Abstract
Influenza viruses of gallinaceous poultry and wild aquatic birds usually have distinguishable receptor-binding properties. Here we used a panel of synthetic sialylglycopolymers and solid-phase receptor-binding assays to characterize receptor-binding profiles of about 70 H7 influenza viruses isolated from aquatic birds, land-based poultry, and horses in Eurasia and America. Unlike typical duck influenza viruses with non-H7 hemagglutinin (HA), all avian H7 influenza viruses, irrespective of the host species, displayed a poultry-virus-like binding specificity, i.e., preferential binding to sulfated oligosaccharides Neu5Acα2-3Galβ1-4(6-O-HSO3)GlcNAc and Neu5Acα2-3Galβ1-4(Fucα1-3)(6-O-HSO3)GlcNAc. This phenotype correlated with the unique amino acid sequence of the amino acid 185 to 189 loop of H7 HA and seemed to be dependent on ionic interactions between the sulfate group of the receptor and Lys193 and on the lack of sterical clashes between the fucose residue and Gln222. Many North American and Eurasian H7 influenza viruses displayed weak but detectable binding to the human-type receptor moiety Neu5Acα2-6Galβ1-4GlcNAc, highlighting the potential of H7 influenza viruses for avian-to-human transmission. Equine H7 influenza viruses differed from other viruses by preferential binding to the N-glycolyl form of sialic acid. Our data suggest that the receptor-binding site of contemporary H7 influenza viruses in aquatic and terrestrial birds was formed after the introduction of their common precursor from ducks to a new host, presumably, gallinaceous poultry. The uniformity of the receptor-binding profile of H7 influenza viruses in various wild and domestic birds indicates that there is no strong receptor-mediated host range restriction in birds on viruses with this HA subtype. This notion agrees with repeated interspecies transmission of H7 influenza viruses from aquatic birds to poultry.
INTRODUCTION
The major natural reservoirs of influenza A viruses are wild aquatic birds of the orders Anseriformes (ducks, geese, and swans) and Charadriiformes (gulls, terns, and waders), which harbor viruses of all 16 hemagglutinin (HA) and 9 neuraminidase (NA) antigenic subtypes currently known. Dabbling ducks (Anatinae), such as mallards and teals, carry almost all subtypes and show particularly high virus isolation rates, suggesting a unique role of these species in the persistence of influenza viruses in nature (for reviews, see references 14, 41, 46, and 64). Occasionally, influenza viruses of aquatic birds infect other birds and mammals, adapt to efficiently replicate and transmit in the new species, and continue to circulate, forming new stable host-specific virus lineages. All known lineages of influenza A viruses in land-based birds and mammals are believed to originate from the viruses of wild aquatic birds. Adaptation of influenza viruses to their sialic acid-containing receptors in a new host species is often required for successful interspecies transmission. Thus, avian influenza viruses bind to receptors containing terminal sialyl-galactosyl residues linked by an α2-3 linkage (Neu5Acα2-3Gal), whereas swine and human viruses bind to receptors which contain terminal α2-6-linked sialyl-galactosyl moieties (Neu5Acα2-6Gal) (10, 18, 27, 32, 35, 44, 47, 50), and a corresponding switch in the receptor specificity of the avian precursor is essential for the emergence of new stable virus lineages in humans and pigs (reviewed in references 1, 25, and 34).
Based on early data (10, 35), it was assumed that all avian influenza viruses have similar receptor-binding specificities and, therefore, that there is no significant receptor-mediated restriction on viral interspecies transmission in birds. The first evidence arguing against this concept was obtained in a study on H5N1 viruses from Hong Kong isolated in 1997, when virus isolates from poultry and humans were found to have a lower receptor binding affinity and a lower neuraminidase activity than closely related viruses of aquatic birds (33). In addition, analysis of the HA and NA sequences of H5 and H7 influenza viruses from various avian species revealed that poultry influenza viruses often differ from duck influenza viruses by additional N-linked glycans at the top of HA and by large deletions in the stalk of NA (2, 33). Furthermore, the H9N2 viruses widely circulating in poultry in Eurasia were found to differ significantly in their receptor specificity from H9N2 viruses of other evolutionary lineages. In particular, these Eurasian poultry influenza viruses displayed good binding to α2-6-linked sialic acids (37, 53). Studies on expression of sialic acids in intestinal and respiratory epithelia of different birds revealed substantial host-specific distinctions, among them, expression of both Neu5Acα2-3Gal- and Neu5Acα2-6Gal-terminated sialyloligosaccharides in chicken and quail, in contrast to ducks, which mainly contain Neu5Acα2-3Gal in their intestinal epithelium (16, 19, 23, 29, 49, 63). Taken together, these findings indicated that influenza viruses perpetuated in different birds can have different receptor specificities owing to distinctions in the sialic acid receptors in these avian species.
Subsequent detailed studies on viral receptor-binding specificity revealed that preferential binding to terminal Neu5Acα2-3Gal disaccharide is shared by the majority of avian viruses; however, viruses adapted to ducks, gulls, and land-based gallinaceous poultry differ in their ability to recognize the subterminal saccharides of Neu5Acα2-3Gal-terminated receptors (reviewed in references 34 and 43). Duck influenza viruses of various HA subtypes (H1 to H5, H9 to H11) preferentially bound to receptors with type 1 and type 3 oligosaccharide sequences, i.e., having the β1-3 linkage between the terminal Neu5Acα2-3Gal moiety and the penultimate sugar residue such as Neu5Acα2-3Galβ1-3GlcNAc (SLec) and Neu5Acα2-3Galβ1-3GalNAcα (STF). Sulfation at the 6-OH group of the subterminal GlcNAc had little effect on binding of duck influenza viruses, whereas fucosylation of this residue reduced the binding significantly (15, 17, 20, 21). In contrast to duck influenza viruses, the H4, H6, H13, and H14 subtype viruses isolated from gulls showed high-avidity binding to fucosylated sialyloligosaccharides Neu5Acα2-3Galβ1-4(Fucα1-3)GlcNAc (SLex) and Neu5Acα2-3Galβ1-3(Fucα1-4)GlcNAc (SLea) (17, 67). Influenza viruses with HA subtypes H5, H7, and H9 are commonly reported in terrestrial gallinaceous poultry (7, 9). The common properties shared by these poultry-adapted viruses are (i) preferential binding to receptors with type 2 sequences having a β1-4 bond between the Neu5Acα2-3Gal moiety and the next sugar residue, such as Neu5Acα2-3Galβ1-4GlcNAc (3′SLN) and (ii) particularly strong binding to the corresponding sulfated analogues Neu5Acα2-3Galβ1-4(6-O-HSO3)GlcNAc (Su-3′SLN) and Neu5Acα2-3Galβ1-4(Fucα1-3)(6-O-HSO3)GlcNAc (Su-SLex) (17, 20, 21).
In the case of H5 and H9 subtypes, virus strains with both duck-virus-like and poultry-virus-like receptor phenotypes were identified in corresponding avian species (17, 20, 21). In contrast to these relatively extensive analyses of H5 and H9 influenza viruses, previous studies on receptor specificity of H7 influenza viruses were limited to a few virus strains from North American and Eurasian poultry. All tested H7 influenza viruses displayed a poultry-virus-like receptor specificity, and some of them showed the ability to bind to human-type receptors (4, 20, 68). To fill the gap in current knowledge on the receptor specificity of H7 influenza viruses, especially those circulating in wild aquatic birds, here we analyzed a large collection of viruses from various host species.
MATERIALS AND METHODS
Viruses.
The viruses were from the repositories of Erasmus Medical Center, Rotterdam, The Netherlands; Istituto Zooprofilattico Sperimentale delle Venezie, Padova, Italy; St. Jude Children's Research Hospital, Memphis, TN; Veterinary Laboratories Agency, Weybridge, Addlestone, United Kingdom; and M. P. Chumakov Institute of Poliomyelitis, Moscow, Russia. Viruses were grown in 10-day-old embryonated chicken eggs. Highly pathogenic viruses were inactivated by treatment with β-propiolactone as described previously (33). The allantoic fluids were clarified by low-speed centrifugation and used in the binding assays without further purification.
