Abstract
In the context of infections with highly pathogenic influenza A viruses, the PB1-F2 protein contributes to virulence and enhances lung inflammation. In contrast, its role in the pathogenesis of seasonal influenza viral strains is less clear, especially in the H1N1 subtype, where strains can have a full-length 87- to 90-amino-acid protein, a truncated 57-amino-acid version, or lack the protein altogether. Toward this, we introduced the full-length 1918 PB1-F2, or prevented PB1-F2 expression, in H1N1 A/USSR/90/77, a seasonal strain that naturally expresses a truncated PB1-F2. All viruses replicated with similar efficiency in ferret or macaque ex vivo lung cultures and elicited similar cytokine mRNA profiles. In contrast, the virus expressing the 1918 PB1-F2 protein caused a delay of proinflammatory responses in ferret blood-derived macrophages, while the PB1-F2 knockout virus resulted in a more rapid response. A similar but less pronounced delay in innate immune activation was also observed in the nasal wash cells of ferrets infected with the 1918 PB1-F2-expressing virus. However, the three viruses did not differ in their virulence or clinical course in ferrets, supporting speculations that PB1-F2 is of limited importance for the pathogenesis of primary viral infection with human seasonal H1N1 viruses.
INTRODUCTION
Most influenza A viruses cause a mild infection of the upper respiratory tract that is usually cleared within 1 to 2 weeks (6); however, certain strains result in more severe disease (12, 29). There is thus considerable interest in identifying and characterizing potential virulence factors. PB1-F2, a small 87- to 90-amino-acid protein encoded by a +1 open reading frame in the PB1 segment, is one of the proposed candidates (2). While the PB1 gene in most avian viruses encodes a full-length PB1-F2, mammalian viruses frequently carry truncated forms due to accumulation of premature stop codons (14). Within the H1N1 subtype, human viruses isolated between 1918 and 1947 express a full-length PB1-F2 protein. In 1950, a stop codon was introduced after amino acid residue 57, resulting in a C-terminally truncated protein (35), and the recently emerged pandemic 2009 H1N1 viruses have a stop codon at residue 11 (9).
The role of PB1-F2 in influenza virus pathogenesis remains controversial. The full-length H1N1 PB1-F2 protein, which carries a mitochondrial targeting sequence in its carboxy-terminal portion, enhances apoptosis and contributes to virulence in mice (2, 34). In contrast, the PB1-F2 protein found in H5N1 viruses does not localize to the mitochondria and has no apoptosis-enhancing effect (1). PB1-F2 also interacts with PB1 and regulates viral polymerase activity in a cell-type- and strain-specific manner (13, 17, 20), and a role of PB1-F2 in the interaction with the cellular antiviral response has been reported. However, while one study observed a PB1-F2-mediated exacerbation of beta interferon (IFN-β) expression in human lung epithelial cells, a different study demonstrated an inhibition of type I IFN signaling that was associated with interaction of PB1-F2 with the mitochondrial antiviral signaling protein (MAVS) (15, 30). Finally, intranasal administration of synthetic full-length or carboxy-terminal portions of A/Brevig Mission/1/1918 (1918) and A/Puerto Rico/8/34 (PR8) PB1-F2 peptides led to recruitment of white blood cells into the lungs and exacerbated secondary bacterial infection in mice (18, 20).
To date, the contribution of PB1-F2 to virulence has been mostly investigated in mice by using mouse-adapted viruses, revealing that the absence of PB1-F2 results in a reduction of virulence and lung inflammation, even though PB1-F2-deleted viruses reach wild-type titers in the lungs (15, 20, 34). Introduction of the H1N1 1918 or the H5N1 A/Hong Kong/156/97 (HK/97) PB1-F2 into mouse-adapted strains revealed that a single amino acid change at position 66 (N66S) was responsible for the observed increased virulence in mice (3, 4, 19). Presence of the HK/97 PB1-F2 also resulted in a delayed onset of type I IFN and proinflammatory cytokine production, leading to more efficient replication and more severe lung pathology (3). In contrast, introduction of a functional PB1-F2 in the 2009 pandemic H1N1 A/California/04/09 (Cal/09) strain had little impact on virulence in mice and ferrets despite an increased induction of proinflammatory cytokines in mice (9).
