Abstract
RNA interference (RNAi) is a critical component of many cellular antiviral responses in plants, invertebrates, and mammals. However, its in vivo role in host protection from the negative-sense RNA virus influenza virus type A (flu) is unclear. Here we have examined the role of RNAi in host defense to flu by analyzing Argonaute 1 and 3 double-knockout mice deficient in components of the RNA-induced silencing complex. Compared to littermate controls, flu-infected double-knockout mice exhibited increased mortality, consistent with more severe alveolitis and pneumonitis. These data indicate that optimal resistance to flu requires Argonaute 1 and/or 3. Enhanced mortality of double-knockout mice was not associated either with increased viral replication or with differential pulmonary recruitment or function of innate and adaptive immune cells. Given the absence of detectable immune defects, our results support the notion that the enhanced flu susceptibility of double-knockout mice arises from an intrinsic impairment in the ability of lung cells to tolerate flu-elicited inflammation.
INTRODUCTION
MicroRNAs (miRNAs) are ∼22-nucleotide gene-regulatory RNAs that modulate cell growth, death, differentiation, and homeostasis, with roles in organismal development, oncogenesis, inflammation, and immunity. Their biogenesis begins in the nucleus with the RNA polymerase II-dependent transcription of primary miRNAs (pri-miRNAs). Subsequently, short stem-loop precursor miRNAs (pre-miRNAs) are cleaved from the body of pri-miRNAs by the RNase III enzyme Drosha and its partner, DGCR8. Upon export to the cytosol, another RNase III enzyme, Dicer, cleaves the hairpin pre-miRNA loops to liberate mature ∼22-nucleotide double-stranded mature miRNAs. One miRNA strand (the passenger) is degraded, and the other (the guide) is loaded onto the Argonaute (Ago) family proteins to form the core of the RNA-induced silencing complex (RISC). RISC-bound guide strands target specific mRNAs through complementary base pairing and repress protein expression through translational inhibition or mRNA degradation (14).
Mammals possess four Ago family members. Of these, only Ago2 exhibits the single-stranded RNA cleavage (or slicer) activity required for small interfering RNA (siRNA). Germ line Ago2 deficiency results in embryonic lethality (18, 21), while conditional Ago2 ablation in hematopoietic cells reveals its requirement for B cell and red blood cell development (22). Much less is known about specific roles for Ago1, Ago3, and Ago4. Biochemical analyses indicate similarity in the small RNAs and mRNAs bound by each Ago family member; thus, the contributions to RNA interference (RNAi) of Ago1, Ago3, and Ago4 could be qualitative or quantitative (2).
RNAi plays a critical role in innate cellular defense against viruses. In plants and invertebrates, viral RNA genomes and mRNAs are targeted for destruction by the stimulated production of siRNAs derived from viral double-stranded RNAs (dsRNAs) (6, 7). Despite considerable effort, analogous viral siRNAs have not been detected within the pool of small RNAs isolated from RNA virus-infected mammalian cells (17, 25, 28). Given the strong type I interferon-based antiviral response initiated by the intracellular dsRNA sensors PKR, RIG-I, and MDA-5, in mammals the siRNAi pathway may have been superseded as a host protection mechanism from RNA viruses (28). Nevertheless, recent studies have begun to reveal examples of cellular miRNAs that target the genomes of RNA viruses. This has been demonstrated for miRNA-32 (miR-32) and primate foamy virus type 1 in 293T cells and miR-24 and miR-93 and vesicular stomatitis virus in mice (15, 23).
Recent in vitro studies suggest a role for the RNAi pathway in cellular protection from influenza A virus (flu). Knockdown of Dicer in Vero cells lacking type I interferon signaling resulted in increased virus production and accelerated apoptosis in flu-infected cells (20). This is consistent with a report that several cellular miRNAs can inhibit flu replication in MDCK cells (29). Further support for a role of miRNAs in the cellular response to flu comes from a deep sequencing analysis of flu-infected chickens, revealing an infection-specific signature of miRNA upregulation in the lung and trachea (29). Similarly, comparison of lungs of mice infected with reconstructed 1918 (r1918) pandemic H1N1 flu and a benign seasonal isolate revealed a 1918 flu-specific miRNA expression profile (16). Interestingly, predicted mRNA targets of these pandemic-specific miRNAs encoded proteins that were enriched for immune response and cell death pathways.
In spite of these recent advances, the in vivo role of the RNAi pathway in the host response to flu remains unknown. Here, we have used viable Ago1- and Ago3-double-knockout (DKO) mice to examine the role of RNAi in the host response to flu. Flu-infected DKO mice exhibited enhanced mortality associated with increased lung pathology. These data indicate that Ago1 and/or Ago3 (and, by extension, the RNAi machinery) is required for optimal resistance to flu.
MATERIALS AND METHODS
Virus.
Influenza virus A/Puerto Rico/8/34 (PR8) was generated with an eight-plasmid reverse genetics system (9). Stocks were propagated no more than twice by allantoic inoculation of 10-day-old embryonated hens' eggs with seed virus diluted 1:106. Virus titers were determined as 50% egg infectious dose (EID50).
Mice.
