Abstract
Higher-order RNA structures in the 5′ untranslated regions (UTRs) of the mouse hepatitis coronavirus (MHV) and bovine coronavirus (BCoV), separate species in the betacoronavirus genus, appear to be largely conserved despite an ∼36% nucleotide sequence divergence. In a previous study, each of three 5′-end-proximal cis-acting stem-loop domains in the BCoV genome, I/II, III, and IV, yielded near-wild-type (wt) MHV phenotypes when used by reverse genetics to replace its counterpart in the MHV genome. Replacement with the BCoV 32-nucleotide (nt) inter-stem-loop fourth domain between stem-loops III and IV, however, required blind cell passaging for virus recovery. Here, we describe suppressor mutations within the transplanted BCoV 32-nt domain that along with appearance of potential base pairings identify an RNA-RNA interaction between this domain and a 32-nt region ∼200 nt downstream within the nonstructural protein 1 (Nsp1)-coding region. Mfold and phylogenetic covariation patterns among similarly grouped betacoronaviruses support this interaction, as does cotransplantation of the BCoV 5′ UTR and its downstream base-pairing domain. Interestingly, cotransplantation of the BCoV 5′ UTR and BCoV Nsp1 coding region directly yielded an MHV wt-like phenotype, which demonstrates a cognate interaction between these two BCoV regions, which in the MHV genome act in a fully interspecies-compliant manner. Surprisingly, the 30-nt inter-stem-loop domain in the MHV genome can be deleted and viral progeny, although debilitated, are still produced. These results together identify a previously undescribed long-range RNA-RNA interaction between the 5′ UTR and Nsp1 coding region in MHV-like and BCoV-like betacoronaviruses that is cis acting for viral fitness but is not absolutely required for viral replication in cell culture.
INTRODUCTION
In positive-strand RNA viruses that replicate in the cytoplasm, genomic 5′-end-proximal RNA structures carry out several functions required for virus reproduction. In coronaviruses, these are thought to include (i) translation initiation, commonly presumed to occur by a canonical cap-dependent 5′-terminal ribosomal entry mechanism, to synthesize the replicase enzymes from open reading frame 1 (13, 14, 31); (ii) signaling an RNA-dependent RNA polymerase template switch during minus-strand synthesis at a heptameric transcription-regulatory sequence (UCUAAAC in the case of mouse hepatitis coronavirus [MHV] and bovine coronavirus [BCoV]) (Fig. 1A and B) for placement of a common leader on subgenomic mRNAs (sgmRNAs) (43, 48, 49, 55); (iii) encoding signals on the 3′ end of minus-strand genomic RNA (the antigenome) for initiating synthesis of plus-strand genomic mRNAs and sgmRNAs (9, 43, 48, 55); (iv) possibly harboring signals that act in trans to initiate synthesis of nascent plus-strand genomes (46); (v) possibly directly influencing initiation of minus-strand synthesis at the 3′ end of the genome (33); and (vi) harboring a genome packaging signal (10, 18). A mechanistic understanding of these events may aid in the development of therapeutic inhibitors of coronavirus replication.
Fig 1.
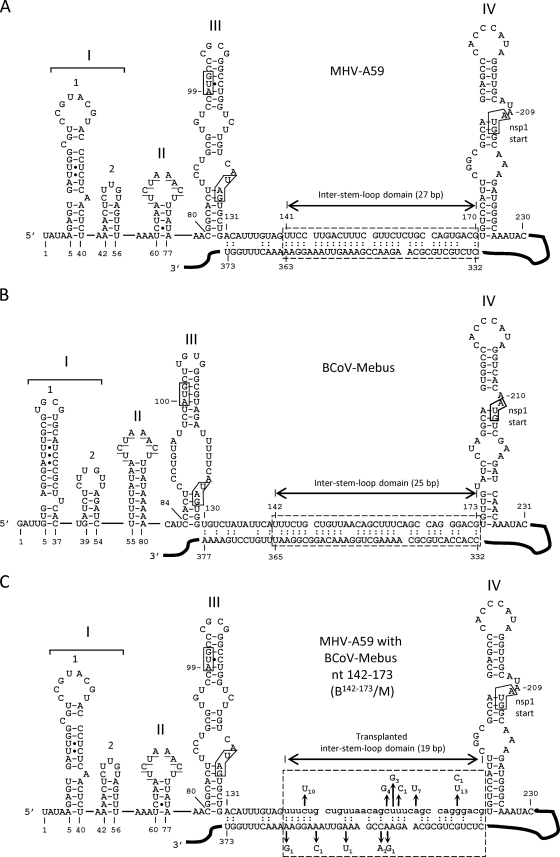
5′ UTRs of MHV-A59 and BCoV-Mebus showing the intra-5′-UTR stem-loops, the inter-stem-loop domain, and a potential long-range RNA-RNA interaction between the 5′ UTR and the Nsp1 coding region. (A) 5′ UTR of MHV showing the Mfold-predicted long-range RNA-RNA interaction (hatched box). (B) 5′ UTR of BCoV showing the Mfold-predicted long-range RNA-RNA interaction (hatched box). (C) 5′ UTR of MHV in which the 32-nt BCoV inter-stem-loop domain (lowercase) replaces the 30-nt MHV counterpart. The hatched box identifies a potential long-range interaction and shows a summary of the potential suppressor mutations in 17 plaque-purified clones described in Table 1. The subscript for each mutation indicates its incidence among the 17 isolates. Note that without suppressor mutations, the number of base pairs within the window is 19, not 27 (as in MHV) or 25 (as in BCoV). Refolding enables the number of base pairs to increase as a function of suppressor mutations (see the text). In all panels, the underlined bases in SLII identify the transcription-regulatory sequence (UCUAAAC), and boxes in SLIII identify the start and stop codons for a short upstream open reading frame. The box in SLIV identifies the start codon for the Nsp1 coding region. Filled circles in SL1, SLII, and SLIII represent non-Watson-and-Crick base pairings, as determined by nuclear magnetic resonance studies (33, 36).
In a previous study (22), we exploited the ∼36% nucleotide (nt) sequence divergence between the 5′ untranslated regions (UTRs) of BCoV and MHV, separate species in the betacoronavirus genus (15) (formerly classified as group 2a coronaviruses), to identify potentially common 5′-end-proximal cis-acting elements for replication of MHV. Despite the relatively high nucleotide sequence divergence, higher-order structures in the 5′-end-proximal genomic regions of these two viruses are largely conserved (Fig. 1A and B) (7, 10, 29, 33, 35, 36, 41, 42), although the nomenclature is not yet uniform, since stem-loops (SLs) II, III, and IV in Fig. 1 are respectively named stem-loops 3, 4 and 5 by the Leibowitz, Giedroc, and Olsthoorn laboratories (10, 29, 33, 35, 36). It was learned that the BCoV SLI/II domain (nt 1 to 84), which contains higher-order structures named SLI (now known to be comprised of stem-loops 1 and 2) (29, 33, 35, 36) and SLII, can functionally replace its MHV counterpart, as can the SLIII domain (nt 85 to 141) and the SLIV domain (nt 174 to 226). For function, SLIV required a 16-nt extension into the nonstructural protein 1 (Nsp1) coding region to form a “long” SLIV (Fig. 1A and B). In each of these substitutions, despite nucleotide differences of 37, 22, and 33%, respectively, the chimeric virus replicated for at least three passages without compensatory suppressor mutations appearing within the 5′-most 1,000 nt and 3′-most 500 nt of the genome. In contrast, a 30-nt inter-stem-loop domain in the MHV 5′ UTR that resides between SLIII and SLIV (nt 141 to 170) could not be replaced by the 32-nt BCoV counterpart (nt 142 to 173) to directly yield viable virus after transfection of the chimeric RNA genome (22). However, the resulting chimera, B142-173/M, produced plaques and viable progeny after two blind cell passages in one of four experimental attempts. These results suggested that there had been compensatory mutations within the surviving chimeras that enabled replication.
