy (r = 0.33; P = 0.39) or posttherapy (r = 0.26; P = 0.49) (see Table S1 in the supplemental material). This is consistent with our prior data showing that sorting small numbers of cells can provide an accurate representation of TCR diversity (51). Taken together, these data provide compelling evidence that the epitope-specific TCR repertoire narrows after initiation of ART, in terms of overall amino acid diversity as well as the number of clonotypes within each response.
Fig 2.
Diversity in the HIV epitope-specific TCR repertoire is reduced after initiation of ART. (A) At pre-ART (unshaded charts) and post-ART (shaded charts) time points, Shannon's entropy (plotted on the y axis) was calculated and shown for amino acid positions within the TRBV CDR3 region (plotted on the x axis) for each of 12 individual epitope-specific T cell responses. Mean entropy for the TCR repertoire is shown in the upper left corner of each chart. The P value for statistical comparison (Wilcoxon matched-pairs test) is shown in the upper right corner of each chart. Pre-ART and post-ART comparisons between independent diversity measurements are shown in panels B to D. (B) Number of distinct clonotypes within the TCR repertoire identified at each time point. (C) Mean Shannon entropy value for each TCR repertoire. (D) Simpson's diversity index for each clonotypic repertoire.
T cell populations undergo memory redistribution and reductions in activation status after initiation of ART.
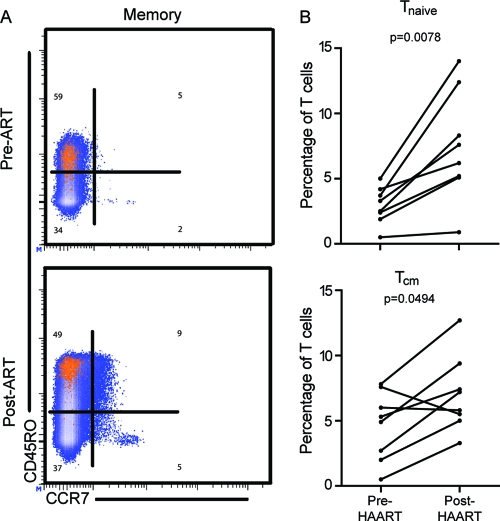
T cell phenotype is governed by environmental signals and is likely influenced by the level of antigen burden. Compared to uninfected controls, T cell populations in individuals with HIV infection are associated with skewed T cell memory differentiation (5) and increased expression levels of CD38 and PD-1 (56). We used expression of CD45RO and CCR7 to measure the memory cell distribution of T cell populations before and after initiation of ART (Fig. 3A). Our longitudinal analysis showed that CCR7+ CD45RO− (Tnaive) and CCR7+ CD45RO+ (Tcm) populations were reconstituted after initiation of ART. Mean Tnaive populations increased 2.5-fold, and Tcm populations increased 1.8-fold (Fig. 3B) (Tnaive P = 0.0078; Tcm P = 0.0494). However, epitope-specific populations did not undergo significant changes in memory phenotype after initiation of ART.
Fig 3.
T cell memory populations are repopulated after initiation of ART. Memory cell subsets were identified based on expression of CD45RO and CCR7. (A) Representative memory cell distributions on CD8+ (blue) and epitope-specific (orange) T cell populations are shown pre-ART and post-ART. (B) Frequencies of CD45RO+ CCR7+ Tcm and CD45RO− CCR7+ Tnaive cell populations as proportions of the overall CD8+ T cell population are increased post-ART.
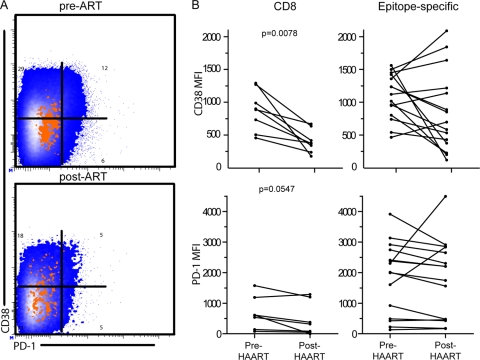
In a similar fashion, we compared post-ART expression of CD38 and PD-1 to pre-ART expression. Our analysis determined a 3.1-fold reduction in CD38 mean fluorescence intensity (MFI) and a 2.2-fold reduction in PD-1 MFI on CD8+ T cell populations (Fig. 4B) (CD38 P = 0.0078; PD-1 P = 0.0547). We evaluated expression of PD-1 and CD38 on epitope-specific T cell populations but did not observe statistically significant changes in the expression of these markers on epitope-specific populations. We next investigated phenotypic differences in these markers on dominant and subdominant clonotypic populations within epitope-specific responses.