Mice were constructed as described previously (S. Cheloufi et al., unpublished data) and maintained on a (B6 × 129)N1B6 background. All experiments were performed with littermate controls. Animals were housed in the St. Jude Children's Research Hospital specific-pathogen-free vivarium. All experiments were conducted under a protocol approved by the St. Jude Children's Research Hospital Animal Care and Use Committee.
Virus infection and sampling.
Mice were anesthetized with 2,2,2-tribromoethanol (Avertin) and infected intranasally with diluted virus (2 × 103 EID50) in 30 μl of endotoxin-free phosphate-buffered saline (PBS). Mice were then weighed and monitored for mortality daily for a period of 14 days or sacrificed at various time points for sampling of the lung lumen by bronchoalveolar lavage (BAL). BAL fluid was collected in 3 1-ml washes with Hanks buffered saline solution (Gibco). After brief centrifugation, BAL fluid supernatant was collected for cytokine expression analysis (see below). BAL fluid cells were resuspended in fluorescence-activated cell sorting (FACS) buffer (PBS with 0.5% fetal bovine serum and 2 mM EDTA); blocked with anti-CD32/CD16 (Fc receptor) monoclonal antibody (MAb; Becton Dickinson); stained with anti-Ly6g fluorescein isothiocyanate (FITC) or anti-Ly6C FITC, allophycocyanin (APC)-labeled anti-CD11c, phycoerythrin (PE)-labeled anti-class II, and anti-CD11b PE-Cy7 (eBioscience) MAbs at 4°C; and then analyzed by flow cytometry. For antigen-specific CD8+ T cell analysis, BAL fluid and spleen cell suspensions were incubated with anti-CD32/CD16 (Fc receptor block), anti-CD8 FITC (BD), anti-CD4 PE-Cy7 (eBioscience), and PE- or APC-conjugated tetramers (DbNP366, DbPA224, KbPB1703, or DbPB1-F26) at 4°C for 30 min. A 10% (wt/vol) lung homogenate was prepared for virus plaque assay in PBS with a Tissue Tearor homogenizer (Fisher Scientific, Pittsburgh, PA) (see below). Blood was collected by terminal cardiac puncture.
Lung pathological analysis and influenza virus immunohistochemistry.
Lungs collected at necropsy were infused with 10% neutral buffered formalin, with additional immersion in fixative for 24 h. Fixed tissues were embedded in paraffin, sectioned at 4 μm, and mounted on positively charged glass slides (Superfrost Plus; Fisher Scientific, Pittsburgh, PA). Slides were stained with hematoxylin-eosin or immunohistochemically for flu antigen. Heat-induced epitope retrieval was performed by heating slides in a BioCare Medical decloaking chamber at 98°C for 30 min in target retrieval buffer (pH 9.0; catalog number S2367; Dako, Carpinteria, CA), followed by a 30-min cooldown period. Slides were placed in Tris-buffered saline–Tween (TBST) buffer (catalog number TA-999TT; Thermo Scientific, Kalamazoo, MI) prior to assay. Endogenous peroxidases were blocked by incubating slides for 5 min in 3% aqueous hydrogen peroxide. Endogenous avidin and biotin (catalog number X0590; Dako, Carpinteria, CA) were blocked (10 min each), followed by blocking for 30 min in Superblock (catalog number 37545ZZ; Thermo Scientific Pierce, Rockford, IL). For detection of antigen, primary goat anti-influenza virus A (H1N1) antibodies (catalog number I7650-05G; US Biologicals, Swampscott, MA) were adsorbed against 3% mouse serum for 24 or 48 h prior to the assay. The primary antibodies were then diluted 1:1,000 in PBS and applied for 1 h to lung sections. After rinsing, the secondary biotinylated donkey antigoat antibodies (catalog number sc-2042; Santa Cruz Biotechnology, Santa Cruz, CA) were used at 1:200 for 30 min. Detection of bound secondary antibodies was performed with horseradish peroxidase-labeled streptavidin (catalog number TS-125-HR; Thermo Scientific, Kalamazoo, MI) with 3,3′ diaminobenzidine (DAB; 5 min; catalog number TA-125-HDX; Thermo Shandon) as chromogenic substrate and a light hematoxylin counterstain (catalog number TA-125-MH; Thermo Scientific, Kalamazoo, MI). Sections were scored blind by a veterinary pathologist (P.V.), and lungs exhibiting 10% or less involvement of flu antigen were excluded from analysis.
Tracheal epithelial cell (TEC) culture and infection.
Mouse tracheas were washed 3 times in PBS with 5 mM dithiothreitol and 1 time with PBS to remove mucous and were incubated overnight at 4°C in protease IV (0.4 mg/ml) in PBS. The cells were dislodged by gentle scraping with a scalpel and dispersed by repeated pipetting. The protease reaction was stopped with the addition of fetal bovine serum to 5%. The cells were briefly centrifuged and resuspended in growth medium (0.5 ml per trachea; 1:1 ratio of Dulbecco modified Eagle medium-Ham's F-12 medium, UltroserG serum substitute, penicillin [100 U/ml], streptomycin, [100 μg/ml], gentamicin [50 μg/ml], kanamycin [100 μg/ml], amphotericin B [2.5 μg/ml] with 5% fetal bovine serum [FBS]). The cells were plated on collagen-fibronectin-coated transwells (Corning) at 250 μl per well with 1 ml growth medium with 5% FBS on the bottom of the transwell. After 2 days, the medium was changed to growth medium without FBS and the cells were cultured until confluent (approximately 5 days). When confluent, the medium from the top of the transwell (apical surface) was removed and the medium at the bottom (basal surface) was changed to differentiation medium (growth medium supplemented with cholera toxin [10 ng/ml; Sigma] and all-trans-retinoic acid [10−7 M; Sigma]). Tight junction integrity was measured with an ohmmeter (World Precision Instruments).
