Abstract
Human noroviruses (family Caliciviridae) are the leading cause of nonbacterial gastroenteritis worldwide. Although Human noroviruses are significant enteric pathogens, there exists no reliable vaccine or therapy to treat infected individuals. To date, attempts to cultivate Human noroviruses within the laboratory have met with little success; however, the related murine norovirus mouse norovirus 1 (MNV-1) has provided an ideal model system to study norovirus replication due to the ease with which the virus is cultivated and the ability to infect a small animal model with this virus. Previously we have identified the association between MNV-1 and components of the host secretory pathway and proposed a role for the viral open reading frame 1 proteins in the replication cycle. Here we describe for the first time a role for cytoskeletal components in early MNV-1 replication events. We show that the MNV-1 utilizes microtubules to position the replication complex adjacent to the microtubule organizing center. Chemical disruption of the microtubule network disperses the sites of MNV-1 replication throughout the cell and impairs production of viral protein and infectious virus. Furthermore, we demonstrate the ability of MNV-1 to redistribute acetylated tubulin to the replication complex and that this association is potentially mediated via the MNV-1 major structural protein, VP1. Transient expression of MNV-1 VP1 exhibited extensive colocalization with both α-tubulin and acetylated tubulin and was observed to alter the distribution of acetylated tubulin in transfected cells. This study highlights the role of the cytoskeleton in early virus replication events and demonstrates the importance of this interaction in establishing the intracellular location of MNV-1 replication complexes.
INTRODUCTION
Caliciviruses (family Caliciviridae) are small (27- to 35-nm), nonenveloped viruses which possess a single-stranded, positive-sense RNA genome of 7 to 8 kb. Of the five genera that have been described within this family (Vesivirus, Lagovirus, Sapovirus, Nebovirus, and Norovirus), only those within the Sapovirus and Norovirus genera have been identified as enteric pathogens of humans. Human noroviruses (HuNoVs) are the major cause of nonbacterial gastroenteritis outbreaks in adults worldwide and account for approximately 50 to 90% of such cases (5, 6, 31, 32). NoV infection is associated with mild to severe illness characterized by diarrhea and/or vomiting, which in extreme cases can lead to dehydration and death (37). Due to the highly contagious nature of NoVs, their low infectious dose, and their high stability within the environment, HuNoVs are significant enteric pathogens.
Due to the inability to cultivate HuNoV and the lack of a small animal model, relatively little is known about NoV biology. As such, very little is known regarding the replication and pathogenesis of NoV, with no reliable treatment or vaccine being available. In 2003, a novel mouse NoV, mouse NoV 1 (MNV-1), was initially isolated, characterized, and used to study the immunological aspects of NoV pathology (28). After this discovery, MNV-1 was discovered to possess a tropism for cells of a mononuclear origin, specifically macrophages and dendritic cells (53), thereby providing a viable tissue culture system for the study of NoV replication.
The NoV genome is protein linked at the 5′ end and polyadenylated at the 3′ end and is organized into three (possibly four) open reading frames (ORFs) (46). ORF1 encodes six nonstructural (NS) proteins which are translated as a single polypeptide (5′-NS1-2–NS3–NS4–NS5–NS6–NS7-3′) and subsequently cleaved by the virally encoded protease (NS6) to produce the mature viral NS proteins (46). The major and minor structural proteins, VP1 and VP2, are encoded by ORF2 and ORF3, respectively, and are translated from a subgenomic positive RNA species to form the virus capsid (4). The 5′ end of ORF2 is the most highly conserved region across all known NoV strains and has been shown to be the site of recombination in this virus (9, 10).
Little is known regarding NoV replication. Previously, we have identified the subcellular localization of the MNV-1 replication complex (RC) and shown that the virus replicates in association with membrane vesicles derived from components of the secretory pathway (21). More recently we have characterized the subcellular localizations of the individual ORF1 proteins, which exhibit distinct localization profiles (20). Currently we know little about the involvement of cytoskeletal components in calicivirus replication. In this study, we show that MNV-1 appears to utilize the host cytoskeleton to establish virus replication proximal to the microtubule organizing center (MTOC). Our results suggest that localization to this site is required for efficient replication. Additionally, we demonstrate that MNV-1 replication induces the redistribution of acetylated tubulin to the site of replication via a potential interaction with VP1.
MATERIALS AND METHODS
Viruses and cells.
RAW264.7 and Vero C1008 cells were grown and maintained in Dulbecco modified Eagle medium (DMEM) (Invitrogen, Australia) supplemented with 5% fetal calf serum (Lonza, Basel, Switzerland), 2 mM Glutamax (Gibco BRL), and penicillin (100 U/ml)-streptomycin (100 μg/ml) (Gibco BRL). Cells were infected with MNV-1 strain CW1 at a multiplicity of infection (MOI) of 5 (i.e., 5 PFU/cell) as previously described (28), and infected cells were maintained in DMEM containing 5% fetal calf serum, 2 mM Glutamax, and penicillin (100 U/ml)-streptomycin (100 μg/ml).
Reagents. (i) Antibodies.
MNV-1-specific guinea pig polyclonal antibodies have been described previously (46). MNV-1-specific rabbit polyclonal antibodies were generously provided by Herbert Virgin (Washington University School of Medicine, St. Louis, MO). Rabbit polyclonal sera raised against MNV-1 NS6 and NS7 were generated and purchased from Invitrogen. Anti-rabbit-, anti-guinea pig-, and anti-mouse-specific IgG–Alexa Fluor 488, 549, and 647 were purchased from Molecular Probes (Invitrogen, Leiden, The Netherlands). Anti-α-tubulin, anti-β-tubulin, anti-γ-tubulin, and antiactin were purchased from Molecular Probes, antivimentin and antipericentrin were purchased from Abcam (Cambridge, MA), mouse anti-acetylated-α-tubulin was purchased from Invitrogen, and anti-double-stranded RNA (anti-dsRNA) (clone J2) was purchased from English & Scientific Consulting Bt. (Hungary).
(ii) Chemicals.
Lipofectamine 2000 transfection reagent was purchased from Invitrogen. Nocodazole (Noz) was purchased from Merck-Calbiochem (Germany). Efficient concentrations of all chemicals were determined by titration to an endpoint where cell viability was maintained and the drug actions were still in effect, using the CytoTox 96 nonradioactive cytotoxicity assay (Promega).
Construction of plasmids.
pcDNA3.1 expression vectors containing the individual ORF1 proteins were constructed as previously described (20). The gene expressing VP1 was similarly cloned into pcDNA3.1(+) using primers purchased from Geneworks (Australia) (forward, 5′-GATATCACCACCATGAGGATGAGTGATGGC-3′; reverse, 5′-ACTAGTTCAATGATGATGATGATGATGTTGTTTGAGCATTCG-3′).
IF analysis.