To determine HA sequences, viral RNA was extracted from virus-containing allantoic fluid, reverse transcribed, and PCR amplified using HA-specific primers. The PCR products were purified using a PCR purification kit (Qiagen) and sequenced using a BigDye Terminator cycle sequencing Ready Reaction kit (Applied Biosystems, Carlsbad, CA).
Receptor-binding assays.
Receptor specificity of the viruses was characterized by determining their binding to soluble synthetic poly-N-(2-hydroxyethyl)acrylamide-based sialylglycopolymers (SGPs) (Lectinity Holding, Inc., Moscow, Russia) (6). The nonlabeled SGPs contained 20 mol% of specific sialyloligosaccharide attached to the 30-kDa polymer. The structures and designations of their oligosaccharide moieties are shown below.
| Neu5Acα2-3Galβ1-4GlcNAcβ | 3′SLN |
| Neu5Gcα2-3Galβ1-4GlcNAcβ | 3′SLN(Gc) |
| Neu5Acα2-3Galβ1-4(6-O-HSO3)GlcNAcβ | Su-3′SLN |
| Neu5Acα2-3Galβ1-4(Fucα1-3)GlcNAcβ | SLex |
| Neu5Acα2-3Galβ1-4(Fucα1-3)(6-O-HSO3)GlcNAcβ | Su-SLex |
| Neu5Acα2-3Galβ1-3GlcNAcβ | SLec |
| Neu5Acα2-3Galβ1-3(6-O-HSO3)GlcNAcβ | Su-SLec |
| Neu5Acα2-3Galβ1-3(Fucα1-4)GlcNAcβ | SLea |
| Neu5Acα2-6Galβ1-4GlcNAcβ | 6′SLN |
The association constants of viral complexes with nonlabeled SGPs were determined in a solid-phase fetuin binding inhibition assay as described in detail previously (20, 36). In brief, 50-μl aliquots of bovine fetuin solution (5 μg/ml) in phosphate-buffered saline (PBS) were incubated in 96-well enzyme-linked immunosorbent assay (ELISA) microplates (Greiner) at 4°C overnight. The plates were washed with water and dried. Viral suspensions were diluted with PBS to hemagglutination titers of 20 to 40. A 50-μl volume of virus solution was added to each well of the fetuin-coated microplates. After incubation at 4°C overnight, the plates were washed with ice-cold washing buffer (0.02% Tween 80–PBS). Serial 2-fold dilutions of SGPs in a solution of PBS containing peroxidase-labeled fetuin, 0.02% Tween 80, 0.2% bovine serum albumin, and 1 μM oseltamivir carboxylate (sialidase inhibitor) were added into the wells (50 μl/well), and the plates were incubated at 4°C for 1 h. After washing, the peroxidase activity in the wells was assayed with tetramethylbenzidine substrate solution. The absorbencies at 450 nm were determined, and the data were transferred to a PC and processed using Microsoft Excel software. The apparent association constants (Kass) of virus complexes with SGPs were calculated for each experimental point within the binding inhibition range of 20% to 80%, and the results were averaged. Receptor-binding assays were repeated three to four times on different days, and the results were averaged.
To facilitate detection of low-avidity binding of the viruses to Neu5Acα2-6Gal-terminated receptors, we used high-molecular-mass polyvalent SGP that contained 20 mol% of 6′SLN and 5 mol% of biotin attached to the 1,500-kDa polymer. A matching biotinylated SGP containing 3′SLN instead of 6′SLN was used as a control. The binding of biotinylated SGPs was determined in a direct solid-phase binding assay (32). The viruses were adsorbed in the well of fetuin-coated plates as described above. Serial 2-fold dilutions of sialylglycopolymers in the reaction buffer (RB; 0.02% Tween 80–0.02% bovine serum albumin–1 μM oseltamivir carboxylate–PBS) were added into the wells (50 μl/well), and the plates were incubated at 4°C for 1 h. After washing, streptavidin-peroxidase solution in RB (1/2,000) was added at 50 μl/well, and the plates were incubated at 4°C for 1 h. After washing, the peroxidase activity in the wells was assayed as described above. The data were converted to Scatchard plots (A450/C versus A450), where C is the concentration of the sialic acid in solution and A450 is the absorbency in the corresponding well. The apparent association constants of virus complexes with SGPs were determined from the slopes of the Scatchard plots.
Analysis of HA amino acid sequences.
The H7 HA sequences were obtained from GenBank through the NCBI Influenza Virus Resource (3) accessed on 21 December 2009. From the 954 available sequences, 705 nonredundant amino acid sequences were selected. After exclusion of short and ambiguous sequences, 665 sequences were left. Nineteen HA sequences were determined in this study (see Table 1 for the accession numbers). All sequences were combined and edited using BioEdit version 7.0.5.32 (24). The H3 numbering system in accord with the alignment of Nobusawa et al. (44) is used throughout the paper. A phylogenetic tree was generated based on the amino acid sequences of the HA1 part using MEGA software version 5 with the neighbor-joining method (61). All ambiguous positions were removed for each sequence pair.
Table 1.
Viruses studied and their receptor-binding phenotypes
| Influenza virus | Accession no.a | Phenotypeb |
|---|---|---|
| Non-H7 duck viruses | ||
| Mallard/Alberta/119/98 (H1N1) | d | |
| Mallard/Ontario/56/76 (H2N3) | d | |
| Mallard/New York/6750/78 (H2N2) | d | |
| Mallard/Alberta/353/88 (H2N3) | d | |
| Mallard/Alberta/205/98 (H2N9) | d | |
| Mallard/Alberta/226/98 (H2N3) | d | |
| Mallard/Alberta/279/98 (H3N8) | d | |
| Mallard/Moscow/3556/08 (H3N2) | d | |
| Mallard/Alberta/47/98 (H4N1) | d | |
| Mallard/Moscow/3661/08 (H4N6) | d | |
| Mallard/Moscow/3641/08 (H11N9) | d | |
| H7 Eurasian poultry viruses | ||
| Chicken/FPV/Rostok/34 (N1) HPAIc | FLAHA7N1 | P$ |
| Turkey/England/647/77 (N7) | AF202247 | p* |
| African starling/England/983/79 (N1) | AF149295 | p |
| Chicken/England/4054/06 (N3) | EF467826 | P |
| Duck/Turkey/55/centinkaya/06 (N1) | N/A | P |
| Chicken/Wales/1306/07 (N2) | EF675618 | p* |
| Chicken/England/1158-11406/08 (N7) | FJ476173 | P |