Here, we describe seven potential suppressor mutations that appeared within the transplanted 32-nt BCoV inter-stem-loop domain and five within a 32-nt partnering domain mapping ∼200 nt downstream within the Nsp1 coding region and demonstrate the importance of these as cognate domains for replication in genetic reconstruction assays. The Nsp1 coding region is the first of 16 in the replicase gene (16, 21), and Nsp1 is an RNA binding protein with several postulated functions in coronavirus replication (5, 6, 23, 27, 28, 32, 39, 47, 56). Surprisingly, the 30-nt inter-stem-loop domain in the 5′ UTR of the MHV genome can be deleted and viral progeny, although debilitated, are still produced. The results together identify a previously undescribed long-range cis-acting RNA-RNA interaction between the 5′ UTR and Nsp1 coding region in the murine and bovine betacoronaviruses that appears to be important for viral fitness but is not absolutely required for virus replication in cell culture.
MATERIALS AND METHODS
Cells.
Delayed brain tumor (DBT) cells (24), mouse L2 cells (19), and baby hamster kidney cells expressing the MHV receptor (BHK-MHVR) (11, 12) were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% defined fetal calf serum (FCS) (HyClone) and 20 μg/ml gentamicin (Invitrogen). Cells were maintained at 37°C with 5% CO2 for all experiments. BHK-MHVR cells were maintained in selection medium containing 0.8 mg/ml Geneticin (G418 sulfate; Invitrogen) (52).
Viruses and GenBank accession numbers.
GenBank accession numbers for virus sequences analyzed are as follows: NC_001846 for MHV-A59; NC_006852 for MHV-JHM; NC_012936 for Parker rat CoV; NC_006577 for human coronavirus (HCoV)-HKU1; NC_010327 for equine coronavirus (ECoV); NC_005147 for HCoV-OC43; NC_007732 for porcine hemagglutinating encephalomyelitis virus (PHEV); FJ415324 for human enteric coronavirus-4408 (HECoV-4408); U00735 for BCoV-Mebus; and NC_004718 for SARS coronavirus, Toronto strain (SCoV-Tor2).
RNA structure prediction and sequence alignments.
The Mfold program of Zuker (http://www.bioinfo.rpi.edu/∼zukerm/) (38, 54) was used for RNA structure predictions. The long-range RNA-RNA base pairing patterns described below were revealed by folding nt 1 to 400 or nt 1 to 500. Sequence alignments were made with Vector NTI suite 8 software (Invitrogen).
Reverse genetics system used for making BCoV/MHV chimeras and methods for analyzing viral progeny.
The reverse genetics system for MHV-A59, infectious clone MHV-A59-1000 (icMHV) (developed by R. Baric and colleagues [17, 52] and kindly provided by R. Baric), was used as previously described in detail for making 5′-end-proximal BCoV/MHV chimeras in the MHV-A59 background (22). Recombinant viruses described here were made by modifying fragment A of MHV-A59 as reported previously (22) but with the appropriate primers for the mutations described below. All procedures for plasmid construction with icMHV DNA, expression of recombinant viral RNA, transfection of cells with infectious recombinant RNA by electroporation, and characterization of mutant progeny by virus titration and growth kinetics were carried out as previously described (22). Plaque morphology was determined on L2 cells after 60 h of growth and after crystal violet staining as described previously (22). Plaque sizes were identified as large (wild type [wt]) if they were ≥2.5 mm, medium if 1.5 to 2.5 mm, or small if ≤1.5 mm in diameter. Representative plaque images were taken with a Nikon digital camera and prepared using Adobe Photoshop CS software.
Genome sequence analysis of chimera progeny.
Routinely, supernatant fluids from cells that first showed cytopathic effect (CPE) (either cells that had been transfected or cells that had been blind-passaged) were collected, and virus contained therein was referred to as virus passage zero (VP0) virus. When 80 to 100% of new DBT cells infected with VP0 virus showed CPE, intracellular RNA was extracted with TRIzol (Invitrogen), and the viral genome was sequenced by RT-PCR for the 5′-end-proximal nt 22 to 1093 and the 3′-terminal 500 nt (excluding the poly(A) tail). VP0 virus was then used to determine plaque morphology, and plaque-purified virus was used as starting material for growth kinetics on DBT cells and sequence analyses.
For analysis of the 5′ nt 22 to 1093 of progeny virus genomes, extracted cellular RNA was reverse transcribed with Superscript II reverse transcriptase (Invitrogen) using primer MHV-1094(+) (5′CGATCAACGTGCCAAGCCACAAGG3′), which binds MHV genomic nt 1094 to 1117, and cDNA was PCR amplified with primers MHV-leader(−) (5′TATAAGAGTGATTGGCGTCCG3′), which binds nt 1 to 21 of the MHV antileader, or BCoV-leader(−) (5′GATTGTGAGCGATTTGCGTGCG3′), which binds nt 1 to 22 of the BCoV antileader, and MHV-1094(+). PCR products were gel purified (QIAEX II; Qiagen) prior to automated sequencing with primers MHV(261–284)(−) (5′CCATGGATGCTTCCGAACGCATCG3′) and MHV(605–623)(+) (5′GTTACACAGGCAGACGCGC3′).
For analysis of the 3′ 500 nt of progeny virus genomes, the same procedure was used except that total RNA was reverse transcribed using primer DI3(+) (5′CGGGATCCGTCGACACGCGTTTTTTTTTTTTTTTTTTTT3′), which binds the poly(A) tail, and the cDNA was PCR amplified with primer MHV(30811–30830)(−) (5′GGATGGTGGTGCAGATGTGG3′), which binds the negative strand of virus nt 30811 to 30830, and primer DI3(+). Gel-purified PCR products were sequenced with primer MHV(30811–30830)(−).
Northern analysis.