To characterize the contribution of PB1-F2 to the virulence of a seasonal H1N1 virus in a naturally susceptible host, we generated recombinant viruses in the A/USSR/90/77 (USSR) background that expressed the 1918 PB1-F2 or that no longer produced the protein. After demonstrating that all viruses replicated with similar efficiency, we characterized the induction of proinflammatory cytokines and apoptosis in ferret and macaque ex vivo lung cultures and ferret blood-derived macrophages. The pathogenesis, cytokine responses, histopathological changes, and extent of apoptosis were then compared in ferrets.
MATERIALS AND METHODS
Cells and viruses.
MDCK cells (ATCC CCL-34) and 293 cells (ATCC CRL-1573) were maintained in Dulbeccos's modified Eagle's medium (DMEM; Invitrogen) supplemented with 5% fetal bovine serum (Invitrogen). All influenza viruses were propagated in MDCK cells cultivated in DMEM with 2 μg/ml of tosyl-phenylalanine chloromethyl ketone–trypsin (DMEM–TPCK; Sigma). Virus titers were quantified by using the limiting dilution method and are expressed as the 50% tissue culture infectious dose per ml (TCID50/ml).
Construction and recovery of recombinant viruses.
To generate a USSR/90/77 (USSR) PB1 gene expressing the 1918 PB1-F2 open reading frame, the necessary nucleotide changes were introduced into the bidirectional PB1 expression plasmid (21) by site-directed mutagenesis, taking care not to alter the PB1 open reading frame. The PB1-F2 knockout (KO) virus was generated by disrupting the start codon and introducing a stop codon after 11 residues, as previously described (20). None of the introduced changes alter the PB1 protein sequence or affect the N40 start codon (32).
Recombinant viruses were recovered as previously described (10). Briefly, semiconfluent 293 cells in a six-well plate were transfected with 0.5 μg of bidirectional expression plasmids containing each of the eight USSR segments by using Lipofectamine 2000 (Invitrogen). Two days after transfection, the cells were trypsinized and transferred onto MDCK cells. The cocultures were maintained in DMEM–TPCK-trypsin until cytopathic effect was observed. The supernatant was then transferred onto fresh MDCK cells to produce viral stocks.
For growth kinetics, 90% confluent MDCK cells were washed twice with phosphate-buffered saline (PBS) and infected at a multiplicity of infection (MOI) of 0.001 in DMEM-TPCK. After 30 min, the medium was changed, cells and supernatant were harvested every 12 h for 2 days, and the virus was titrated by using the limiting dilution method.
Ex vivo lung cultures.
Lung slice cultures were generated from three ferrets (Mustela putorius furo) and one cynomolgus macaque (Macaca fascicularis) based on a previously published protocol (26). Briefly, lungs were inflated by infusion with a 1:1 solution of DMEM–F-12 (Invitrogen) and 0.8% low-melting-point agarose (Fisher Scientific) at 50°C. After incubation on ice for 30 min, 3-mm-thick sections were cut. Lung slices were infected with 3 × 104 TCID50 of the recombinant viruses and placed in a 1:1 solution of DMEM–F-12 and DMEM with antibiotics. At 24 and 48 h postinfection, the supernatant and the lungs were harvested for titration, immunohistochemistry, and RNA extraction.
Milliplex analysis.
Cultured macaque lung slices were homogenized in 400 μl of radioimmunoprecipitation assay (RIPA) buffer (1% Triton X-100, 1 mM dithiothreitol, 150 mM NaCl, 20 mM Tris [pH 7.5], 10% glycerol) in a bead mill homogenizer (Tissue Lyser; Qiagen), and debris was pelleted by centrifugation at 14,000 × g for 20 min at 4°C. The protein concentration of the supernatant was determined by the Bradford assay (Thermo Scientific), and 12.5 μg of the cell lysate was used for each reaction mixture. Concentrations of active caspase-3, glyceraldehyde-3-phosphate dehydrogenase, and cleaved poly(ADP-ribose) polymerase (PARP) were determined using the 3-Plex apoptosis signaling kit (EMD Millipore), while the concentrations of IFN-γ, interleukin-1β (IL-1β), IL-6, IL-8, IL-10, IL-17, monocyte chemoattractant protein 1 (MCP-1), macrophage inflammatory protein 1α (MIP-1α), MIP-1β, granulocyte-macrophage colony-stimulating factor (GM-CSF), and tumor necrosis factor alpha (TNF-α) were quantified with the nonhuman cytokine Milliplex map kit (EMD Millipore) following the manufacturer's instructions. Plates were read with a Bio-Plex 200 system (Bio-Rad).