| Turkey/Italy/977/99 (N1) | CY024754 | p |
| Turkey/Italy/1081/99 (N1) | AF364135 | P |
| Turkey/Italy/1082/99 (N1) | AF364136 | p |
| Turkey/Italy/2379/99 (N1) | AF364148 | p* |
| Turkey/Italy/2715/99 (N1) | AF364150 | p* |
| Turkey/Italy/2732/99 (N1) | CY025181 | p* |
| Turkey/Italy/3185/99 (N1) | AF364153 | p* |
| Turkey/Italy/3560/99 (N1) | AF364158 | P* |
| Turkey/Italy/4294/99 (N1) | AF364162 | P* |
| Turkey/Italy/5079/99 (N1) HPAI | AF364169 | P |
| Ostrich/Italy/984/00 (N1) HPAI | DQ991343 | P |
| Ostrich/Italy/2332/00 (N1) HPAI | DQ991312 | P |
| Turkey/Italy/2984/00 (N1) HPAI | CY021533 | P |
| Turkey/Italy/4426/99 (N1) | GU052992 | p* |
| Turkey/Italy/1351/01 (N1) | CY021421 | p |
| Turkey/Italy/8912/02 (N3) | CY020605 | P |
| Turkey/Italy/251/03 (N3) | CY020589 | p |
| Turkey/Italy/3620/03 (N3) | CY021357 | P |
| Turkey/Italy/4608/03 (N3) | CY021485 | p* |
| Turkey/Italy/4479/04 (N3) | CY020581 | p |
| H7 North American poultry viruses | ||
| Turkey/Minnesota/1200/80 (N3) | CY014778 | P* |
| Turkey/Minnesota/1/88 (N9) | CY014786 | P |
| Turkey/Virginia/4529/02 (N2) | N/A | p* $ |
| Avian/New York/273874/03 (N?) | N/A | p* $ |
| Chicken/NJ/294508-12/04 (N2) | EU743253 | p* $ |
| Chicken/Delaware/296763/04 (N?) | N/A | p* $ |
| H7 viruses isolated from humans | ||
| England/268/96 (N7) | AF028020 | P |
| Netherlands/219/2003 (N7) HPAI | AY338459 | P$ |
| Netherlands/230/2003 (N7) HPAI | EPI319937 | P$ |
| Netherlands/231/2003 (N7) HPAI | EPI319940 | P$ |
| New York/107/03 (N2) | EU587368 | p* $ |
| H7 Eurasian aquatic bird viruses | ||
| Duck/Hong Kong/293/78 (N2) | U20461 | P* |
| Mallard/Netherlands/12/00 (N3) | CY014718 | P |
| Mallard/Netherlands/9/05 (N7) | CY077008 | p |
| Mallard/Netherlands/22/07 (N1) | CY043840 | P |
| Mallard/Sweden/56/02 (N7) | AY999977 | P |
| Mallard/Sweden/82/02 (N7) | AY999978 | P |
| Mallard/Sweden/85/02 (N7) | AY999979 | P |
| Mallard/Sweden/87/02 (N7) | AY999980 | P |
| Mallard/Sweden/91/02 (N9) | AY999981 | P* |
| Mallard/Sweden/92/02 (N7) | AY999982 | P |
| Mallard/Sweden/94/02 (N7) | AY999984 | P |
| Mallard/Sweden/102/02 (N7) | AY999986 | P |
| Mallard/Sweden/105/02 (N7) | AY999989 | P |
| Mallard/Sweden/106/02 (N7) | AY999990 | P |
| Mallard/Sweden/107/02 (N7) | AY999991 | P |
| Mallard/Sweden/64/03 (N7) | CY096611+ | P |
| Mallard/Sweden/65/03 (N7) | CY096612+ | P |
| Redknot/Sweden/1/04 (N8) | CY096610+ | P |
| Mallard/Italy/33/01 (N3) | AY586411 | P |
| Mallard/Italy/440-8/05 (N7) | EPI167284+ | P |
| Mallard/Italy/497-29/06 (N7) | EPI167286+ | P |
| Mallard/Italy/497-35/06 (N7) | EPI167287+ | P |
| Shoveler/Italy/2698-27/06 (N7) | EPI167289+ | P |
| Shoveler/Italy/377-6/06 (N7) | EPI167285+ | P |
| Mallard/Italy/1336/07 (N3) | EPI167295+ | P* |
| Mallard/Italy/6104-14/07 (N3) | EPI167300+ | P |
| Teal/Italy/794-3/08 (N1) | EPI167298+ | P |
| H7 North American aquatic bird virues | ||
| Mallard/Alberta/279/77 (N3) | CY005976 | P |
| Ruddy turnstone/NJ/65/85 (N3) | CY005928 | P |
| Green-winged teal/Alberta/228/85 (N3) | CY005978 | P |
| Ruddy turnstone/DE/2378/88 (N7) | CY005980 | P |
| Redknot/NJ/325/89 (N7) | CY005981 | P |
| Mallard/Alberta/34/01 (N1) | CY005983 | P |
| Laughing gull/DE/22/02 (N3) | CY099313+ | p* |
| Laughing gull/DE/42/06 (N3) | EU030984 | p* |
| Mallard/Alberta/243/06 (N3) | CY099314+ | P* |
| Ruddy turnstone/DE/294/06 (N3) | N/A | p* |
| Ruddy turnstone/DE/108/07 (N3) | CY036775 | P* |
| Seal/Massachusetts/1/80 (N7) | K00429 | p |
| H7 equine viruses | ||
| Equine/Detroit/3/64 (N7) | CY099312+ | E7 |
| Equine/Lexington/1/66 (N7) | X62556 | E7 |
| Equine/Cordoba/5/76 (N7) | CY099311+ | E7 |
| Equine/Santiago/1/77 (N7) | AY383756 | E7 |
| H3 equine viruses | ||
| Equine/Miami/63 | E3 | |
| Equine/Tennessee/5/86 | E3 | |
| Equine/Kentucky/2/86 | E3 |
Data represent GenBank or EpiFlu accession numbers for the nucleotide sequences. Data for non-H7 HAs are not included. N/A, not available; “+,” sequences determined in this study.
Data represent distinctive patterns of virus binding to a panel of SGPs (see typical examples shown in Fig. 2 to 4; see also Table S1 in the supplemental material). d, see Fig. 2A; P, see Fig. 2B; p, see Fig. 2C; E7 and E3, see Fig. 3. “*,” virus binding to 6′SLN (Fig. 4B and C); “$,” data from the previous study (20).
HPAI, highly pathogenic avian influenza viruses with multibasic cleavage site between HA1 and HA2. FPV, fowl plague plague virus.
Molecular models.
Modeling was performed using atomic coordinates of the H7 HA complex with pentasaccharide LSTa determined previously (52). The molecular models were generated using PyMOL 1.4 (Schrödinger, LLC).
RESULTS
Selection of viruses for the study.
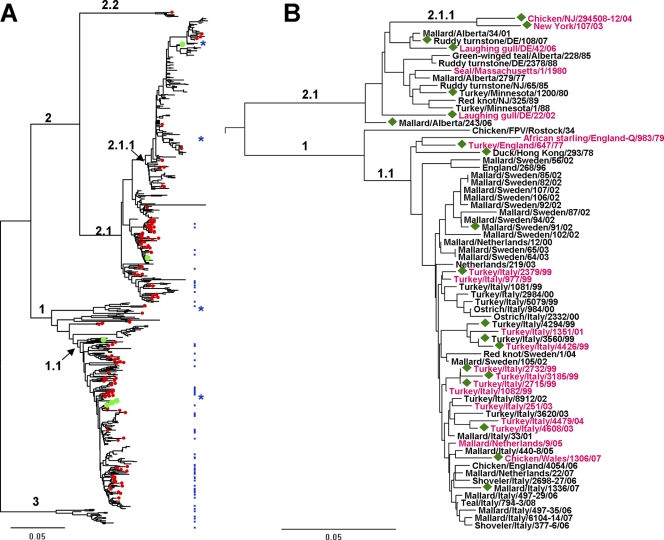
To explore the natural diversity of viruses with H7 HA and to select representative viruses for the receptor-binding studies, we analyzed HA sequences available in GenBank together with a few newly determined sequences of viruses from our repositories. The phylogenetic relationship between the H7 influenza viruses is shown in Fig. 1A. They are separated into avian viruses of the Eastern Hemisphere (clade 1) and Western Hemisphere (clade 2) and viruses of the equine H7 lineage (clade 3), which is roughly equidistant from either avian group. A few available sequences of South American viruses (clade 2.2) are well separated from the North American viruses (clade 2.1). The majority of sequences of viruses isolated in the Western Hemisphere are represented by those of viruses isolated in Europe and Asia after 1970 (clade 1.1). Four less-represented lineages include historical Eurasian isolates from the first half of the 20th century and African, Australian, and Pakistani viruses (not labeled on the tree).
Fig 1.