Northern analyses were done as previously described (25). Briefly, freshly confluent DBT cells in 25-cm2 flasks (∼4 × 106 cells) were infected with wt or chimeric viruses at an MOI of 0.01 PFU/cell. At 20 h postinfection, intracellular RNA was TRIzol extracted, and 1/10 of the total RNA from one 25-cm2 flask (∼60 μg RNA total per 25-cm2 flask) was resolved by electrophoresis in a 1.0% agarose-formaldehyde gel at 150 V for 4 h. RNA was transferred to a Hybond N+ nylon membrane (Amersham Biosciences) by vacuum blotting for 3 h followed by UV cross-linking. After prehybridization of the membrane with NorthernMax prehybridization/hybridization buffer (Ambion) at 55°C for 4 h, the blot was probed at 55°C overnight with 20 pmol (∼4 × 105 cpm/pmol) of the 5′-end γ-32P-labeled 3′-UTR-specific oligonucleotide MHV(31094–31122)(+) (5′CAGCAAGACATCCATTCTGATAGAGAGTG3′), which binds MHV genomic nt 31094 to 31122. Probed blots were exposed to Kodak XAR-5 film at −80°C for imaging, and images were prepared using Adobe Photoshop CS software.
RESULTS
Identification of seven mutations within the transplanted inter-stem-loop domain and five within the Nsp1 coding domain that potentially reconstitute base-pairing between the two domains in phenotypic revertants.
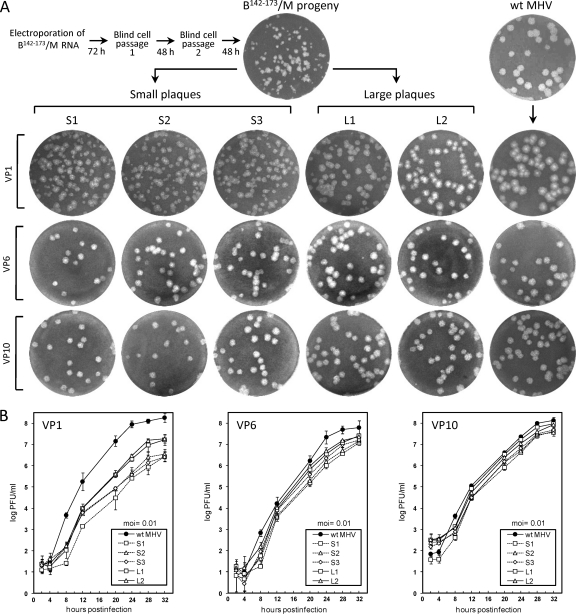
It was demonstrated previously that the 30-nt inter-stem-loop domain mapping between SLIII and SLIV in the MHV 5′ UTR behaves as a species-specific cis-acting replication element, since the 32-nt homologue from the BCoV genome was essentially functionally incompatible with the MHV genome, as shown by the chimera B142–173/M (Fig. 1C) (22). Nucleotides 131 to 140, the 5′-terminal 10 nt of the entire sequence between SLIII and SLIV, were not included as part of the inter-stem-loop domain here or previously (22), because by some predictions it makes up part of SLIII in the MHV 5′ UTR (29). Likewise, BCoV nt 130 to 141 were not included as part of the transplanted 32-nt inter-stem-loop domain. B142–173/M chimeric RNA containing the 32-nt transplanted BCoV domain during three transfection experiments was found to yield no viable progeny after four blind cell passages, but in a fourth trial, a mixture of small and wt-like large plaques were found after two blind cell passages (Fig. 2A, VP0 plaques). From this, six small plaques and 11 large plaques were selected, and each was passaged 10 times in DBT cells. Note that virus causing smaller plaques developed larger plaques and near-wt growth kinetics with passaging (Fig. 2). To determine what potential suppressor mutations might have caused the phenotypic changes within the transplanted 5′ UTR, the adjacent Nsp1 coding domain, and the 3′ UTR, viral genomic RNA isolated from cells infected at virus passages 1, 6, and 10 was sequenced through these regions. No 3′-end-proximal changes were found, and nucleotide changes within the 5′-end-proximal nt 22 through 1093 are summarized in Table 1.
Fig 2.
Phenotypes of representative plaque-purified progeny from cells electroporated with chimeric B142–173/M RNA. (A) Plaque appearances after culturing for 60 h on L2 cells. Isolated small plaques (S1 to S3 out of 6 total) and large plaques (L1 and L2 out of 11 total) were serially passaged 10 times on DBT cells. (B) Growth kinetics for three isolated small plaques (hatched lines) and two isolated large plaques (solid lines) following their selection at viral passages 1, 6, and 10.
Table 1.
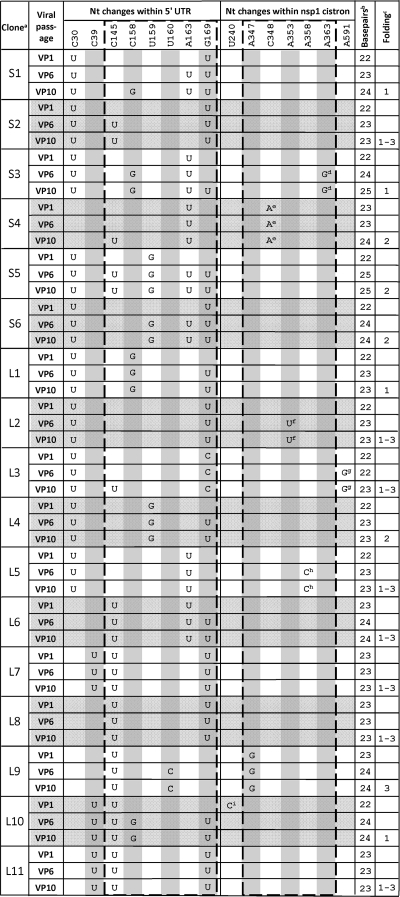
Potential compensatory mutations within the 5′ UTR and Nsp1 coding region of progeny from the B142-173/M chimera
Plaque purified clones were selected at virus passage 0 (VP0) and identified as small (S) or large (L).
Number of base pairs within the base-pairing window (identified as positions 145 to 169 in the 5′ UTR and 347 to 363 in the Nsp1 cistron).
See Fig. 3 for descriptions of the folding patterns.
Causes K52E.
Causes P47T.
Causes K48N.
Causes M128V.
Causes K50T.
Causes F11L.
The most striking result was a clustering of potential suppressor mutations within the transplanted 32-nt BCoV domain and within a 32-nt region mapping ∼200 nt downstream within the MHV Nsp1 coding region (summarized in Table 1 and schematically depicted in Fig. 1C and 3A). It was also striking that whereas there were fewer predicted base pairings initially between the two regions in B142–173/M RNA (19 bp) (Fig. 1C) than in Mfold-predicted wt MHV RNA (27 bp) (Fig. 1A), when predicted foldings included the putative suppressor mutations at virus passage 10 (Table 1), the number of base pairs within the window increased from 19 (Fig. 1C and 3A) to 22 to 25 (Table 1 and Fig. 3B). For example, when the combined suppressor mutations in S1 and S2 at VP10 were used, Mfold predicted the first potential folding pattern (potential folding 1) in Fig. 3B. Base pairings of 22 to 25 in revertants compare with 27 and 25 bp, respectively, for wt MHV and wt BCoV (Fig. 1A and B). These observations together led us to hypothesize that the suppressor mutations may reveal a specific long-range RNA-RNA interaction between the 5′ UTR and the Nsp1 coding region that is important for virus replication.
