Abstract
Routine serodiagnosis of herpes simplex virus (HSV) infections is currently performed using recombinant glycoprotein G (gG) antigens from herpes simplex virus 1 (HSV-1) and HSV-2. This is a single-antigen test and has only one diagnostic application. Relatively little is known about HSV antigenicity at the proteome-wide level, and the full potential of mining the antibody repertoire to identify antigens with other useful diagnostic properties and candidate vaccine antigens is yet to be realized. To this end we produced HSV-1 and -2 proteome microarrays in Escherichia coli and probed them against a panel of sera from patients serotyped using commercial gG-1 and gG-2 (gGs for HSV-1 and -2, respectively) enzyme-linked immunosorbent assays. We identified many reactive antigens in both HSV-1 and -2, some of which were type specific (i.e., recognized by HSV-1- or HSV-2-positive donors only) and others of which were nonspecific or cross-reactive (i.e., recognized by both HSV-1- and HSV-2-positive donors). Both membrane and nonmembrane virion proteins were antigenic, although type-specific antigens were enriched for membrane proteins, despite being expressed in E. coli.
INTRODUCTION
Herpes simplex virus 1 (HSV-1) and HSV-2 cause significant human morbidity. HSV-2 is the causative agent of most recurrent genital herpes lesions and is sexually transmitted. Infections are often asymptomatic, and most infected individuals are unaware of the infection, yet HSV-2 is associated with an increased risk of HIV acquisition (33) and an increased risk during pregnancy of spontaneous abortion, premature birth, and perinatal herpes (12, 13). Unawareness of HSV-2 infection is also a major contributing factor to transmission to uninfected partners (64, 65). In contrast, HSV-1 is usually transmitted during childhood and is found to be associated predominantly with orolabial infections (cold sores). Also, HSV-1 infection of the eye (ocular herpes) is the most common cause of infectious corneal blindness in industrialized countries. Both HSV-1 and HSV-2 establish lifelong latent infections within the dorsal root and trigeminal ganglia and are characterized by periodic reactivation and virus shedding from mucocutaneous epithelium. Owing to the different natural histories and outcomes of HSV-1 and -2 infections, accurate diagnosis of the HSV type is important for patient management and prognosis and controlling potential transmission. For example, knowing the specific HSV type can help the patient take appropriate precautions to prevent transmission of the disease to others. In particular, the identification of unrecognized HSV-2 infection can be used to carefully monitor virus shedding during pregnancy and minimize the risk of perinatal infection.
Laboratory tests for HSV infection include virus culture, virus neutralization, PCR, and serological tests. Virus culture is considered the “gold standard” in the early stages of a primary infection. However, it is less sensitive during the healing stage of infection or during recurrent infections. Culture testing may also report false negatives if the sample is not collected and transported properly to maintain virus viability. PCR is more sensitive than virus culture (21, 81) but is not easily performed as a routine point-of-care test. Serological testing is the method of choice for a point-of-care test because of the simplicity of antibody tests and because of the limitations of the other approaches.
HSV-1 and HSV-2 show close amino acid sequence homology and therefore exhibit extensive antigenic cross-reactivity (51). In the past, this has hampered the discrimination between HSV-1 and -2 infections using serological approaches. However, the discovery that US4/glycoprotein G (gG) from HSV-1 and -2 (termed gG-1 and gG-2, respectively) were serologically distinct (56) led to the development of type-specific serological (TSS) tests which discriminate between HSV-1 and -2 antibodies (2, 3). Several TSS tests have been developed, but only those based on US4/gG show acceptable specificity and are FDA approved. The test has acceptable sensitivity in convalescent-phase individuals but is less sensitive in the early stages of infection (5, 6), a limitation that may be overcome by measurement of IgM or the discovery of another antigen(s) that detects IgG during acute infection. Moreover, single-antigen-based tests are prone to false negatives when infected individuals lack gG-specific antibodies in their profile (74). Western blots of whole virions can also discriminate between HSV-1 and HSV-2 infections and are more sensitive than gG-based tests (5). Virus neutralization by complement-fixing antibody is negative during the acute state of infection and less sensitive than gG-based tests such as the immunodot assay (5). Neither virus neutralization nor Western blotting is routinely available as a point-of-care test.
In addition to diagnostics, there has been considerable interest in the development of subunit vaccines against HSV. Subunit vaccines are inherently safer than conventional inactivated or attenuated live vaccines. The choice of vaccine targets for testing has been heavily influenced by prior defined antibody targets, and several glycoproteins (gD, gB, gC, and gE) show promise in animal challenge models (14, 30, 50, 55, 57, 59, 73). However, the outcomes of clinical trials have been disappointing (19, 77) but not too surprising, given that the current generation of subunit vaccines is based on only a very small number of antigens defined in the preproteomic era. New antigens, particularly those associated with cases of naturally occurring (asymptomatic) immunity, are needed (17, 23).
HSV encodes >80 different open reading frames (ORFs), including approximately 16 glycoproteins and other membrane-associated proteins. In recent years, new tools have been developed to enable the serological response to a pathogen to be profiled at the proteome level. In the study described in this report, we have used a proteome microarray platform to profile the antibody response in >90 characterized serum samples. The aim was to provide an overall picture of the antibody response. Although we see variation between individuals, the most commonly recognized antigens were structural proteins. Importantly, several HSV type-common and type-specific antigens that may be used to improve the sensitivity and specificity of existing tests and open up opportunities for vaccine development were discovered.
MATERIALS AND METHODS
Sera.
Blood samples were collected from patients attending the University of California, Irvine (UCI), Medical Center for diagnosis and treatment of herpesvirus infections. All samples were collected as part of an immunological study with consent and under local institutional review board (IRB) approval (UCI IRB protocol 2009-6963). For the present study, the serum fraction was separated from each blood sample and stored at −80°C prior to use. All sera were assayed by FDA-approved commercial HSV-1 and HSV-2 enzyme-linked immunosorbent assays (ELISAs; HerpeSelect; Focus Diagnostics) according to the manufacturer's instructions. All infections were latent, and no acute cases were examined. Sera representing the healthy adult population in the same geographical location as the patients (Orange County, CA) were collected by the UCI General Clinical Research Center (GCRC) with consent and under local IRB approval (UCI IRB protocol 2007-5896).
Construction of HSV-1 and -2 proteome microarrays.
Proteome microarrays were fabricated essentially as described previously (24, 54) by PCR amplification of coding sequences in genomic DNA and insertion of amplicons into a T7 expression vector by homologous recombination, followed by expression in coupled transcription-translation in vitro (IVTT) and direct printing onto microarrays. Gene sequences for PCR primer design were obtained from NCBI (GenBank accession numbers NC_001806 and NC_001798 for HSV-1 strain 17 and HSV-2 strain HG52, respectively). The gene nomenclature used is as published in the curated Oral Pathogen Genome Sequence Databases (Oralgen) at the Los Alamos National Laboratory (http://www.oralgen.lanl.gov/). Template DNA was a generous gift from Dale Carpenter and Steve Wechsler, UCI Department of Ophthalmology. HSV-1 strain 17 DNA was supplied as 5 overlapping genomic fragments cloned into cosmids (22). HSV-2 strain 333 DNA was prepared from virion-extracted DNA or purchased from ATCC.
Primers used for PCR amplification contained 20 bp nucleotides specific for each gene with an extension of 20 bp complementary to ends of linear pXT7 vector at the 5′ ends (24, 54). The genomes of herpes simples viruses are GC rich (68% for HSV-1 and 70% for HSV-2). For PCR, genes were amplified using AccuPrime GC-rich DNA polymerase (catalog no. 12337-016; Invitrogen) or 2× Phusion High-Fidelity PCR master mix with GC buffer (catalog no. F-532S; Finnzymes/Thermo Scientific) with addition of dimethyl sulfoxide (DMSO; final concentration, 2%) and 8 ng/μl bovine serum albumin (BSA), using touchdown PCR with cycling conditions of initial denaturation at 98°C/1 min, followed by 20 cycles of 98°C/10 s, 68°C/20 s with decremental temperature of 0.5°C/cycle, and 72°C for 30 s/kb, followed by 20 cycles of 98°C/10 s, 58°C/20 s, and 72°C/30 s/kb. In vivo homologous recombination takes place between the PCR product and pXT7 vector in competent Escherichia coli DH5α cells. The recombinant plasmids were isolated from this culture using a QIAprep 96 Turbo kit (Qiagen). All recombinant plasmids were confirmed as containing the insert by quality control PCR (QC-PCR), in which a band of expected size was amplified from the recombinant using the same primers used in the original PCR. Plasmids that generated strong hits on the array (see below) were also confirmed by sequencing.