Fig 4.
CD8+ T cell activation is reduced after ART, while epitope-specific changes are variable. T cell activation was measured by surface expression of CD38 and PD-1 molecules. (A) Representative plots are shown for CD8+ (blue) and epitope-specific (orange) populations. (B) Expression of CD38 and PD-1 as measured by MFI was reduced on CD8+ T cell populations after initiation of ART.
PD-1 expression patterns on epitope-specific clonotypes remain stable after initiation of ART.
Expression of PD-1 is closely related to antigen exposure and T cell exhaustion in several infection systems (10, 24, 43), and we have previously shown that PD-1 is expressed more highly on dominant clonotypes within epitope-specific T cell populations (13). Having demonstrated clonotypic remodeling after initiation of ART, we were interested in determining whether initiation of ART would influence the relationship between clonotypic dominance and PD-1 expression.
Epitope-specific T cell clonotypes were identified using tetramer reagents and anti-TRBV antibodies, and PD-1 expression was determined by colabeling these populations with anti-PD-1 antibodies. As previously reported, we observed a distinct pattern of PD-1 expression on dominant and subdominant clonotypes within HIV epitope-specific T cell populations whereby dominant clonotypes within epitope-specific populations expressed higher levels of PD-1 than did corresponding subdominant clonotypes (Fig. 5A). This pattern of higher PD-1 expression on dominant clonotypes remained statistically significant post-ART (Fig. 5B). We noted no significant relationship between clonotypic dominance and expression of CD38, CD127, or CD57 (data not shown). These results indicate that while overall immune activation may decrease on the parent CD8+ T cell population after initiation of ART, the environmental signals which influence PD-1 expression on epitope-specific clonotypes remain intact.
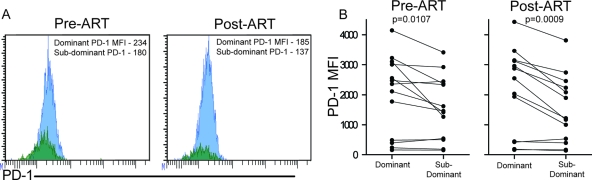
Fig 5.
Dominant clonotypes express higher levels of PD-1 than subdominant clonotypes before and after ART. PD-1 expression was measured on dominant and subdominant clonotypes within epitope-specific responses before and after initiation of ART. (A) Representative histograms show PD-1 expression on dominant (blue) and subdominant (green) clonotypes before and after ART. The MFI value is shown in the upper right corner of each histogram for each clonotypic population. (B) PD-1 expression levels on dominant and subdominant clonotypes of 13 epitope-specific responses were compared at pre-ART and post-ART time points (Wilcoxon matched-pairs test).
DISCUSSION
Clonotypes within the epitope-specific T cell receptor repertoire recognize viral epitopes with an exquisite specificity (22), and antigen exposure almost certainly drives their phenotypic qualities (9). Our data here demonstrate that after initiation of ART, the magnitudes of epitope-specific T cell responses contract and the TCR repertoires of these epitope responses narrow, as measured by the number of clonotypes and by the amino acid diversity within the repertoire. Despite this repertoire narrowing, clonotypic dominance within the TCR repertoire is most often maintained, and dominant T cell receptor clonotypes remain PD-1high compared to subdominant clonotypes. While other studies have noted phenotypic changes in T cell populations (16, 44, 57) and contraction of epitope-specific responses (11, 17, 21), this is the first report, to our knowledge, to assess diversity within the TCR repertoire and the clonotypic phenotype before and after initiation of ART.