Fig 3.
Potential suppressor mutation-induced base pairings in progeny of the B142–173/M chimera. (A) Summary of potential suppressor mutations and their positions within the inter-stem-loop and downstream partnering domains that appeared in the small and large plaque isolates after 10 serial passages (depicted as in Fig. 1C). (Top) Base positions (in parentheses) for the BCoV sequence (lowercase) use BCoV numbering. Base positions elsewhere use MHV numbering. (Bottom) Summary of base changes at the named positions. The subscript numbers indicate the number of occurrences in 17 total isolates, shown in Table 1. (B) Three potential folding patterns as suggested by Mfold that together maximize the base pairings enabled by the suppressor mutations. Any of the three potential foldings can accommodate revertants S2, L2, L3, L5, L6, L7, L8, and L11, and others are accommodated as indicated. For revertants depicted in Table 1, base pairings within the base-pairing window at passage 10 increased from 19 to 22, 23, 24, or 25 (as summarized in the last two columns of Table 1).
Mfold predicts base pairing between the inter-stem-loop domain and a region within the Nsp1 coding region of several MHV-like and BCoV-like betacoronaviruses.
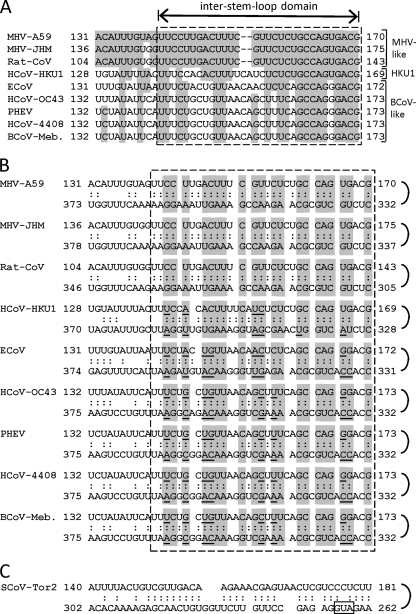
After a potential base pairing between the two domains was first noted, as described above, Mfold patterns that might identify base pairings for the long-range RNA-RNA interaction were sought by folding nt 1 to 400 or nt 1 to 500 of additional fully sequenced MHV-like and BCoV-like betacoronaviruses. Interestingly, base pairing between the inter-stem-loop domains (Fig. 4A) and respective partnering domains were readily found for the MHV-like, HCoV-HKU1, and BCoV-like coronaviruses (Fig. 4B) that revealed 25 to 27 potential base pairings within the 30- to 32-nt window. Figure 4B also depicts the phylogenetic covariation of sequences that maintain base pairings within the virus group. The Mfold and covariation results, therefore, appear to be consistent with a functional role for the base pairing as postulated above.
Fig 4.
Base pairings between the inter-stem-loop and partnering domains for 10 betacoronaviruses. (A) Alignment of the inter-stem-loop domains for MHV-like, HCoV-HKU1, and BCoV-like betacoronaviruses. Shading identifies bases that are in common with MHV-A59. The inter-stem-loop domains are identified with a hatched box. (B) Base pairings as predicted by Mfold. Shaded areas identify regions of base pairing common to the whole group. Underlined bases within the shaded areas are variant bases relative to those in MHV-A59 that maintain base pairing at the same position. Note that the number of base pairings within the inter-stem-loop domain for any given virus ranges from 25 to 27. (C) Potential 5′-end-proximal long-range RNA-RNA interaction derived by Mfold analysis of the 5′ region of the SCoV-Tor2 genome. This folding prediction is identical to that named for stem-loop 5 by Chen and Olsthoorn (10). The AUG start codon for the Nsp1 coding region is identified with a hatched box.
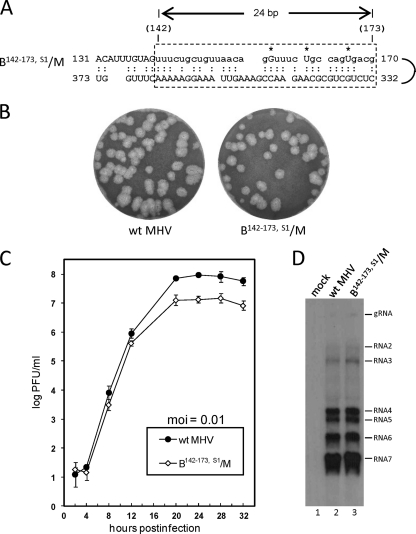
In vitro mutagenesis to form base pairing in the B142–173/M chimera with C158G, A163U, and G169U (thus recapitulating reverted virus S1 at passage 10) enabled immediate wt-like progeny after transfection of the chimeric genome.
To directly test the hypothesis that base-pairing between the inter-stem-loop and the Nsp1 partnering domains imparts fitness to the debilitated chimera, C158G, A163U, and G169U, the three putative suppressor mutations in isolate S1 at passage 10 (Table 1), were used to prepare chimera B142–173,S1/M (Fig. 5A), which was then tested by transfection. Whereas B142–173/M required two blind cell passages before the appearance of CPE (Fig. 2A), B142–173,S1/M developed CPE within 24 h posttransfection (hpt). Moreover, progeny from VP0 virus developed large plaques (Fig. 5B), had no additional putative suppressor mutations within the sequenced regions, had growth kinetics similar to that of wt MHV but with a 10-fold-lower final titer (107 PFU/ml versus 108 PFU/ml for wt MHV) (Fig. 5C), and had a wt-like sgmRNA profile as determined by Northern analysis (Fig. 5D).
Fig 5.
Testing of B142–173,S1/M chimeric RNA for replication. (A) B142–173/M RNA was made to contain C158G, A163U, and G169U (*) in order to mimic reverted virus S1 at virus passage 10. The predicted base-pairing pattern of B142–173,S1/M long-range RNA-RNA interaction is shown. BCoV sequence is indicated by lowercase letters. (B) Plaque patterns of wt MHV and B142–173,S1/M at VP1. (C) Growth kinetics of wt MHV-A59 and B142–173,S1/M at VP1. (D) Northern blot showing genomic and sgmRNA patterns for uninfected cells (lane 1), wt-MHV-infected cells (lane 2), and B142–173,S1/M virus-infected cells (lane 3). The radiolabeled probe was an oligonucleotide complementary to a 26-nt region within the MHV 3′ UTR.
These results, therefore, are consistent with the hypothesis that base pairing between the inter-stem-loop domain and the Nsp1 coding domain contributes to the replicating ability and fitness of MHV in cell culture.
Preplaced suppressor mutations within the Nsp1 coding region of the otherwise B142–173/M chimeric RNA hastened the appearance of phenotypic revertants and suppressor mutations within the 5′-UTR-mapping inter-stem-loop domain.