Viral titer determination.
Virus titers in lung homogenates and apical conditioned medium from TEC cultures were determined by plaque assay on Madin-Darby canine kidney (MDCK) cells. Nearly confluent 9.6-cm2 MDCK cell monolayers were infected with 10-ml aliquots of six 10-fold serial dilutions of lung homogenates for 1 h at 37°C and then washed 3 times in PBS before adding 3 ml of minimal essential medium (MEM) containing 0.9% agarose and 1 mg/ml l-1-tosylamido-2-phenylethyl chloromethyl ketone (TPCK; Worthington)-treated trypsin. Cultures were incubated at 37°C in 5% CO2 for 72 h, and plaques were visualized with crystal violet. For determination of viral titers in TEC cultures, apical medium was diluted 10-fold and 100 μl of diluent was used to infect nearly confluent MDCK cells in 96-well plates containing 100 μl MEM with 1 mg/ml TPCK-treated trypsin. Cells were incubated for 72 h and stained with crystal violet to score viability.
Antigen-specific ELISA.
Microtiter plates were coated with purified PR8 whole virus in PBS overnight at 4°C. Serum samples were serially diluted 10-fold and incubated for 1 h at room temperature. Influenza virus-specific IgG1, IgG2a, IgG3, and IgM were detected with alkaline phosphatase-conjugated antimouse secondary antibodies (Southern Biotech) at a 1:1,000 dilution in PBS plus 0.5% bovine serum albumin. The substrate p-nitrophenyl phosphate (Sigma-Aldrich) was added, and plates were incubated at room temperature for 60 min. Optical density values were determined at 405 nm on a 96-well enzyme-linked immunosorbent assay (ELISA) reader (Molecular Devices).
RESULTS
It has been difficult to study the physiological role of the RNAi pathway in infectious disease because null mutations in its major components (Drosha, Dicer, and Ago2) lead to early embryonic lethality (3, 5, 18). However, mice lacking Ago1 or combined knockouts including Ago3 and Ago4 are viable (Cheloufi et al., unpublished), permitting analysis of the impact of RNAi impairment on the host response to infectious agents. Reasoning that compound Ago mutants would display more severe RNAi defects than single mutants, we focused our analysis on Ago1/Ago3-DKO mice. Indeed, consistent with a perturbation in an RNAi-dependent process required for pre- or postnatal development, the frequency at weaning of DKO pups born from heterozygote (Het; Ago1+/− Ago3+/−) intercrosses was sub-Mendelian (15%; Table 1). The incomplete penetrance of this phenotype suggested a compensatory role of Ago2 and/or Ago4. Necropsy did not reveal any abnormal phenotypes in adult DKO mice, suggesting that, despite lacking a function that contributes to survival to weaning, DKO mice may serve as a useful model to study the in vivo role of RNAi in the host response to flu.
Table 1.
DKO mice are born at sub-Mendelian frequency
| Genotype | Number |
|
|---|---|---|
| Expected | Observed | |
| WT | 37.5 | 45 |
| Het | 75 | 79 |
| DKO | 37.5 | 22 |
| Total | 150 | 150 |
To determine whether Ago1/3 is required for normal immune system development, we performed immunophenotypic analysis of central and peripheral lymphoid organs of DKO and wild-type (WT) littermate control mice. DKO mice exhibited normal cellularity of the thymus, peripheral lymph nodes, spleen, and bone marrow (see Fig. S1 in the supplemental material). The numbers of CD4 T and CD8 T, NKT, and nTreg cells in the thymus were similar between DKO and WT mice, indicating normal thymic development (see Fig. S1 in the supplemental material). Similarly, in peripheral lymphoid organs these T cell lineages were maintained at WT levels in DKO mice. We detected a marginal increase in the number of NK cells in both the spleen and lymph nodes of DKO mice (WT mice = 1.2 × 106 and DKO mice = 1.9 × 106 [P = 0.003, Student's t test] and WT mice = 6.24 × 103 and DKO mice = 1.4 × 104 [P = 0.009, Student's t test], respectively). B cell development in the bone marrow and spleen of DKO mice appeared normal across all Hardy fraction developmental stages (see Fig. S2 in the supplemental material). Finally, myeloid development appeared normal, with dendritic cells (DCs), neutrophils, and macrophages occurring in similar numbers in DKO and WT mouse spleens (see Fig. S1 in the supplemental material).