Raw264.7 cell monolayers on coverslips were infected with MNV-1 at an MOI of 5 and incubated at 37°C for the time periods specified. Alternately, Vero C1008 cell monolayers were transfected with pcDNA3.1 expression plasmids encoding the MNV-1 proteins as specified by the manufacturer. The cells were subsequently washed with phosphate-buffered saline (PBS), fixed with 4% paraformaldehyde (ProSciTech, Kirwan, Australia) for 10 min at 20°C, and permeabilized with 0.1% Triton X-100 for immunofluorescence (IF) analysis as previously described (20, 21). Alternatively, cell monolayers were fixed with 4% paraformaldehyde for 10 min and permeabilized with 100% methanol for 5 to 10 min before being washed with PBS. Primary and secondary antibodies were incubated with blocking buffer (PBS containing 1% bovine serum albumin [BSA]) and washed with PBS containing 0.1% BSA–0.1% Tween 20 between incubation steps. After a final wash with PBS, coverslips were drained and mounted onto glass slides with a quick-dry mounting medium (United Biosciences, Brisbane, Australia) before visualization on either a Zeiss LSM 510 Meta confocal microscope or a Leica confocal microscope. Images were collected and collated for publication using Adobe Photoshop software.
Western blotting (WB).
MNV-1-infected or uninfected cells were lysed in 1× COP buffer (10 mM Tris-HCl [pH 8.2], 150 mM NaCl, 5 mM EDTA, 1% NP-40, 0.5 mM phenylmethylsulfonyl fluoride [PMSF], 5 μg/ml leupeptin, sodium fluoride, sodium orthovanadate), and proteins were separated on 4 to 12% precast Tris-Bis polyacrylamide gels (Invitrogen). Proteins were subsequently transferred to a Hybond-ECL nitrocellulose membrane (Amersham Biosciences Corp, Piscataway, NJ) using the Bio-Rad wet-transfer blotting module. The membrane was subsequently blocked with 5% skim milk powder (Diploma) or 5% BSA in PBS containing 0.1% Tween 20 at room temperature. Incubation with the appropriate antibodies was performed in blocking buffer on a rotating wheel overnight at 4°C. The bound antibodies were subsequently visualized with species-specific IgG conjugated to Alexa Fluor 488, 647, or 680 (Invitrogen) or IRDye 800CW (Rockland Inc.), using the Li-Cor Storm or Li-Cor Odyssey scanner. The resulting images were digitally scanned and processed in Adobe Photoshop for publication.
Plaque assay.
Plaque assays were performed in 6-well plates by seeding 1 × 106 RAW264.7 cells and the next day infecting cell monolayers with dilutions of virus-containing culture fluids for 60 min. Cells were overlaid with 2 ml/well of medium containing 70% DMEM, 2.5% fetal bovine serum (FBS), 15 mM sodium hydrogen carbonate, 5 U penicillin-streptomycin, 25 mM HEPES, 2 mM Glutamax, and 0.35% low-melting-point agarose and were incubated for 2 days at 37°C with 5% CO2. Cells were fixed by adding 1 ml/well of 4% formaldehyde directly onto the overlay and incubating for 30 min at room temperature. Cells were rinsed with water and stained with 0.2% crystal violet for 20 min, and plaques were counted.
qPCR.
RNA extraction and quantitative PCR (qPCR) conditions have been described previously (1). Briefly, 2.65 × 106 Raw264.7 cells were seeded into 6-well plates and the next day were infected with MNV-1 at an MOI of 5. Cell lysates were collected at 12 h postinfection (hpi), and RNA was extracted for qPCR analysis. cDNA was generated from RNA samples using SuperScript III reverse transcriptase (RT) (Invitrogen). Briefly, 1 to 2 μg of RNA, 2 pmol of each reverse primer (see below), and 10 nmol of deoxynucleoside triphosphates (dNTPs) were added to a PCR tube and made up to 13 μl with diethyl pyrocarbonate–Milli-Q H2O (DEPC-MQ H2O). Quantitation of samples by qPCR was performed as follows. To each well of a 96-well PCR rack was added 200 nM forward (F) and reverse (R) primers (MNV1.RT.F, TTGTTGGCATCAAGGACACCTG; MNV1.RT.R, TGGATGGAATGAAGGGCTCC; mGAPDH.RT.F, CGTCCCGTAGACAAAATGGT; mGAPDH.RT.R, TCAATGAAGGGGTCGTTGAT), 12.5 μl iQ SYBR green mix (Bio-Rad Laboratories), 5 μl RT, and 2.5 μl DEPC-MQ H2O. qPCR was performed using the iCycler iQ real-time PCR detection system (Bio-Rad Laboratories). Relative quantitative expression and fold induction were then calculated and plotted using GraphPad Prism 5.0 (GraphPad Software).
RESULTS
The MNV-1 RC localizes around the MTOC and induces alterations in microtubule morphology.
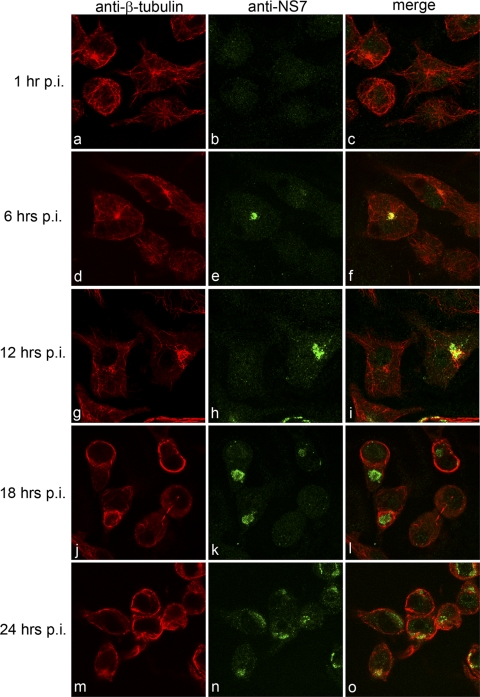
We have previously shown that MNV-1 replication is associated with components of the endocytic and secretory pathways (21). To extend our analyses further, we investigated whether cytoskeletal components also contribute to MNV-1 replication. To undertake this study, MNV-1-infected Raw264.7 cells were fixed for IF analysis at 1, 6, 12, 18, and 24 h postinfection (hpi) and dual labeled with antibodies against the MNV-1 RC (NS6-7 or NS7) and β-tubulin, vimentin, or α-actin and then analyzed by confocal microscopy (Fig. 1 to 3). An involvement of microtubules was investigated in cells labeled for β-tubulin, where the MNV-1 RC was consistently observed to localize proximate to the microtubule organizing center (MTOC) as early as 6 hpi (Fig. 1d to f). In addition, we observed significant alterations in microtubule morphology from 12 hpi, which were characterized by the formation of thick microtubule bundles. The observed changes in microtubule morphology became more apparent at later time points during infection (i.e., 18 and 24 hpi), where β-tubulin was observed to label prominently around the periphery of infected cells (Fig. 1j to o).
Fig 1.
The MNV-1 RC localizes around the MTOC and induces alterations in microtubule morphology. Raw264.7 cells were infected with MNV-1 at an MOI of 5, fixed at 1, 6, 12, 18, and 24 hpi, and labeled for the RC (NS7) (green) and β-tubulin (red). The MNV-1 RC was observed to localize to a perinuclear location proximal to the MTOC at 6 hpi and to induce alterations in the morphology of microtubules, namely, the formation of microtubule bundles, at 12, 18, and 24 hpi.