Differentiation of ferret blood-derived macrophages.
Peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll (GE Healthcare) gradient centrifugation at 400 × g for 40 min. Adherent monocytes were isolated by seeding PBMCs in 12-well plates overnight, followed by a careful medium change, and macrophage differentiation was induced with 5 ng/ml of human GM-CSF (R&D Systems). After 6 days, the cells were infected with 3 × 104 TCID50 of the recombinant viruses and harvested at 8, 16, and 24 h postinfection for RNA extraction and terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) staining.
Animal experiments.
All animal studies were approved by the Institutional Care and Use committee of the INRS-Institut Armand-Frappier. Only ferrets seronegative for circulating influenza virus strains were used. Groups of at least 6 animals were infected intranasally with 105 TCID50 of the respective recombinant virus. Body temperature was recorded every 30 min using intraperitoneal implants (DSI) starting 1 day before infection. Clinical signs were monitored daily for the first 4 days and every second day thereafter. The clinical score was established according to a scale of 0 to 3 scale, with 0 representing the physiological range as previously described (21). Nasal washes were collected daily for the first 4 days and at day 6, two animals were sacrificed on days 2 and 4 postinfection, and nasal turbinates and lungs were harvested. Data shown for rUSSR were obtained from 8 animals infected in the context of an earlier study (21).
Immunohistochemistry and titration.
One-third of the nasal turbinate and lung tissue samples were formalin fixed, paraffin embedded, and cut into 5-μm sections. The slides were deparaffinized and rehydrated following standard immunohistochemistry protocols and hematoxylin and eosin (H&E) stained. For the detection of infected cells by immunohistochemistry, the slides were microwaved for 15 min in 10 mM sodium citrate (pH 6) solution (Fisher Scientific) for antigen retrieval. After blocking with a 1:50 dilution of rabbit serum in PBS for 30 min, an influenza virus-specific antiserum (OBT1551; AbD Serotec) was added for 90 min at room temperature. The slides were washed three times with PBS, and a biotinylated secondary antibody was added for 30 min, followed by 30 min of incubation with Alexa Fluor 488-labeled streptavidin. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI), and coverslips were mounted with Mowiol (Calbiochem).
For titration, tissues were weighed and homogenized in DMEM with a bead mill homogenizer (Tissue Lyser; Qiagen). Cell debris was removed by centrifugation, and the supernatant was titrated by the limiting dilution method. Titers are expressed as the TCID50/g.
TUNEL assay.
The number of apoptotic cells was determined by TUNEL staining, using the in situ cell death detection kit TMR red (Roche) as previously described (24). Briefly, cells were fixed with 4% paraformaldehyde (Sigma) in PBS, permeabilized with 0.1% Triton X-100 in 0.1% sodium citrate, and incubated for 1 h at 37°C with the TUNEL reaction mixture. Slides were washed twice with PBS and counterstained with DAPI. For infected macrophages, DAPI- and TUNEL-positive cells were counted in each well, and the percentage of apoptotic cells was calculated. Results are expressed as the fold change compared to noninfected cells.
RNA extraction and cytokine mRNA quantification by real time reverse transcription-PCR (RT-PCR) analysis.
Immediately after collection, 140 μl of nasal wash was mixed with 560 μl of AVL buffer (QIAamp viral RNA minikit; Qiagen) and stored at −80°C. Lung ex vivo culture samples were placed in RNALater (Qiagen) and stored at −80°C. For RNA isolation, samples were homogenized with a bead mill homogenizer (Tissue Lyser; Qiagen) in 350 μl of RLT buffer (RNeasy RNA extraction kit) supplemented with 1% β-mercaptoethanol. The RNA from the nasal washes, macrophages, and tissues was extracted following the manufacturer instructions for the corresponding kit.
Ferret cytokine mRNA was quantified as previously described (21). Briefly, 10 ng of sample RNA was used for each reaction mixture. The genes encoding type I IFN, IFN-γ, TNF-α, IL-1β, -6, -8, and CCL5 (GenBank accession number AB513129.1) were cloned into pCR2.1-TOPO based on previously published sequences (22, 27, 28) and in vitro transcribed using T7 polymerase (New England BioLabs). Serial dilutions of the standards and samples were analyzed by real-time RT-PCR using the QuantiTect SYBR green RT-PCR kit (Qiagen). The gene copy numbers were calculated by extrapolating data from the standard curve.