To determine whether the rarer putative suppressor mutations within the Nsp1 coding region are sufficient for phenotypic reversion, the mutations A353U and A363G were separately placed into the MHV genome along with the wt BCoV inter-stem-loop domain and tested. With B142–173,A353U/M RNA, CPE appeared within 48 hpt and VP0 virus generated wt-like plaques, retained the A353U mutation, and also contained G169U within the transplanted domain (data not shown). With B142–173,A363G/M, CPE appeared within 48 hpt and VP0 virus generated wt-like plaques, retained the A363G mutation, and also contained G169U within the transplanted domain (data not shown). These results show that the A353U and A363G mutations each alone may not suffice as suppressor mutations but likely hastened the appearance of additional suppressor mutations within the transplanted pyrimidine-rich domain.
With the exception of the of a C30U mutation, single potential suppressor mutations occurring outside the long-range base-pairing windows, namely, C39U, U240C, and A591G, were not tested as suppressors (Table 1). When the C30U mutation alone was transplanted along with the BCoV inter-stem-loop domain into the MHV background, viable virus appeared, but so did C145 and G169 within the window. This result suggested that C30U may not alone function as a suppressor but may by some unknown mechanism hasten the appearance of suppressor mutations. This phenomenon was not explored further.
The BCoV 5′ UTR and the (nearly) entire BCoV Nsp1 coding region function together in the MHV background to yield wt MHV-like progeny in cell culture.
The evidence showing that a virus species-specific interaction between the inter-stem-loop domain in the 5′ UTR and its partnering domain in the Nsp1 coding region is required for virus replication led us to directly test a bidirectional matching of the two domains. Since it had been shown previously that the functional BCoV SLIV alone in the MHV genome extends 16 nt into the Nsp1 coding region (22), the full BCoV 5′ UTR in addition to the 16-nt extension (i.e., nt 1 to 226) was used for testing with the remaining MHV Nsp1 coding region in the MHV background. For this, B1–226/M (Fig. 6A) was tested but was found to be nonviable over three blind cell passages from three separate electroporation attempts (Fig. 6B) (22), probably because of the incompatibility between the inter-stem-loop domain (from BCoV) and the Nsp1 partnering domain (from MHV) described above. Likewise, no viable progeny were generated when a chimera was tested in which the entire MHV 5′ UTR plus its 16-nt extension into its Nsp1 coding region was used with the adapted BCoV Nsp1 coding region (i.e., one lacking the first 16 nt and last 42 nt) in the MHV background to form B227–906/M (Fig. 6A and B). Note that for this construct, the MHV 3′ 42 nt of the Nsp1 coding region were retained to keep the region compatible with the MHV papain-like protease that separates Nsp1 and Nsp2 (17). However, when a construct in which the BCoV 5′ UTR containing the 30-nt MHV inter-stem-loop domain was tested in the MHV background as in B1-141,174-226/M (Fig. 6A), CPE appeared within 48 hpt, and progeny from electroporated RNA (VP0) yielded medium plaques, a peak titer of 1.0 × 106 PFU/ml, and a full set of viral sgmRNAs (Fig. 6B to D). These results together demonstrated a requirement for compatibility between the 5′-UTR-mapping inter-stem-loop domain and its cognate partnering domain within the Nsp1 coding region. Conversely, when a construct with the entire MHV 5′ UTR but with the BCoV 32-nt inter-stem-loop domain and the BCoV Nsp1 coding region in the MHV background, B142-173,227-906/M (Fig. 6A), was tested, CPE was observed within 48 hpt, and viable progeny (VP0) producing large plaques, a titer of 1.0 × 106 PFU/ml, and a full set of viral sgmRNAs (Fig. 6B to D) were obtained. This construct, therefore, also demonstrated a compatibility requirement between the 5′ UTR-mapping inter-stem-loop domain and its cognate partnering domain within the Nsp1 coding region.
Fig 6.
Requirement for compatibility between the 5′-UTR-located inter-stem-loop and downstream partnering domains for MHV replication. (A) For viability, the chimeric virus must contain the MHV inter-stem-loop domain within the BCoV 5′ UTR when in the MHV background (as in B1–141,174-226/M), the BCoV inter-stem-loop domain and BCoV Nsp1 coding region when in the MHV background (as in B142–173,227-906/M), or the BCoV 5′ UTR and BCoV Nsp1 coding region when in the MHV background (as in B1–906/M). Note that the in-frame C-terminal fusion site for the Nsp1 coding region occurs between BCoV nt 906 and MHV nt 909. (B) Plaque phenotypes for the progeny from the variant constructs described for panel A. (C) Growth kinetics of the progeny depicted in B. (D) Northern analyses of the progeny depicted in for panel B.
These results are fully consistent with the notion that functional base pairing between the two domains is needed for virus replication. Still, the compatibility in these constructs was not perfect, since the titer of progeny was 100-fold lower than that for wt MHV. Interestingly, when the entire BCoV 5′-proximal region of the genome through the Nsp1 coding region to nt 906 was tested in construct B1-906/M, CPE was found within 24 hpt and viral progeny produced plaques, growth kinetics, a final titer, and a sgmRNA profile that were indistinguishable from those of wt MHV (Fig. 6A to D). These results further demonstrate a requirement for compatibility between the 5′ UTR and cognate Nsp1 coding region for BCoV and MHV when used in the MHV background for replication and fitness studies in cell culture. Without a reverse genetics system for BCoV, we could not apply the reciprocal test for this finding.
The 30-nt inter-stem-loop domain in the 5′ UTR is not an absolute requirement for MHV replication in cell culture.
Since in MHV, distinct regions in the 5′ UTR, including SLIII and the loop region of SLIV (Fig. 1), and a large region of the 3′ UTR have been shown to be dispensable for virus replication in cell culture (20, 22, 50), this test was applied to the 30-nt inter-stem-loop domain in MHV. Surprisingly, a deletion of nt 141 to 170 in the MHV genome (Fig. 7A) was not lethal, but rather, CPE appeared after one blind cell passage following transfection of the mutated genomic transcript, and progeny showing wt-like large plaques and a final growth titer of 1.0 × 106 PFU/ml were obtained (Fig. 7B and C). Interestingly, an A649C mutation (H147P) was found in the Nsp1 coding region of all progeny, and G516A (V103I), C583U (T125I), and A880C (E224A) mutations were found in some but not all progeny at VP10 (Fig. 7C). Furthermore, a UCUAA sequence was found inserted within the upstream transcription-regulatory sequence of all progeny by VP10 (Fig. 7A and C). These results together with the observation of very poor viral growth when the BCoV inter-stem-loop domain replaced the homologous native MHV domain demonstrate that the inter-stem-loop domain is required for optimal viral fitness but is not absolutely required for viral replication in cell culture.
Fig 7.
Deletion of the 30-nt inter-stem-loop domain in MHV leads to viable but debilitated progeny. (A) Schematic depiction of the MHV 5′ UTR with a deleted inter-stem-loop region (Δ141–170) and the inserted UCUAA element. The UCUAA insertion could theoretically have occurred following nt 59, 64, or 69 (illustrated as the latter). (B) Plaque patterns for plaque clones 1 to 3 at virus passages 1 and 10. (C) Genotypes and virus titers for the viruses depicted in panel B.