For array fabrication, purified minipreparations of DNA were expressed in an E. coli-based IVTT expression system (Expressway cell-free expression system; Invitrogen). Twenty-microliter reaction mixtures (comprising 4.8 μl E. coli lysate as a source of ribosomes, 4.0 μl reaction mixture containing T7 RNA polymerase, 5.2 μl amino acid mixture, 2 μl buffer, and 4 μl plasmid DNA) were set up in sealed 384-well plates and incubated for 16 h on a platform shaker at 250 rpm at 24°C. A protease inhibitor cocktail (Cømplete; Roche) and Tween 20 to a final concentration of 0.05% were then added prior to printing. The expressed protein reactions were printed in singlicate without further purification onto 8-pad nitrocellulose-coated Oncyte Nova slides (Grace Bio-Labs) using an OmniGrid Accent 100 microarray printer (Genomic Solutions) in a 1-by-4 subarray format. Each subarray included multiple negative-control spots comprising mock IVTT expression reaction mixtures lacking DNA template. Each subarray also included positive-control spots of four serial dilutions of a mixture of mouse, rat, and human IgG and two serial dilutions of human IgM. Together, these positive and negative controls are used to normalize the data from different arrays (see below). Also included were four serial dilutions of purified recombinant Epstein-Barr virus nuclear antigen-1 (EBNA-1; DevaTal Inc.), which is recognized by the majority of humans and which serves as a useful guide to serum quality.
To monitor the protein expression in each spot, we used antibodies against the N-terminal poly-His (clone His-1; Sigma) and the C-terminal hemagglutinin (HA; clone 3F10; Roche) tags engineered into each protein. Arrays were first blocked for 30 min in protein array blocking buffer (Whatman) at room temperature and then probed for 1 h with antitag antibodies diluted 1/1,000 in blocking buffer. The slides were then washed 3 times in Tris-buffered saline (TBS) containing 0.05% (vol/vol) Tween 20 (T-TBS) and incubated in biotinylated secondary antibodies (Jackson ImmunoResearch). After washing the slides 3 times in T-TBS, bound antibodies were detected by incubation with streptavidin-conjugated SureLight P-3 (Columbia Biosciences). The slides were then washed 3 times in T-TBS, followed by washing 3 times in TBS, and dipped in distilled water prior to air drying by brief centrifugation. Slides were scanned in a Perkin Elmer ScanArray confocal laser scanner, and data were acquired using ScanArrayExpress software.
For probing with human sera, samples were diluted to 1/100 in 1× protein array blocking buffer supplemented with E. coli lysate (Antigen Discovery, Inc.) at a final concentration of 10 mg/ml protein to block anti-E. coli antibodies and incubated at 37°C for 30 min with constant mixing. Meanwhile, the arrays were blocked with protein array blocking buffer for 30 min. The blocking buffer was removed, and the arrays were probed with the pretreated sera overnight at 4°C with gentle rocking. The slides were then washed 3 times in T-TBS and incubated in biotinylated anti-human IgG Fc (Jackson ImmunoResearch) diluted 1/200 in protein array blocking buffer. After washing the slides 3 times each in T-TBS and TBS, bound antibodies were visualized as described above. Control arrays were similarly probed, except that the primary antibody was replaced by protein array blocking buffer supplemented with E. coli lysate. To minimize any potential variability between probing and scanning performed on different days or between different batches of arrays, the data shown are derived from sera that were all probed at the same time using the same batch of arrays (i.e., from the same print run). These data are representative of two separate experiments.
Array data analysis and statistical treatment.
Raw data were collected as the mean pixel signal intensity data for each spot. Raw data from each donor were subtracted from the average of the negative-control spots (IVTT reaction mixtures lacking plasmid template) and then log transformed and normalized against the negative-control spots (IVTT lacking DNA template) using the VSN package in the statistical environment R from the Bioconductor suite (http://Bioconductor.org/). Antigens were defined to be seroreactive when the log-normalized signals were above a cutoff determined for each donor as the mean plus 2 standard deviations (SDs) of the negative-control spots. Pairs of donor populations were compared using a Bayes-regularized t test adapted from Cyber-T software for use with protein arrays (7, 29, 38, 53, 78, 79), and P values were corrected for multiple test conditions using the Benjamini-Hochberg (BH) method (10). Positive antigens were classified as type specific or cross-reactive according to significance (BH P [pBH] values, <0.05 and ≥0.05, respectively). Receiver operator characteristic (ROC) analyses were performed with log-normalized array data for single antigens by testing signals for each donor as the threshold cutoff to discriminate HSV-1 and HSV-2 infection. The area under the curve (AUC) was used as a relative measure of each antigen's ability to discriminate between HSV-1 and -2 infections. For frequency of recognition (seroprevalence) analysis, a cutoff was defined for each antigen on the array using the log-normalized data and set as the average signal plus 3 SDs of the ELISA-defined seronegative population. The numbers of individuals above the cutoff in each of the seropositive groups were determined and expressed as a percentage. For graphic representations, log-normalized data were retransformed to a scale approximating the raw data using 2 to the exponent of the log value.
Enrichment analysis.
An enrichment analysis was performed to determine if there were structural or functional properties of HSV proteins that favored their recognition by the antibody response. Proteins in HSV-1 (strain 17) were grouped using the gene ontology (GO) components defined by uniprot.org. For each group, the numbers of proteins represented on the array (i.e., the proteome) and the numbers that were seroreactive were determined as percentages of the respective totals, and the ratio was used to determine enrichment. A group with a ratio of >1.5 was considered to be enriched relative to the proteome; that is, proteins of that type were overrepresented among the antibody targets compared with the number that would be expected by random recognition. P values for enrichment statistical analysis were calculated using Fisher's exact test in the R environment.
Purified protein ELISAs.
ELISAs were developed essentially as described previously (36). To facilitate expression in E. coli BL21 cells, HSV glycoproteins were engineered without signal sequences and transmembrane domains by redesigning PCR primers. Transformants were colony selected, and a single sequence-verified clone was used for protein expression. Proteins were autoinduced in Magic medium (Invitrogen), and after overnight incubation at 37°C, cells were lysed in BugBuster reagent (Novagen) and recombinant proteins were purified from the insoluble fraction as SDS-solubilized inclusion bodies and/or the soluble fraction using nickel chelate columns (nickel-nitrilotriacetic acid His Bind; Novagen), as described previously (36). Optimal antigen-coating concentrations were first determined by dilution experiments with control sera. The 90 serum samples used for array probing were then assayed at a dilution of 1/150 in casein blocking buffer (Blocker; Thermo Scientific). Bound antibodies were detected with anti-human IgG secondary antibody conjugated to horseradish peroxidase (Bethyl Labs), followed by tetramethylbenzidine developer (SureBlue reserve; KPL, Gaithersburg, MD), and optical densities (ODs) were measured at 450 nm.
RESULTS
Production of HSV-1 and HSV-2 protein microarrays.
DNA with high G+C content has a higher melting temperature and is inherently more difficult to amplify by PCR and sequence. For PCR, several rounds were required to obtain a full ORF library, each employing slightly different conditions. In the first round of PCR, in which the bulk of the amplicons were obtained, AccuPrime GC-rich DNA polymerase (Invitrogen) was used. Those that failed to amplify were then subjected to a second round of PCR using Phusion DNA polymerase and GC buffer (Finnzymes), and the amplification of remaining ORFs in a third round was attempted using the same buffer modified by the addition of BSA and DMSO.
The DNA genomes of HSV-1 and -2 both comprise two unique coding segments, termed short and long (US and UL, respectively), each of which is flanked by repeat elements. The HSV-1 genome is 152,260 bp and has an overall base composition 68.3% G+C (see reference 60 and papers cited therein). The UL segment is 107,943 bp and encodes ORFs UL1 to UL56. UL15 (5,794 bp) contains an intron producing exons 1 and 2 (1,094 and 1,243 bp, respectively), which were amplified and cloned separately. The shorter US segment encodes ORFs US1 to US12. The HSV-2 genome is 154,746 bp with a G+C content of 70.4% (26). The genome encodes UL1 to UL56 and US1 to US12. In addition, we cloned UL26.5, UL49.5, and US8.5 from both HSV-1 and -2 (see Table S1 in the supplemental material). UL36 (9,495 bp) was successfully amplified from HSV-2 only, although the plasmid failed to express detectable protein. We also cloned repeat region intermediate early (IE) genes RL1/ICP34.5, RL2/ICP0, and RS1/ICP4 from HSV-1 and -2 (which are additional to IE genes UL54/ICP27 and US12/ICP47 and putative IE genes UL29/ICP8 and UL39/ICP6). The LAT genes of HSV-1 and 2 were successfully amplified and cloned, but details will be described elsewhere.