Statistical analysis.
The statistical analyses were performed using GraphPad Prism 5.0a. P values were calculated by individually comparing the different groups using an unpaired Mann-Whitney test.
RESULTS
Characterization of recombinant viruses with altered PB1-F2 proteins.
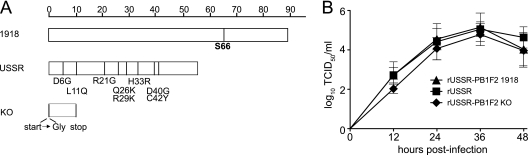
There is considerable variation in the length of PB1-F2 open reading frame within the H1N1 subtype, ranging from a full-length 87- to 90-amino-acid protein for viruses isolated before 1950, to a truncated protein of 57 amino acids in later strains, including USSR, to a complete deletion in the recently emerged 2009 pandemic H1N1 viruses (9). To evaluate the contribution of PB1-F2 to the virulence of H1N1 viruses, we introduced the 1918 PB1-F2 open reading frame in the seasonal USSR strain by site-directed mutagenesis, carefully avoiding changes to the USSR PB1 protein sequence. In addition, a virus lacking PB1-F2 was produced by mutation of the start codon and introduction of a stop codon after 11 amino acids (20). The 1918 and USSR PB1-F2 proteins shared 86% amino acid identity. However, the shorter USSR PB1-F2 lacks the C-terminal domain, which contains a mitochondrial targeting sequence, mediates PB1 binding, promotes inflammation, and interferes with type I IFN signaling (2, 17, 20, 30). In addition to a full-length C-terminal domain, the 1918 PB1-F2 protein has a serine at position 66 (Fig. 1A), which is associated with increased pathogenicity of H5N1 and H1N1 viruses in mice (4). None of the other diverging residues has been associated with virulence or specific functions (Fig. 1A). While direct detection of PB1-F2 was not possible due to the lack of USSR PB1-F2-specific antibodies, the sequence of the PB1 segments was confirmed for each virus. Growth kinetics of recombinant and parental viruses in MDCK cells were not significantly different, even though replication of the PB1-F2 KO virus was slightly delayed (Fig. 1B), confirming previous reports that PB1-F2 slightly enhances the viral polymerase activity (20).
Fig 1.
Generation of recombinant viruses. (A) Schematic representation of the different PB1-F2 proteins. Amino acid sequences were aligned using the Megalign 8.0.2 software. Each PB1-F2 protein is represented by a bar, and residues that differ between the 1918 and USSR PB1-F2 proteins are indicated below the USSR protein. The serine at position 66, which has been associated with increased virulence, is highlighted in bold. (B) Growth kinetics of the recombinant viruses. MDCK cells were infected at an MOI of 0.001, and the supernatant was harvested at the indicated time points. Each value represents the mean of three independent experiments performed in triplicate; error bars indicate the standard deviations.
Infection efficacy and cytokine profiles in ferret and macaque ex vivo lung cultures are PB1-F2 independent.
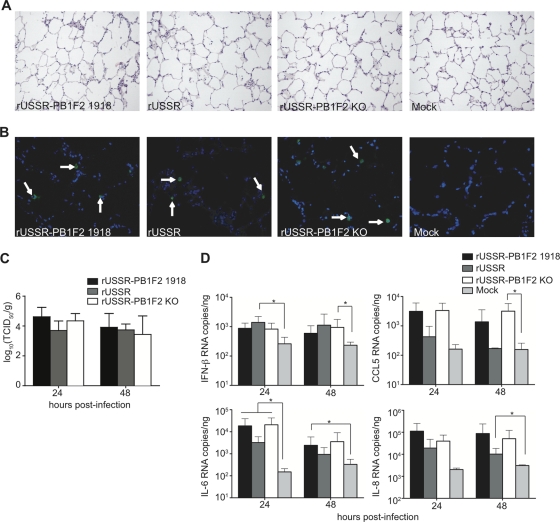
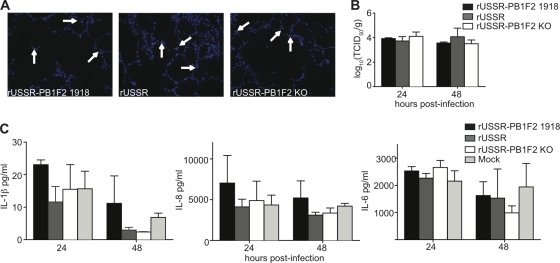
The presence of a full-length PB1-F2 protein has been associated with increased lung pathology and inflammation in mice (3, 19). To directly compare the contribution of PB1-F2 to infection efficacy and inflammatory responses in the lungs of naturally susceptible host species, we established ferret and macaque ex vivo lung cultures. Lungs from healthy animals were inflated with a mixture of DMEM–F-12 medium and 0.8% low-melting-point agarose to preserve the lung anatomy and structure and cut in slices of approximately 3-mm thickness. The lung slices maintained a typical pulmonary parenchyma with alveolar septa of normal thickness and uniformly expanded alveoli without areas of atelectasis or overinflated regions (Fig. 2A). Since viability started to decrease on day 3 after isolation (data not shown), samples were analyzed 24 and 48 h after infection with 3 × 104 TCID50 of the recombinant viruses. At these time points, no differences in frequency and morphology of infected cells were detected (Fig. 2B and data not shown), and all viruses reached similar titers in tissue homogenates (Fig. 2C).
