Abstract
We analyzed the nuclear trafficking ability of Gag proteins from six retroviral genera. Contrary to a previous report, human immunodeficiency virus type 1 (HIV-1) Gag showed no propensity to cycle through the nucleus. The only Gag protein that displayed CRM1-dependent nuclear cycling was that of Rous sarcoma virus (RSV). Surprisingly, this cycling could be eliminated without compromising infectivity by replacing the RSV Gag N-terminal matrix (MA) domain with HIV MA.
TEXT
The retroviral structural protein, Gag, from several viral genera has been reported to be present in minor amounts in the nucleus of infected cells (3, 11, 14, 17). These viruses include Rous sarcoma virus (RSV), human immunodeficiency virus type 1 (HIV-1), murine leukemia virus (MLV), and prototypic foamy virus (PFV). For RSV, the mechanism of this trafficking has been carefully studied. RSV Gag contains two nuclear localization sequences (NLSs), one in the matrix (MA) domain and one in the nucleocapsid (NC) domain. These NLSs bind to the nuclear import receptors importin-11 and importin-alpha, respectively (1, 5). RSV Gag also contains a nuclear export sequence (NES) in the p10 domain that interacts with the CRM1 nuclear export machinery (14, 16). At steady state, very little RSV Gag is located in the nucleus, but treatment of cells with the CRM1 inhibitor leptomycin B (LMB) results in a striking accumulation of Gag in the nucleus within minutes of treatment (14). It has been proposed that the nuclear trafficking of RSV Gag is required for proper packaging of the genomic RNA (4, 5, 12). In support of this hypothesis, RSV Gag mutants that do not traffic through the nucleus fail to properly package their RNA genome and are noninfectious (2, 4, 15, 16). However, another report suggests that RSV Gag does not play a role in the export of unspliced genomic RNA (9).
For HIV-1, according to a single report (3), the MA domain contains both an NLS and an NES. An HIV-1 Gag mutant known as M4 that contains two point mutations in MA (K18A, R22G) was reported to be severely replication deficient, apparently due to loss of genomic RNA packaging. According to the data in that study, wild-type HIV-1 Gag was exclusively cytoplasmic, but HIV-1 M4 Gag accumulated in the nucleus. Further, a green fluorescent protein (GFP)-tagged HIV-1 MA was also expressed exclusively in the cytoplasm but accumulated in the nucleus upon LMB treatment (3).
For PFV, it was recently reported that cell division was required for nuclear Gag accumulation, that PFV Gag does not have a functional NLS, and that the nuclear trafficking of PFV Gag was not essential for infectivity (10). The report of MLV Gag nuclear accumulations did not determine the mechanism of transport (11).
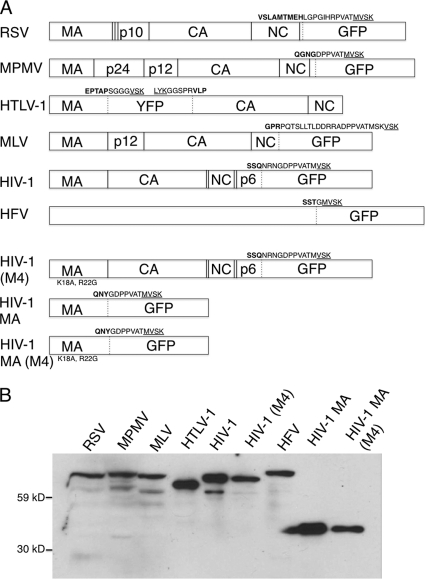
Because it appeared from these publications that nuclear trafficking of retroviral Gag proteins might be a common feature among retroviruses, we chose to assess the nuclear trafficking characteristics of fluorescently tagged Gag proteins from each of the major retroviral genera (Fig. 1A). These viruses included an alpharetrovirus (RSV) (18), a betaretrovirus (Mason-Pfizer monkey virus [MPMV]), a gammaretrovirus (MLV), a deltaretrovirus (human T cell leukemia virus type 1 [HTLV-1]), a lentivirus (HIV-1) (6), and a spumaretrovirus (human foamy virus [HFV] GagGFP). Plasmid constructs expressing each of these tagged Gag proteins were transfected into 293FT cells (Invitrogen) using Fugene6 and analyzed by Western blotting to confirm appropriate protein expression using an antibody against GFP (Fig. 1B). Next, the transfections were repeated in glass-bottom dishes (Mattek), and cells were imaged the next day by epifluorescence. None of the cells other than a portion of the HFV Gag-expressing cells displayed significant nuclear accumulation of Gag. To determine if any of the Gag proteins were being exported from the nucleus by the CRM1 pathway, we treated the cells with LMB (10 ng/ml) and imaged the cells 30 min later (Fig. 2). As expected (14), RSV Gag was found to be predominantly in the nucleus after treatment. None of the other retroviral Gag proteins demonstrated increased nuclear accumulation after LMB treatment, although MPMV Gag began to lose nuclear exclusion in some cells (Fig. 2). By 4 h posttreatment, the majority of MPMV Gag-expressing cells had lost nuclear exclusion, but even after 24 h there was no accumulation in the nucleus like was observed with RSV Gag. Each transfection was also repeated in the HeLa-derived cell line TZM-bl cells, with identical results (not shown). It should be noted that this assay would show only nuclear accumulation if Gag contained both a nuclear import sequence and a CRM1-dependent export sequence.