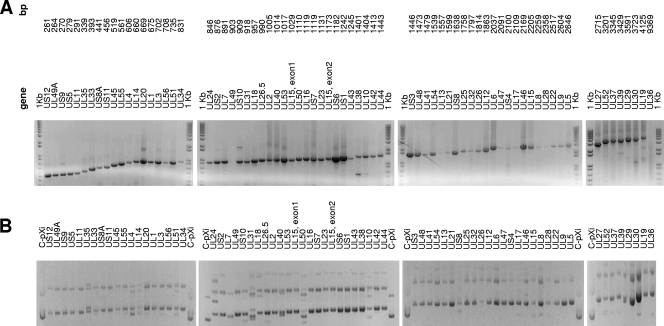
Amplicons were inserted onto the T7 expression vector pXi by homologous recombination using our standard methodology (24, 54). Gels showing the HSV-2 PCR amplicon library and corresponding DNA minipreparations are shown in Fig. 1A and B, respectively, and are representative of those for HSV-1 (data not shown). Recombinants were first verified using QC-PCR, in which the recombinant is checked for an insert of the correct size using the original ORF-specific PCR primers. Where possible, verification by sequencing was preferred, although some plasmids were difficult to sequence owing to high G+C content.
Fig 1.
Construction of expressible HSV ORFeome by PCR and recombination cloning. (A) Gel showing PCR amplicon library arranged by predicted size. (B) Gel showing corresponding DNA minipreparations after recombination cloning of PCR amplicons into expression plasmid pXi. All plasmids are circular/nonlinearized. C-pXi, control (nonrecombinant) pXi plasmid. Results for HSV-2 are shown. Results for HSV-1 were similar (data not shown).
Verified plasmids were expressed in 16-h IVTT reactions and printed, and the arrays were probed with antipolyhistidine and anti-HA epitope tags to evaluate expression of each of the proteins. A total of 99% of HSV-1 proteins and 97% of HSV-2 proteins printed were reactive with one or both of the antitag antibodies, and the remaining 1% and 3% were negative for both. The double negatives were HSV-1 UL54 and HSV-1 UL19 and UL39 and were also nonreactive with immune sera.
Despite repeated attempts, we were unable to produce seroreactive HSV-2 US4 (gG), as will be seen below. A PCR product of the expected size (2,100 bp) was obtained and successfully inserted into the pXi expression vector by homologous recombination, and an insert of the correct size was amplified by QC-PCR. Sequencing confirmed the presence of a full-length insert of the correct sequence, although plasmids with truncated inserts were also present. The IVTT product of this plasmid preparation was not recognized by HSV-2 immune sera. Extensive individual colony selection was then performed, although only truncated genes were obtained, and none were reactive with immune sera.
Overview of HSV-1 and HSV-2 antibody profiles.
Sera from a total of 90 patients attending the UCI Medical Center were used to probe chips displaying the HSV-1 and HSV-2 protein microarrays. The serological status of each patient was determined using commercially available ELISAs (HerepeSelect 1 and 2 IgG), and the statuses were as follows: seronegative (n = 47), HSV-1 positive only (n = 32), HSV-2 positive only (n = 6), and HSV-1/HSV-2 double positive (n = 5). In addition, 21 randomly selected healthy individuals from the same geographical location as the patient samples were probed, giving 5 donor groups.
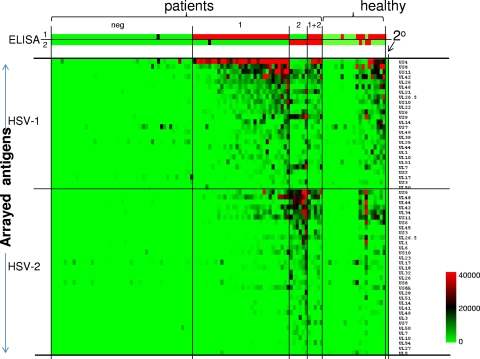
The heat map, shown in Fig. 2, provides an overview of the reactivity of all these samples. Only those antigens that were seroreactive are shown, i.e., where the average signal intensity of one or more of the 5 donor groups was above a cutoff defined as C plus 2 SDs (where C is the average of the IVTT control spots lacking DNA template). By this criterion, 26 different HSV-1 antigens and 33 different HSV-2 antigens were seroreactive. The heat map shows that the seronegative samples are also minimally reactive against HSV-1 or -2 antigens on the array, whereas the samples from the seropositive donors showed elevated reactivity to many HSV antigens, as expected. Overall, the HSV-1-seropositive donor samples (defined by commercial ELISA) reacted preferentially with HSV-1 antigens on the array and the HSV-2-seropositive donor samples reacted preferentially with HSV-2 antigens. However, both patient groups showed reactivity to one or more antigens from the reciprocal virus type, and this reactivity is likely to represent cross-reactivity. For example, the heat map indicates that HSV-2-seropositive donors react to HSV-1 antigens US9 and UL7. Both antigens are components of the HSV-2 profile and conserved between the viruses, and the antibodies may therefore be cross-reactive. Indeed, these donors react well to both the HSV-1 and HSV-2 orthologs of US9, consistent with the notion of cross-reactivity. Responses to the orthologs of UL7 are stronger for the HSV-1 version, although it is important to stress that any differences in reactivity against HSV-1 and -2 orthologs of a given antigen may reflect the protein concentrations of each spot, which are not uniform in this type of protein array. Reactivity by the HSV-1/HSV-2 double-positive donors appeared to be equally distributed between the HSV-1 and HSV-2 antigens. The percentages for seroprevalence of each reactive antigen are presented below in the seroprevalence analysis.
Fig 2.
Heat map overview of the HSV-1 and HSV-2 antibody profiles of human sera. Columns correspond to sera used to probe the array, and rows are arrayed antigens. Sera were serotyped using HerpeSelect-1 and -2 IgG ELISAs (Focus Diagnostics), as shown at the top, and used as the reference for sample categorization. The patient sera were thus classified into the groups seronegative (neg; n = 47), HSV-1 seropositive only (1; n = 32), HSV-2 seropositive only (2; n = 6), and HSV-1 and -2 seropositive (1 + 2; n = 5). For comparison, sera from a healthy population (healthy; n = 21) were probed. Only those antigens that were reactive against sera from the HSV-1- or HSV-2-seropositive populations are shown. An antigen was defined to be reactive when the average signal intensity for a donor population was greater than the mean plus 2 SDs of the control spots consisting of IVTT reaction mixtures lacking DNA template (C + 2 SDs). The HSV-1 antigens are ranked by descending average signal of the HSV-1-seropositive population, and the HSV-2 antigens are similarly ranked for the HSV-2-positive population. The sera are also ranked from left to right within each group by increasing sum of the signals. The heat map was generated from log-normalized data that were retransformed to approximate raw values, and the signal was converted into a color as shown in the legend. For the ELISA data, a positive signal is shown in red, borderline in black, and negative in green.
Identification of seroreactive antigens in HSV-1 and HSV-2.
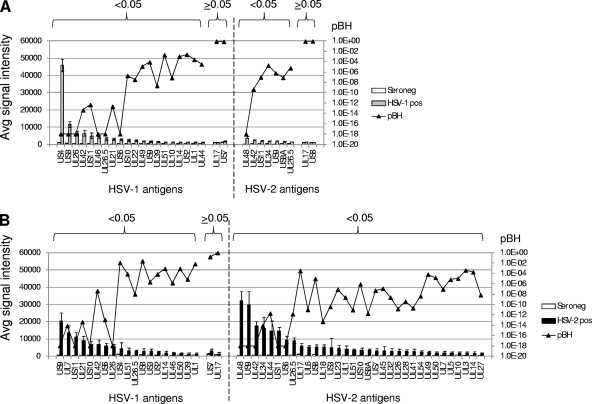
The various signals for a particular antigen probed with different sera (i.e., reading left to right across the heat map in Fig. 2) are directly correlated with antibody titers, since the concentration of a given protein is constant between different arrays. Thus, to identify antigens diagnostic of infection, we derived P values by comparing seropositive and seronegative donors using Bayesian t tests (Fig. 3). When comparing the HSV-1-seropositive and -seronegative groups, 22 HSV-1 antigens were seroreactive, of which 20 were significantly different (Fig. 3A). Of these, glycoproteins G and E (US4 and US8, respectively) gave the two highest signals. The HSV-1-seropositive population also recognized 9 HSV-2 antigens, of which 7 were significant. When comparing the HSV-2-seropositive and -seronegative groups, 33 HSV-2 antigens were reactive, of which all were significant (Fig. 3B). Interestingly, the HSV-2-seropositive population also cross-reacted to 19 HSV-1 antigens. This could be a consequence of the relatively small sample size of HSV-2-seropositive donors. However, if confirmed on a larger sample set, this disparate pattern of cross-reactivity in HSV-1 and -2 infections may reflect the natural history of infection, such as the route of entry or the preferred site of infection. None of the samples were from known acute cases, thereby ruling out a role for acute verses latent infection in the observed reactivity between HSV-1 and -2 infections.