Fig 2.
Infection and cellular responses in ferret ex vivo lung cultures. (A and B) H&E (A) and immunohistochemical (B) staining of lung sections. Lungs slices were harvested 24 h after infection with 3 × 104 TCID50 of the respective virus, paraffin embedded, and H&E stained or stained with a polyclonal anti-influenza virus antibody and counterstained with DAPI. Arrows indicate infected cells. Pictures were taken at 200× or 400× magnification. (C) Viral titers in the homogenized lung slices, expressed as TCID50/g of tissue. (D) Cytokine responses induced by the recombinant viruses. Infected lung slices were harvested at the indicated time points, total RNA was extracted, and 10 ng was used in each reaction mixture. The copy number was extrapolated from results with an in vitro-transcribed mRNA of known concentration. Means and standard deviations, based on three independent experiments performed in duplicate, are shown.
To determine if PB1-F2 alters the induction or magnitude of type I IFNs and proinflammatory cytokines, we generated cytokine mRNA profiles from the lung homogenates. An upregulation of IFN-β, IL-6, IL-8, and CCL5 mRNAs was observed at 24 or 48 h postinfection (Fig. 2D), with USSR generally inducing a weaker but not statistically significantly different response, while IFN-α, IFN-γ, and IL-1β mRNA expression was not induced (data not shown). Since full-length PB1-F2 with the mitochondrial targeting sequence in its carboxy terminus has been associated with increased apoptosis (2, 33), we performed TUNEL assays on paraffin-embedded lung slices. However, only a few apoptotic cells were observed 24 h or 48 h after infection, and there was no difference between the viruses (data not shown).
Since mRNA levels do not always reflect cytokine protein levels, we repeated the experiment in ex vivo lung cultures from cynomolgus macaques, for which sensitive protein-based cytokine assays are available. Consistent with the results obtained in ferret cultures, no differences in infection and replication efficiency were observed for the three recombinant viruses (Fig. 3A and B). The Milliplex cytokine profiles in lung homogenates revealed only a weak induction of IL-8 and IL-1β at 48 h after infection, with USSR again resulting in lower levels. However, protein concentrations did not differ significantly between the viruses and mock-infected control lysates (Fig. 3A). Cytokines that are mainly produced by immune cells, such as MCP-1, MIP-1α, MIP-1β, GM-CSF, IL-10, and IL-17, were not induced in our cultures (data not shown). Furthermore, protein concentrations of active caspase-3 and cleaved PARP were also independent of the virus used, suggesting that PB1-F2 has no impact on the induction of apoptosis in ex vivo lung slice cultures (data not shown). Taken together, these results indicate that PB1-F2 alone has little effect on infection efficiency or local cytokine responses in isolated lung tissue.
Fig 3.
Infection and cytokine profiles in macaque lung ex vivo cultures. (A) Detection of infected cells in paraffin sections from tissue harvested 24 h after infection was performed as previously described. Pictures were taken at a 400× magnification, and arrows indicate infected cells. (B) Titers of homogenized lung slices were determined by using the limiting dilution method and are expressed as TCID50/g of tissue. (C) Quantification of IL-1β, IL-8, and IL-6 production at 24 and 48 h postinfection. Tissues were homogenized in RIPA buffer, and 12.5 μg of each sample was analyzed in a bioplex assay. Values represent the means of three samples, and error bars indicate the standard deviations.