To determine whether T and B cells from DKO mice were functionally impaired, we activated cells in vitro and measured the proliferation rate and cytokine production. CD4 T cell proliferation and cytokine production appeared normal in DKO mice (see Fig. S3 in the supplemental material). Similarly, B cell proliferation appeared normal (see Fig. S3 in the supplemental material). Taken together, these data indicate that the immune system develops normally in DKO mice. Thus, with baseline immune parameters intact, DKO mice appeared to be a suitable model for investigating the in vivo role of RNAi in the host response to flu.
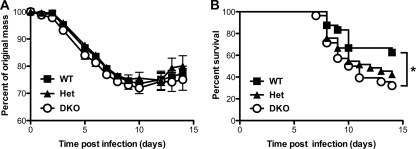
To determine whether DKO mice were able to mount a normal protective response to flu, we infected DKO, Het, and WT littermate controls intranasally with 2,000 EID50 of the virulent A/Puerto Rico/8/34 laboratory strain (H1N1 strain PR8) and monitored morbidity (weight loss) and mortality (Fig. 1). Over the 14-day infection course, DKO mice exhibited a similar pattern of weight loss as their WT littermate controls (Fig. 1A). However, by day 14 (d14) postinfection, DKO mice exhibited a striking 50% increase in mortality compared to WT littermate controls (68% versus 38%; P = 0.03, Mantel-Cox test), with Het mice exhibiting an intermediate phenotype. Thus, normal host resistance to the lethal effect of flu exhibits a dose-dependent requirement for Ago1 and/or Ago3.
Fig 1.
Susceptibility of WT and DKO mice to influenza virus A infection. WT (n = 25), Het (n = 34), and DKO (n = 29) mice were infected intranasally with 2,000 EID50 of PR8. Mice were monitored daily for weight loss (A) and survival (B). *, P < 0.04, log-rank Mantel-Cox test. Data are from three independent experiments (mean and SEM).
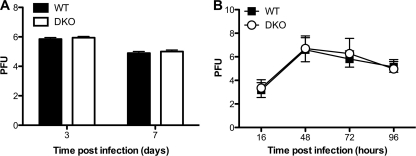
Several studies demonstrate that host miRNAs and virally derived siRNAs can regulate viral replication (28). In the case of flu, both Dicer and specific cellular miRNAs have been reported to inhibit viral replication in Vero and MDCK cells, respectively (20, 29). Therefore, we hypothesized that increased viral replication was the underlying cause of the enhanced mortality exhibited by DKO mice. To test this idea, we prepared whole-lung homogenates from mice 3 and 7 days postinfection and performed MDCK plaque assays to measure viral titers. We detected no difference in viral titers in the lungs of WT and DKO mice, suggesting that in vivo the host-protective role of Ago1 and/or Ago3 does not involve suppression of viral replication (Fig. 2A). Similar results were obtained in experiments comparing infected primary tracheal epithelial cell (TEC) cultures from WT and DKO mice (Fig. 2B).
Fig 2.
Viral replication in flu-infected WT and DKO mouse lungs and TEC cultures. (A) Mice were infected intranasally with 2,000 EID50 of PR8. Lungs were isolated at d3 (WT mice, n = 15; DKO mice, n = 16) and at d7 (WT mice, n = 18; DKO mice, n = 15). Viral titers in the lungs were determined by plaque-forming assay with MDCK cells. Data are from three independent experiments (mean and SEM). (B) WT and DKO mouse TECs were infected with PR8 at a multiplicity of infection of 0.01 in triplicate. Apical supernatants were collected at 16, 48, 72, and 96 h postinfection and assessed for viral titer by plaque-forming assay with MDCK cells. Data are from three independent experiments (mean and SEM).
Another possible explanation for the increased mortality of DKO mice is immune response impairment. To determine if there was an early defect in type I interferon production, we measured the beta interferon concentration in BAL fluid at d3 postinfection and found no difference between WT and DKO mice (see Fig. S4 in the supplemental material). For a more comprehensive analysis, we used the Milliplex Map mouse 32-plex cytokine kit (Millipore) and the Luminex multiplex bead analyzer to assess the levels of 32 additional cytokines in BAL fluid at d7 postinfection. Again, we found no significant difference in cytokine expression (see Fig. S4 in the supplemental material). Although these data cannot rule out aberrant expression of cytokines not included in the 32-plex assay, they do suggest that airway cytokine levels are normal in flu-infected DKO mice and indicate that their enhanced mortality was not due to cytokine storm.
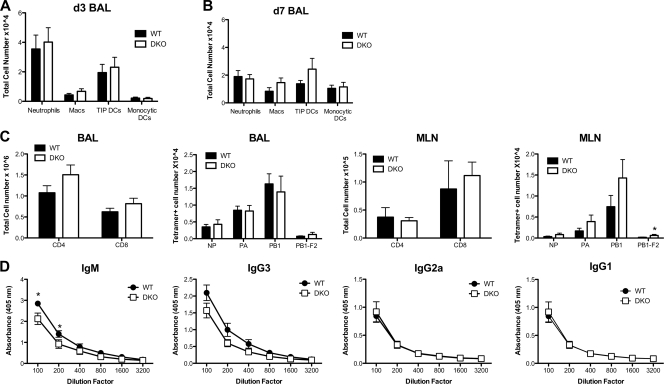
Neutrophils, tumor necrosis factor (TNF)-inducible nitric oxide synthase (iNOS)-producing dendritic cells (TIP DCs), and macrophages have been implicated as key players in the protective host response to flu infection (1, 31). To test whether the recruitment of these myeloid cells to flu-infected lungs was abnormal in DKO mice, we measured their abundance in BAL fluid at d3 and d7 postinfection. Over the 4-day time course, the numbers of neutrophils decreased, macrophages exhibited a marginal increase, TIP DCs remained unchanged, and monocytic DCs increased (Fig. 3A and B). Despite these temporal changes, we detected no significant difference in the abundance of each cell type between DKO mice and WT controls, suggesting that abnormal myeloid cell recruitment did not underlie the enhanced mortality of flu-infected DKO mice.