Fig 3.
Comparisons between HSV-1- or HSV-2-seropositive and -seronegative donors. Histograms show average array signals ± SEMs of seronegative, HSV-1-seropositive, and HSV-2-seropositive donors. Only the seroreactive antigens (>C + 2 SDs) are shown. The responses to each antigen by HSV-1-seropositive donors (A) and HSV-2-seropositive donors (B) were compared to those by the seronegative donors by t tests, and the Benjamini-Hochberg-corrected P (pBH) values (shown overlaid onto the histograms) were used to classify the antigens into significant and nonsignificant responses (pBH values, <0.05 and ≥0.05, respectively).
Identification of HSV type-specific antibodies.
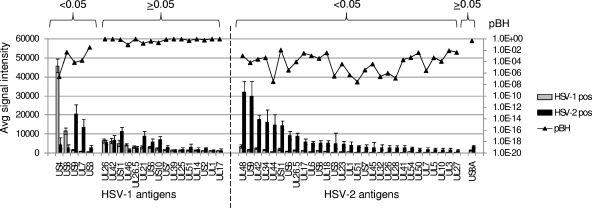
One of the main aims of this study was to define additional potential HSV type-specific antigens. For this we compared the HSV-1-seropositive and HSV-2-seropositive donors by t tests (Fig. 4). Five HSV-1 antigens were discriminatory, although only US4 and US8 gave stronger signals with the HSV-1-seropositive donors. The remaining three significant antigens (US9, UL7, and US3) were not strongly recognized by HSV-1-seropositive donors but were recognized by cross-reactive antibodies from the HSV-2-seropositive population. The remaining reactive HSV-1 antigens were nondiscriminatory, i.e., were recognized equally well by both HSV-1- and HSV-2-infected populations. The HSV-2 antigens, in contrast, were mostly type specific, i.e., were recognized only by the HSV-2-seropositive population.
Fig 4.
Comparisons between HSV-1- and HSV-2-seropositive donors. Data are as described in the legend to Fig. 3, except that HSV-1- and HSV-2-seropositive donors were compared.
ROC and seroprevalence analysis.
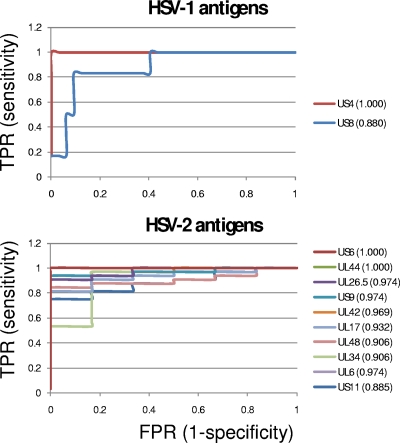
In Table 1 are listed the reactive antigens with BH-corrected P values from t tests comparing HSV-1- and HSV-2-seropositive donors (Fig. 4). Antigens with pBH values of <0.05 (indicated by asterisks) are considered discriminatory between HSV-1- and HSV-2-seropositive populations. In addition to the t test, we also used (i) ROC analysis to determine the sensitivity and specificity of individual antigens and AUC as a measure of discrimination (Fig. 5) and (ii) frequency-of-recognition (seroprevalence) analysis of each antigen within each population. For the latter, a cutoff was defined for each antigen as the mean plus 3 SDs of the seronegative population, and the number of donors above the cutoff was represented as a percentage of the population. Overall, there was good concordance between the pBH and the single-antigen AUC. Using pBH and AUC as criteria, HSV-1 antigens that were the best discriminators were US4 and US8. The majority of HSV-1-reactive antigens, however, were recognized by both HSV-1- and HSV-2-seropositive populations. Although not type specific, such antigens may have utility for broad diagnosis of HSV infection without a need for discriminating between HSV-1 and -2. For a specific test, only US4/gG showed acceptable specificity (AUC = 1). US8/gE showed inferior sensitivity and specificity (AUC = 0.88). In contrast, there were several potential discriminatory HSV-2 antigens with AUC values of >0.9, including US6, UL44, UL26.5, US9, and UL42. US6 and UL44 appear to be perfect classifiers, with AUCs of 1.
Table 1.
Discriminatory and nondiscriminatory antigensa
| ORF | Product description | % reactivity |
pBH | ||||
|---|---|---|---|---|---|---|---|
| Negative | HSV-1 | HSV-2 | HSV-1 + -2 | Controls | |||
| HSV1_US4 | Glycoprotein G, gG | 2 | 94 | 0 | 80 | 19 | 2.5E−07* |
| HSV1_US8 | Glycoprotein E, gE | 0 | 25 | 0 | 20 | 33 | 4.9E−03* |
| HSV1_US9 | Envelope protein | 0 | 19 | 100 | 100 | 19 | 7.4E−05* |
| HSV1_UL7 | Virion protein | 0 | 13 | 100 | 20 | 14 | 1.7E−04* |
| HSV1_US3 | Serine/threonine protein kinase cellular homolog, KR1 | 2 | 9 | 33 | 0 | 19 | 3.3E−02* |
| HSV1_UL26 | Serine protease, VP40, self-cleaves to form VP21 and VP24 | 2 | 91 | 100 | 100 | 52 | 9.6E−01 |
| HSV1_UL42 | DNA polymerase accessory protein, VPAP | 0 | 38 | 50 | 40 | 43 | 9.6E−01 |
| HSV1_US11 | RNA-binding tegument protein, DNB | 0 | 66 | 100 | 100 | 33 | 7.6E−01 |
| HSV1_UL46 | Tegument protein, VP11/12 | 2 | 81 | 33 | 100 | 52 | 9.3E−02 |
| HSV1_UL26.5 | Capsid scaffolding protein, ICP35, VP22a | 0 | 47 | 17 | 20 | 38 | 9.6E−01 |
| HSV1_UL21 | Tegument protein | 0 | 53 | 83 | 80 | 14 | 1.7E−01 |
| HSV1_US6 | Glycoprotein D, gD | 0 | 63 | 83 | 100 | 48 | 3.9E−01 |
| HSV1_US10 | Tegument protein | 2 | 56 | 100 | 80 | 33 | 2.2E−01 |
| HSV1_US7 | Glycoprotein I, gI | 0 | 3 | 0 | 0 | 0 | 7.9E−01 |
| HSV1_UL39 | Ribonucleotide reductase large-subunit cellular homolog, ICP6, RIR1 | 0 | 41 | 33 | 80 | 29 | 9.6E−01 |
| HSV1_UL25 | DNA packaging virion protein | 0 | 3 | 0 | 0 | 0 | 9.3E−01 |
| HSV1_UL51 | Tegument protein | 0 | 6 | 33 | 0 | 14 | 5.0E−01 |
| HSV1_UL14 | Minor tegument protein | 0 | 25 | 17 | 20 | 14 | 9.3E−01 |
| HSV1_US2 | Tegument protein | 0 | 9 | 17 | 0 | 10 | 5.9E−01 |
| HSV1_UL1 | Virion glycoprotein L, gL | 0 | 16 | 33 | 0 | 14 | 9.8E−01 |
| HSV1_UL17 | Virion packaging protein | 2 | 0 | 0 | 0 | 0 | 9.6E−01 |
| HSV2_UL48 | Tegument protein, VP16 | 0 | 75 | 100 | 100 | 29 | 1.1E−03* |
| HSV2_US9 | Envelope protein | 0 | 28 | 100 | 100 | 29 | 7.1E−05* |
| HSV2_UL42 | DNA polymerase accessory protein | 0 | 47 | 100 | 100 | 43 | 2.6E−04* |
| HSV2_UL34 | Membrane-associated virion protein? | 0 | 19 | 88 | 80 | 38 | 5.1E−04* |
| HSV2_UL44 | Glycoprotein C, gC | 0 | 0 | 88 | 20 | 10 | 3.3E−08* |
| HSV2_US11 | RNA-binding tegument protein (DNB) | 0 | 56 | 100 | 100 | 33 | 1.2E−02* |
| HSV2_US6 | Glycoprotein D, gD | 0 | 9 | 100 | 40 | 24 | 3.3E−06* |
| HSV2_UL26.5 | Capsid scaffolding protein, ICP35, VP22a | 0 | 6 | 88 | 60 | 14 | 9.3E−05* |
| HSV2_UL17 | Capsid protein | 0 | 0 | 0 | 0 | 5 | 3.0E−03* |
| HSV2_UL6 | Virion protein | 0 | 6 | 75 | 20 | 10 | 1.1E−03* |
| HSV2_US8 | Glycoprotein E, gE | 0 | 0 | 25 | 0 | 5 | 3.9E−04* |
| HSV2_UL18 | Capsid protein, VP23 | 0 | 31 | 100 | 40 | 24 | 3.7E−03* |
| HSV2_US3 | Nuclear phosphoprotein cellular homolog | 0 | 0 | 75 | 40 | 0 | 3.0E−07* |
| HSV2_UL23 | Thymidine kinase, ICP36 | 0 | 0 | 50 | 20 | 5 | 3.9E−05* |
| HSV2_UL1 | Glycoprotein L, gL | 0 | 3 | 63 | 0 | 5 | 4.7E−07* |
| HSV2_UL51 | Tegument protein | 0 | 0 | 88 | 0 | 5 | 2.8E−08* |
| HSV2_US7 | Glycoprotein I, gI | 0 | 0 | 63 | 60 | 10 | 3.6E−06* |
| HSV2_UL45 | Membrane protein type II | 0 | 0 | 63 | 0 | 10 | 3.9E−05* |
| HSV2_UL32 | Cleavage and packaging protein | 0 | 0 | 63 | 20 | 5 | 2.5E−07* |