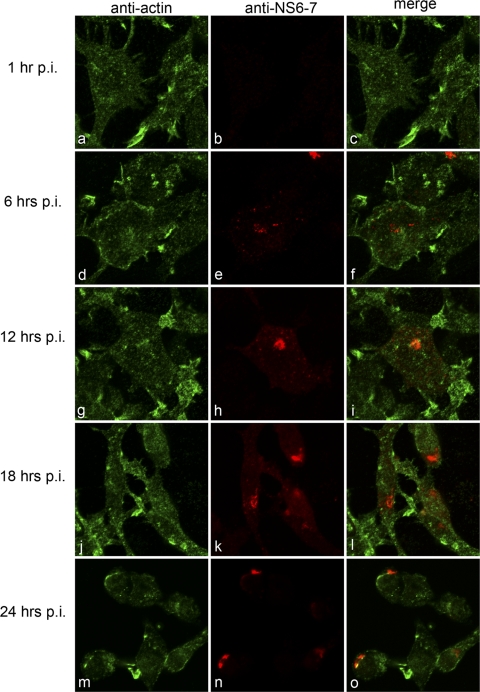
Fig 3.
MNV-1 does not associate with actin during replication. Raw264.7 cells were infected with MNV-1 at an MOI of 5, fixed at 1, 6, 12, 18, and 24 hpi, and labeled for the RC (NS6-7) (red) and α-actin (green). No colocalization was observed between MNV-1 and actin at any of the time points analyzed. Minor changes in actin morphology were observed at 18 and 24 hpi, which may be attributed to the induction of apoptosis by MNV-1.
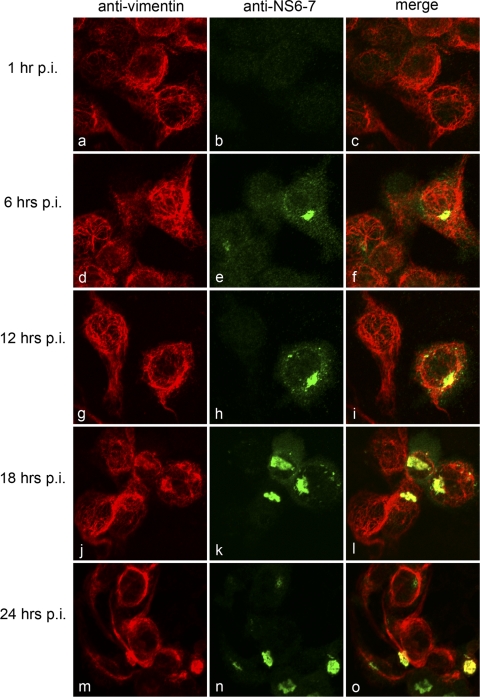
A potential role for intermediate filaments was investigated in cells costained for vimentin, where we observed some colocalization at 12, 18, and 24 hpi (Fig. 2g to o). The distribution of vimentin did appear to overlap with that of MNV-1 NS6-7 in the perinuclear region, close to the MTOC (Fig. 2). This may represent a localization to aggresomes, which have previously been proposed to support the replication of African swine fever virus (ASFV) (17). However, unlike ASFV and aggresome formation, the vimentin staining did not appear to surround the MNV-1 RC but rather colocalized with it. Additionally, the distribution of vimentin within MNV-1-infected cells remained relatively unchanged until very late in the replication cycle (i.e., 24 hpi) (Fig. 2m to o), when a cytopathic effect was evident. In contrast, we observed relatively little association between the MNV-1 RC and actin during the course of infection (Fig. 3). However, we observed some colocalization of NS6-7 with actin at later time points (particularly at 24 hpi [Fig. 3m to o), when there was a more punctuate distribution actin. This alteration may be a result of the induction of apoptosis, as this process is known to affect the morphology of cytoskeletal components and is upregulated during MNV-1 infection at later time points (7, 12, 14, 16, 33).
Fig 2.
MNV-1 induces alterations in the distribution and morphology of vimentin. Raw264.7 cells were infected with MNV-1 at an MOI of 5, fixed at 1, 6, 12, 18, and 24 hpi, and labeled for the RC (NS6-7) (green) and vimentin (red). MNV-1 was observed to colocalize with vimentin at 12 hpi and to induce alterations in the distribution and morphology of vimentin at 18 and 24 hpi.
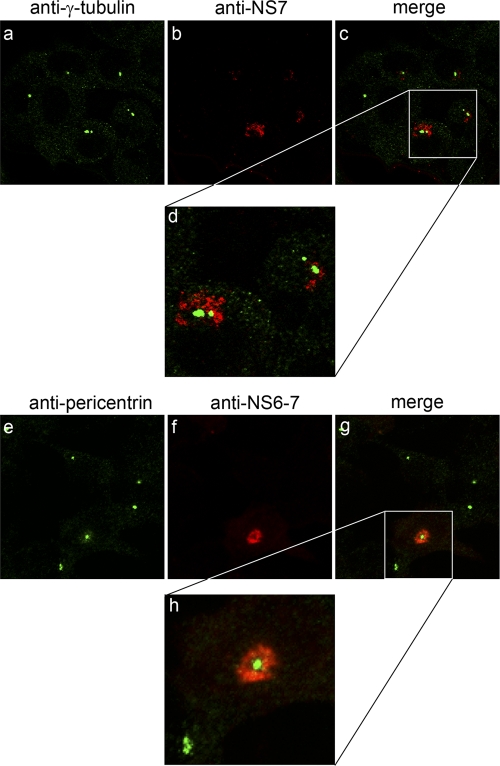
In order to further characterize the interaction between the MNV-1 RC and the MTOC, we investigated the localization of two MTOC components, γ-tubulin and pericentrin, during the course of infection. MNV-1-infected Raw264.7 cells were fixed at 1, 6, 12, 18, and 24 hpi and dual labeled with antibodies against either NS6-7 or NS7 and γ-tubulin or pericentrin, (Fig. 4 and data not shown). At 12 hpi the MNV-1 RC was clearly observed to localize around both γ-tubulin- and pericentrin-labeled structures (Fig. 4). Interestingly, while the localization of the MNV-1 RC proximal to the MTOC was consistently observed throughout the course of infection, we observed no direct colocalization between the MNV-1 RC and either γ-tubulin or pericentrin at time points when there was optimal virus replication, suggesting that the anchoring of the RC to the perinuclear region of the cell is not dependent on a direct association between the RC and the MTOC. Some colocalization was observed at the later time points; however, these cells were undergoing severe cytopathic effects, so the true impact of this association is difficult to resolve. Importantly, we observed no alteration in the localization of either γ-tubulin or pericentrin during the course of infection (Fig. 4 and data not shown), nor did we observe any ablation in the labeling of these components. This would suggest that the localization and function of these components are not overtly affected during MNV-1 replication.
Fig 4.
MNV-1 localizes proximal to the MTOC during replication. Raw264.7 cells were infected with MNV-1 at an MOI of 5, fixed at 12 hpi, and labeled for the RC (NS7/NS6-7) (red) and pericentrin or γ-tubulin (green). MNV-1 was observed to localize proximal to the MTOC but did not exhibit any colocalization with either pericentrin or γ-tubulin. The morphology and distribution of pericentrin and γ-tubulin did not alter during the course of infection.