Fig 3.
Characterization of immune responses in WT and DKO mice. (A and B) Mice were infected intranasally with 2,000 EID50 of PR8 (WT mice, n = 15; DKO mice, n = 16). BAL fluid was collected at d3 (A) and d7 (B) postinfection; cells were analyzed by FACS; and cell numbers for neutrophils (CD11b+, Gr1+, major histocompatibility complex class II negative), macrophages (Macs; CD11b+, Gr1+, major histocompatibility complex class II positive), TNF-iNOS-producing DCs (TIP DCs; Ly6Chigh, CD11b+, CD11c+), and monocytic DCs (CD11b+, CD11c+) were enumerated. (C) Mice were infected intranasally with 2,000 EID50 of PR8, and BAL fluid was collected at d7 postinfection (WT mice, n = 18; DKO mice, n = 15). Cells from BAL fluid and MLN were analyzed by FACS, and total numbers of CD4+ T cells, CD8+ CTLs, and tetramer-positive CTLs were enumerated. *, P < 0.05, Student's t test. (D) PR8-specific serum antibody titers at d7 postinfection (WT mice, n = 11; DKO mice, n = 9) were determined by ELISA. *, P < 0.02, two-way analysis of variance. Data in panels A to C are from three independent experiments (mean and SEM).
CD8+ cytotoxic T lymphocytes (CTLs) are a critical component of the host protective adaptive immune response to flu. Therefore, we measured the abundance of immunodominant flu peptide-specific CTLs and CD4+ T cells in BAL fluid and draining mediastinal lymph node (MLN) at d7 postinfection (Fig. 3C). We observed no significant difference in total CD4+ T cell numbers in BAL fluid or MLN between DKO and WT mice. The same was true for total or flu peptide-specific CD8+ T cell numbers, with one minor exception. Compared to WT controls, DKO mice exhibited a small, but significant, increase in the number of PB1-F2-specific CTLs in BAL fluid (WT mice = 173, DKO mice = 628; P < 0.05, Student's t test) but not MLN, although the overall contribution of this specificity to the CD8 response was marginal. Thus, aberrant T cell responses appear to be unlikely to account for the enhanced mortality of flu-infected DKO mice.
As B cells and flu-specific antibody production are also known to play a key role in adaptive immune responses to flu (34), we examined serum immunoglobulin levels in flu-infected mice. At d7 postinfection, DKO mice exhibited WT levels of serum anti-flu IgG3, IgG1, and IgG2a and a minor 2-fold reduction in IgM (Fig. 3D). These data suggest that DKO mice are not defective in antibody class switching and that defects in B cell immunoglobulin secretion are unlikely to account for the enhanced mortality of flu-infected DKO mice. Overall, these data suggest that immune response impairment is unlikely to be a major cause of the increased mortality of flu-infected DKO mice.
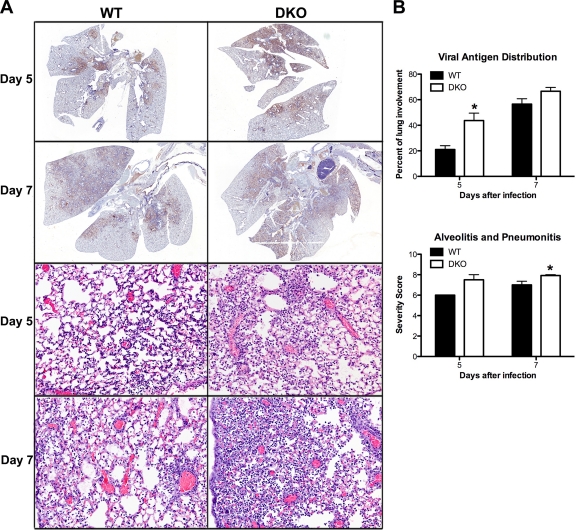
To explore a nonimmunological basis for the elevated mortality of DKO mice, we performed histological analysis of lungs at d5 and d7 postinfection. Lung sections were stained with hematoxylin-eosin to assess lesion severity and immunohistochemically with anti-flu (H1N1) antibody to determine flu antigen distribution. Compared to WT littermate controls, Ago1/3-DKO mouse lungs exhibited a significant increase in viral antigen distribution at d5 postinfection (Fig. 4A, top four panels, and B, top graph; P = 0.04, Student's t test). This correlated with a significant increase of the combined histological score for alveolitis and pneumonitis in DKO mice relative to WT mice at d7 postinfection (Fig. 4A, bottom four panels, and B, bottom graph; P = 0.02, Student's t test). Thus, DKO mice exhibit a transient increase in viral protein dissemination preceding the onset of enhanced alveolitis that is likely the proximal cause of their elevated mortality.