| HSV2_UL26 | Serine protease, self-cleaves to form VP21 and VP24 (VP40) | 0 | 6 | 63 | 20 | 14 | 9.7E−07* |
| HSV2_UL28 | DNA cleavage and packaging protein, ICP18.5 | 0 | 0 | 38 | 20 | 10 | 1.3E−07* |
| HSV2_UL41 | Virion host shutoff protein, cellular homolog, VHS | 0 | 19 | 88 | 60 | 14 | 2.1E−04* |
| HSV2_UL54 | Regulates and transports RNA, IE63, VMW63, ICP27, IE63 | 0 | 9 | 50 | 60 | 5 | 5.7E−04* |
| HSV2_UL50 | Deoxyuridine triphosphatase (dUTPase) | 2 | 0 | 13 | 0 | 0 | 3.9E−03* |
| HSV2_UL7 | Virion protein | 0 | 0 | 50 | 20 | 5 | 2.8E−06* |
| HSV2_UL5 | Component of DNA helicase-primase complex, cellular homolog, HELI | 0 | 3 | 13 | 0 | 5 | 5.1E−04* |
| HSV2_UL10 | Tegument protein | 0 | 0 | 63 | 0 | 0 | 1.2E−04* |
| HSV2_UL3 | Nuclear phosphoprotein | 0 | 0 | 13 | 20 | 0 | 8.6E−03* |
| HSV2_UL27 | Glycoprotein B, gB, VP7 | 0 | 3 | 75 | 0 | 5 | 4.3E−03* |
| HSV2_US8A | ? | 0 | 56 | 88 | 40 | 33 | 4.6E−01 |
Percentages shown are those in each patient group reactive for individual HSV-1 and HSV-2 antigens, using a cutoff defined by the mean plus 3 SDs of the seronegative population. Patient groups were defined as described in the legend to Fig. 2, and the antigens are listed in the same order as shown in Fig. 4 for comparison. Antigens that discriminate HSV-1- and HSV-2-seropositive patients (i.e., pBH < 0.05 when the two populations were compared by t tests) are indicated (*). ORF product descriptions were obtained from Uniprot (http://www.uniprot.org/) and Oralgen (http://www.oralgen.lanl.gov/).
Fig 5.
ROC plots for discriminatory HSV-1 and HSV-2 antigens. For HSV-1 antigens, only those that gave stronger signals with HSV-1-seropositive donors are shown. For HSV-2 antigens, only the top 10 are shown, although 29 antigens were discriminatory (Fig. 4). Antigens are ranked by AUCs, shown in parentheses. TPR, true-positive rate; FPR, false-positive rate.
As a rule, the seroprevalence analysis shows agreement with significance tests. For example, HSV-2 UL44/gC was the most specific single antigen, being recognized by 88% and 0% of HSV-2- and HSV-1-seropositive donors, respectively. Other glycoproteins, notably, HSV-2 UL1/gL and US7/gI, also gave good discrimination. There were also some exceptions, such as HSV-2 UL42 and UL48, which, although they had high AUCs (0.97 and 0.91, respectively), were recognized by a large percentage of HSV-1-seropositive donors when using the threshold defined.
Enrichment analysis.
The identities of the reactive antigens listed in Table 1 show that a large number of structural proteins were reactive, notably, glycoproteins such as US4/gG, US6/gD, US8/gE, UL10/gM, UL22/gH, and UL44/gC. This is consistent with the notion that the antibody response was biased toward surface-located viral proteins. We therefore performed an enrichment analysis for the HSV-1 antibody profile. Each protein encoded by HSV-1 (strain 17) was assigned one or more gene ontology (GO) component classifiers according to the database at www.uniprot.org. The percentage of the total number of genes assigned to each GO component present in the proteome and in the seroreactive antigens was determined, and the ratio was used to determine the fold enrichment. The analysis revealed 12 proteins on the array that were assigned the GO component virion membrane, of which 9 were seroreactive. This represents a 1.63-fold enrichment, although it failed to reach significance. Tegument proteins were not enriched in the seroreactive antigen set.
Recombinant protein ELISAs.
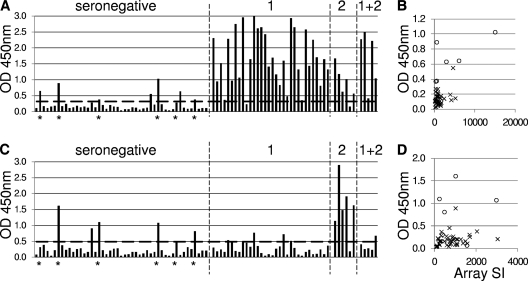
One aim of this study was the identification of potential serodiagnostic antigens. To begin to address this, we expressed candidates revealed by microarray analysis in BL21 cells that could discriminate between HSV-1-seropositive and HSV-2-seropositive individuals. Glycoproteins lacking hydrophobic sequences (leader sequences and transmembrane domains) were reengineered, and sequence-verified clones were used for expression. Data for two glycoproteins, HSV-1 US8/gE and HSV-2 UL44/gC, purified by nickel chelate affinity chromatography, are presented in Fig. 6A and C. The ELISAs were used to assay the same serum sample collection used to probe the arrays (as described in the legend to Fig. 2). Both antigens showed excellent performance for discriminating seronegative from seropositive donors. Thus, HSV-1 US8/gE showed 97% sensitivity and 83% specificity, whereas HSV-2 UL44/gC showed 83% sensitivity and 85% specificity (Table 2). Within the seronegative populations (as defined by the FocuSelect ELISA) were 6 individuals that also responded to one or both antigens. Most of these individuals also gave high signals to the corresponding antigen on the array, as illustrated in the scatter plots in Fig. 6B and D. When these equivocal samples were removed, there were strong improvements to the sensitivity and/or specificity of the ELISAs to detect infection (Table 2). Importantly, on the ELISA, HSV-2 UL44/gC also retained excellent discrimination between the HSV-1- and HSV-2-seropositive donors (97% specificity, AUC = 0.870). The HSV1 US8/gE protein did not discriminate as well (67% specificity, AUC = 0.604).
Fig 6.
ELISA data using two purified HSV antigens. (A and B) HSV-1 US8/gE; (C and D) HSV-2 UL44 gC. Sera are as described in the legend to Fig. 2, with serological status shown in panels A and C. Diagnoses for individuals marked with an asterisk were considered equivocal. Horizontal dashed line, average plus 2 SDs of the seronegative population minus the 6 equivocal samples. Scatter plots in panels B and D show ELISA ODs and corresponding microarray signal intensity (SI) for the seronegative population only. The six equivocal seronegatives are indicated by the open circles.
Table 2.
Receiver operator characteristic analysis of ELISA performancea
| ELISA | Comparison | Sensitivity (%) | Specificity (%) | AUC |
|---|---|---|---|---|
| HSV-1 US8/gE | Seronegative vs HSV-1 seropositive | 97 | 83 | 0.955 |
| HSV-1 US8/gE | Seronegative (minus equivocal) vs HSV-1 seropositive | 100 | 98 | 0.976 |
| HSV-1 US8/gE | HSV-1 seropositive vs HSV-2 seropositive | 69 | 67 | 0.604 |
| HSV-2 UL44/gC | Seronegative vs HSV-2 seropositive | 83 | 85 | 0.780 |
| HSV-2 UL44/gC | Seronegative (minus equivocal) vs HSV-2 seropositive | 83 | 98 | 0.894 |
| HSV-2 UL44/gC | HSV-1 seropositive vs HSV-2 seropositive | 83 | 97 | 0.870 |
See Fig. 6 for definition of equivocal samples (n = 6) in the seronegative population. AUC, area under the curve. The AUC of a perfect classifier is 1.0, whereas an antigen that shows no discriminatory properties has an AUC of 0.5.