PB1-F2 modulates cytokine responses in ferret blood-derived macrophages.
Previous studies proposed that the PB1-F2-mediated induction of apoptosis and cytokine responses may be immune cell specific (2, 25). We thus compared cytokine profiles in ferret blood-derived macrophages infected with the recombinant viruses. All viruses resulted in similar NP mRNA levels (Fig. 4A), confirming that there were no differences in infection efficiency. While the virus lacking PB1-F2 resulted in an upregulation of IL-1β, Il-6, and IL-8 mRNA levels, indicative of a proinflammatory response, rUSSR only displayed a mild and delayed response, and rUSSR-PB1F2 1918 viruses prevented all immune activation (Fig. 4B). None of the recombinant viruses induced IFN-α, IFN-γ, or CCL5 expression (Fig. 4B and data not shown), indicating that PB1-F2 modulates the induction of certain proinflammatory cytokines in ferret macrophages. The corresponding analysis of protein-based cytokine profiles in macaque blood-derived macrophages was not possible due to insufficient protein concentrations.
Fig 4.
Cytokine responses in ferret blood-derived macrophages. (A and B) Copy numbers of influenza virus NP RNA (A) and of the proinflammatory cytokines IL-1β, IL-6, IL-8, and IFN-α (B) at 16 and 24 h postinfection. Blood-derived macrophages were infected with 3 × 104 TCID50 of the indicated viruses. Copy numbers were quantified by real-time RT-PCR using 10 ng of total RNA. Means and standard deviations, based on three independent experiments performed in duplicate, are shown. (C) Induction of apoptosis in ferret blood-derived macrophages. The cells were infected with the recombinant viruses, and TUNEL staining was performed after 8 h or 16 h. Values represent the means of at least two independent experiments performed in duplicate. Statistical significance was determined using an unpaired Mann-Whitney test. *, P < 0.05; **, P < 0.01.
Exposure to the 1918 PB1-F2 peptide induced apoptosis in a murine macrophage cell line, but this effect was lost when the peptide was expressed in the context of PR8 (18). To investigate the effect of PB1-F2 during infection of ferret blood-derived macrophages, we determined the increase of TUNEL-positive apoptotic cells associated with each virus. After 8 h, there were around twice as many apoptotic cells in rUSSR-PB1F2 1918-infected cultures than in cultures infected with the parental USSR or the PB1-F2 KO virus, while the levels of apoptosis after 16 h were similar for all samples (Fig. 4C), demonstrating that PB1-F2 proteins carrying a mitochondrial localization signal more rapidly induce apoptosis in ferret blood-derived macrophages.
PB1-F2 does not affect the virulence of a seasonal H1N1 strain in ferrets.
To evaluate the contribution of PB1-F2 to the virulence of a seasonal influenza virus strain in a naturally susceptible host, groups of 6 to 8 ferrets were infected intranasally with 105 TCID50 of different recombinant viruses. All animals developed a fever that peaked between days 1 and 2 postinfection and rapidly declined (data not shown). The different viruses caused an overall similar disease, with moderate clinical signs indicative of an upper and lower respiratory tract infection (Fig. 5A). Some of the rUSSR-PB1F2 1918 virus-infected animals developed more severe clinical signs between days 2 and 4 postinfection, with an increased quantity of seromucous nasal exudates compared to the other groups. However, none of these differences was statistically significant.
Fig 5.
Pathogenesis and virulence of the recombinant viruses in ferrets. (A and B) Clinical scores (A) and titers obtained for nasal turbinates and lung samples (B). Ferrets were infected intranasally with 105 TCID50 of the indicated virus (n = 8 for rUSSR-PB1-F2 1918; n = 6 for rUSSR-PB1-F2 KO). For rUSSR, data from 8 animals infected with USSR during an earlier study (21) are shown. See Materials and Methods. Clinical scores represent the sum of the activity and respiratory signs of disease, scored as outlined in Material and Methods. At days 2 and 4 postinfection, two ferrets of each group were sacrificed, and nasal turbinates and the lower right lung lobe were harvested and homogenized for tissue titration or fixed for histopathological analysis. The number of days postinfection is indicated on the x axis, and clinical scores and virus titers, expressed as TCID50/g, are plotted on the respective y axes. Each symbol represents one animal. (C) Histopathological changes in the lungs. The lower right lobes of ferrets sacrificed at day 2 postinfection were hematoxylin and eosin stained, and pictures were taken at a 400× magnification.