Evolutionary relationships of H7 HAs. Phylogenetic trees for the amino acid sequences of the HA1 protein were inferred by the neighbor-joining method using MEGA software version 5 (61). The scale bars represent 0.05 units of amino acid substitutions per site. (A) The tree is based on 665 sequences available from GenBank and 19 sequences determined in this study. Numbered branches include viruses from the Eastern Hemisphere (clade 1) and the Western Hemisphere (clade 2); viruses from North America (clade 2.1) and South America (clade 2.2); viruses isolated in Europe and Asia after 1970 (clade 1.1); North American poultry influenza viruses with an eight-amino-acid deletion in the receptor-binding site (clade 2.1.1); and H7 equine influenza viruses (clade 3). Red dots mark viruses isolated from wild and domestic aquatic birds (mainly ducks). Green dots mark viruses isolated from humans. Blue dots depict viruses tested for receptor binding properties in this study; blue stars show four viruses characterized previously (20). (B) Tree for the viruses tested in this study and in reference 20. The strain names are colored in accord with the viral receptor-binding specificity: black, typical poultry-virus-like phenotype (Fig. 2B); purple, atypical poultry-virus-like phenotype (Fig. 2C). Green diamonds depict viruses that bind to 6′SLN. The numbering of the clades is the same as in panel A.
To cover the major lineages of H7 influenza viruses, we studied viruses isolated from (i) North American ducks and shorebirds (clade 2.1), (ii) European ducks and poultry (clade 1.1), and (iii) horses (clade 3) (see Fig. 1 and Table 1). Representatives of the largest group of contemporary North American poultry influenza viruses with amino acid deletion 221 to 228 in the HA receptor-binding site (RBS) (58) (Fig. 1; clade 2.1.1) were not included, as their receptor-binding properties were characterized previously (20). To choose specific viral strains for the study, we analyzed amino acid sequences of the HA globular head (amino acid positions 90 to 250; see Fig. S1 in the supplemental material) and selected strains that differed by amino acid substitutions in the vicinity of the RBS. As a result, there was some bias in the selected virus panel toward the viruses with atypical sequences, because many European duck and poultry influenza viruses with identical HA sequences were represented in our panel by a single isolate. In addition to the viruses with known HA sequences, we tested a few nonsequenced viruses (Table 1). For the comparison with H7 influenza viruses, 11 representative duck influenza viruses of other subtypes and three H3 subtype equine influenza viruses were tested.
Receptor-binding properties of avian H7 influenza viruses.
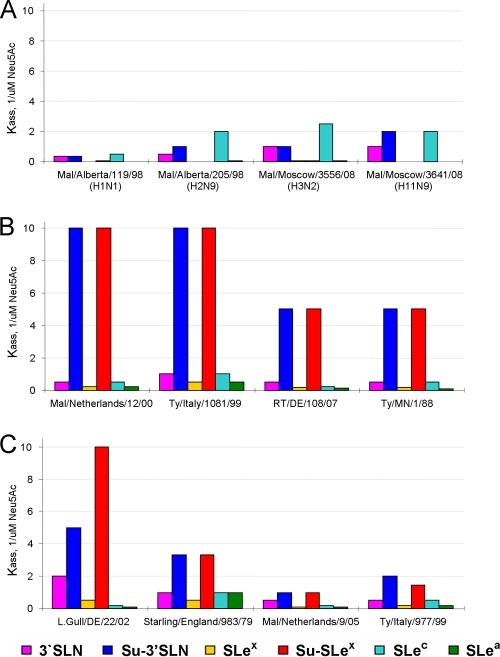
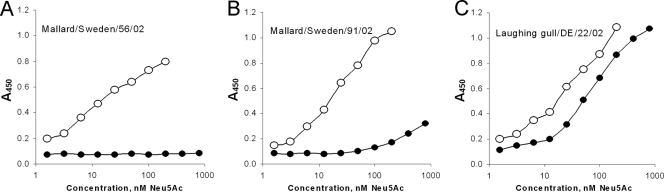
The receptor-binding characteristics were studied using a panel of seven sialylglycoproteins with terminal Neu5Acα2-3Gal moiety and distinctive structures of penultimate sugar residues (see Materials and Methods for structures and abbreviations of SGPs). The values of association constants of viral complexes with SGPs for all tested viruses are presented in Table S1 in the supplemental material. Based on these data, a specific receptor-binding phenotype (d, P, or p) was assigned to each virus (Table 1) as illustrated in Fig. 2. Viruses shown in Fig. 2A (phenotype d) displayed maximal binding avidity for Neu5Acα2-3Galβ1-3GlcNAc-containing SGP (SLec) and did not bind to fucosylated analogues SLex, Su-SLex, and SLea. Sulfation of the receptor moiety had relatively little or no effect on the binding avidity. This binding pattern was shared by all 11 tested duck influenza viruses with H1, H2, H3, H4, and H11 HAs.
Fig 2.
Examples of receptor-binding profiles of avian influenza viruses. Association constants of viral complexes with nonlabeled Neu5Ac2-3Gal-containing sialylglycopolymers were determined using a binding inhibition assay. Colors depict the sialyoligosaccharide moiety of the SGP. (A) Duck influenza viruses with non-H7 HA (receptor-binding phenotype d). (B) Viruses representing typical binding phenotype (P) of H7 influenza viruses with a Kass[Su-SLex]/Kass[3′SLN] ratio equal to or higher than 10 (see Table S1 in the supplemental material). (C) H7 influenza viruses with atypical binding phenotypes (p) (Kass[Su-SLex]/Kass[3′SLN] ratio between 1 and 8).
All tested H7 influenza viruses displayed another binding pattern. Virus strains represented in Fig. 2B (phenotype P) showed comparable levels of binding avidity to 5 SGPs (3′SLN, SLex, SLec, Su-SLec, and SLea), indicating that neither the type of linkage between Gal and the penultimate GlcNAc nor fucosylation of GlcNAc significantly affects the binding. The most prominent feature of these viruses was their strong and equal binding to two SGPs that contained sulfated sialyloligosaccharide moieties, Su-3′SLN and Su-SLex. The binding avidity for these sulfated SGPs was 10 to 20 times higher than that for corresponding nonsulfated analogues 3′SLN and SLex. As can be seen from Table 1, Table S1 in the supplemental material, and Fig. 1B, phenotype P is typically observed in the H7 influenza viruses tested in this study irrespective of their isolation place (Europe or North America) and host species (wild aquatic birds or poultry).
The viruses represented in Fig. 2C show the same distinctive H7-virus-like binding preference for sulfated receptors. However, their binding pattern is less pronounced. In particular, these viruses show a smaller difference in their binding to sulfated and nonsulfated SGPs compared with the typical H7 influenza viruses represented in Fig. 2B. For brevity, the viruses with such deviations from the typical H7 P phenotype are designated “atypical” (phenotype p). They were present in various lineages of H7 influenza viruses, with some prevalence among the viruses isolated from poultry (Table 1 and Fig. 1B).
Importantly, none of the tested H7 influenza viruses showed a receptor-binding profile characteristic of wild duck influenza viruses with other HA subtypes (Table 1 and previous reports [17, 20]).
Recognition of the N-glycolyl species of sialic acid.
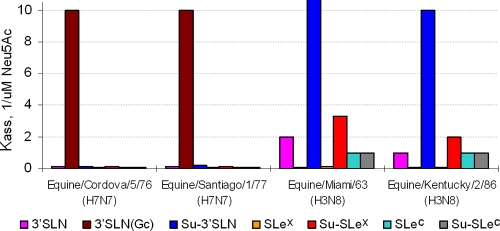
The N-glycolylneuraminic acid (Neu5Gc) does not seem to be present in birds, but it is found in all mammals except humans (54). To assess viral recognition of Neu5Gc, we used SGP with a Neu5Gcα2-3Galβ1-4GlcNAc moiety [3′SLN(Gc)]. Control non-H7 duck influenza viruses did not bind to 3′SLN(Gc), whereas all representative avian H7 influenza viruses tested showed detectable binding (see Table S1 in the supplemental material). However, the avian H7 influenza viruses bound to 3′SLN(Gc) 10 to 50 times less avidly than they bound to the corresponding Neu5Ac-containing analog (3′SLN) and 100 to 200 times less avidly than to their highest-avidity receptor, Su-SLex. The equine H7N7 viruses showed a strikingly different binding pattern. They bound to 3′SLN(Gc) with markedly higher avidity than any of the Neu5Ac-containing SGPs tested (Fig. 3; see also Table S1 in the supplemental material). Three equine H3N8 viruses were tested for a comparison. These viruses bound to 3′SLN(Gc) much less efficiently than to the Neu5Ac-containing counterpart 3′SLN and, especially, than to the highest-affinity receptor, Su-3′SLN.