DISCUSSION
The development of HSV type-specific serological (TSS) tests based on the gG-1 and gG-2 (US4) glycoproteins has had a major impact on the management of HSV infection. The test is particularly useful for diagnosis of genital herpes, where clinical symptom-based diagnosis is inaccurate. The identification of unrecognized HSV-2 infection can be used to more carefully monitor virus shedding during pregnancy. TSS tests are also useful for monitoring the partners of individuals with HSV or HIV infections. The antigenic differences between gG-1 and gG-2 that underlie type specificity are not fully understood. It is conventional to express recombinant gG-1 and gG-2 for serological tests in eukaryotic cells to ensure glycosylation. However, the same antigens expressed in E. coli also show good type specificity (41, 83), indicating that there are type-specific antigenic differences in the amino acid backbone that are independent of glycosylation. Alignment of the gG-1 and gG-2 ORFs shows that the gG-2 ORF is 1,460 nucleotides longer, with the smaller gG-1 protein predicted to align with the C-terminal portion of gG-2 (61). Both genes have also undergone drift by accumulating separate point mutations, additions, and deletions (61). Epitope mapping studies of gG-2 reveal type-specific domains not only in the unique N-terminal portion, as expected, but also in the shared C-terminal region (34).
US4/gG, the basis of current TSS tests, was discovered in the preproteomic era. There is every reason to expect that other antigens can be used to discriminate HSV-1 and -2 infections and increase the sensitivity of the test. Indeed, UL44/gC has recently been shown to be as specific as US4/gG for type-specific tests (74), indicating that gG-1 and gG-2 are not the only HSV proteins known to induce type-specific antibodies. The only rational way to approach this problem is to test individual antigens in order to identify those that are antigenically distinct. We believe that the study described here provides the first proteome-wide study of the humoral response to human herpes simplex virus 1 and 2.
Owing to the small volumes of sera required to probe arrays and the ease with which they can be screened in a high-throughput fashion, the array platform is ideal for seroepidemiological studies against hundreds of pathogen antigens simultaneously (20, 31, 49, 52). Seroprevalence of HSV is influenced by age, sexual activity, and ethnicity, and a much larger study would be required to estimate seroprevalence in the United States properly using the array platform. Nevertheless, it is useful to determine how the data presented here compare with estimates of seroprevalence by gG-based approved assays. Recent studies estimate that seroprevalences in the United States are about 60% for HSV-1 (75, 82) and 16 to 22% for HSV-2 (9, 16, 32, 82). By array, the best type-specific antigens are US4/gG for HSV-1 and UL44/gC and US6/gD for HSV-2. These antigens indicate seroprevalences in the Orange County, CA, general population of ∼19% for HSV-1 and 10 to 24% for HSV-2. We were unable to produce seroreactive HSV-2 US4/gG (gG-2), although it is anticipated that this antigen would also provide sensitive detection of HSV-2 infection. The reason(s) for this lack of gG-2 reactivity is not established but may relate to insufficient antigen expression or a requirement for recognition, such as antigen folding or posttranslational modification, that is not met by IVTT.
Overall, the array data confirm and extend our understanding of HSV serology. Specifically, we confirm that UL44/gC shows type-specific antigenicity (27, 74) and warrants further investigation as a candidate antigen for the development of improved tests. We also confirm the type specificity of US4/gG, at least for HSV-1. A number of other proteins are known to be the targets of antibodies generated during infection. For example, Western blots of whole virions reveal several major bands (4, 5). The identities of some can be made with antigen-specific monoclonal antibodies, as is the case with gG (5). However, it is not known at this time how the bands identified by Western blotting correlate with those identified by array screening. Our proteome-wide screening approach adds to this database additional antigens that have not been described before.
The importance of antibody in mediating immunity to HSV infection is confirmed in passive transfer experiments in animal models (35, 39, 58, 62, 68, 72). Immune sera are known to have broad reactivity to several antigens, including different envelope glycoproteins, tegument proteins, and capsid proteins (25, 63). Of the ∼11 glycoproteins on the envelope of HSV that mediate viral attachment and entry, several reports (mainly concerning US6/gD, UL27/gB, and UL44/gC) show these to be the targets of virus-neutralizing antibodies in vitro (1, 8, 50) and able to engender protection in vivo (14, 19, 57, 59), which may be mediated by complement fixation and/or antibody-dependent cell-mediated cytotoxic mechanisms (28, 47, 48, 80). In the context of HSV serology, it is conventional to focus on membrane proteins, and this has proved a successful approach for engendering antibody-mediated protection with subunit vaccines (44, 76). Unlike vaccines, the choice of ideal antigens for serodiagnostic applications appears to be less dependent on membrane proteins. Capsid and tegument proteins are not normally surface exposed, so the presence of antibodies to these antigens (Table 1) may reflect their exposure to the adaptive immune system in other ways, perhaps when the integrity of virus particles is compromised, such as by complement-mediated disruption of the membrane.
It has been suggested that the most important T cell epitopes in HSV may be in tegument or other noncapsid proteins (15, 37, 42, 45), although many CD4 and CD8 T cell epitopes have recently been identified in the envelope glycoproteins UL27/gB and US6/gD (11, 17, 18, 43, 46, 66, 67, 71). Tegument proteins such as those encoded by HSV-2 UL41, UL46/VP11/12, UL47/VP13/14, UL48/VP16, and UL49 and IE proteins such as RL2/ICP0 and RS1/ICP4 have also been identified as major targets for effector T cells (15, 37, 42, 45). Of these, the arrays detect strong antibody signals against US6/gD (from both HSV-1 and -2) and UL48 of HSV-2, both of which are CD4 target antigens. The overlap between the targets of antibody and CD4 cells rather than CD8 cells is consistent with findings in vaccinia virus (40, 69, 70).
Overall, we found that sera from HSV-1-seropositive donors were specific for HSV-1 antigens on the array, whereas the sera from HSV-2-seropositive donors appeared to be reactive against HSV-1 and -2 antigens. Given the high degree of phylogenetic relatedness of HSV-1 and -2, it is perhaps surprising that the antibody profiles of HSV-1 and -2 revealed in this study are not more alike. Part of the explanation is technical and is perhaps related to differential levels of expression or correct folding of the HSV-1 and -2 orthologs of each protein. Nevertheless, the asymmetry of recognition may also reflect biological differences in HSV-1 and HSV-2 infections, such as the route of entry or site of primary infection. None of the HSV-1- or HSV-2-seropositive donors had acute infections, thereby ruling out a role for acute infection versus latency in the observed differences. Previous serological analyses of HSV-1 and -2 infections have alluded to an asymmetrical antibody response (4). While the microarray platform represents a useful first-pass screen of the proteome, any conclusions about differential recognition must be confirmed on other platforms using purified proteins of known concentration. A similar strategy may help identify antigens with other useful diagnostic properties. For example, provided that the sera used for screening are well characterized, it may be possible to identify antigens that discriminate between current (acute) infections and previous (latent) infection, between symptomatic and asymptomatic latent infections, or between different routes of entry, such as by the oropharyngeal versus genital routes of HSV-1 acquisition.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dale Carpenter and Steve Wechsler, UCI Department of Ophthalmology, for gifts of HSV DNA.
This work was supported by NIH grants R44AI058365 (to D.H.D.), U01AI078213 (to P.L.F.), and EY14900 and EY019896 (to L.B.), grants from The Discovery Eye Foundation and The Henry L. Guenther Foundation, and a Research to Prevent Blindness Challenge grant (to L.B.).