DISCUSSION
Here, we report the discovery that for the bovine coronavirus and mouse hepatitis coronavirus, separate species in the (formerly group 2a) betacoronavirus genus, there exists a functional interdependence between their respective 5′ UTRs and Nsp1 coding regions for optimal virus growth in cell culture and that part of that interdependence is a long-range (∼200-nt) RNA-RNA interaction. Furthermore, it was observed that whereas only some parts of the BCoV 5′ UTR and Nsp1 coding region can be transplanted into the MHV genome to directly yield viable virus and some parts cannot (22), when the entire BCoV 5′ UTR and Nsp1 coding region are transplanted together as a cognate unit, nearly fully fit MHV wt-like progeny are directly obtained, and there appear to be no interspecies constraints on replication. These results therefore show that the BCoV cis-acting RNA elements in the 5′ UTR and Nsp1 coding region, although evolutionarily divergent (∼36% nucleotide divergence in the 5′ UTR, ∼40% nucleotide divergence in the Nsp1 coding region, and ∼53% amino acid divergence in the Nsp1 protein), are successfully recognized as a cognate unit and utilized by the MHV proteins for genome replication and packaging with apparently no detrimental effect. The long-range RNA-RNA interaction function for replication also explains, at least in part, the previously reported cis-replication requirement for the 5′-terminal part of the Nsp1 coding region in BCoV defective interfering (DI) RNA (7, 8) and the presence of this region in all known naturally occurring group 1 and group 2 coronavirus DI RNAs (3, 4). They possibly also explain, in part, the requirement for this region in the MHV genome for virus replication (5).
Is the identified long-range RNA-RNA interaction a common feature of coronaviruses? In a recent comprehensive prediction of coronavirus 5′-end-proximal higher-order structures based predominantly on Mfold and phylogenetic and covariation analyses by Chen and Olsthoorn (10), seven substantial stem-loops (1 through 5, and unnamed 6 and 7) were described for the 5′-terminal 340 nt of group 2a coronaviruses that, for BCoV, are essentially identical to the seven stem-loops depicted in Fig. 1B (SL1, -2, -II, -III, and -IV) and in references 23 (SLV) and 7 (SLVI) (sequence differences are due to the different strains of BCoV used, BCoV-Mebus versus BCoV-Ent). It is of interest that for both BCoV and MHV, stem-loops V and VI (equivalent to stem-loops 6 and 7 in reference 10; also data not shown) map entirely within the Nsp1 coding region and (except for possibly a short region as noted below) lie between the upstream and downstream partnering domains in the long-range RNA-RNA interaction. A possibly analogous long-range RNA-RNA interaction is predicted for each of the group 2b, 2c, and 2d coronaviruses as well, but in these cases, in contrast to the group 2a coronaviruses, the long-distance upstream and downstream base-pairing partners are contained entirely within the stem of SL5 (equivalent to SLIV in Fig. 1; see also Fig. 4C, and see Fig. 3B in reference 10). SL5 for groups 2b, 2c, and 2d coronaviruses also harbors a large terminal loop that, in turn, contains three smaller stem-loops with primarily hexameric (UUYCGU) terminal loops (10). That is, for the group 2b SCoV-like coronaviruses (harboring an 80-nt terminal loop), the group 2c BtCoV-HKU5-1-like coronaviruses (harboring a 94-nt terminal loop), and the group 2d BtCoV-HKU9-1-like coronaviruses (harboring a 55-nt terminal loop), there is a long-range RNA-RNA interaction, but the upstream portion is nearly entirely within the 5′ UTR and the downstream portion is confined to the 5′-terminal 29 to 113 nt of the Nsp1 coding region. In a pattern similar to that of the group 2b through 2d coronaviruses, long-range RNA-RNA interactions that might also be analogous are predicted within SL5 of the group 1a TGEV-Purdue-like coronaviruses (with a 106-nt terminal loop), the group 1b HCoV-229E-inf-1-like coronaviruses (with a 100-nt terminal loop), the group 1c PEDV-CV777-like coronaviruses (with a 116-nt terminal loop), and the group 1d GId BtCoV-1A-like coronaviruses (with a 114-nt terminal loop). That is, in the group 1 coronaviruses, the upstream portion of the long-range interaction and the terminal loop map entirely within the 5′ UTR and the downstream portion is confined to the 5′-terminal 9 to 13 nt of the Nsp1 coding region (10). Intriguingly, three small stem-loops containing mostly the UUYCGU loop, like those in the group 2b through 2d coronaviruses, make up the large terminal loop of SL5 in the group 1 coronaviruses (10, 42). Within the group 3 coronaviruses, no base pairing between the 5′ UTR and open reading frame 1 coding region was noted (10). For coronaviruses other than MHV and BCoV, the functional significance of the specific long-range RNA-RNA interaction, to our knowledge, has not been experimentally examined; however, the large terminal loop in SL5 of the group 2b through 2d and group 1 coronaviruses is a postulated genome packaging signal (10, 18).
One interesting feature of the BCoV 72-nt SLV and 30-nt SLVI that map within the loop of the long-range RNA-RNA interaction is that each functions as a higher-order cis-replication element for DI RNA in virus-infected cells based on analysis of synonymous structure-disrupting and compensatory structure-restoring mutations (7, 23). In experiments concurrent with those described above, attempts were made to analyze the cis-acting role of MHV SLV and SLVI in viral replication. For this, however, structure-disrupting synonymous mutations were found available in only the MHV 83-nt SLV (nt 233 through 315) in which mutations C242U and C239U as a pair in the left strand and G305A and G308A as a pair in the right strand were separately made and tested. These mutations altered SLV ΔG from −32.5 kcal/mol to −26.7 and −26.9 kcal/mol, respectively, but resulted in no changes in progeny plaque size or replication kinetics (data not shown). Hence, the potential role of MHV SLV and SLVI in virus replication has not yet been determined. In a retrospective analysis relevant to the long-range RNA-RNA interaction, however, we noted that the downstream 9 nt of SLVI in BCoV (nt 332 to 340) (7) are also part of the downstream base-pairing window in the long-range RNA-RNA interaction (Fig. 1B). That is, the mutations that destabilized the lower stem of SLVI in BCoV DI RNA (C333G and C336U) (7) would also have decreased base pairing in the long-range RNA-RNA interaction from 25 to 24 (Fig. 1B). Thus, either disruption (i.e., that in SLVI or that in the long-range RNA-RNA interaction) may have been the lesion that blocked DI RNA replication. This arrangement may suggest a possible functional switching between two alternate structures. A similar arrangement involving the four 3′-terminal nucleotides in MHV SLVI (nt 332 to 335) is also possible (data not shown).