After initiation of ART, we noted narrowed diversity within the epitope-specific TCR repertoire. We analyzed the TCR repertoire for 14 epitopes of 8 HIV-positive individuals before and after initiation of ART, using a variety of methodologies. The first method involved simply counting the number of clonotypes identified by sequencing. The second method used Simpson's diversity index (55) to assess the number of clonotypes in a response and, importantly, also accounted for the frequency of each clonotype as a part of the whole population. The third method used calculations to determine entropy (H) by assessing the variability of amino acid substitutions within the TCRβ CDR3 region expressed in a given epitope-specific TCR repertoire. Each independent methodology indicated with statistical significance that the clonotypic TCR repertoire narrows after initiation of ART.
Epitope-specific T cell populations decreased in relative frequency after suppression of viral replication under ART. Presumably, and as suggested previously (11, 17, 21), these contractions resulted from reductions in antigen exposure and corresponding decreases in antigen-mediated T cell expansion. Within lower-frequency epitope-specific responses, however, dominance within the clonotypic hierarchy was largely maintained after ART, suggesting that the features of the environment and of the immune response responsible for maintaining the clonotypic hierarchy are independent of absolute antigen levels. Various lines of research support the theory that TCR avidity for antigen is the driving factor in establishing and maintaining dominance within the TCR repertoire (23, 40), although clonotypic dominance may result from deletion of high-avidity clonotypes from the repertoire (27).
If avidity is a primary biophysical determinant of dominance within the repertoire, then clonotypes with higher or lower avidity would have different relative capacities to sense and respond to their cognate antigens regardless of the absolute levels of circulating antigen (4). Indeed, highly avid clonotypes may contribute to natural control of viremia in vivo (3). In two epitope-specific TCR repertoires, we observed dramatic shifts in dominance where the frequency of the pre-ART dominant clonotype contracted to much lower levels post-ART and previously subdominant clonotypes increased in frequency. If the dominant clonotype had been generated in response to a viral variant epitope and after suppression of viremia another epitope sequence became predominant, even at low levels, we might expect a different clonotype to establish dominance in the TCR repertoire. Our group has shown the dynamic nature of the HIV epitope-specific T cell receptor repertoire over the course of chronic infection (30), and these data suggest that individual T cell clonotypes likely respond to the prevalence of circulating HIV variants (22, 30, 51). Since T cell clonotypes expand to differing degrees in vivo, we reasoned that this might be reflected in the phenotype of these cells. We recently demonstrated that within epitope-specific populations, individual clonotypes may differ markedly in their memory maturation phenotype but not in the expression of other markers, such as CD57, indicating a degree of senescence that is maintained across clonotypes (33). Our most recent work showed that dominant clonotypes express a PD-1high CD127low surface phenotype and recognize dominant circulating viral sequences, whereas subdominant clonotypes are better able to recognize and suppress epitope variants (13). Another study, by van Bockel et al., found that epitope-specific clonotypes are persistent over time in vivo and may contribute to the recognition and suppression of viral variants (54). Taken together, the data suggest that individual T cell clonotypes may preferentially contribute to recognition of discrete virus populations and that a diverse T cell receptor repertoire may provide more effective control of viral replication. Future studies should be directed at evaluating epitope sequence variation in the context of ART-mediated viral suppression, as well as a further characterization of the mechanisms that maintain epitope-specific clonotypic hierarchies when antigen levels are reduced.
We assessed the cell surface phenotype on various cell populations. Activation and memory profiles on CD8+ T cells clearly changed after initiation of ART. CD38 and PD-1 expression was reduced on CD8+ T cells, perhaps providing some evidence that T cells are less prone to apoptosis after initiation of ART (38); however, no pattern of overall reduction was apparent for epitope-specific populations in our study. In line with other studies indicating that activation levels are strong prognostic indicators of disease progress and viral load (16, 44), reductions in activation were a positive indicator of clinical improvement in these individuals. We noted consistent redistributions within CD8+ T memory cell subsets that indicated increases in the frequencies of Tnaive and Tcm populations after initiation of ART. These results are consistent with previously reported data (6, 12) showing that ART alters the phenotype of T cells such that they more closely resemble those found in uninfected individuals. The changes we noted in the CD8+ T cell population support our hypothesis that these individuals underwent significant immune remodeling after initiation of ART.