Footnotes
Published ahead of print 8 February 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Adamiak B, et al. 2010. Human antibodies to herpes simplex virus type 1 glycoprotein C are neutralizing and target the heparan sulfate-binding domain. Virology 400:197–206 [DOI] [PubMed] [Google Scholar]
- 2. Ashley RL. 2001. Sorting out the new HSV type specific antibody tests. Sex. Transm. Infect. 77:232–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ashley RL, Eagleton M, Pfeiffer N. 1999. Ability of a rapid serology test to detect seroconversion to herpes simplex virus type 2 glycoprotein G soon after infection. J. Clin. Microbiol. 37:1632–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ashley RL, Militoni J. 1987. Use of densitometric analysis for interpreting HSV serologies based on Western blot. J. Virol. Methods 18:159–168 [DOI] [PubMed] [Google Scholar]
- 5. Ashley RL, Militoni J, Lee F, Nahmias A, Corey L. 1988. Comparison of Western blot (immunoblot) and glycoprotein G-specific immunodot enzyme assay for detecting antibodies to herpes simplex virus types 1 and 2 in human sera. J. Clin. Microbiol. 26:662–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ashley-Morrow R, Krantz E, Wald A. 2003. Time course of seroconversion by HerpeSelect ELISA after acquisition of genital herpes simplex virus type 1 (HSV-1) or HSV-2. Sex. Transm. Dis. 30:310–314 [DOI] [PubMed] [Google Scholar]
- 7. Baldi P, Long AD. 2001. A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics 17:509–519 [DOI] [PubMed] [Google Scholar]
- 8. Banks T, et al. 1989. A major neutralizing domain maps within the carboxyl-terminal half of the cleaved cytomegalovirus B glycoprotein. J. Gen. Virol. 70(Pt 4):979–985 [DOI] [PubMed] [Google Scholar]
- 9. Bauer GR, Khobzi N, Coleman TA. 2010. Herpes simplex virus type 2 seropositivity and relationship status among U.S. adults age 20 to 49: a population-based analysis. BMC Infect. Dis. 10:359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57:289–300 [Google Scholar]
- 11. BenMohamed L, et al. 2003. Identification of novel immunodominant CD4+ Th1-type T-cell peptide epitopes from herpes simplex virus glycoprotein D that confer protective immunity. J. Virol. 77:9463–9473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brown ZA, et al. 1997. The acquisition of herpes simplex virus during pregnancy. N. Engl. J. Med. 337:509–515 [DOI] [PubMed] [Google Scholar]
- 13. Brown ZA, et al. 2003. Effect of serologic status and cesarean delivery on transmission rates of herpes simplex virus from mother to infant. JAMA 289:203–209 [DOI] [PubMed] [Google Scholar]
- 14. Cantin EM, et al. 1987. Expression of herpes simplex virus 1 glycoprotein B by a recombinant vaccinia virus and protection of mice against lethal herpes simplex virus 1 infection. Proc. Natl. Acad. Sci. U. S. A. 84:5908–5912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Carmack MA, et al. 1996. T cell recognition and cytokine production elicited by common and type-specific glycoproteins of herpes simplex virus type 1 and type 2. J. Infect. Dis. 174:899–906 [DOI] [PubMed] [Google Scholar]
- 16. CDC 2010. Seroprevalence of herpes simplex virus type 2 among persons aged 14–49 years—United States, 2005–2008. MMWR Morb. Mortal. Wkly. Rep. 59:456–459 [PubMed] [Google Scholar]
- 17. Chentoufi AA, et al. 2008. Asymptomatic human CD4+ cytotoxic T-cell epitopes identified from herpes simplex virus glycoprotein B. J. Virol. 82:11792–11802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chentoufi AA, et al. 2008. HLA-A*0201-restricted CD8+ cytotoxic T lymphocyte epitopes identified from herpes simplex virus glycoprotein D. J. Immunol. 180:426–437 [DOI] [PubMed] [Google Scholar]
- 19. Corey L, et al. 1999. Recombinant glycoprotein vaccine for the prevention of genital HSV-2 infection: two randomized controlled trials. Chiron HSV Vaccine Study Group. JAMA 282:331–340 [DOI] [PubMed] [Google Scholar]
- 20. Crompton PD, et al. 2010. A prospective analysis of the Ab response to Plasmodium falciparum before and after a malaria season by protein microarray. Proc. Natl. Acad. Sci. U. S. A. 107:6958–6963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cullen AP, Long CD, Lorincz AT. 1997. Rapid detection and typing of herpes simplex virus DNA in clinical specimens by the Hybrid Capture II signal amplification probe test. J. Clin. Microbiol. 35:2275–2278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cunningham C, Davison AJ. 1993. A cosmid-based system for constructing mutants of herpes simplex virus type 1. Virology 197:116–124 [DOI] [PubMed] [Google Scholar]
- 23. Dasgupta G, Chentoufi AA, Nesburn AB, Wechsler SL, BenMohamed L. 2009. New concepts in herpes simplex virus vaccine development: notes from the battlefield. Expert Rev. Vaccines 8:1023–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Davies DH, et al. 2005. Profiling the humoral immune response to infection by using proteome microarrays: high-throughput vaccine and diagnostic antigen discovery. Proc. Natl. Acad. Sci. U. S. A. 102:547–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Deshpande SP, Kumaraguru U, Rouse BT. 2000. Dual role of B cells in mediating innate and acquired immunity to herpes simplex virus infections. Cell. Immunol. 202:79–87 [DOI] [PubMed] [Google Scholar]
- 26. Dolan A, Jamieson FE, Cunningham C, Barnett BC, McGeoch DJ. 1998. The genome sequence of herpes simplex virus type 2. J. Virol. 72:2010–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dolter KE, Goins WF, Levine M, Glorioso JC. 1992. Genetic analysis of type-specific antigenic determinants of herpes simplex virus glycoprotein C. J. Virol. 66:4864–4873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dubin G, Socolof E, Frank I, Friedman HM. 1991. Herpes simplex virus type 1 Fc receptor protects infected cells from antibody-dependent cellular cytotoxicity. J. Virol. 65:7046–7050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Durbin BP, Hardin JS, Hawkins DM, Rocke DM. 2002. A variance-stabilizing transformation for gene-expression microarray data. Bioinformatics 18(Suppl 1):S105–S110 [DOI] [PubMed] [Google Scholar]
- 30. Eisenberg RJ, et al. 1985. Synthetic glycoprotein D-related peptides protect mice against herpes simplex virus challenge. J. Virol. 56:1014–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Felgner PL, et al. 2009. A Burkholderia pseudomallei protein microarray reveals serodiagnostic and cross-reactive antigens. Proc. Natl. Acad. Sci. U. S. A. 106:13499–13504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fleming DT, et al. 1997. Herpes simplex virus type 2 in the United States, 1976 to 1994. N. Engl. J. Med. 337:1105–1111 [DOI] [PubMed] [Google Scholar]
- 33. Freeman EE, et al. 2006. Herpes simplex virus 2 infection increases HIV acquisition in men and women: systematic review and meta-analysis of longitudinal studies. AIDS 20:73–83 [DOI] [PubMed] [Google Scholar]
- 34. Grabowska A, et al. 1999. Identification of type-specific domains within glycoprotein G of herpes simplex virus type 2 (HSV-2) recognized by the majority of patients infected with HSV-2, but not by those infected with HSV-1. J. Gen. Virol. 80(Pt 7):1789–1798 [DOI] [PubMed] [Google Scholar]
- 35. Hayashida I, et al. 1982. Mechanism of antibody-mediated protection against herpes simplex virus infection in athymic nude mice: requirement of Fc portion of antibody. Microbiol. Immunol. 26:497–509 [DOI] [PubMed] [Google Scholar]
- 36. Hermanson G, et al. 2012. Measurement of antibody responses to modified vaccinia virus Ankara (MVA) and Dryvax® using proteome microarrays and development of recombinant protein ELISAs. Vaccine 30:614–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hosken N, et al. 2006. Diversity of the CD8+ T-cell response to herpes simplex virus type 2 proteins among persons with genital herpes. J. Virol. 80:5509–5515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Huber W, von Heydebreck A, Sultmann H, Poustka A, Vingron M. 2002. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 18(Suppl 1):S96–S104 [DOI] [PubMed] [Google Scholar]
- 39. Incorvaia C, et al. 1995. Effects of the passive transfer of anti-gB antibodies in a rabbit model of HSV-1-induced keratitis. Ophthalmologica 209:340–345 [DOI] [PubMed] [Google Scholar]
- 40. Jing L, et al. 2008. An extremely diverse CD4 response to vaccinia virus in humans is revealed by proteome-wide T-cell profiling. J. Virol. 82:7120–7134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kakkanas A, et al. 1995. Escherichia coli expressed herpes simplex virus gG1 and gG2 proteins in ELISA and immunoblotting assays. Intervirology 38:346–351 [DOI] [PubMed] [Google Scholar]
- 42. Koelle DM, Abbo H, Peck A, Ziegweid K, Corey L. 1994. Direct recovery of herpes simplex virus (HSV)-specific T lymphocyte clones from recurrent genital HSV-2 lesions. J. Infect. Dis. 169:956–961 [DOI] [PubMed] [Google Scholar]