Fig 3.
Examples of receptor-binding profiles of equine influenza viruses with H7 and H3 HA. Association constants of viral complexes with nonlabeled sialylglycopolymers were determined using a binding inhibition assay.
Binding to Neu5Acα2-6Gal-containing receptor analogue.
Although most avian influenza viruses do not bind to α2-6-linked sialic acid, we and others previously noticed that a few tested H7 poultry influenza viruses showed weak but significant binding to Neu5Acα2-6Gal-containing receptors (4, 20, 68). To facilitate detection of viral low-avidity interactions with α2-6-linked receptors, we assayed binding of high-molecular-mass 6′SLN-containing biotinylated SGP (6′SLN-biot) in a direct binding assay. Figure 4 illustrates the spectrum of binding patterns observed. All non-H7 duck influenza viruses, all equine influenza viruses, and many H7 avian viruses showed no binding to 6′SLN-biot (Fig. 4A; see also Table S1 in the supplemental material). Yet a significant fraction of H7 avian viruses displayed low (Fig. 4B) to pronounced (Fig. 4C) binding to 6′SLN-biot; such binding, however, was always weaker than the binding to the matching 3′SLN-containing counterpart (3′SLN-biot). As can be concluded from Fig. 1B, Table 1, and Table S1 in the supplemental material, the H7 avian viruses that were capable of binding to α2-6-linked sialic acid belonged to different evolutionary lineages and were isolated from both poultry and wild aquatic birds.
Fig 4.
Examples of virus binding to Neu5Acα2-3Gal- and Neu5Acα2-6Gal-containing biotinylated SGPs in a direct binding assay. The viruses displayed comparable levels of binding to 3′SLN-containing SGPs (open circles); two of them (panels B and C) also bound to 6′SLN-containing SGPs (closed circles).
Analysis of HA sequences.
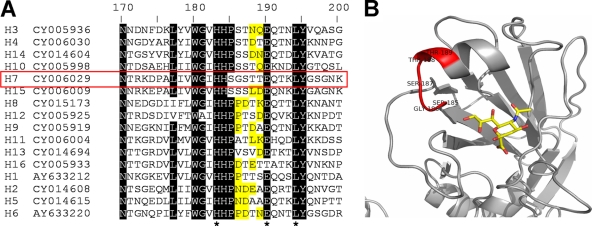
Our data revealed several distinctions in the receptor binding profiles of H7 influenza viruses and duck influenza viruses with non-H7 HA, such as enhanced binding of H7 influenza viruses to sulfated receptors Su-3′SLN and Su-SLex, their ability to bind fucosylated sialyloligosaccharides, and their occasional binding to α2-6-linked sialic acids. To understand the molecular basis of these distinctions, we first compared amino acid sequences of H7 HA with avian HAs of other subtypes, focusing on amino acids forming the receptor-binding domain (amino acids 90 to 250) (Fig. 5; see also Fig. S1 in the supplemental material). The H7 HA and the phylogenetically close H15 HA (51) differed from all other avian HAs by the substitution of Pro to Ser in conserved position 185 of the RBS (Fig. 5). Residues in positions 185 to 189 of the H7 HA have small side chains (typically, Ser-Gly-Ser-Thr-Thr), whereas other avian viruses have, in addition to Pro185, at least one large amino acid in these positions. These distinctions in the sequence of the 185 to 189 loop of H7 HA could affect the structure of the “left” side of its RBS.
Fig 5.
Structure of the 185 to 189 peptide loop of H7 HA. (A) Partial amino acid sequences of the 16 HA subtypes. Amino acids in the region from 185 to 189 with bulky side chains are highlighted in yellow. H7 HA is indicated with a red box. GenBank accession numbers for the sequences are shown next to the subtype. The top line shows H3 numbering. Stars in the bottom line depict amino acid residues that interact with sialic acid in the HA-receptor complex. The sequences are listed according to their homology; amino acids conserved among at least 14 of 16 sequences are shaded. (B) Location of the 185 to 189 loop (colored in red) on the model of H7 HA complex with pentasaccharide LSTa (52). Only the sialic acid moiety of LSTa is shown (stick model).
Inefficient binding of duck influenza viruses to the fucosylated receptors SLex, SLea, and Su-SLex seems to depend at least partially on steric clashes of the fucose moiety with the subtype-conserved bulky amino acid in position 222 of the HA (17, 20). Apparently, there are no such clashes in the case of the H7 HAs, which display similar binding avidities for the fucosylated receptors SLea and SLex and their nonfucosylated counterparts SLec and 3′SLN, respectively (Fig. 2; see also Table S1 in the supplemental material). Our previous studies on H5N1/97 avian and H3N8 equine influenza viruses suggested that enhanced binding of these viruses to sulfated receptors with type 2 cores (Su-3′SLN and Su-SLex) can be explained by favorable electrostatic interactions of the sulfate moiety with the positively charged side chain of lysine or arginine in position 193 of the HA (17, 20, 21). The same mechanism may be responsible for the strong binding of H7 influenza viruses to Su-3′SLN and Su-SLex, as all these viruses have Lys193 (see Fig. S1 in the supplemental material).
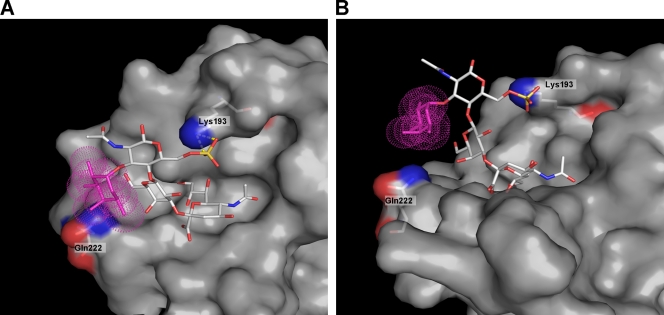
To test our hypotheses about atomic interactions of the H7 HA with the sulfate and fucose moieties of the receptors, we modeled the positioning of the Su-SLex analogue in the RBS by the use of the crystal structure of the H7 HA complex with the nonfucosylated and nonsulfated pentasaccharide LSTa (52). As illustrated in Fig. 6, the modeling predicted formation of an ionic bond between the sulfo group of Su-SLex and Lys193 and a concomitant lack of sterical conflicts between fucose and its closest counterpart in the RBS, glutamine in position 222.
Fig 6.
Molecular model of H7 HA complexed with sialyoligosaccharide Su-SLex. The model represents the crystal structure of the H7 HA complex with LSTa (52), in which bound sialyloligosaccharide was modified to generate Su-SLex by the use of Pymol. The HA is shown as a gray molecular surface. Lys193 and Gln222 are highlighted and labeled. The fucose residue of Su-SLex is depicted by purple dots; the sulfo group is shown in orange-red. Panels A and B show different views of the same model and illustrate formation of and ionic bonds between the sulfo group and Lys193 and the lack of sterical conflicts between fucose and Gln222.