Inasmuch as the 5′ UTR-mapping 30-nt inter-stem-loop component of the long-range RNA-RNA interaction can be deleted without killing MHV replication (Fig. 7), the question of what role(s) the long-range RNA-RNA interaction might play that could be considered optional for virus survival arises. Currently, we can only speculate from known functions of 5′-end-proximal cis-acting replication elements in coronaviruses and other plus-strand RNA viruses (see the introduction). Based on available data, it is plausible that the long-range RNA-RNA interaction plays a role in one or both of two potential regulatory pathways. First, it might regulate an optional noncanonical translation initiation site in a manner that ensures viral genome translation under conditions of cell stress when canonical translation initiation mechanisms are largely repressed (1, 26, 30, 44). This role is suggested by the demonstration of a potential internal ribosomal entry site on group 3 coronavirus genomic RNA that needs yet to be demonstrated in other coronavirus groups (34) and by robust translation of MHV RNAs during global inhibition of cellular mRNA translation (2, 40, 45). Such an alternate mechanism of translation initiation has been shown to function on some capped and polyadenylated viral and cellular mRNAs, to be influenced by double-stranded RNA regions near the translation start codon, and to be under the control of trans-acting proteins (1, 26, 30, 44).
A second potential regulatory pathway for the long-range RNA-RNA interaction could be maintenance of a proper acceptor orientation for the RNA-dependent RNA polymerase (RdRp) template switching during generation of minus-strand templates for subgenomic mRNA synthesis (43, 49, 55). This is suggested by the appearance of the UCUAA component within the UCUAAAC intergenic transcriptional regulatory sequence following deletion of the 30-nt inter-stem-loop region (Fig. 7). Addition of the UCUAA element in the same region was also found when stem-loop 4 (referred to as SLIII in Fig. 1A) was deleted, which led to the suggestion that stem-loop 4 functions as a required spacer element for sgmRNA synthesis (50). In earlier studies, the UCUAA element was also found to spontaneously appear at this site in some MHV strains in which its occurrence positively correlated with enhanced transcription (37, 51, 53). Regulation of mRNA synthesis is also suggested by the increased synthesis of sgmRNAs when amino acid-changing mutations were made in the (then unknown) downstream partnering domain of the MHV long-range interaction during the analysis of Nsp1 function by Brockway and Denison (5). For Nsp1 functional analysis in MHV-A59, 13 amino acids within the amino-terminal third of Nsp1 were changed to alanine by reverse genetics and tested for viability. Although only some of the mutations were found to be lethal, two tested simultaneously, E46A and K48A, made by A346C, A351G, and A352C mutations, were not lethal but inexplicably caused an ∼20% increase in overall viral RNA synthesis (5). Although these three mutations which map within the long-range base-pairing window would have decreased the base pairings only within the window from 27 to 25, there might have been enough RNA rearrangement to affect upstream RdRp template switching and an increase in subgenomic mRNA synthesis. Alternatively, the amino acid changes could have caused alternate template switching behavior by an entirely different unknown mechanism.
From the present study, there is no clear evidence to suggest that MHV behavior was altered as a function of amino acid changes arising from suppressor mutations. However, in no case was an amino acid-changing mutation found in the absence of other potential suppressor mutations, making it unfeasible to assess the effects of single amino acid changes alone. Of the five suppressor mutations within the downstream Nsp1 coding region, four (S3, S4, L2, and L5) led to amino acid changes within Nsp1 (K52E, P47T, K48N, and K50T, respectively) (Table 1), but progeny from each of these made large plaques (Fig. 2A and data not shown), and two, S3 and L2, had near-wt growth kinetics at passage 10 (Fig. 2B). No evaluation of viral RNA levels was made for any of these, however.
ACKNOWLEDGMENTS
We thank Ralph Baric for the MHV-A59-1000 reverse genetics system.
This work was supported by Public Health Service grant AI014367 from the National Institute of Allergies and Infectious Diseases and by funds from the University of Tennessee Center of Excellence in Livestock Diseases and Human Health.
Footnotes
Published ahead of print 15 February 2012
REFERENCES
- 1. Balvay L, Soto Rifo R, Ricci EP, Decimo D, Ohlmann T. 2009. Structural and functional diversity of viral IRESes. Biochim. Biophys. Acta 1789:542–557 [DOI] [PubMed] [Google Scholar]
- 2. Bechill J, Chen Z, Brewer JW, Baker SC. 2008. Coronavirus infection modulates the unfolded protein response and mediates sustained translational repression. J. Virol. 82:4492–4501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brian DA, Baric RS. 2005. Coronavirus genome structure and replication. Curr. Top. Microbiol. Immunol. 287:1–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brian DA, Spaan WJM. 1997. Recombination and coronavirus defective interfering RNAs. Semin. Virol. 8:101–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brockway SM, Denison MR. 2005. Mutagenesis of the murine hepatitis virus nsp1-coding region identifies residues important for protein processing, viral RNA synthesis, and viral replication. Virology 340:209–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brockway SM, Lu XT, Peters TR, Dermody TS, Denison MR. 2004. Intracellular localization and protein interactions of the gene 1 protein p28 during mouse hepatitis virus replication. J. Virol. 78:11551–11562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brown CG, Nixon KS, Senanayake SD, Brian DA. 2007. An RNA stem-loop within the bovine coronavirus nsp1 coding region is a cis-acting element in defective interfering RNA replication. J. Virol. 81:7716–7724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chang RY, Brian DA. 1996. cis requirement for N-specific protein sequence in bovine coronavirus defective interfering RNA replication. J. Virol. 70:2201–2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chang RY, Hofmann MA, Sethna PB, Brian DA. 1994. A cis-acting function for the coronavirus leader in defective interfering RNA replication. J. Virol. 68:8223–8231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen SC, Olsthoorn RC. 2010. Group-specific structural features of the 5′-proximal sequences of coronavirus genomic RNAs. Virology 401:29–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen W, Baric RS. 1996. Molecular anatomy of mouse hepatitis virus persistence: coevolution of increased host cell resistance and virus virulence. J. Virol. 70:3947–3960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen W, Madden VJ, Bagnell CR, Jr., Baric RS. 1997. Host-derived intracellular immunization against mouse hepatitis virus infection. Virology 228:318–332 [DOI] [PubMed] [Google Scholar]
- 13. Chen Y, et al. 2009. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc. Natl. Acad. Sci. U. S. A. 106:3484–3489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Decroly E, et al. 2008. Coronavirus nonstructural protein 16 is a cap-0 binding enzyme possessing (nucleoside-2′O)-methyltransferase activity. J. Virol. 82:8071–8084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. de Groot RJ, Ziebuhr J, Poon LL, Woo PC, et al. 2008. Proposal to divide the family Coronaviridae into subfamilies, Coronavirinae and Torovirinae, and rationalize the classification of species. http://talk.ictvonline.org/files/ictv_official_taxonomy_updates_since_the_8th_report/m/vertebrate-2008/1230.aspx