- 43. Koelle DM, et al. 2001. CD8 CTL from genital herpes simplex lesions: recognition of viral tegument and immediate early proteins and lysis of infected cutaneous cells. J. Immunol. 166:4049–4058 [DOI] [PubMed] [Google Scholar]
- 44. Koelle DM, Corey L. 2003. Recent progress in herpes simplex virus immunobiology and vaccine research. Clin. Microbiol. Rev. 16:96–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Koelle DM, et al. 1994. Antigenic specificities of human CD4+ T-cell clones recovered from recurrent genital herpes simplex virus type 2 lesions. J. Virol. 68:2803–2810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Koelle DM, et al. 2003. Immunodominance among herpes simplex virus-specific CD8 T cells expressing a tissue-specific homing receptor. Proc. Natl. Acad. Sci. U. S. A. 100:12899–12904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kohl S, Sigouroudinia M, Engleman EG. 1999. Adhesion defects of antibody-mediated target cell binding of neonatal natural killer cells. Pediatr. Res. 46:755–759 [DOI] [PubMed] [Google Scholar]
- 48. Kunder SC, Wu L, Morahan PS. 1993. Role of NK cells in immunomodulator-mediated resistance to herpesvirus infection. Antiviral Res. 21:103–118 [DOI] [PubMed] [Google Scholar]
- 49. Kunnath-Velayudhan S, et al. 2010. Dynamic antibody responses to the Mycobacterium tuberculosis proteome. Proc. Natl. Acad. Sci. U. S. A. 107:14703–14708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Langenberg AG, et al. 1995. A recombinant glycoprotein vaccine for herpes simplex virus type 2: safety and immunogenicity [corrected]. Ann. Intern. Med. 122:889–898 [DOI] [PubMed] [Google Scholar]
- 51. Lasky LA, Dowbenko DJ. 1984. DNA sequence analysis of the type-common glycoprotein-D genes of herpes simplex virus types 1 and 2. DNA 3:23–29 [DOI] [PubMed] [Google Scholar]
- 52. Liang L, et al. 2011. Identification of potential serodiagnostic and subunit vaccine antigens by antibody profiling of toxoplasmosis cases in Turkey. Mol. Cell. Proteomics 10:M110.00691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Long AD, et al. 2001. Improved statistical inference from DNA microarray data using analysis of variance and a Bayesian statistical framework. Analysis of global gene expression in Escherichia coli K12. J. Biol. Chem. 276:19937–19944 [DOI] [PubMed] [Google Scholar]
- 54. Luevano M, et al. 2010. High-throughput profiling of the humoral immune responses against thirteen human papillomavirus types by proteome microarrays. Virology 405:31–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Manservigi R, et al. 1990. Protection from herpes simplex virus type 1 lethal and latent infections by secreted recombinant glycoprotein B constitutively expressed in human cells with a BK virus episomal vector. J. Virol. 64:431–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Marsden HS, Buckmaster A, Palfreyman JW, Hope RG, Minson AC. 1984. Characterization of the 92,000-dalton glycoprotein induced by herpes simplex virus type 2. J. Virol. 50:547–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. McClements WL, Armstrong ME, Keys RD, Liu MA. 1996. Immunization with DNA vaccines encoding glycoprotein D or glycoprotein B, alone or in combination, induces protective immunity in animal models of herpes simplex virus-2 disease. Proc. Natl. Acad. Sci. U. S. A. 93:11414–11420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. McDermott MR, Brais LJ, Evelegh MJ. 1990. Mucosal and systemic antiviral antibodies in mice inoculated intravaginally with herpes simplex virus type 2. J. Gen. Virol. 71(Pt 7):1497–1504 [DOI] [PubMed] [Google Scholar]
- 59. McDermott MR, Graham FL, Hanke T, Johnson DC. 1989. Protection of mice against lethal challenge with herpes simplex virus by vaccination with an adenovirus vector expressing HSV glycoprotein B. Virology 169:244–247 [DOI] [PubMed] [Google Scholar]
- 60. McGeoch DJ, et al. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69(Pt 7):1531–1574 [DOI] [PubMed] [Google Scholar]
- 61. McGeoch DJ, Moss HW, McNab D, Frame MC. 1987. DNA sequence and genetic content of the HindIII l region in the short unique component of the herpes simplex virus type 2 genome: identification of the gene encoding glycoprotein G, and evolutionary comparisons. J. Gen. Virol. 68(Pt 1):19–38 [DOI] [PubMed] [Google Scholar]
- 62. McKendall RR. 1985. IgG-mediated viral clearance in experimental infection with herpes simplex virus type 1: role for neutralization and Fc-dependent functions but not C′ cytolysis and C5 chemotaxis. J. Infect. Dis. 151:464–470 [DOI] [PubMed] [Google Scholar]
- 63. McKendall RR, Pettit M, Woo W. 1988. The immunoglobulin response to individual HSV-1 viral polypeptides: kinetics of the response during primary and secondary experimental infection with herpes simplex virus. J. Med. Microbiol. 25:59–66 [DOI] [PubMed] [Google Scholar]
- 64. Mertz GJ, Benedetti J, Ashley R, Selke SA, Corey L. 1992. Risk factors for the sexual transmission of genital herpes. Ann. Intern. Med. 116:197–202 [DOI] [PubMed] [Google Scholar]
- 65. Mertz GJ, et al. 1988. Transmission of genital herpes in couples with one symptomatic and one asymptomatic partner: a prospective study. J. Infect. Dis. 157:1169–1177 [DOI] [PubMed] [Google Scholar]
- 66. Mikloska Z, Cunningham AL. 1998. Herpes simplex virus type 1 glycoproteins gB, gC and gD are major targets for CD4 T-lymphocyte cytotoxicity in HLA-DR expressing human epidermal keratinocytes. J. Gen. Virol. 79(Pt 2):353–361 [DOI] [PubMed] [Google Scholar]
- 67. Mikloska Z, Kesson AM, Penfold ME, Cunningham AL. 1996. Herpes simplex virus protein targets for CD4 and CD8 lymphocyte cytotoxicity in cultured epidermal keratinocytes treated with interferon-gamma. J. Infect. Dis. 173:7–17 [DOI] [PubMed] [Google Scholar]
- 68. Morrison LA, Zhu L, Thebeau LG. 2001. Vaccine-induced serum immunoglobulin contributes to protection from herpes simplex virus type 2 genital infection in the presence of immune T cells. J. Virol. 75:1195–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Moutaftsi M, et al. 2007. Vaccinia virus-specific CD4+ T cell responses target a set of antigens largely distinct from those targeted by CD8+ T cell responses. J. Immunol. 178:6814–6820 [DOI] [PubMed] [Google Scholar]
- 70. Moutaftsi M, et al. 2010. Uncovering the interplay between CD8, CD4 and antibody responses to complex pathogens. Future Microbiol. 5:221–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Posavad CM, et al. 2003. T cell immunity to herpes simplex viruses in seronegative subjects: silent infection or acquired immunity? J. Immunol. 170:4380–4388 [DOI] [PubMed] [Google Scholar]
- 72. Raizman MB, Foster CS. 1988. Passive transfer of anti-HSV-1 IgG protects against stromal keratitis in mice. Curr. Eye Res. 7:823–829 [DOI] [PubMed] [Google Scholar]
- 73. Rooney JF, Wohlenberg C, Cremer KJ, Moss B, Notkins AL. 1988. Immunization with a vaccinia virus recombinant expressing herpes simplex virus type 1 glycoprotein D: long-term protection and effect of revaccination. J. Virol. 62:1530–1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Scheper T, et al. 2010. The glycoproteins C and G are equivalent target antigens for the determination of herpes simplex virus type 1-specific antibodies. J. Virol. Methods 166:42–47 [DOI] [PubMed] [Google Scholar]
- 75. Schillinger JA, et al. 2004. National seroprevalence and trends in herpes simplex virus type 1 in the United States, 1976–1994. Sex Transm. Dis. 31:753–760 [DOI] [PubMed] [Google Scholar]
- 76. Stanberry LR. 2004. Clinical trials of prophylactic and therapeutic herpes simplex virus vaccines. Herpes 11(Suppl 3):161A–169A [PubMed] [Google Scholar]
- 77. Stanberry LR, et al. 2002. Glycoprotein-D-adjuvant vaccine to prevent genital herpes. N. Engl. J. Med. 347:1652–1661 [DOI] [PubMed] [Google Scholar]
- 78. Sundaresh S, et al. 2006. Identification of humoral immune responses in protein microarrays using DNA microarray data analysis techniques. Bioinformatics 22:1760–1766 [DOI] [PubMed] [Google Scholar]
- 79. Sundaresh S, et al. 2007. From protein microarrays to diagnostic antigen discovery: a study of the pathogen Francisella tularensis. Bioinformatics 23:i508–i518 [DOI] [PubMed] [Google Scholar]
- 80. Vollstedt S, et al. 2004. Interplay between alpha/beta and gamma interferons with B, T, and natural killer cells in the defense against herpes simplex virus type 1. J. Virol. 78:3846–3850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Wald A, et al. 1997. Frequent genital herpes simplex virus 2 shedding in immunocompetent women. Effect of acyclovir treatment. J. Clin. Invest. 99:1092–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Xu F, et al. 2006. Trends in herpes simplex virus type 1 and type 2 seroprevalence in the United States. JAMA 296:964–973 [DOI] [PubMed] [Google Scholar]
- 83. Zandi K, et al. 2007. Production of recombinant gG-1 protein of herpes simplex virus type 1 in a prokaryotic system in order to develop a type-specific enzyme-linked immunosorbent assay kit. FEMS Immunol. Med. Microbiol. 50:319–323 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.