Fig 1.
Fluorescently tagged retroviral Gag constructs. (A) RSV construct (RSV.3 h.GFP) was obtained from John Wills, HTLV-1 construct (mCMVREM-MAYFP) from David Derse, HIV-1 construct from Marilyn Resh, and HFV (pcHFV/gfp) construct from Maxine Linial. MLV GagCFP was obtained from Alan Rein and subcloned into a GFP vector. All other clones were generated using standard cloning procedures. Amino acid sequences at junctions are shown. Bold letters represents Gag sequences, underlined letters represent GFP/YFP sequences. (B) Vectors were transfected into 293FT cells, and a Western blot analysis was performed on the cell lysates using an antibody against GFP.
Fig 2.
LMB sensitivity of retroviral Gag proteins. (A) 293FT cells were transfected with the Gag constructs illustrated in Fig. 1. Cells were imaged the next day. LMB-treated cells received 10 ng/ml LMB for 30 min prior to imaging. (B) Distribution of cellular phenotypes. The y axis is the percentage of cells in each treatment with the various distributions. Cells were scored as nuclear if there was a noticeable accumulation of protein in the nucleus. Cells were scored as cytoplasmic if the protein was enriched in the cytoplasm or plasma membrane. Cells were scored as nuclear and cytoplasmic if the protein was equally distributed between the nucleus and cytoplasm.
The absence of HIV Gag accumulation after LMB treatment was unexpected, given the apparently unambiguous data in the single relevant publication. To further probe the CRM1-dependent trafficking of HIV-1 Gag, we generated an HIV-1 MA-GFP expression construct, as well as HIV GagGFP and MA-GFP constructs with the M4 mutations in MA exactly as previously described (3). Surprisingly, we saw no nuclear accumulation of any of these proteins with or without LMB treatment (Fig. 2).
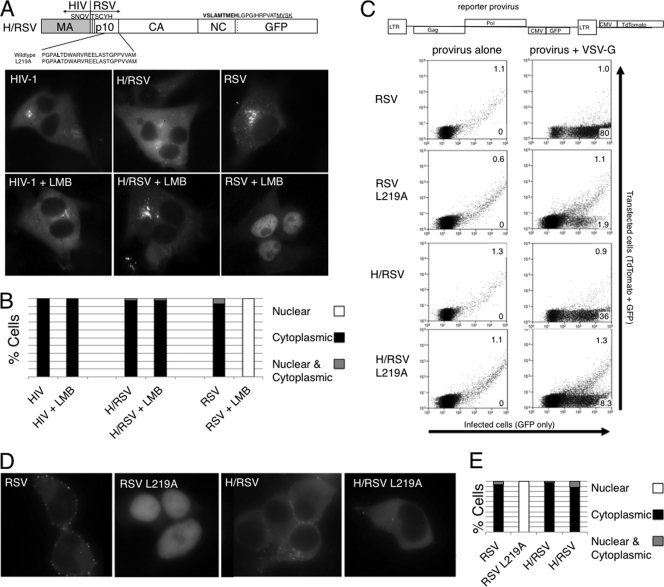
Because RSV MA contains an NLS, but in our experiments HIV MA does not, we tested whether an RSV Gag chimera that contained HIV MA in place of RSV MA (H/RSV) would continue to traffic through the nucleus. This construct was generated in the context of RSV GagGFP and tested as described above (Fig. 3A and B). At steady state, H/RSV GagGFP was found predominantly in the cytoplasm, and LMB treatment did not lead to significant nuclear accumulation.
Fig 3.