While the changes we observed in activation and memory phenotypes on CD8+ T cells after initiation of ART have been reported previously for other cohorts, and while Yamamoto et al. reported reductions in negative regulatory surface molecules on T cell populations (57), the measurement of phenotypic changes after initiation of ART on epitope-specific clonotypes has not been reported. Recent findings from our group established a relationship between dominance in the epitope-specific TCR repertoire and a PD-1high CD127low phenotype (13). By assessing epitope-specific clonotypic T cell populations before and after initiation of ART, we noted that dominant clonotypes had a PD-1high phenotype compared to subdominant clonotypes, which had been established during chronic infection and which endured after initiation of ART. We did not note statistically significant relationships between clonotypic dominance and the expression of CD38, CD127, or CD57 or memory cell distribution before or after initiation of ART. The absence of these relationships conforms with results from a previous study of chronic HIV infection in the absence of ART that also found no relationships between clonotypic dominance and CD57 expression or memory cell distribution of HIV-specific CD8+ T cells (33).
Our finding with regard to the maintenance of the PD-1high phenotype on dominant HIV epitope-specific clonotypes after ART is supported by data from other model systems and from parent T cell populations (57). If dominance is established as a result of TCR avidity, and if PD-1 expression is predicated on antigen sensing, as suggested by other groups for both the lymphocytic choriomeningitis virus (LCMV) and simian immunodeficiency virus (SIV) models as well as for naturally controlled HIV infection (4, 9, 47), then we might expect PD-1 expression patterns on epitope-specific T cell clonotypes to remain consistent despite a large decrease in the level of HIV antigen after the start of ART. In this manner, PD-1 expression might represent the degree of cognate antigen recognition rather than absolute levels of generalized antigen exposure. Furthermore, considering our observations that PD-1 and CD38 expression patterns were not reduced universally on epitope-specific populations or on their clonotypic constituents, it is likely that these cells continue to receive some level of antigen stimulation even after the dramatic antigen reductions seen after initiation of ART. Indeed, the increased expression of PD-1 and CD38 we noted on a few epitope-specific populations may suggest that these responses actually saw relative increases in antigen exposure after initiation of ART. Despite years of study, the dynamics of antigen exposure and the mechanisms that define clonotypic diversity and phenotype within epitope-specific populations after initiation of ART have not been well characterized. A more detailed understanding of these mechanisms could inform our expectations of immune recovery and capacity for epitope-specific T cell populations in the context of viral suppression mediated primarily by ART.
Our study focused on 14 epitope-specific populations in 8 individuals before and after initiation of ART. Operating within the constraints of subject and sample availability and clonotypic repertoire composition, we necessarily performed our comparisons between single time points before and after initiation of ART and assessed several epitope-specific responses within short periods after initiation of ART. In the majority of subjects, we observed gross immunological changes, such as contraction of responses or reductions in CD38 expression on CD8+ T cell populations, after initiation of ART. However, had we assessed clonotypic composition or phenotype at later time points, we may have observed even more profound changes compared to pre-ART time points. This sort of longitudinal study to further characterize the epitope-specific clonotypic repertoire and cell surface phenotype after initiation of ART could provide a context for epitope-specific immune responses. As a matter of necessity due to the contraction of epitope-specific responses after initiation of ART, we sorted fewer epitope-specific T cells at post-ART time points (see Table S1 in the supplemental material). However, there was no relationship between the number of cells sorted and diversity at either pre- or post-ART time points. It is therefore unlikely that the reduced TCR diversity we observed post-ART was simply an artifact of the sorting process, and we have previously published data to support this analysis (51). More detailed analyses of epitope-specific TCR repertoire dynamics may be possible as deep sequencing techniques become more widespread (45, 55). However, current methods for deep sequencing of TCR repertoires rely either on a very limited TRBV family primer set (55) or on multiplex PCR with a set of primers representing each TRBV family (45). These methods will need to be compared to the current state-of-the-art method, i.e., 5′-anchored PCR, which has been shown to be correlated highly with anti-TRBV antibody staining in defining epitope-specific TCR repertoires (13).