Inhibition of microtubule dynamics perturbs steps in the MNV-1 replication cycle.
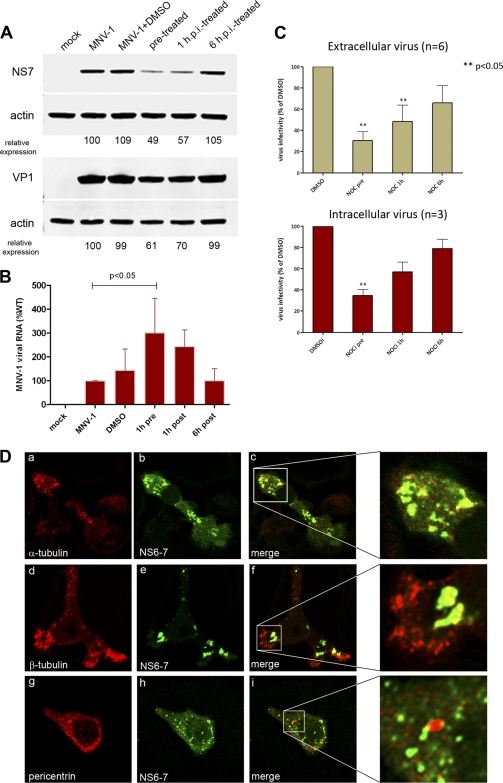
Based on the observation above that the MNV-1 RC localizes around the MTOC early in infection, we proposed that the virus utilizes microtubules to traffic from the periphery of the cell to the site of replication, which resides proximal to the MTOC. In order to determine at what stage during the MNV-1 replication cycle the microtubule network is important (i.e., entry, replication, or egress), we utilized the biochemical properties of nocodazole (Noz), a cytotoxic agent known to inhibit microtubule polymerization. An effective concentration of 10 μM Noz, which inhibited microtubule polymerization but maintained cell viability, was used to treat Raw264.7 cells either prior to infection, 1 hpi, or 6 hpi The pretreatment was to determine if there were major effects on the entry process, the 1-h treatment was used to determine effects after entry (a time period identified by Perry et al. [39]), and the 6 h treatment was used to determine effects after the initial rounds of MNV-1 replication. Immunoblots of MNV-1-infected cell lysates harvested at 12 hpi were probed with antibodies against the ORF1 protein NS7 and the ORF2 protein VP1 and analyzed for changes in the level of viral protein expression (Fig. 5A). In samples treated with Noz prior to and immediately after infection, we observed a significant decrease in the level of viral protein expression compared to those in untreated samples and samples treated with Noz at 6 hpi (Fig. 5A). We observed an equally significant reduction for the ORF1 protein NS3 (data not shown), suggesting that this was a global effect on viral protein translation. Interestingly, we when assessed the effect on viral RNA replication, we observed a large increase in the intracellular accumulation of viral RNA during the pre- and posttreatments (Fig. 5B). The most significant increase in viral RNA occurred at 1 h pretreatment, which coincided with an associated decrease in protein production. When we examined the effect of Noz on production of both intracellular and extracellular virus, we observed a significant decrease in the Noz-treated samples prior to or at 1 hpi (Fig. 5B). It appeared that Noz dually affected the production of intra- and extracellular virus, suggesting that it is not solely an issue of release of the virus particles from infected cells (Fig. 5C). Interestingly, again the most significant decrease in virus production was associated with the increase in viral RNA accumulation. In order to explore the role of microtubules further, we analyzed the effect of Noz treatment on the localization of the MNV-1 RC (Fig. 5D). Raw264.7 cells were treated with Noz prior to infection and then infected with MNV-1. Cells were then fixed at 12 hpi, colabeled for NS6-7 and either α-tubulin, β-tubulin, or pericentrin, and analyzed by confocal microscopy (Fig. 5D). In untreated cells infected with MNV-1, the RC was observed predominantly as a single large, dense body which localized to a perinuclear position, as shown in Fig. 4d to f. However, in cells treated with Noz, this perinuclear localization was disrupted and sites of replication were observed as multiple foci distributed randomly throughout the cytoplasm (Fig. 5D). This disruption is particularly evident when comparing the staining patterns of NS6-7 and pericentrin in untreated and Noz-treated cells (compare Fig. 4h with inset in Fig. 5D, panel i).
Fig 5.
Disruption of microtubules inhibits MNV-1 replication. Raw264.7 cells were infected with MNV-1 at an MOI of 5 and treated with nocodazole (Noz) at 1 h prior to infection, 1 hpi, and 6 hpi. (A) Lysates were collected at 12 hpi and analyzed by immunoblotting using antisera against NS7, VP1, and actin. Relative expression was quantitated using actin as the internal control and Quantity One software. (B) Production of viral RNA was assessed by real-time qPCR. The relative amounts of viral RNA are expressed as a percentage compared to the MNV-1 untreated sample. Error bars represent standard errors of the means (SEM), and a one-way analysis of variance (ANOVA) was performed to derive the statistical significance (n = 6). (C) Tissue culture fluid was collected at 12 hpi and analyzed for the production of infectious virus by plaque assay. For virus titer analyses, intracellular virus (n = 3) and extracellular virus (n = 6) were measured by plaque assay, and the titers are expressed as a percentage compared to the dimethyl sulfoxide (DMSO)-treated control. Error bars represent SEM, and a one-way ANOVA was performed to derive the statistical significance. (D) RAW264.7 cells were treated with Noz prior to infection were fixed at 12 hpi and analyzed by IF for the distribution of the MNV-1 RC (NS6-7) and pericentrin, α-tubulin, or β-tubulin. Noz treatment inhibited localization of the MNV-1 RC proximal to the MTOC and resulted in the distribution of the RC throughout the cytoplasm.
Thus, we suggest that disruption of microtubule dynamics alters the intracellular site of viral replication (Fig. 5D), leading to an accumulation of viral RNA (Fig. 5B) at a distinct site, and it is not efficiently utilized or accessible for translation (Fig. 5A) and is not efficiently packaged into newly formed virions (Fig. 5C). These data imply that the establishment and positioning of the MNV-1 RC proximal to the MTOC are microtubule dependent and are critical for efficient viral protein translation and virus assembly.
Acetylated tubulin is redistributed to the MNV-1 RC during replication.