Fig 4.
Lung histology of flu-infected WT and DKO mice. WT and DKO mice were infected intranasally with 2,000 EID50 of PR8. Lungs were harvested for histopathological analysis at d5 (WT mice, n = 5; DKO mice, n = 8) and d7 (WT mice, n = 19; DKO mice, n = 14) postinfection. (A) Lung sections from d5 and d7 postinfection were immunohistochemically stained for flu antigen (top four panels, brown). Representative micrographs are shown. Magnification, ×5. Lung sections from d5 and d7 were stained with hematoxylin-eosin (bottom four panels). Representative micrographs are shown. Magnification, 200×. (B) Sections were scored blindly for flu antigen distribution (% involvement; top four panels) and the combined score for severity of alveolitis and interstitial pneumonitis (bottom four panels). Severity scoring criteria for lung lesions is the following: 0, no lesions; 1, minimal, focal to multifocal, inconspicuous; 2, mild, multifocal, conspicuous; 3, moderate, multifocal, prominent; 4, marked, diffuse or coalescing, lobar; 5, severe, diffuse consolidation, multilobar.
DISCUSSION
Our data support a physiologically important role for Ago1 and/or Ago3 in mammalian host protection from the negative-sense RNA virus influenza A. In principle, the enhanced susceptibility to flu exhibited by DKO mice could arise from either qualitative or quantitative deficiencies in the RNAi machinery. Qualitatively, Ago1 and/or Ago3 may mediate nonredundant aspects of the cellular RNAi machinery, such as binding of unique subsets of mature miRNAs or siRNAs or delivery of unique effector functions. Such unique aspects, in turn, could play a critical role in determining host susceptibility to flu. However, at this point, evidence in support of such a qualitative view of the role of different Ago family members (apart from Ago2) is lacking in the literature.
In contrast, evidence does exist to support a quantitative view of the contribution of different Ago proteins to RNAi, insofar as Ago1, -3, and -4 have been shown to bind a similar profile of miRNAs and mRNAs (2). Also, they have been found individually to be nonessential in embryonic stems cells (30; Cheloufi et al., unpublished). In addition, early Xenopus embryos ectopically overexpressing different Ago proteins are able to support RNAi function during early embryogenesis (19). Finally, our data demonstrate that embryonic and immune system development can occur in the absence of Ago1 and Ago3. Thus, DKO mice may suffer a global impairment in the efficiency but not necessarily the specificity of small RNA pathways. Such a quantitative impairment is consistent with the incompletely penetrant developmental block to weaning and the enhanced susceptibility to flu infection exhibited by DKO mice. This view finds further support in the dose-dependent requirement for Ago1/3 in host protection from flu-dependent mortality. Thus, our data support a model in which Ago1 and/or Ago3 contributes quantitatively to the delivery of critical functions required for host resistance to flu. However, they do not rule out further contributions from unique qualitative aspects of Ago1 and/or Ago3 function.
Regardless of whether loss of Ago1/3 function leads to qualitative or quantitative impairment of RNAi, it is instructive to consider the ways that the Argonautes could be promoting host resistance to flu. It is possible that flu infection induces the production of viral siRNAs and that these require Ago1/3 to target the destruction of viral mRNAs and/or viral genomic segments. However, this is highly unlikely. First, despite extensive efforts of multiple groups, viral siRNAs have not been detected in virus-infected mammalian cells, including those infected with flu (19). Second and most importantly, we did not detect increased viral titers in the lungs of infected DKO mice.
Another possibility is that the absence of Ago1/3 impacts a subset of cellular miRNAs whose functions contribute to flu resistance. Recent evidence provides support for this idea. The virulent r1918 H1N1 flu induces a distinct miRNA profile compared to a nonvirulent seasonal flu isolate, suggesting that the regulation of miRNA expression can contribute to flu pathogenesis (16). Since miRNAs can impact immune cell function, it is possible that a critical antiflu function of innate and/or adaptive immune cells could be defective in Ago1/3-DKO mice. For example, miR-146a and miR-146b control both interleukin-1β-induced inflammation and RIG-I-dependent type I interferon production via a negative-feedback loop resulting in downregulation of IRAK1 and TRAF6 (10, 24). In addition, the r1918 H1N1 flu-specific miRNA profile includes elevated expression of miR-200a and miR-223 (16). MiR-200a targets IFNAR1 and STAT2 and miR-223 represses CREB activity, consistent with roles in regulating the type I interferon response. Moreover, miR-223-deficient mice develop neutrophilic lung inflammation, suggesting a role as a key regulator of granulocyte production and inflammation (12). Despite these precedents, flu-infected DKO mice did not display altered cytokine (including type I interferon) expression in or recruitment defects of innate immune cells (including neutrophils) to BAL fluid. Although flu-infected DKO mice did exhibit a decrease in flu-specific serum IgM and an increase in flu-specific CTLs in the lung, in both cases the changes were very modest and unlikely to account for elevated mortality. Although we cannot rule out the possibility of undiscovered immune cell defects in DKO mice, it appears unlikely that impaired immune function can account for the enhanced susceptibility of DKO mice to flu infection.