With their expression of identical TCR repertoires, HIV epitope-specific clonotypes represent the fundamental unit of the T cell response to HIV (19, 22, 30, 39, 46, 50). Overall, the results we report here indicate that despite dramatic decreases in viral load, increases in CD4 count, and decreases in systemic activation, the factors which govern clonotypic expansion and phenotype within epitope-specific T cell responses remain intact at some level. While TCR avidity may be a key component, it is likely not a singular determinant of clonotypic repertoire diversity or phenotype. A more complete understanding of how epitope-specific clonotypes are maintained over time in the presence and absence of their cognate antigens is of great interest as we seek to elicit effective, durable T cell responses via vaccination.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grant AI39966 (S.A.K.) and by the Vanderbilt-Meharry Center for AIDS Research (AI54999). Flow cytometry acquisition and analysis were performed at the NIH-funded Vanderbilt-Meharry CFAR Immunopathogenesis Core Facility.
Footnotes
Published ahead of print 18 January 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Addo MM, et al. 2007. Fully differentiated HIV-1 specific CD8+ T effector cells are more frequently detectable in controlled than in progressive HIV-1 infection. PLoS One 2:e321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Addo MM, et al. 2003. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load. J. Virol. 77:2081–2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Almeida JR, et al. 2007. Superior control of HIV-1 replication by CD8+ T cells is reflected by their avidity, polyfunctionality, and clonal turnover. J. Exp. Med. 204:2473–2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Almeida JR, et al. 2009. Antigen sensitivity is a major determinant of CD8+ T-cell polyfunctionality and HIV-suppressive activity. Blood 113:6351–6360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Appay V, et al. 2002. Memory CD8+ T cells vary in differentiation phenotype in different persistent virus infections. Nat. Med. 8:379–385 [DOI] [PubMed] [Google Scholar]
- 6. Autran B, et al. 1997. Positive effects of combined antiretroviral therapy on CD4+ T cell homeostasis and function in advanced HIV disease. Science 277:112–116 [DOI] [PubMed] [Google Scholar]
- 7. Betts M, et al. 2001. Analysis of total human immunodeficiency virus (HIV)-specific CD4+ and CD8+ T-cell responses: relationship to viral load in untreated HIV infection. J. Virol. 75:11983–11991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Betts M, et al. 2006. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood 107:4781–4789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Blattman JN, Wherry EJ, Ha SJ, van der Most RG, Ahmed R. 2009. Impact of epitope escape on PD-1 expression and CD8 T-cell exhaustion during chronic infection. J. Virol. 83:4386–4394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brown KE, Freeman GJ, Wherry EJ, Sharpe AH. 2010. Role of PD-1 in regulating acute infections. Curr. Opin. Immunol. 22:397–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Casazza JP, Betts MR, Picker LJ, Koup RA. 2001. Decay kinetics of human immunodeficiency virus-specific CD8+ T cells in peripheral blood after initiation of highly active antiretroviral therapy. J. Virol. 75:6508–6516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Champagne P, et al. 2001. Skewed maturation of memory HIV-specific CD8 T lymphocytes. Nature 410:106–111 [DOI] [PubMed] [Google Scholar]
- 13. Conrad JA, et al. 2011. Dominant clonotypes within HIV-specific T cell responses are programmed death-1high and CD127low and display reduced variant cross-reactivity. J. Immunol. 186:6871–6885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Day CL, et al. 2006. PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature 443:350–354 [DOI] [PubMed] [Google Scholar]
- 15. Fidler S, et al. 2002. Virological and immunological effects of short-course antiretroviral therapy in primary HIV infection. AIDS 16:2049–2054 [DOI] [PubMed] [Google Scholar]
- 16. Giorgi JV, et al. 1999. Shorter survival in advanced human immunodeficiency virus type 1 infection is more closely associated with T lymphocyte activation than with plasma virus burden or virus chemokine coreceptor usage. J. Infect. Dis. 179:859–870 [DOI] [PubMed] [Google Scholar]