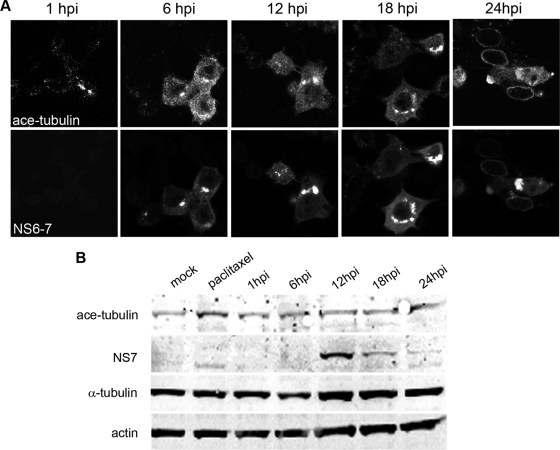
Cellular modification of microtubules by adaptor proteins such as microtubule-associated proteins (MAPs) and posttranslational processes allows for the alteration of the properties of microtubules, thereby changing their functional characteristics. Posttranslational acetylation of microtubules is associated with an increase the stability of microtubules and is characterized by the formation of microtubule bundles (49). As we observed the formation of similar structures in MNV-1-infected cells (Fig. 1), we sought to further analyze the effect of MNV-1 replication on the formation and localization of acetylated microtubules. Raw264.7 cells were infected at an MOI of 5, and cells and cell lysates were then collected for IF or WB analysis, respectively, at 1, 6, 12, 18, and 24 hpi For IF, cells were dual labeled with antibodies against NS6-7 and acetylated α-tubulin and analyzed by confocal microscopy (Fig. 6A). Cell lysates were also analyzed by Western blotting using antisera against acetylated α-tubulin, NS7, and actin (Fig. 6B). The distribution of acetylated tubulin labeling was observed to be altered in MNV-1-infected cells, and it was seen to be recruited to the RC (Fig. 6A). Interestingly, when quantified by WB, the level of acetylated tubulin was not observed to increase significantly at any time point during infection (Fig. 6B). This would suggest that while the level of acetylation of microtubules remains unaltered in infected cells, MNV-1 is able to redistribute stable microtubules to the site of replication.
Fig 6.
MNV-1 replication induces the redistribution of acetylated tubulin to the RC. (A) Raw264.7 cells were infected with MNV-1 at an MOI of 5, fixed at 1, 6, 12, 18, and 24 hpi, and labeled for the RC (NS6-7) and acetylated (ace) α-tubulin. Acetylated tubulin was observed to localize with the MNV-1 RC in infected cells. (B) Immunoblots of cell lysates collected at 1, 6, 12, 18, and 24 hpi were also analyzed for changes in the level of acetylated tubulin. The level of acetylated tubulin was observed to remain unchanged, as was the overall level of α-tubulin and actin. A paclitaxel-treated sample which showed an increase in tubulin acetylation was used as a control.
MNV-1 major structural protein VP1 associates with and alters the distribution of acetylated tubulin.
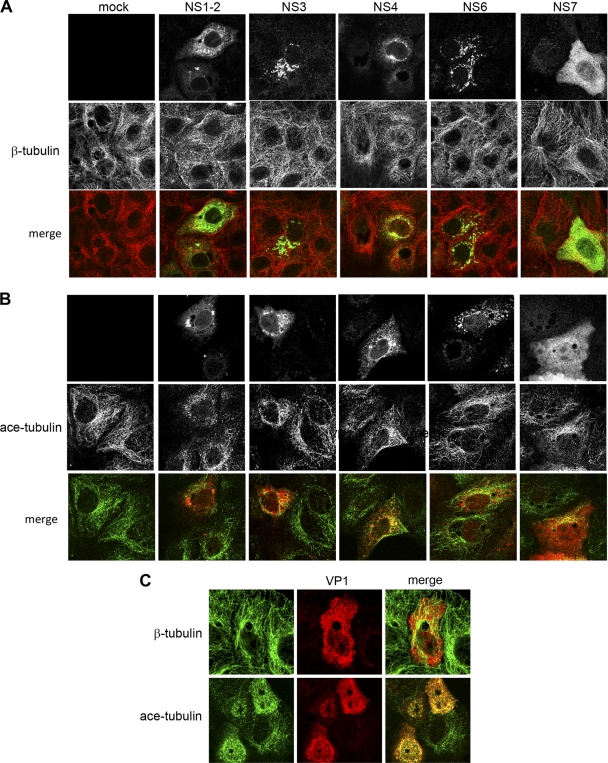
In order to determine how MNV-1 potentially interacts with microtubules, we analyzed the morphology and distribution of microtubules in cells transfected with individual MNV-1 proteins. Vero C1008 cells were transfected with MNV-1 ORF1 proteins as described previously (20) and here. At 24 h posttransfection (hpt), cells were fixed and dual labeled with antisera against β-tubulin or acetylated α-tubulin and antisera specific for the individual viral proteins and then analyzed by confocal microscopy (Fig. 7A). In cells transfected with MNV-1 ORF1 proteins, we observed some minor colocalization between the MNV-1 ORF1 proteins and β-tubulin; however, the vast majority of the labeling was not coincident (Fig. 7A). In addition, we did not detect any significant alterations in β-tubulin distribution or morphology in these cells. However, in ORF1-transfected cells labeled for acetylated tubulin, we observed some minor colocalization between acetylated tubulin and both NS3 and NS4, although various degrees of colocalization were observed with all ORF1 proteins, albeit most likely not significant (Fig. 7B). As other viruses which are known to interact with microtubules do so via structural proteins, we expanded our analysis to include the MNV-1 structural proteins VP1 and VP2. Unfortunately, we were unable to express VP2 under the conditions utilized in this study. When expressed in Vero cells, VP1 was observed to be distributed throughout the cytoplasm and within the nucleus and was present as very small punctuate structures (Fig. 7C). In cells colabeled with β-tubulin, we did not observe any significant colocalization of VP1 with microtubules (Fig. 7C). However, in cells labeled for acetylated tubulin we observed extensive colocalization with VP1. Additionally, we observed considerable redistribution of acetylated tubulin in VP1-expressing cells, including the absence of distinct filamentous acetylated microtubules which are typically seen in mock cells. This would suggest that VP1 may redistribute acetylated tubulin within transfected cells in a manner that prevents assembly of this component into distinct filamentous structures.
Fig 7.
MNV-1 VP1 localizes with and induces the redistribution of acetylated tubulin. (A and B) Vero C1008 cells were transfected with MNV-1 ORF1 proteins, fixed at 24 hpt, and analyzed by IF using antisera specific for each ORF1 protein and β-tubulin (A) (red) and acetylated tubulin (ace-tubulin) (B) (green). Only minor amounts of the ORF1 proteins analyzed were observed to colocalize with either β-tubulin or ace-tubulin. (C) VP1-transfected cells were also analyzed by IF using antisera specific for VP1 and β-tubulin or ace-tubulin (green). In contrast to the ORF1 proteins, VP1 exhibited extensive colocalization with acetylated tubulin and was observed to alter the distribution of acetylated tubulin in transfected cells.
A proportion of the MNV-1 major structural protein VP1 associates with the MNV-1 RC during infection.