To identify amino acid substitutions responsible for the deviation of some H7 influenza viruses from the typical receptor-binding phenotype, we analyzed HA sequences of H7 influenza viruses tested in this study (see Fig. S1 in the supplemental material).The majority of atypical European viruses shared substitution G186V. Furthermore, three out of five atypical North American strains had mutation G186A or G186E. This correlation agreed with our hypothesis about the important role played by residues 185 to 189 in the structure and distinctive receptor-binding profile of H7 HA. Substitutions G205E (Turkey/Italy/977/99), S227T (Laughing gull/Delaware/42/06), and K193R (Seal/Massachusetts/1/80) and deletion 221 to 228 (NewYork/107/03 and Chicken/NJ/294508-12/04) also correlated with atypical binding phenotypes. Substitutions in these positions are known to affect receptor-binding characteristics of influenza viruses (4, 11, 15, 38, 57, 60, 66, 68). Given the significant evolutionary divergence of HAs of H7 poultry and equine influenza viruses, it was impossible to predict specific amino acid substitutions responsible for the preferential binding of equine influenza viruses to the N-glycolyl form of sialic acid. Substitutions in the 130 and 220 loops of the HA RBS seem to be the most plausible candidates for the differences in binding specificity.
N-linked glycans in the proximity of the RBS can affect viral receptor-binding characteristics (for reviews, see references 34 and 55). The viruses in our panel differed by potential glycosylation sites at Asn133 and Asn158; these sites are known to be glycosylation competent (62). The presence or absence of these sites did not clearly correlate with the binding phenotype of the virus, although we noticed that 7 of 11 viruses with the site at Asn158 showed binding to 6′SLN.
DISCUSSION
We previously found that duck influenza viruses with various divergent HA subtypes (H1 to H5, H9 to H11) shared receptor binding specificity, suggesting that these viruses either evolved from a common duck ancestor or acquired duck-virus-like receptor specificity during their prolonged circulation in ducks (20, 34). The H5N1 and H9N2 stable virus lineages in gallinaceous poultry originated from viruses of aquatic birds (7, 9, 48) and have distinctive poultry-virus-like receptor specificity (17, 20). These findings indicate that the duck-to-poultry adaptation of H5 and H9 influenza viruses altered their receptor specificity.
The influenza viruses with H7 HA subtypes were isolated from a variety of aquatic and land-based birds. The viruses were associated with poultry outbreaks of avian influenza (“fowl plaque”) of low and high pathogenicity in both hemispheres and occasionally infected mammals, including humans (reviewed in references 5, 9, and 65). Interestingly, little genetic diversity has been observed between H7 influenza viruses isolated from wild aquatic and land-based domestic birds in the same geographical area (2, 8, 42), suggesting their relatively frequent and perhaps unrestricted interspecies transmission. Given the importance of H7 influenza viruses with respect to animal and human health and their apparent expanded host range, we characterized receptor specificity of H7 influenza viruses isolated from different host species and representing almost all currently known evolutionary lineages (Fig. 1 and Table 1).
The equine H7 influenza viruses differed from all other H7 influenza viruses by their clear preference for the N-glycolyl form of sialic acid, which is the major sialic acid species expressed in the horse tracheal epithelium (59). The ability of H3 and H7 influenza viruses to bind both Neu5Ac and Neu5Gc was noticed previously (17, 18, 59). The receptor-binding assay used in this study shows that H7 equine influenza viruses bind Neu5Gc significantly better than Neu5Ac, whereas H3 equine influenza viruses display the opposite binding preference. Because Neu5Gc is absent in birds and humans (54), one can assume that equine H7 influenza viruses were strongly adapted to their host and that their receptor specificity could have restricted their transmission to other species.
All non-equine H7 influenza viruses displayed relatively homogeneous receptor-binding properties. The key feature of these viruses was their enhanced avidity for sulfated receptor sequences with type 2 cores (Su-3′SLN and Su-SLex), most likely owing to ionic interactions of the negatively charged sulfate with a conserved positively charged amino acid in position 193 (Lys or Arg). In addition, most H7 influenza viruses tolerated fucosylation of the third sugar in both type 1 and type 2 sequences (SLea and SLex). These features, individually or in combination, are known hallmarks of poultry-adapted viruses with H5 and H9 HAs that differentiate these viruses from viruses in ducks (17, 20, 21). An independent acquisition by H5, H7, and H9 poultry influenza viruses of a strong binding to Su-SLex suggests that this property provides the viruses with a selective replicative advantage in poultry. In mammals, SLex and its sulfated forms, among them Su-SLex, are well-known naturally occurring sequences recognized by E-, P-, and L-selectins (12, 39, 56). SLex and related molecules are constitutively expressed glycan sequences typical for endothelial cells and leukocytes, and their levels can increase manyfold during the inflammation process (12). Given that both selectins and their ligands are evolutionary conserved molecules, one can predict expression of Su-SLex on avian endothelial cells. Interestingly, three previous reports described a strong HA-dependent tropism of H5 and H7 poultry influenza viruses to endothelial cells of chicken embryo cells (13) and to cultured human pulmonary microvascular endothelial cells (45, 69). Thus, enhanced binding of poultry influenza viruses to Su-SLex and other sulfated and fucosylated sialyloligosaccharide sequences could have emerged as an adaptive change required for viral endotheliotropism in birds, a well-known feature of highly pathogenic poultry influenza viruses with H5 and H7 HA (28).
In is believed that influenza A viruses in land-based birds and mammals originate from the viruses of wild aquatic birds. Remarkably, we found that none of the H7 influenza viruses isolated from North American and Eurasian ducks and shorebirds tested in this study showed a binding phenotype typical of the duck influenza viruses with non-H7 HAs. Because all H7 influenza viruses, irrespective of the host of origin, share the unique structure of the 190 loop and distinctive poultry-virus-like binding phenotype, one can speculate that structural and functional features of the RBS of contemporary H7 influenza viruses were formed after the introduction of their common precursor from ducks to a new host, presumably, gallinaceous poultry. The original H7 duck influenza virus seems to have become extinct; alternatively, the H7 HA could have evolved from a non-H7 HA subtype after its duck-to-poultry transmission.
Efficient circulation of H7 influenza viruses in ducks despite their poultry-virus-like receptor specificity could be explained by a relatively good binding of these viruses to the “duck-type” receptor sequence SLec (see Fig. 2; see also Table S1 in the supplemental material). In contrast, the H9 poultry-adapted viruses have almost completely lost binding to this sequence (20). The ability of the progenitor H7 subtype virus to replicate in both ducks and poultry without major changes in the receptor specificity could have contributed to its fast expansion.
Although distinctive receptor-binding properties of the H7 HA permit virus replication in various avian species without significant changes in the RBS and receptor specificity, adaptive changes in the neuraminidase seem to be essential for the duck-to-poultry adaptation. Thus, deletions in the stalk of the NA that affect its catalytic activity are commonly observed in H5 and H7 poultry influenza viruses but not in their putative duck precursors (2, 8, 31, 33). Emergence of the NA stalk deletions as the first essential change in the duck influenza viruses during their experimental adaptation to poultry was confirmed in several independent studies (22, 26, 30, 40). In addition to deletions in the NA, HA mutations leading to emergence of new glycosylation sites near the RBS of H5 and H7 influenza viruses correlate with virus isolation from poultry. These mutations do not seem to emerge immediately after the virus transmission to poultry (2, 33). They rather correlate with the acquisition of the multibasic cleavage site by the virus and could thus be a consequence of the extended tissue tropism of the virus such as the ability to replicate in endothelial cells (13). We have tested only a few HPAI H7 influenza viruses, and their receptor-binding phenotypes were similar to phylogenetically close low-pathogenic strains (Table 1). Further studies are needed to understand the role of NA deletions and enhanced HA glycosylation in virus replication and pathogenicity in poultry.