An HIV/RSV Gag chimera does not require the CRM1 export sequence. (A) Amino acid sequence at the junction sequence is shown. LMB treatment was performed as described in the legend to Fig. 2. (B) Distribution of cells from panel A, as described in the legend to Fig. 2. (C) Illustration of the RSV reporter construct. DF1 cells were transfected with 1,600 ng of provirus along with 400 ng of filler DNA or 400 ng of VSV-G. Cells were collected 2 to 3 days posttransfection and analyzed on a MoFloXDP flow cytometer. Percentages of cells transfected (expressing TdTomato and GFP) are shown in the upper right corners, and percentages of cells infected (expressing GFP only) are shown in the lower right corners. (D) Cells were transfected and imaged 6 to 8 h later to avoid possible nuclear translocation during cell division. (E) Distribution of cells from panel D.
To test if this chimeric protein is capable of generating infectious particles, the chimeric gene was introduced into an RSV provirus. Because the major splice donor for RSV is located in MA, we were not able to generate a chimera capable of producing both Gag and Env. Therefore, an Env-defective provirus was used and a functional viral glycoprotein was provided in trans from a separate plasmid. To monitor infectivity, we used a two-color flow cytometry system described previously (7). For this, we engineered a plasmid that contained a GFP cassette under the control of a cytomegalovirus (CMV) promoter within the RSV provirus in place of Env and a red fluorescent protein cassette (TdTomato) also under CMV control in the plasmid backbone (Fig. 3C). Cells transfected with the provirus express both red and green. However, if infectious particles are generated, the newly infected cells express green only, because only the GFP expression cassette is incorporated into the viral genome. The chicken fibroblast cell line DF1 was transfected with this provirus alone or in combination with a vesicular stomatitis virus glycoprotein (VSV-G) expression vector, and the cells were collected for analysis 2 to 3 days later. Infectivity was measured as the ratio of infected (green only) cells to transfected (red and green) cells (Fig. 3C, Table 1).
Table 1.
Proviral infectivity
| Virus | No. of infected cells/transfected cell |
||
|---|---|---|---|
| Experiment 1a | Experiment 2 | Experiment 3 | |
| RSV | 77.4 | 6.62 | 16.65 |
| RSV L219A mutant | 1.8 | 0.22 | 0.60 |
| H/RSV | 40.0 | 2.96 | 5.52 |
| H/RSV L219A mutant | 6.5 | 0.66 | 0.61 |
The raw data from experiment 1 are shown in Fig. 3.
Because unlike RSV Gag the H/RSV Gag chimera does not cycle through the nucleus in a CRM1-dependent fashion, we did not expect it to be capable of generating infectious particles. To our surprise, the H/RSV chimera was still fully capable of generating infectious particles, with the infectivity reduced only a few fold compared with that of the wild type. This is the first known example of an RSV Gag protein that does not display significant CRM1-dependent nuclear trafficking but remains capable of producing infectious particles.
If nuclear trafficking of the chimeric RSV Gag is not required for infectivity, then the CRM1-dependent NES in p10 should be dispensable in this context. Unfortunately, the NES in RSV p10 overlaps an important structural domain that is required for proper assembly (8, 13, 15). Because both functions have been probed extensively, we were able to predict which amino acids are required for each function. The position most likely to abolish the NES function without completely destroying structural function is the leucine 21 amino acid residues before the p10/CA cleavage site. When this amino acid was mutated to alanine (L219A), the NES activity was abolished (15), but mutation of the same amino acid to methionine did not abolish viral assembly in an in vitro assembly assay (8). We therefore introduced this L219A mutation into the RSV and H/RSV GagGFP constructs and into the proviral construct (Fig. 3D and E). As expected, the wild-type RSV GagGFP was cytoplasmic at steady state, but the GagGFP L219A mutant was exclusively nuclear. In contrast, H/RSV GagGFP was predominantly cytoplasmic both with and without the L219A mutation. Although there was no nuclear accumulation of the H/RSV GagGFP L219A mutant, a fraction of the cells did not display complete nuclear exclusion. It is not clear whether this was a result of cell division or very weak nuclear import.
Next, we tested the NES-deficient mutants for infectivity (Fig. 3C, Table 1). As expected, the L219A mutation dramatically reduced wild-type RSV infectivity (an average 33-fold drop; Table 1). The effect on the H/RSV chimera was less dramatic, reducing infectivity by 6.5-fold on average. It is not possible to know with certainty if this drop was due to an effect on the NES or to changes in the overlapping structural domain. This experiment was repeated three times, and the H/RSV chimera on average was 2.5-fold less infectious than wild-type RSV when the NES was intact but 2.5-fold more infectious than wild-type RSV when the NES was inactivated (Table 1). Because the H/RSV chimera and the L219A mutation undoubtedly cause additional changes to the viral life cycle, besides affecting the NLS and NES functions, we cannot conclude that the nuclear trafficking of Gag does not contribute to genome packaging efficiency. However, because the nontrafficking Gag proteins generate infectious particles, we can conclude that bulk CRM1-dependent nuclear trafficking of Gag is not absolutely required for genome packaging and infectivity.