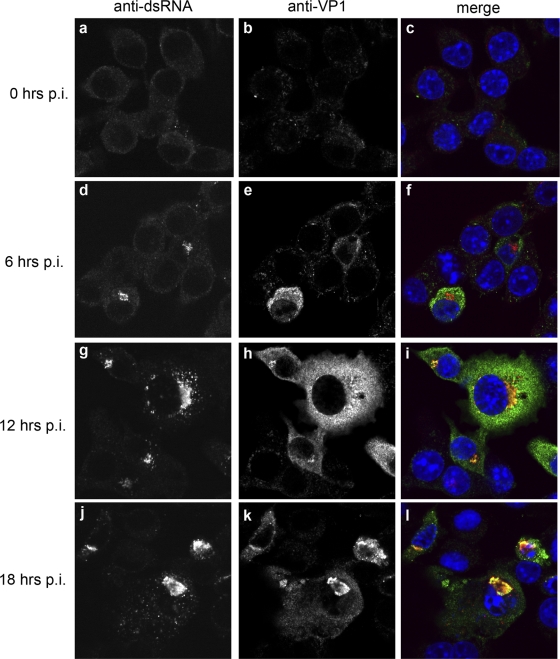
To extend these studies, we wished to determine whether the MNV-1 VP1 protein localized to the RC. RAW264.7 cells were infected as described above and IF analysis performed over the course of infection (Fig. 8). As can be observed, the VP1 proteins were associated with the MNV-1 RC throughout the duration of infection; however, only a small proportion of the VP1 protein colocalized with the RC. At the peak time of infection (i.e., 12 hpi), the VP1 distribution was observed to be diffuse throughout the cytoplasm (Fig. 8d to i), and it became more perinuclear, and colocalized with the RC, at the later time points (Fig. 8j–l).
Fig 8.
A proportion of MNV-1 VP1 protein associates with the RC during infection. A time course infection shows localization of the MNV-1 structural protein VP1 with the RC. Raw264.7 cells were infected with MNV-1 at an MOI of 5 and fixed at 0, 6, 12, and 18 hpi. Cells were labeled with antisera against the RC (dsRNA) and the MNV-1 capsid (VP1). A proportion of the MNV-1 VP1 was observed to localize within the RC at all time points analyzed, in addition to there being some diffuse labeling within the cytoplasm.
DISCUSSION
The cytoskeleton plays a vital role in a myriad of cellular functions, including cell architecture, movement, signaling, and intracellular trafficking (18, 29, 35). Accordingly, many viruses have evolved mechanisms which enable them to exploit these structures to aid in their replication (30, 41, 42). Here we show that MNV-1 appears to utilize microtubules during the early stages of replication to establish localization/positioning of the RC proximal to the MTOC. Previously we have characterized the subcellular localization of the MNV-1 RC and shown that replication is associated with significant changes in cellular organelle morphology and distribution (21). In addition, we have showed that the transient expression of the MNV-1 ORF1 proteins NS1-2 and NS4 displays localization patterns consistent with what we observed during virus infection (20). In this study, we have further defined the subcellular localization of the MNV-1 RC and shown it to localize to a site juxtaposed to the MTOC (Fig. 1 and 4). Localization of the RC to this site was dependent on apparent microtubule integrity, as nocodazole treatment led to dispersion of the RC within the cytoplasm (Fig. 5). Furthermore, treatment with nocodazole early during infection resulted in a significant inhibition of production of viral protein and infectious particles (Fig. 5), strongly suggesting that localization of MNV-1 to the MTOC is required for optimal virus replication. Interestingly, while disruption of microtubules inhibited localization of the MNV-1 RC to the MTOC, virus replication was still detected in the periphery of cells and presented as discrete foci (Fig. 5D), and in fact it appeared that viral genome replication was enhanced. Currently we can only speculate that this apparent increase in intracellular viral RNA may be attributable to the fact that this RNA does not appear to be accessible for translation or packaging into newly formed virions. Therefore, our conclusion is that the localization of the RC proximal to the MTOC is a crucial step in the virus life cycle to promote the production of infectious particles. Currently we are attempting to elucidate the mechanism and potential coupling between viral RNA replication, protein translation, and virus assembly.
While localization of the MNV-1 RC proximal to the MTOC was maintained throughout infection, IF analysis revealed no direct association between MNV-1 and host proteins residing within the MTOC, namely, γ-tubulin or pericentrin, nor did the morphology and distribution of these proteins appear to be affected (Fig. 4). The apparent absence of a direct interaction between MNV-1 and the MTOC suggests that the virus may utilize an alternative strategy for maintaining its positioning around the MTOC. Tethering of the RC to other cellular components that localize to this region (e.g., the Golgi apparatus and nuclear envelope) seems plausible, particularly in light of our recent observations that several MNV-1 ORF1 proteins associate with these organelles (namely, NS1-2 with the endoplasmic reticulum [ER] and NS4, with endosomes) (20). However, as many of these organelles are dynamic in nature, with their components continually being trafficked from their resident compartment and recycled back again, it is also possible that some other mechanism is required to keep the RC anchored to this region.
In addition to investigating the role of microtubules in MNV-1 replication, we analyzed the effect of replication on posttranslational acetylation of tubulin (Fig. 6 and 7). In MNV-1-infected cells we observed the redistribution of acetylated tubulin to the RC but did not detect any appreciable change in the level of acetylated tubulin (Fig. 6A and B). Further analysis of transfected cells revealed an association between acetylated tubulin and VP1. Redistribution of acetylated tubulin by VP1 (Fig. 7B), but not the ORF1 proteins (Fig. 6), suggests that interaction of the MNV-1 RC with microtubules is potentially mediated via VP1. We have observed that VP1 is consistently found within the MNV-1 RC during the course of infection as well as diffuse within the cytoplasm (Fig. 8).
Herpes simplex virus, African swine fever virus, and hepatitis E virus have been shown to modulate acetylation of microtubules (13, 25, 27, 55). However, in contrast to MNV-1, these viruses were observed to stimulate increased tubulin acetylation. Conversely, other viruses, such as poliovirus and foot-and-mouth disease virus, have been shown to disrupt microtubule formation via cleavage of cytoskeletal components which stabilize microtubules (3, 23, 24). Why the recruitment of acetylated tubulin to the RC would contribute to MNV-1 replication still remains unclear. One possible explanation is that sequestration of acetylated tubulin may serve to inhibit anterograde trafficking and signal transduction. The molecular motor kinesin-1 traffics intracellular cargo in an anterograde manner and has been shown to bind with greater affinity to acetylated microtubules (8, 43). Additionally, Giustiniani and colleagues have demonstrated that microtubule acetylation promotes the association of the chaperone Hsp90 with microtubules in addition to promoting the signaling of the proapoptotic proteins p53 and protein kinase B (PKB) (15). In addition to regulating signal transduction via interactions with PKB, acetylated microtubules also regulate the function of ATPases which are involved in signal transduction (reviewed in reference 2). In light of this, it seems feasible that sequestration of acetylated tubulin by MNV-1 would have some effect not only on intracellular trafficking of key molecules such as cytokines but also on key signaling pathways that may interact with the microtubule network.
In addition to illustrating a role for microtubules in MNV-1 replication, our results also indicate a role for actin in early stages of replication, though the exact role of the actin cytoskeleton in the MNV-1 replication cycle still remains unclear. While our initial observation of infected cells revealed an association with and morphological changes in microtubules, only minor alterations in actin morphology were observed until much later during the replication cycle (Fig. 3). Interestingly, we observed that cytochalasin D treatment did not appear to significantly inhibit the production of infectious virus particles, yet drug treatment prior to infection did appear to inhibit localization of the RC to the MTOC (data not shown). Two previous reports have suggested roles for actin in the replication cycle of Caliciviruses; however, the observations are conflicting. Perry et al. (38) observed that treatment with cytochalasin D enhanced MNV-1 infection, whereas Stuart and Brown (48) observed that feline calicivirus infection was impaired. In each case, though, only virus-positive cells were enumerated and effects on virus replication were not quantitated.