- 16. Denison M, Perlman S. 1987. Identification of putative polymerase gene product in cells infected with murine coronavirus A59. Virology 157:565–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Denison MR, et al. 2004. Cleavage between replicase proteins p28 and p65 of mouse hepatitis virus is not required for virus replication. J. Virol. 78:5957–5965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Escors D, Izeta A, Capiscol C, Enjuanes L. 2003. Transmissible gastroenteritis coronavirus packaging signal is located at the 5′ end of the virus genome. J. Virol. 77:7890–7902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gaertner DJ, Winograd DF, Compton SR, Paturzo FX, Smith AL. 1993. Development and optimization of plaque assays for rat coronaviruses. J. Virol. Methods 43:53–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Goebel SJ, Miller TB, Bennett CJ, Bernard KA, Masters PS. 2007. A hypervariable region within the 3′ cis-acting element of the murine coronavirus genome is nonessential for RNA synthesis but affects pathogenesis. J. Virol. 81:1274–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gorbalenya AE, Enjuanes L, Ziebuhr J, Snijder EJ. 2006. Nidovirales: evolving the largest RNA virus genome. Virus Res. 117:17–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Guan BJ, Wu HY, Brian DA. 2011. An optimal cis-replication stem-loop IV in the 5′ untranslated region of the mouse coronavirus genome extends 16 nucleotides into open reading frame 1. J. Virol. 85:5593–5605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gustin KM, Guan BJ, Dziduszko A, Brian DA. 2009. Bovine coronavirus nonstructural protein 1 (p28) is an RNA binding protein that binds terminal genomic cis-replication elements. J. Virol. 83:6087–6097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hirano N, Fujiwara K, Matumoto M. 1976. Mouse hepatitis virus (MHV-2). Plaque assay and propagation in mouse cell line DBT cells. Jpn. J. Microbiol. 20:219–225 [PubMed] [Google Scholar]
- 25. Hofmann MA, Sethna PB, Brian DA. 1990. Bovine coronavirus mRNA replication continues throughout persistent infection in cell culture. J. Virol. 64:4108–4114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Holcik M, Sonenberg N. 2005. Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 6:318–327 [DOI] [PubMed] [Google Scholar]
- 27. Kamitani W, Huang C, Narayanan K, Lokugamage KG, Makino S. 2009. A two-pronged strategy to suppress host protein synthesis by SARS coronavirus Nsp1 protein. Nat. Struct. Mol. Biol. 16:1134–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kamitani W, et al. 2006. Severe acute respiratory syndrome coronavirus nsp1 protein suppresses host gene expression by promoting host mRNA degradation. Proc. Natl. Acad. Sci. U. S. A. 103:12885–12890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kang H, Feng M, Schroeder ME, Giedroc DP, Leibowitz JL. 2006. Putative cis-acting stem-loops in the 5′ untranslated region of the severe acute respiratory syndrome coronavirus can substitute for their mouse hepatitis virus counterparts. J. Virol. 80:10600–10614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Komar AA, Hatzoglou M. 2011. Cellular IRES-mediated translation: the war of ITAFs in pathophysiological states. Cell Cycle 10:229–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lai MM, Stohlman SA. 1981. Comparative analysis of RNA genomes of mouse hepatitis viruses. J. Virol. 38:661–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Law AH, Lee DC, Cheung BK, Yim HC, Lau AS. 2007. Role for nonstructural protein 1 of severe acute respiratory syndrome coronavirus in chemokine dysregulation. J. Virol. 81:416–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li L, et al. 2008. Structural lability in stem-loop 1 drives a 5′ UTR-3′ UTR interaction in coronavirus replication. J. Mol. Biol. 377:790–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu DX, Xu HY, Lim KP. 1998. Regulation of mRNA 1 expression by the 5′-untranslated region (5′-UTR) of the coronavirus infectious bronchitis virus (IBV). Adv. Exp. Med. Biol. 440:303–311 [DOI] [PubMed] [Google Scholar]
- 35. Liu P, et al. 2009. Mouse hepatitis virus stem-loop 2 adopts a uYNMG(U)a-like tetraloop structure that is highly functionally tolerant of base substitutions. J. Virol. 83:12084–12093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu P, et al. 2007. A U-turn motif-containing stem-loop in the coronavirus 5′ untranslated region plays a functional role in replication. RNA 13:763–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Makino S, Lai MM. 1989. Evolution of the 5′-end of genomic RNA of murine coronaviruses during passages in vitro. Virology 169:227–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mathews DH, Sabina J, Zuker M, Turner DH. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911–940 [DOI] [PubMed] [Google Scholar]
- 39. Narayanan K, et al. 2008. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J. Virol. 82:4471–4479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Raaben M, Groot Koerkamp MJ, Rottier PJ, de Haan CA. 2007. Mouse hepatitis coronavirus replication induces host translational shutoff and mRNA decay, with concomitant formation of stress granules and processing bodies. Cell Microbiol. 9:2218–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Raman S, Bouma P, Williams GD, Brian DA. 2003. Stem-loop III in the 5′ untranslated region is a cis-acting element in bovine coronavirus defective interfering RNA replication. J. Virol. 77:6720–6730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Raman S, Brian DA. 2005. Stem-loop IV in the 5′ untranslated region is a cis-acting element in bovine coronavirus defective interfering RNA replication. J. Virol. 79:12434–12446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sawicki SG, Sawicki DL, Siddell SG. 2007. A contemporary view of coronavirus transcription. J. Virol. 81:20–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Spriggs KA, Bushell M, Willis AE. 2010. Translational regulation of gene expression during conditions of cell stress. Mol. Cell 40:228–237 [DOI] [PubMed] [Google Scholar]
- 45. Tahara SM, et al. 1994. Coronavirus translational regulation: leader affects mRNA efficiency. Virology 202:621–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vogt DA, Andino R. 2010. An RNA element at the 5′-end of the poliovirus genome functions as a general promoter for RNA synthesis. PLoS Pathog. 6:e1000936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wathelet MG, Orr M, Frieman MB, Baric RS. 2007. Severe acute respiratory syndrome coronavirus evades antiviral signaling: role of nsp1 and rational design of an attenuated strain. J. Virol. 81:11620–11633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wu HY, Brian DA. 2010. Subgenomic messenger RNA amplification in coronaviruses. Proc. Natl. Acad. Sci. U. S. A. 107:12257–12262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wu HY, Ozdarendeli A, Brian DA. 2006. Bovine coronavirus 5′-proximal genomic acceptor hotspot for discontinuous transcription is 65 nucleotides wide. J. Virol. 80:2183–2193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yang D, Liu P, Giedroc DP, Leibowitz J. 2011. Mouse hepatitis virus stem-loop 4 functions as a spacer element required to drive subgenomic RNA synthesis. J. Virol. 85:9199–9209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yokomori K, Banner LR, Lai MM. 1991. Heterogeneity of gene expression of the hemagglutinin-esterase (HE) protein of murine coronaviruses. Virology 183:647–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yount B, Denison MR, Weiss SR, Baric RS. 2002. Systematic assembly of a full-length infectious cDNA of mouse hepatitis virus strain A59. J. Virol. 76:11065–11078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang X, Lai MM. 1994. Unusual heterogeneity of leader-mRNA fusion in a murine coronavirus: implications for the mechanism of RNA transcription and recombination. J. Virol. 68:6626–6633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zuniga S, Sola I, Alonso S, Enjuanes L. 2004. Sequence motifs involved in the regulation of discontinuous coronavirus subgenomic RNA synthesis. J. Virol. 78:980–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zust R, et al. 2007. Coronavirus non-structural protein 1 is a major pathogenicity factor: implications for the rational design of coronavirus vaccines. PLoS Pathog. 3:e109. [DOI] [PMC free article] [PubMed] [Google Scholar]