Since the imaging assay used to analyze Gag trafficking is qualitative, it is not possible to exclude that a small percentage of Gag molecules traffic through the nucleus. Nor can it be excluded that there is trafficking that does not utilize the CRM1 export pathway. However, our data provide no evidence that nuclear trafficking is a critical step in the production of infectious viral particles for any of the retroviruses tested.
ACKNOWLEDGMENTS
We thank John Wills for the RSV GagGFP plasmid, Gisela Heideker and David Derse for the HTLV plasmid, Alan Rein for the MLV plasmid, Marilyn Resh for the HIV-1 plasmid, and Maxine Linial for the HFV plasmid.
This work was supported by U.S. Public Health Service grant AI73098.
Footnotes
Published ahead of print 8 February 2012
REFERENCES
- 1. Butterfield-Gerson KL, Scheifele LZ, Ryan EP, Hopper AK, Parent LJ. 2006. Importin-beta family members mediate alpharetrovirus Gag nuclear entry via interactions with matrix and nucleocapsid. J. Virol. 80:1798–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Callahan EM, Wills JW. 2003. Link between genome packaging and rate of budding for Rous sarcoma virus. J. Virol. 77:9388–9398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dupont S, et al. 1999. A novel nuclear export activity in HIV-1 matrix protein required for viral replication. Nature 402:681–685 [DOI] [PubMed] [Google Scholar]
- 4. Garbitt-Hirst R, Kenney SP, Parent LJ. 2009. Genetic evidence for a connection between Rous sarcoma virus Gag nuclear trafficking and genomic RNA packaging. J. Virol. 83:6790–6797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gudleski N, Flanagan JM, Ryan EP, Bewley MC, Parent LJ. 2010. Directionality of nucleocytoplasmic transport of the retroviral Gag protein depends on sequential binding of karyopherins and viral RNA. Proc. Natl. Acad. Sci. U. S. A. 107:9358–9363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hermida-Matsumoto L, Resh MD. 2000. Localization of human immunodeficiency virus type 1 Gag and Env at the plasma membrane by confocal imaging. J. Virol. 74:8670–8679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jorgenson RL, Vogt VM, Johnson MC. 2009. Foreign glycoproteins can be actively recruited to virus assembly sites during pseudotyping. J. Virol. 83:4060–4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Joshi SM, Vogt VM. 2000. Role of the Rous sarcoma virus p10 domain in shape determination of Gag virus-like particles assembled in vitro and within Escherichia coli. J. Virol. 74:10260–10268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. LeBlanc JJ, Uddowla S, Abraham B, Clatterbuck S, Beemon KL. 2007. Tap and Dbp5, but not Gag, are involved in DR-mediated nuclear export of unspliced Rous sarcoma virus RNA. Virology 363:376–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mullers E, Stirnnagel K, Kaulfuss S, Lindemann D. 2011. Prototype foamy virus Gag nuclear localization: a novel pathway among retroviruses. J. Virol. 85:9276–9285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nash MA, Meyer MK, Decker GL, Arlinghaus RB. 1993. A subset of Pr65gag is nucleus associated in murine leukemia virus-infected cells. J. Virol. 67:1350–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Parent LJ. 2011. New insights into the nuclear localization of retroviral Gag proteins. Nucleus 2:92–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Phillips JM, Murray PS, Murray D, Vogt VM. 2008. A molecular switch required for retrovirus assembly participates in the hexagonal immature lattice. EMBO J. 27:1411–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Scheifele LZ, Garbitt RA, Rhoads JD, Parent LJ. 2002. Nuclear entry and CRM1-dependent nuclear export of the Rous sarcoma virus Gag polyprotein. Proc. Natl. Acad. Sci. U. S. A. 99:3944–3949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Scheifele LZ, Kenney SP, Cairns TM, Craven RC, Parent LJ. 2007. Overlapping roles of the Rous sarcoma virus Gag p10 domain in nuclear export and virion core morphology. J. Virol. 81:10718–10728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Scheifele LZ, Ryan EP, Parent LJ. 2005. Detailed mapping of the nuclear export signal in the Rous sarcoma virus Gag protein. J. Virol. 79:8732–8741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schliephake AW, Rethwilm A. 1994. Nuclear localization of foamy virus Gag precursor protein. J. Virol. 68:4946–4954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Spidel JL, et al. 2004. Lysines close to the Rous sarcoma virus late domain critical for budding. J. Virol. 78:10606–10616 [DOI] [PMC free article] [PubMed] [Google Scholar]