One possible explanation for these seemingly disparate observations may be found in the close interrelation between the microtubule and actin networks. While the roles of microtubules and actin in intracellular trafficking have typically been treated as separate, it has become increasingly apparent that there is significant overlap in the function and regulation of these two networks. Correspondingly, several proteins (WAFL and mDia), including molecular motors (MyoVA), have been demonstrated to associate with both the microtubule and actin networks to coordinate intracellular trafficking and other functions, such as cell movement (11, 19, 22, 40, 44, 45, 50). Additionally, a number of viruses, including Semliki forest virus, vaccinia virus, and respiratory syncytial virus, have been shown to utilize both microtubules and actin in a coordinated manner to facilitate replication (26, 34, 47). In light of this, we propose that while actin does not appear to be necessary for MNV-1 entry, it is likely required for transition of endocytosed virus onto microtubule tracks postentry. However, it also cannot be excluded that delivery of a host factor to the MTOC, facilitating viral transcription and/or translation, is equally required by MNV-1 for efficient replication and tethering of the MNV-1 RC within this area.
An alternate explanation for the localization of MNV-1 around the MTOC may be that MNV-1 replicates in association with aggresomes or aggresome-like structures (17, 36, 52). Aggresomes form in response to an accumulation of misfolded or aberrant proteins and localize around or adjacent to the MTOC (54). Similarities between aggresomes and the RCs of viruses which also localize proximal to the MTOC have prompted the speculation that these viruses may associate with these structures to facilitate their replication (17, 36, 51, 52). While the MNV-1 RC may share similarities with aggresomes, the lack of certain structural components characteristic of aggresomes, such as the formation of a vimentin cage, suggests that this may not be the case.
In the present study, we have further characterized the subcellular localization of the MNV-1 RC and shown that it localizes proximal to the MTOC. Additionally, we have demonstrated for the first time a role for microtubules in the replication of a calicivirus. While it is clear that localization proximal to the MTOC is crucial for efficient MNV-1 replication, we are still attempting to understand how this contributes to virus replication and how this affects cellular responses. Further understanding of this association between MNV-1 and the cytoskeleton will provide greater insights into the replication and pathogenesis of the Caliciviridae.
ACKNOWLEDGMENTS
This research was supported by research grants to J.M.M. from the National Health and Medical Research Council of Australia (grant 1010327) and from the U.S. National Institutes of Health (grants AI054483 and AI065982).
We thank Kim Green, Slava Sosnovtsev, and Herbert Virgin for generously providing antibodies. We also thank Rebecca Ambrose for help and advice with the qPCR.
Footnotes
Published ahead of print 1 February 2012
REFERENCES
- 1. Ambrose RL, Mackenzie JM. 2011. West Nile virus differentially modulates the unfolded protein response to facilitate replication and immune evasion. J. Virol. 85:2723–2732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arce CA, Casale CH, Barra HS. 2008. Submembraneous microtubule cytoskeleton: regulation of ATPases by interaction with acetylated tubulin. FEBS J. 275:4664–4674 [DOI] [PubMed] [Google Scholar]
- 3. Armer H, et al. 2008. Foot-and-mouth disease virus, but not bovine enterovirus, targets the host cell cytoskeleton via the nonstructural protein 3C(pro). J. Virol. 82:10556–10566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Asanaka M, et al. 2005. Replication and packaging of Norwalk virus RNA in cultured mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 102:10327–10332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Best SM, et al. 2005. Inhibition of interferon-stimulated JAK-STAT signaling by a tick-borne flavivirus and identification of NS5 as an interferon antagonist. J. Virol. 79:12828–12839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Blanton LH, et al. 2006. Molecular and epidemiologic trends of caliciviruses associated with outbreaks of acute gastroenteritis in the United States, 2000–2004. J. Infect. Dis. 193:413–421 [DOI] [PubMed] [Google Scholar]
- 7. Bok K, Prikhodko VG, Green KY, Sosnovtsev SV. 2009. Apoptosis in murine norovirus-infected RAW264.7 cells is associated with downregulation of survivin. J. Virol. 83:3647–3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bulinski JC. 2007. Microtubule modification: acetylation speeds anterograde traffic flow. Curr. Biol. 17:R18–R20 [DOI] [PubMed] [Google Scholar]
- 9. Bull RA, et al. 2005. Norovirus recombination in ORF1/ORF2 overlap. Emerg. Infect. Dis. 11:1079–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bull RA, Tanaka MM, White PA. 2007. Norovirus recombination. J. Gen. Virol. 88:3347–3359 [DOI] [PubMed] [Google Scholar]
- 11. Caufour PS, et al. 2001. Construction, characterization and immunogenicity of recombinant yellow fever 17D-dengue type 2 viruses. Virus Res. 79:1–14 [DOI] [PubMed] [Google Scholar]
- 12. Diallo M, Thonnon J, Traore-Lamizana M, Fontenille D. 1999. Vectors of Chikungunya virus in Senegal: current data and transmission cycles. Am. J. Trop. Med. Hyg. 60:281–286 [DOI] [PubMed] [Google Scholar]
- 13. Elliott G, O'Hare P. 1998. Herpes simplex virus type 1 tegument protein VP22 induces the stabilization and hyperacetylation of microtubules. J. Virol. 72:6448–6455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Furmanski M. 1999. Unlicensed vaccines and bioweapon defense in World War II. JAMA 282:822. [PubMed] [Google Scholar]
- 15. Giustiniani J, et al. 2009. Tubulin acetylation favors Hsp90 recruitment to microtubules and stimulates the signaling function of the Hsp90 clients Akt/PKB and p53. Cell. Signal. 21:529–539 [DOI] [PubMed] [Google Scholar]
- 16. Grzanka A, Grzanka D, Orlikowska M. 2003. Cytoskeletal reorganization during process of apoptosis induced by cytostatic drugs in K-562 and HL-60 leukemia cell lines. Biochem. Pharmacol. 66:1611–1617 [DOI] [PubMed] [Google Scholar]
- 17. Heath CM, Windsor M, Wileman T. 2001. Aggresomes resemble sites specialized for virus assembly. J. Cell Biol. 153:449–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Herrmann H, Strelkov SV, Burkhard P, Aebi U. 2009. Intermediate filaments: primary determinants of cell architecture and plasticity. J. Clin. Invest. 119:1772–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huang JD, et al. 1999. Direct interaction of microtubule- and actin-based transport motors. Nature 397:267–270 [DOI] [PubMed] [Google Scholar]