Given the absence of detectable immune defects in DKO mice, we favor the notion that their elevated mortality arises from a defect in pulmonary tolerance to flu infection. Indeed, evidence for nonimmune tissue tolerogenic mechanisms of disease susceptibility has been mounting (11, 26, 27). An excellent recent example is of hemeoxygenase-1 (HO-1), found to regulate the ability of mice to tolerate Plasmodium chabaudi chabaudi malaria by limiting the amount of toxic heme that accumulates in the liver. In contrast, HO-1 has no impact on parasite burden, demonstrating that its beneficial effects are achieved via enhancing tissue tolerance, rather than modulation of the anti-P. chabaudi chabaudi immune response (26, 27). One possible mechanism of pulmonary tolerance to flu infection could be the prevention of cytotoxic viral protein buildup by an Ago1/3-dependent miRNA pathway. The enhanced viral protein dissemination evident in d5 flu-infected DKO mouse lung sections may reflect the perturbation of such a mechanism and is consistent with evidence that in MDCK cells miR-323, miR-491, and miR-654 can repress translation of flu genes encoding PB1 and PB1-F2 (29) and the small but significant elevation in the number of PB1-F2-specific CD8+ CTLs detected in DKO mice. Arguing against this mechanism is the comparable viral titer and protein epitope abundance (as measured by immunohistochemistry; data not shown) in primary cultures of DKO mouse compared to WT mouse flu-infected tracheal epithelial cells. Therefore, if elevated viral protein expression underlies enhanced viral protein dissemination in DKO mouse lungs, it must occur in a cell type-specific fashion.
Another possible pulmonary flu tolerance mechanism postulates that Ago1/3-dependent miRNAs are required in pathways that promote the general resistance of lung cells to inflammation-related toxic effectors, resulting in a given level of inflammatory response inflicting greater damage in DKO than WT mouse lungs. In support of such a mechanism, a growing number of miRNAs have been shown to play important roles in lung tissue homeostasis (32), including miR-155 and miR-26a (32), let-7 (13), miR-29 (8), and miR-15 and miR-16 (4, 33). Viral titer, controlled by the net contribution of multiple cellular and viral processes, measures the abundance of functional viral particles and is thus a poor surrogate for total viral protein level. Nevertheless, comparable viral titers in WT and DKO mouse lungs together with comparable viral epitope abundance in flu-infected WT and DKO mouse TECs suggest that elevated viral protein dissemination in DKO mouse lungs may not reflect elevated viral protein abundance. Indeed, as immunohistochemistry is sensitive both to the abundance and to the accessibility of target epitopes, it is possible that the immunohistochemical evidence of enhanced viral protein dissemination in DKO mouse lungs reflects elevated cellular damage-induced viral protein release to the extracellular space, where antibody epitopes may be more easily accessed. The capacity to clear cellular debris (including released viral proteins) would increase over time with accumulating phagocytic cell recruitment to the lungs, providing an explanation for why enhanced viral dissemination was evident at d5 but not d7 postinfection—in inverse temporal correlation with the appearance of enhanced pneumonitis/alveolitis. Indeed, there was a trend toward greater macrophage and monocytic DC abundance in d7 but not d5 BAL fluid from DKO mice compared to WT mice. It will be important to explore whether Ago1/3-dependent flu-induced miRNAs that can suppress expression of specific flu proteins exist and the potential of such targeted proteins, when overexpressed, to induce cytotoxicity in DKO mouse alveolar pneumocytes. Further, if Ago1/3 deficiency leads to a general state of lung fragility, DKO mice may exhibit enhanced susceptibility to pulmonary inflammatory diseases, whether of infectious origin or not. Thus, it will be important to explore whether DKO mice exhibit enhanced susceptibility to noninfectious pulmonary inflammatory diseases such as experimental allergic asthma.
Supplementary Material
ACKNOWLEDGMENTS
We thank Helen Beere and Joonsoo Kang for constructive comments on the manuscript, Linda Chung for help with genotyping, and the Cellular Immunology Core, Flow Cytometry Core, Animal Resources Center, and Veterinary Pathology Core Facilities of St. Jude Children's Research Hospital for technical support.
This work was supported by ALSAC and the St. Jude Children's Infection Defense Center (M.B.) and by NIH RO1GM062534 (G.J.H.).