Similarly to most avian viruses, many H7 influenza viruses do not bind Neu5Acα2-6Gal-terminated receptors. The potential structural reasons underlying the lack of binding of the human-type receptors by the typical H7 influenza virus A/Turkey/Italy/02 were previously discussed (52). Since 1994, the H7 influenza viruses circulating in North American live bird markets have acquired an eight-amino-acid deletion in the RBS (positions 221 to 228) (see Fig. 1; see also Fig. S1 in the supplemental material). The viruses of this lineage maintained preferential binding to Neu5Acα2-3Gal-terminated sequences and acquired weak-to-moderate binding to 2-6-linked sialic acids (4, 20, 68). Structural analysis revealed that Arg220 and Arg229 compensate for the deletion and interact with receptor analogues (68). Importantly, we found in this study that not only H7 poultry influenza viruses of a lineage with the HA deletion but H7 aquatic bird and poultry influenza viruses from many other evolutionary lineages displayed detectable binding to human-type receptor analogue 6′SLN (see Fig. 1B). We speculate that the peculiar structure of the 185 to 189 loop located close to the 221 to 228 loop reduces HA conflicts with the Neu5Acα2-6Gal-terminated receptor sequences and that various additional substitutions within and in the vicinity of the RBS allow binding to Neu5Acα2-6Gal. It is not clear whether this property provides the virus with some selective advantage or merely reflects occasional HA variations. Whatever is the case, the frequently emerging ability of the H7 influenza viruses to bind to 2-6-linked sialic acids can facilitate transmission of these viruses to humans.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the European Union FP6 project FLUPATH. Parts of the study were funded by the Von Behring-Roentgen-Stiftung (M.N.M.), the Deutsche Forschungsgemeinschaft SFB 593 (H.-D.K.), NIAID-NIH contracts HHSN266200700010C (R.A.M.F.) and HHSN266200700005C (R.G.W.), NIAID-NIH Division of Intramural Research (V.J.M.), research grant “Molecular and Cell Biology” from the Presidium of the Russian Academy of Sciences (N.V.B.), and research grant 11-04-00517 from the Russian Foundation for Basic Research (A.S.G.).
We thank John Skehel (MRC National Institute for Medical Research, London, United Kingdom) and Rupert Russell (University of St Andrews, St. Andrews, United Kingdom) for atomic coordinates of the H7 HA complex with LSTa and for critical comments on the manuscript.
Footnotes
Published ahead of print 15 February 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Baigent SJ, McCauley JW. 2003. Influenza type A in humans, mammals and birds: determinants of virus virulence, host-range and interspecies transmission. Bioessays 25:657–671 [DOI] [PubMed] [Google Scholar]
- 2. Banks J, et al. 2001. Changes in the haemagglutinin and the neuraminidase genes prior to the emergence of highly pathogenic H7N1 avian influenza viruses in Italy. Arch. Virol. 146:963–973 [DOI] [PubMed] [Google Scholar]
- 3. Bao Y, et al. 2008. The influenza virus resource at the National Center for Biotechnology Information. J. Virol. 82:596–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Belser JA, et al. 2008. Contemporary North American influenza H7 viruses possess human receptor specificity: implications for virus transmissibility. Proc. Natl. Acad. Sci. U. S. A. 105:7558–7563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Belser JA, Bridges CB, Katz JM, Tumpey TM. 2009. Past, present, and possible future human infection with influenza virus A subtype H7. Emerg. Infect. Dis. 15:859–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bovin NV. 1998. Polyacrylamide-based glycoconjugates as tools in glycobiology. Glycoconj. J. 15:431–446 [DOI] [PubMed] [Google Scholar]
- 7. Brown IH, et al. 2006. Recent epidemiology and ecology of influenza A viruses in avian species in Europe and the Middle East. Dev. Biol. (Basel) 124:45–50 [PubMed] [Google Scholar]
- 8. Campitelli L, et al. 2004. Interspecies transmission of an H7N3 influenza virus from wild birds to intensively reared domestic poultry in Italy. Virology 323:24–36 [DOI] [PubMed] [Google Scholar]
- 9. Capua I, Alexander DJ. 2009. Avian influenza infection in birds: a challenge and opportunity for the poultry veterinarian. Poult. Sci. 88:842–846 [DOI] [PubMed] [Google Scholar]
- 10. Connor RJ, Kawaoka Y, Webster RG, Paulson JC. 1994. Receptor specificity in human, avian, and equine H2 and H3 influenza virus isolates. Virology 205:17–23 [DOI] [PubMed] [Google Scholar]
- 11. Daniels PS, et al. 1987. The receptor-binding and membrane-fusion properties of influenza virus variants selected using anti-haemagglutinin monoclonal antibodies. EMBO J. 6:1459–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Feizi T, Galustian C. 1999. Novel oligosaccharide ligands and ligand-processing pathways for the selectins. Trends Biochem. Sci. 24:369–372 [DOI] [PubMed] [Google Scholar]
- 13. Feldmann A, Schafer MK, Garten W, Klenk HD. 2000. Targeted infection of endothelial cells by avian influenza virus A/FPV/Rostock/34 (H7N1) in chicken embryos. J. Virol. 74:8018–8027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fouchier RA, Munster VJ. 2009. Epidemiology of low pathogenic avian influenza viruses in wild birds. Rev. Sci. Tech. 28:49–58 [DOI] [PubMed] [Google Scholar]
- 15. Gambaryan A, et al. 2006. Evolution of the receptor binding phenotype of influenza A (H5) viruses. Virology 344:432–438 [DOI] [PubMed] [Google Scholar]
- 16. Gambaryan A, Webster R, Matrosovich M. 2002. Differences between influenza virus receptors on target cells of duck and chicken. Arch. Virol. 147:1197–1208 [DOI] [PubMed] [Google Scholar]
- 17. Gambaryan A, et al. 2005. Receptor specificity of influenza viruses from birds and mammals: new data on involvement of the inner fragments of the carbohydrate chain. Virology 334:276–283 [DOI] [PubMed] [Google Scholar]
- 18. Gambaryan AS, et al. 2005. Receptor-binding properties of swine influenza viruses isolated and propagated in MDCK cells. Virus Res. 114:15–22 [DOI] [PubMed] [Google Scholar]
- 19. Gambaryan AS, et al. 2003. Differences between influenza virus receptors on target cells of duck and chicken and receptor specificity of the 1997 H5N1 chicken and human influenza viruses from Hong Kong. Avian Dis. 47:1154–1160 [DOI] [PubMed] [Google Scholar]
- 20. Gambaryan AS, et al. 2008. 6-Sulfo sialyl Lewis X is the common receptor determinant recognized by H5, H6, H7 and H9 influenza viruses of terrestrial poultry. Virol. J. 5:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gambaryan AS, et al. 2004. H5N1 chicken influenza viruses display a high binding affinity for Neu5Acalpha2-3Galbeta1-4(6-HSO3)GlcNAc-containing receptors. Virology 326:310–316 [DOI] [PubMed] [Google Scholar]
- 22. Giannecchini S, et al. 2010. Molecular adaptation of an H7N3 wild duck influenza virus following experimental multiple passages in quail and turkey. Virology 408:167–173 [DOI] [PubMed] [Google Scholar]
- 23. Guo CT, et al. 2007. The quail and chicken intestine have sialyl-galactose sugar chains responsible for the binding of influenza A viruses to human type receptors. Glycobiology 17:713–724 [DOI] [PubMed] [Google Scholar]
- 24. Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95–98 [Google Scholar]
- 25. Horimoto T, Kawaoka Y. 2001. Pandemic threat posed by avian influenza A viruses. Clin. Microbiol. Rev. 14:129–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hossain MJ, Hickman D, Perez DR. 2008. Evidence of expanded host range and mammalian-associated genetic changes in a duck H9N2 influenza virus following adaptation in quail and chickens. PLoS One 3:e3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ito T, et al. 1998. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. J. Virol. 72:7367–7373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Klenk HD. 2005. Infection of the endothelium by influenza viruses. Thromb. Haemost. 94:262–265 [DOI] [PubMed] [Google Scholar]
- 29. Kuchipudi SV, et al. 2009. Differences in influenza virus receptors in chickens and ducks: implications for interspecies transmission. J. Mol. Genet. Med. 3:143–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li J, Cardona CJ. 2010. Adaptation and transmission of a wild duck avian influenza isolate in chickens. Avian Dis. 54:586–590 [DOI] [PubMed] [Google Scholar]