- 20. Hyde JL, Mackenzie JM. 2010. Subcellular localization of the MNV-1 ORF1 proteins and their potential roles in the formation of the MNV-1 replication complex. Virology 406:138–148 [DOI] [PubMed] [Google Scholar]
- 21. Hyde JL, et al. 2009. Mouse norovirus replication is associated with virus-induced vesicle clusters originating from membranes derived from the secretory pathway. J. Virol. 83:9709–9719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ishizaki T, et al. 2001. Coordination of microtubules and the actin cytoskeleton by the Rho effector mDia1. Nat. Cell Biol. 3:8–14 [DOI] [PubMed] [Google Scholar]
- 23. Joachims M, Etchison D. 1992. Poliovirus infection results in structural alteration of a microtubule-associated protein. J. Virol. 66:5797–5804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Joachims M, Harris KS, Etchison D. 1995. Poliovirus protease 3C mediates cleavage of microtubule-associated protein-4. Virology 211:451–461 [DOI] [PubMed] [Google Scholar]
- 25. Jouvenet N, Monaghan P, Way M, Wileman T. 2004. Transport of African swine fever virus from assembly sites to the plasma membrane is dependent on microtubules and conventional kinesin. J. Virol. 78:7990–8001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kallewaard NL, Boxena AL, Crowe JE. 2005. Cooperativity of actin and microtubule elements during replication of respiratory syncytial virus. Virology 331:73–81 [DOI] [PubMed] [Google Scholar]
- 27. Kannan H, Fan S, Patel D, Bossis I, Zhang YJ. 2009. The hepatitis E virus open reading frame 3 product interacts with microtubules and interferes with their dynamics. J. Virol. 83:6375–6382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Karst SM, Wobus CE, Lay M, Davidson J, Virgin HWIV. 2003. STAT1-dependent innate immunity to a Norwalk-like virus. Science 299:1575–1578 [DOI] [PubMed] [Google Scholar]
- 29. Lee TYJ, Gotlieb AI. 2003. Microfilaments and microtubules maintain endothelial integrity. Microsc. Res. Tech. 60:115–125 [DOI] [PubMed] [Google Scholar]
- 30. Leopold PL, Pfister KK. 2006. Viral strategies for intracellular trafficking: motors and microtubules. Traffic 7:516–523 [DOI] [PubMed] [Google Scholar]
- 31. Lopman BA, et al. 2003. Viral gastroenteritis outbreaks in Europe, 1995-2000. Emerg. Infect. Dis. 9:90–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Maguire AJ, Green J, Brown DWG, Desselberger U, Gray JJ. 1999. Molecular epidemiology of outbreaks of gastroenteritis associated with small round-structured viruses in East Anglia, United Kingdom, during the 1996-1997 season. J. Clin. Microbiol. 37:81–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mashima T, Naito M, Tsuruo T. 1999. Caspase-mediated cleavage of cytoskeletal actin plays a positive role in the process of morphological apoptosis. Oncogene 18:2423–2430 [DOI] [PubMed] [Google Scholar]
- 34. Newsome TP, Scaplehorn N, Way M. 2004. Src mediates a switch from microtubule- to actin-based motility of vaccinia virus. Science 306:124–129 [DOI] [PubMed] [Google Scholar]
- 35. Nogales E. 2001. Structural insights into microtubule function. Annu. Rev. Biophys. Biomol. Struct. 30:397–420 [DOI] [PubMed] [Google Scholar]
- 36. Nozawa N, Yamauchi Y, Ohtsuka K, Kawaguchi Y, Nishiyama Y. 2004. Formation of aggresome-like structures in herpes simplex virus type 2-infected cells and a potential role in virus assembly. Exp. Cell Res. 299:486–497 [DOI] [PubMed] [Google Scholar]
- 37. Patel MM, Hall AJ, Vinje J, Parashar UD. 2009. Noroviruses: a comprehensive review. J. Clin. Virol. 44:1–8 [DOI] [PubMed] [Google Scholar]
- 38. Perry JW, Taube S, Wobus CE. 2009. Murine norovirus-1 entry into permissive macrophages and dendritic cells is pH-independent. Virus Res. 143:125–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Perry JW, Wobus CE. 2010. Endocytosis of murine norovirus 1 into murine macrophages is dependent on dynamin II and cholesterol. J. Virol. 84:6163–6176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Petrasek J, Schwarzerova K. 2009. Actin and microtubule cytoskeleton interactions. Curr. Opin. Plant Biol. 12:728–734 [DOI] [PubMed] [Google Scholar]
- 41. Ploubidou A, Way M. 2001. Viral transport and the cytoskeleton. Curr. Opin. Cell Biol. 13:97–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Radtke K, Dohner K, Sodeik B. 2006. Viral interactions with the cytoskeleton: a hitchhiker's guide to the cell. Cell. Microbiol. 8:387–400 [DOI] [PubMed] [Google Scholar]
- 43. Reed NA, et al. 2006. Microtubule acetylation promotes kinesin-1 binding and transport. Curr. Biol. 16:2166–2172 [DOI] [PubMed] [Google Scholar]
- 44. Rodionov V, Yi J, Kashina A, Oladipo A, Gross SP. 2003. Switching between microtubule- and actin-based transport systems in melanophores is controlled by cAMP levels. Curr. Biol. 13:1837–1847 [DOI] [PubMed] [Google Scholar]
- 45. Rodriguez OC, et al. 2003. Conserved microtubule-actin interactions in cell movement and morphogenesis. Nat. Cell Biol. 5:599–609 [DOI] [PubMed] [Google Scholar]
- 46. Sosnovtsev SV, et al. 2006. Cleavage map and proteolytic processing of the murine norovirus nonstructural polyprotein in infected cells. J. Virol. 80:7816–7831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Spuul P., Balistreri G., Kaariainen L., Ahola T. 2010. Phosphatidylinositol 3-kinase-, actin-, and microtubule-dependent transport of Semliki Forest virus replication complexes from the plasma membrane to modified lysosomes. J. Virol. 84:7543–7557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Stuart AD, Brown TDK. 2006. Entry of feline calicivirus is dependent on clathrin-mediated endocytosis and acidification in endosomes. J. Virol. 80:7500–7509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Takemura R, et al. 1992. Increased microtubule stability and alpha-tubulin acetylation in cells transfected with microtubule-associated proteins MAP1B, MAP2 or TAU. J. Cell Sci. 103:953–964 [DOI] [PubMed] [Google Scholar]
- 50. Viklund IM, et al. 2009. WAFL, a new protein involved in regulation of early endocytic transport at the intersection of actin and microtubule dynamics. Exp. Cell Res. 315:1040–1052 [DOI] [PubMed] [Google Scholar]
- 51. Wileman T. 2006. Aggresomes and autophagy generate sites for virus replication. Science 312:875–878 [DOI] [PubMed] [Google Scholar]
- 52. Wileman T. 2007. Aggresomes and pericentriolar sites of virus assembly: cellular defense or viral design? Annu. Rev. Microbiol. 61:149–167 [DOI] [PubMed] [Google Scholar]
- 53. Wobus CE, et al. 2004. Replication of norovirus in cell culture reveals a tropism for dendritic cells and macrophages. PLoS Biol. 2:e432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wojcik C, DeMartino GN. 2003. Intracellular localization of proteasomes. Int. J. Biochem. Cell Biol. 35:579–589 [DOI] [PubMed] [Google Scholar]
- 55. Xuan CH, et al. 2007. Regulation of microtubule assembly and stability by the transactivator of transcription protein of Jembrana disease virus. J. Biol. Chem. 282:28800–28806 [DOI] [PubMed] [Google Scholar]