Footnotes
Published ahead of print 8 February 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Aldridge JR, JR, et al. 2009. TNF/iNOS-producing dendritic cells are the necessary evil of lethal influenza virus infection. Proc. Natl. Acad. Sci. U. S. A. 106:5306–5311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beitzinger M, Peters L, Zhu JY, Kremmer E, Meister G. 2007. Identification of human microRNA targets from isolated argonaute protein complexes. RNA Biol. 4:76–84 [DOI] [PubMed] [Google Scholar]
- 3. Bernstein E, et al. 2003. Dicer is essential for mouse development. Nat. Genet. 35:215–217 [DOI] [PubMed] [Google Scholar]
- 4. Calin GA, Croce CM. 2006. MicroRNA signatures in human cancers. Nat. Rev. Cancer 6:857–866 [DOI] [PubMed] [Google Scholar]
- 5. Chong MM, et al. 2010. Canonical and alternate functions of the microRNA biogenesis machinery. Genes Dev. 24:1951–1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Deddouche S, et al. 2008. The DExD/H-box helicase Dicer-2 mediates the induction of antiviral activity in Drosophila. Nat. Immunol. 9:1425–1432 [DOI] [PubMed] [Google Scholar]
- 7. Deleris A, et al. 2006. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science 313:68–71 [DOI] [PubMed] [Google Scholar]
- 8. Fabbri M, et al. 2007. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc. Natl. Acad. Sci. U. S. A. 104:15805–15810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hoffmann E, Krauss S, Perez D, Webby R, Webster RG. 2002. Eight-plasmid system for rapid generation of influenza virus vaccines. Vaccine 20:3165–3170 [DOI] [PubMed] [Google Scholar]
- 10. Hou J, et al. 2009. MicroRNA-146a feedback inhibits RIG-I-dependent type I IFN production in macrophages by targeting TRAF6, IRAK1, and IRAK2. J. Immunol. 183:2150–2158 [DOI] [PubMed] [Google Scholar]
- 11. Jamieson AM, Yu S, Annicelli CH, Medzhitov R. 2010. Influenza virus-induced glucocorticoids compromise innate host defense against a secondary bacterial infection. Cell Host Microbe 7:103–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Johnnidis JB, et al. 2008. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature 451:1125–1129 [DOI] [PubMed] [Google Scholar]
- 13. Johnson CD, et al. 2007. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 67:7713–7722 [DOI] [PubMed] [Google Scholar]
- 14. Kim VN, Han J, Siomi MC. 2009. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 10:126–139 [DOI] [PubMed] [Google Scholar]
- 15. Lecellier CH, et al. 2005. A cellular microRNA mediates antiviral defense in human cells. Science 308:557–560 [DOI] [PubMed] [Google Scholar]
- 16. Li Y, et al. 2010. MicroRNA expression and virulence in pandemic influenza virus-infected mice. J. Virol. 84:3023–3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lin J, Cullen BR. 2007. Analysis of the interaction of primate retroviruses with the human RNA interference machinery. J. Virol. 81:12218–12226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu J, et al. 2004. Argonaute2 is the catalytic engine of mammalian RNAi. Science 305:1437–1441 [DOI] [PubMed] [Google Scholar]
- 19. Lund E, Sheets MD, Imboden SB, Dahlberg JE. 2011. Limiting Ago protein restricts RNAi and microRNA biogenesis during early development in Xenopus laevis. Genes Dev. 25:1121–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Matskevich AA, Moelling K. 2007. Dicer is involved in protection against influenza A virus infection. J. Gen. Virol. 88:2627–2635 [DOI] [PubMed] [Google Scholar]
- 21. Meister G, et al. 2004. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell 15:185–197 [DOI] [PubMed] [Google Scholar]
- 22. O'Carroll D, et al. 2007. A Slicer-independent role for Argonaute 2 in hematopoiesis and the microRNA pathway. Genes Dev. 21:1999–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Otsuka M, et al. 2007. Hypersusceptibility to vesicular stomatitis virus infection in Dicer1-deficient mice is due to impaired miR24 and miR93 expression. Immunity 27:123–134 [DOI] [PubMed] [Google Scholar]
- 24. Perry MM, et al. 2008. Rapid changes in microRNA-146a expression negatively regulate the IL-1beta-induced inflammatory response in human lung alveolar epithelial cells. J. Immunol. 180:5689–5698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pfeffer S, et al. 2005. Identification of microRNAs of the herpesvirus family. Nat. Methods 2:269–276 [DOI] [PubMed] [Google Scholar]
- 26. Sansonetti PJ, Medzhitov R. 2009. Learning tolerance while fighting ignorance. Cell 138:416–420 [DOI] [PubMed] [Google Scholar]
- 27. Seixas E, et al. 2009. Heme oxygenase-1 affords protection against noncerebral forms of severe malaria. Proc. Natl. Acad. Sci. U. S. A. 106:15837–15842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Skalsky RL, Cullen BR. 2010. Viruses, microRNAs, and host interactions. Annu. Rev. Microbiol. 64:123–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Song L, Liu H, Gao S, Jiang W, Huang W. 2010. Cellular microRNAs inhibit replication of the H1N1 influenza A virus in infected cells. J. Virol. 84:8849–8860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Su H, Trombly MI, Chen J, Wang X. 2009. Essential and overlapping functions for mammalian Argonautes in microRNA silencing. Genes Dev. 23:304–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Thomas PG, et al. 2009. The intracellular sensor NLRP3 mediates key innate and healing responses to influenza A virus via the regulation of caspase-1. Immunity 30:566–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tomankova T, Petrek M, Kriegova E. 2010. Involvement of microRNAs in physiological and pathological processes in the lung. Respir. Res. 11:159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Volinia S, et al. 2006. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. U. S. A. 103:2257–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Waffarn EE, Baumgarth N. 2011. Protective B cell responses to flu—no fluke! J. Immunol. 186:3823–3829 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.