To compare viral replication in the upper and lower respiratory tracts, two ferrets of each group were sacrificed at days 2 and 4 postinfection and the nasal turbinates and lungs were harvested. Similar titers were found for the different viruses in both the upper and lower respiratory tracts (Fig. 5B and C), indicating that PB1-F2 does not influence replication efficacy in ferrets, thus reproducing results obtained in mice (9, 20, 34). Histological examination of H&E-stained lung sections from animals sacrificed on day 2 after infection revealed discrete changes characterized by thickening of the alveolar walls, partial loss of the epithelial layer in the bronchi, and recruitment of inflammatory cells that seemed slightly more pronounced in rUSSR-PB1F2 1918-infected and rUSSR-infected animals (Fig. 5D). TUNEL assays were performed on the lung slides, and no differences in the number of apoptotic cells were observed (data not shown).
The presence of the 1918 PB1-F2 is associated with a stronger induction of IL-1β but lower levels of IFN-α and IL-6 in vivo.
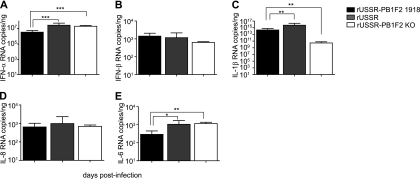
Previous studies in vitro and in mice demonstrated that PB1-F2 influences the proinflammatory response and inhibits type I IFN production (19, 30). To determine if PB1-F2 had a similar effect in the context of an infection of a naturally susceptible host with a seasonal strain, we generated cytokine profiles from nasal wash cell mRNA at different times over the course of the infection. One day after infection, the virus with the 1918 PB1-F2 induced the weakest IFN-α response, with 10-fold lower expression levels than the two other viruses (P < 0.001) (Fig. 6A). However, there was no difference in IFN-β production (Fig. 6B). We next analyzed the expression of the proinflammatory cytokines IL-1β, IL-8, and IL-6. The presence of PB1-F2 resulted in significantly higher IL-1β mRNA levels, but the response associated with the 1918 PB1-F2 was significantly lower than that induced by rUSSR (P < 0.01) (Fig. 6C). No difference in IL-8 mRNA levels was observed (Fig. 6D), while the 1918 PB1-F2 virus induced weaker IL-6 expression than rUSSR and rUSSR-PB1F2 KO (P < 0.05 and P < 0.01, respectively) (Fig. 6E). At later time points, differences between the groups were less pronounced, most likely reflecting the overall mild course of disease (data not shown). Similar cytokine profiles and kinetics were observed in cells from bronchoalveolar lavages (data not shown), indicating that the 1918 PB1-F2 reduces the initial type I IFN and proinflammatory cytokine responses in the upper and lower respiratory tracts.
Fig 6.
Comparison of cytokine mRNA expression in ferret nasal washes. Copy numbers of the type I IFNs IFN-α (A) and IFN-β (B) and the proinflammatory cytokines IL-1β (C), IL-8 (D), and IL-6 (E) at day 1 postinfection were quantified by real-time RT-PCR. Total RNA was extracted from nasal wash cells, and 10 ng was used for each reaction mixture. The copy number was extrapolated from in vitro-transcribed mRNA of known concentration. Each value represents the mean of at least 6 animals, and error bars reflect the standard deviations. Statistical significance was determined using an unpaired Mann-Whitney test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
DISCUSSION
Since its discovery as an accessory viral protein, PB1-F2 has been extensively investigated. In vitro, PB1-F2 induces apoptosis and modulates proinflammatory cytokine and type I IFN expression (2, 15, 16, 30), and studies of mice with infected with mouse-adapted viruses have revealed increased innate immune activation and lung inflammation in the presence of PB1-F2 from virulent strains (4, 19). Here, we investigated the role of PB1-F2 in the context of a seasonal human influenza virus strain in a naturally susceptible host. While PB1-F2 had little impact on the replication and cellular responses in ex vivo lung cultures, the presence of the full-length 1918 PB1-F2 protein resulted in a delay of proinflammatory responses in ferret blood-derived macrophages and to a lesser extent in the nasal wash cells of infected ferrets. However, neither the presence of the full-length 1918 PB1-F2 nor its deletion altered the course or severity of disease in ferrets, indicating that the contribution of PB1-F2 to the pathogenesis of primary viral infection with seasonal influenza viruses is minor.