- 17. Gray CM, et al. 1999. Frequency of class I HLA-restricted anti-HIV CD8+ T cells in individuals receiving highly active antiretroviral therapy (HAART). J. Immunol. 162:1780–1788 [PubMed] [Google Scholar]
- 18. Hunt PW, et al. 2003. T cell activation is associated with lower CD4 T cell gains in human immunodeficiency virus-infected patients with sustained viral suppression during antiretroviral therapy. J. Infect. Dis. 187:1534–1543 [DOI] [PubMed] [Google Scholar]
- 19. Iglesias MC, et al. 2011. Escape from highly effective public CD8+ T-cell clonotypes by HIV. Blood 118:2138–2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jin X, et al. 1999. Dramatic rise in plasma viremia after CD8+ T cell depletion in simian immunodeficiency virus-infected macaques. J. Exp. Med. 189:991–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kalams SA, et al. 1999. Levels of human immunodeficiency virus type 1-specific cytotoxic T-lymphocyte effector and memory responses decline after suppression of viremia with highly active antiretroviral therapy. J. Virol. 73:6721–6728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kalams SA, et al. 1996. T cell receptor usage and fine specificity of human immunodeficiency virus 1-specific cytotoxic T lymphocyte clones: analysis of quasispecies recognition reveals a dominant response directed against a minor in vivo variant. J. Exp. Med. 183:1669–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kedzierska K, La Gruta NL, Davenport MP, Turner SJ, Doherty PC. 2005. Contribution of T cell receptor affinity to overall avidity for virus-specific CD8+ T cell responses. Proc. Natl. Acad. Sci. U. S. A. 102:11432–11437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Keir ME, Francisco LM, Sharpe AH. 2007. PD-1 and its ligands in T-cell immunity. Curr. Opin. Immunol. 19:309–314 [DOI] [PubMed] [Google Scholar]
- 25. Koup R, et al. 1994. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J. Virol. 68:4650–4655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lefranc MP, et al. 2003. IMGT unique numbering for immunoglobulin and T cell receptor variable domains and Ig superfamily V-like domains. Dev. Comp. Immunol. 27:55–77 [DOI] [PubMed] [Google Scholar]
- 27. Lichterfeld M, et al. 2007. Selective depletion of high-avidity human immunodeficiency virus type 1 (HIV-1)-specific CD8+ T cells after early HIV-1 infection. J. Virol. 81:4199–4214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu Z, et al. 1997. Elevated CD38 antigen expression on CD8+ T cells is a stronger marker for the risk of chronic HIV disease progression to AIDS and death in the Multicenter AIDS Cohort Study than CD4+ cell count, soluble immune activation markers, or combinations of HLA-DR and CD38 expression. J. Acquir. Immune Defic. Syndr. 16:83–92 [DOI] [PubMed] [Google Scholar]
- 29. Maecker HT, Trotter J. 2006. Flow cytometry controls, instrument setup, and the determination of positivity. Cytometry A 69A:1037–1042 [DOI] [PubMed] [Google Scholar]
- 30. Meyer-Olson D, et al. 2006. Fluctuations of functionally distinct CD8+ T-cell clonotypes demonstrate flexibility of the HIV-specific TCR repertoire. Blood 107:2373–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Meyer-Olson D, et al. 2003. Analysis of the TCR beta variable gene repertoire in chimpanzees: identification of functional homologs to human pseudogenes. J. Immunol. 170:4161–4169 [DOI] [PubMed] [Google Scholar]
- 32. Meyer-Olson D, et al. 2004. Limited T cell receptor diversity of HCV-specific T cell responses is associated with CTL escape. J. Exp. Med. 200:307–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Meyer-Olson D, et al. 2010. Clonal expansion and TCR-independent differentiation shape the HIV-specific CD8+ effector-memory T-cell repertoire in vivo. Blood 116:396–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Migueles SA, et al. 2002. HIV-specific CD8+ T cell proliferation is coupled to perforin expression and is maintained in nonprogressors. Nat. Immunol. 3:1061–1068 [DOI] [PubMed] [Google Scholar]