- 31. Li J, Zu DH, Cardona CJ, Miller J, Carpenter TE. 2011. Emergence and genetic variation of neuraminidase stalk deletions in avian influenza viruses. PLoS One 6:e14722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Matrosovich M, et al. 2000. Early alterations of the receptor-binding properties of H1, H2, and H3 avian influenza virus hemagglutinins after their introduction into mammals. J. Virol. 74:8502–8512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Matrosovich M, Zhou N, Kawaoka Y, Webster R. 1999. The surface glycoproteins of H5 influenza viruses isolated from humans, chickens, and wild aquatic birds have distinguishable properties. J. Virol. 73:1146–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Matrosovich MN, Gambaryan AS, Klenk H-D. 2008. Receptor specificity of influenza viruses and its alteration during interspecies transmission, p 134–155 In Klenk H-D, Matrosovich MN, Stech J. (ed), Avian influenza, vol 27 Karger, Basel, Switzerland [Google Scholar]
- 35. Matrosovich MN, et al. 1997. Avian influenza A viruses differ from human viruses by recognition of sialyloligosaccharides and gangliosides and by a higher conservation of the HA receptor-binding site. Virology 233:224–234 [DOI] [PubMed] [Google Scholar]
- 36. Matrosovich MN, et al. 1993. Probing of the receptor-binding sites of the H1 and H3 influenza A and influenza B virus hemagglutinins by synthetic and natural sialosides. Virology 196:111–121 [DOI] [PubMed] [Google Scholar]
- 37. Matrosovich MN, Krauss S, Webster RG. 2001. H9N2 influenza A viruses from poultry in Asia have human virus-like receptor specificity. Virology 281:156–162 [DOI] [PubMed] [Google Scholar]
- 38. Medeiros R, Naffakh N, Manuguerra JC, van der Werf S. 2004. Binding of the hemagglutinin from human or equine influenza H3 viruses to the receptor is altered by substitutions at residue 193. Arch. Virol. 149:1663–1671 [DOI] [PubMed] [Google Scholar]
- 39. Mitoma J, et al. 2007. Critical functions of N-glycans in L-selectin-mediated lymphocyte homing and recruitment. Nat. Immunol. 8:409–418 [DOI] [PubMed] [Google Scholar]
- 40. Mundt E, et al. 2009. Replication and pathogenesis associated with H5N1, H5N2, and H5N3 low-pathogenic avian influenza virus infection in chickens and ducks. Arch. Virol. 154:1241–1248 [DOI] [PubMed] [Google Scholar]
- 41. Munster VJ, et al. 2007. Spatial, temporal, and species variation in prevalence of influenza A viruses in wild migratory birds. PLoS Pathog. 3:e61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Munster VJ, et al. 2005. Mallards and highly pathogenic avian influenza ancestral viruses, northern Europe. Emerg. Infect. Dis. 11:1545–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nicholls JM, Chan RW, Russell RJ, Air GM, Peiris JS. 2008. Evolving complexities of influenza virus and its receptors. Trends Microbiol. 16:149–157 [DOI] [PubMed] [Google Scholar]
- 44. Nobusawa E, et al. 1991. Comparison of complete amino acid sequences and receptor-binding properties among 13 serotypes of hemagglutinins of influenza A viruses. Virology 182:475–485 [DOI] [PubMed] [Google Scholar]
- 45. Ocaña-Macchi M, et al. 2009. Hemagglutinin-dependent tropism of H5N1 avian influenza virus for human endothelial cells. J. Virol. 83:12947–12955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Olsen B, et al. 2006. Global patterns of influenza a virus in wild birds. Science 312:384–388 [DOI] [PubMed] [Google Scholar]
- 47. Paulson JC. 1985. Interactions of animal viruses with cell surface receptors, p 131–219 In Conn M. (ed), The receptors, vol. 2 Academic Press, Orlando, FL [Google Scholar]
- 48. Peiris JS, de Jong MD, Guan Y. 2007. Avian influenza virus (H5N1): a threat to human health. Clin. Microbiol. Rev. 20:243–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pillai SP, Lee CW. 2010. Species and age related differences in the type and distribution of influenza virus receptors in different tissues of chickens, ducks and turkeys. Virol. J. 7:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rogers GN, D'Souza BL. 1989. Receptor binding properties of human and animal H1 influenza virus isolates. Virology 173:317–322 [DOI] [PubMed] [Google Scholar]
- 51. Röhm C, Zhou N, Suss J, Mackenzie J, Webster RG. 1996. Characterization of a novel influenza hemagglutinin, H15: criteria for determination of influenza A subtypes. Virology 217:508–516 [DOI] [PubMed] [Google Scholar]
- 52. Russell RJ, Stevens DJ, Haire LF, Gamblin SJ, Skehel JJ. 2006. Avian and human receptor binding by hemagglutinins of influenza A viruses. Glycoconj. J. 23:85–92 [DOI] [PubMed] [Google Scholar]
- 53. Saito T, et al. 2001. Characterization of a human H9N2 influenza virus isolated in Hong Kong. Vaccine 20:125–133 [DOI] [PubMed] [Google Scholar]
- 54. Schauer R, Srinivasan GV, Coddeville B, Zanetta JP, Guerardel Y. 2009. Low incidence of N-glycolylneuraminic acid in birds and reptiles and its absence in the platypus. Carbohydr. Res. 344:1494–1500 [DOI] [PubMed] [Google Scholar]
- 55. Schulze IT. 1997. Effects of glycosylation on the properties and functions of influenza virus hemagglutinin. J. Infect. Dis. 176(Suppl. 1):S24–S28 [DOI] [PubMed] [Google Scholar]
- 56. Sperandio M, Gleissner CA, Ley K. 2009. Glycosylation in immune cell trafficking. Immunol. Rev. 230:97–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Stevens J, et al. 2008. Recent avian H5N1 viruses exhibit increased propensity for acquiring human receptor specificity. J. Mol. Biol. 381:1382–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Suarez DL, Garcia M, Latimer J, Senne D, Perdue M. 1999. Phylogenetic analysis of H7 avian influenza viruses isolated from the live bird markets of the Northeast United States. J. Virol. 73:3567–3573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Suzuki Y, et al. 2000. Sialic acid species as a determinant of the host range of influenza A viruses. J. Virol. 74:11825–11831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Suzuki Y, Kato H, Naeve CW, Webster RG. 1989. Single-amino-acid substitution in an antigenic site of influenza virus hemagglutinin can alter the specificity of binding to cell membrane-associated gangliosides. J. Virol. 63:4298–4302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tamura K, et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 24:1596–1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wagner R, Wolff T, Herwig A, Pleschka S, Klenk HD. 2000. Interdependence of hemagglutinin glycosylation and neuraminidase as regulators of influenza virus growth: a study by reverse genetics. J. Virol. 74:6316–6323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wan H, Perez DR. 2006. Quail carry sialic acid receptors compatible with binding of avian and human influenza viruses. Virology 346:278–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. 1992. Evolution and ecology of influenza A viruses. Microbiol. Rev. 56:152–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Webster RG, Hulse DJ. 2004. Microbial adaptation and change: avian influenza. Rev. Sci. Tech. 23:453–465 [DOI] [PubMed] [Google Scholar]
- 66. Yamada S, et al. 2006. Haemagglutinin mutations responsible for the binding of H5N1 influenza A viruses to human-type receptors. Nature 444:378–382 [DOI] [PubMed] [Google Scholar]
- 67. Yamnikova SS, et al. 2003. Differences between HA receptor-binding sites of avian influenza viruses isolated from Laridae and Anatidae. Avian Dis. 47(Suppl. 3):1164–1168 [DOI] [PubMed] [Google Scholar]
- 68. Yang H, Chen LM, Carney PJ, Donis RO, Stevens J. 2010. Structures of receptor complexes of a North American H7N2 influenza hemagglutinin with a loop deletion in the receptor binding site. PLoS Pathog. 6:e1001081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zeng H, et al. 2012. Human pulmonary microvascular endothelial cells support productive replication of highly pathogenic avian influenza viruses: possible involvement in the pathogenesis of human H5N1 virus infection. J. Virol. 86:667–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.