- 35. Ogg G, et al. 1999. Decay kinetics of human immunodeficiency virus-specific effector cytotoxic T lymphocytes after combination antiretroviral therapy. J. Virol. 73:797–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Oxenius A, et al. 2000. Early highly active antiretroviral therapy for acute HIV-1 infection preserves immune function of CD8+ and CD4+ T lymphocytes. Proc. Natl. Acad. Sci. U. S. A. 97:3382–3387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Petrovas C, et al. 2006. PD-1 is a regulator of virus-specific CD8+ T cell survival in HIV infection. J. Exp. Med. 203:2281–2292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Petrovas C, et al. 2009. Differential association of programmed death-1 and CD57 with ex vivo survival of CD8+ T cells in HIV infection. J. Immunol. 183:1120–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Price DA, et al. 2009. Public clonotype usage identifies protective Gag-specific CD8+ T cell responses in SIV infection. J. Exp. Med. 206:923–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Price DA, et al. 2005. Avidity for antigen shapes clonal dominance in CD8+ T cell populations specific for persistent DNA viruses. J. Exp. Med. 202:1349–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Price DA, et al. 2004. T cell receptor recognition motifs govern immune escape patterns in acute SIV infection. Immunity 21:793–803 [DOI] [PubMed] [Google Scholar]
- 42. Rehr M, et al. 2008. Emergence of polyfunctional CD8+ T cells after prolonged suppression of human immunodeficiency virus replication by antiretroviral therapy. J. Virol. 82:3391–3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Riley J. 2009. PD-1 signaling in primary T cells. Immunol. Rev. 229:114–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Robbins GK, et al. 2009. Incomplete reconstitution of T cell subsets on combination antiretroviral therapy in the AIDS Clinical Trials Group protocol 384. Clin. Infect. Dis. 48:350–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Robins HS, et al. 2009. Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood 114:4099–4107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rudd BD, et al. 2010. Diversity of the CD8+ T cell repertoire elicited against an immunodominant epitope does not depend on the context of infection. J. Immunol. 184:2958–2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Salisch NC, et al. 2010. Inhibitory TCR coreceptor PD-1 is a sensitive indicator of low-level replication of SIV and HIV-1. J. Immunol. 184:476–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schmitz JE, et al. 1999. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science 283:857–860 [DOI] [PubMed] [Google Scholar]
- 49. Shannon CE, Weaver W. 1948. The mathematical theory of communication. Bell Syst. Tech. J. 27:379–423 [Google Scholar]
- 50. Simons BC, Kalams SA. 2007. The potential role of epitope-specific T-cell receptor diversity in the control of HIV replication. Curr. Opin. HIV AIDS 2:177–182 [DOI] [PubMed] [Google Scholar]
- 51. Simons BC, et al. 2008. Despite biased TRBV gene usage against a dominant HLA B57-restricted epitope, TCR diversity can provide recognition of circulating epitope variants. J. Immunol. 181:5137–5146 [DOI] [PubMed] [Google Scholar]
- 52. Smith MZ, et al. 2008. Limited maintenance of vaccine-induced simian immunodeficiency virus-specific CD8 T-cell receptor clonotypes after virus challenge. J. Virol. 82:7357–7368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Stewart JJ, et al. 1997. A Shannon entropy analysis of immunoglobulin and T cell receptor 1. Mol. Immunol. 34:1067–1082 [DOI] [PubMed] [Google Scholar]
- 54. van Bockel DJ, et al. 2011. Persistent survival of prevalent clonotypes within an immunodominant HIV Gag-specific CD8+ T cell response. J. Immunol. 186:359–371 [DOI] [PubMed] [Google Scholar]
- 55. Venturi V, Kedzierska K, Turner SJ, Doherty PC, Davenport MP. 2007. Methods for comparing the diversity of samples of the T cell receptor repertoire. J. Immunol. Methods 321:182–195 [DOI] [PubMed] [Google Scholar]
- 56. Vollbrecht T, et al. 2010. Impact of changes in antigen level on CD38/PD-1 expression on HIV-specific CD8 T cells in chronic, untreated HIV-1 infection. J. Med. Virol. 82:358–370 [DOI] [PubMed] [Google Scholar]
- 57. Yamamoto T, et al. 2011. Surface expression patterns of negative regulatory molecules identify determinants of virus-specific CD8+ T-cell exhaustion in HIV infection. Blood 117:4805–4815 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.