PB1-F2 delays proinflammatory responses in macrophages.
In mice, introduction of the entire 1918 PB1-F2, or the N66S mutation alone, into the context of the mouse-adapted H1N1 WSN/33 strain resulted in an increased production of proinflammatory cytokines at later infection stages (3, 4, 19). However, further characterization of the N66S mutant revealed decreased type I IFN production during the initial phase of the infection (3, 5, 30). Comparison of the PB1-F2-associated cellular responses in different cell types revealed a reduced type I IFN and proinflammatory response in blood-derived macrophages and dendritic cells, while no differences were observed in epithelial cells (25). We also did not observe any effect of PB1-F2 on the cytokine profile in ferret and macaque lung ex vivo cultures, confirming that the contribution of the protein to the induction of an inflammatory response in lung cells is limited. In ferret blood-derived macrophages, the absence of PB1-F2 resulted in a strong induction of IL-1β, IL-6, and IL-8, while the differences in proinflammatory responses between the truncated 57-residue USSR PB1-F2 and the full-length 1918 protein with a serine at position 66 were modest, suggesting that this residue is less important in the context of a seasonal virus. However, the 1918 PB1-F2 protein was consistently associated with the lowest responses, reproducing the overall tendencies reported for WSN/33 viruses in murine cells (25). Our study thus provides further evidence for an immune cell-specific activity of PB1-F2 in mammals.
PB1-F2 contributes little to the primary viral pathogenesis of seasonal influenza viruses.
In mice, which are only susceptible to mouse-adapted or highly pathogenic influenza viruses, PB1-F2 deletion or the presence of asparagine at position 66 results in reduced disease severity, while the change of position 66 to serine increases mortality (3, 18, 19, 34). In contrast, results obtained in ferrets are less clear-cut. A recombinant seasonal H1N1 virus carrying the 1918 PB1 segment led to efficient spread to and replication in the lung (31), implicating PB1-F2 as a contributing factor, while restoration of a full-length PB1-F2 in an H1N1 pandemic 2009 virus regardless of position 66 did not increase virulence (9). Consistent with the latter report, we did not detect significant differences in the disease severity, lung pathology, and viral load in the upper or lower respiratory tracts of ferrets infected with the different USSR-based PB1-F2 mutant viruses, indicating that PB1-F2 alone does not influence the clinical disease caused by seasonal influenza viruses. However, its contribution to disease severity in the context of viral or bacterial coinfections underlying a large proportion of severe influenza cases (7, 8) remains to be determined. While the mouse studies indicate a role of PB1-F2 in the primary viral pathogenesis of highly virulent influenza viruses in mammals, the extent of its contribution has to be validated ferrets or macaques, which more closely reproduce the disease seen in human patients.
Only the 1918 PB1-F2 protein modulates local cytokine responses in vivo.
An early delay in innate immune activation has been observed in lung tissues of mice infected with a sublethal mouse-adapted virus carrying the PB1-F2 N66S mutation (3), supporting a growing body of evidence from in vitro studies that PB1-F2 delays innate immune activation (5, 30). Here we observed that only the N66S-carrying 1918 PB1-F2 protein was associated with inhibition of IFN-α, IL-1β, and IL-6 induction in ferret nasal wash cells at day 1 after infection, providing further evidence for an immunomodulatory role of full-length PB1-F2 proteins with this mutation. However, in contrast to the results obtained in blood-derived macrophages, where absence of PB1-F2 led to 10- to 100-fold increases in cytokine mRNAs, the differences associated with the 1918 PB1-F2 in vivo, although statistically significant, were only around 3- to 5-fold higher. By demonstrating that PB1-F2 contributes little to innate immune activation and overall inflammatory responses in the context of a primary seasonal influenza virus infection, where infection of macrophages and other immune cells is rare, our results support an increasing body of evidence that this protein is not a major virulence factor in mammals. However, the observed PB1-F2-mediated delayed activation of inflammatory cells represents a possible mechanism of virulence enhancement in the context of infections with already highly pathogenic strains or coinfections with several pathogens, which result in an increased influx of immune cells (11, 23).
ACKNOWLEDGMENTS
We thank all laboratory members for continuing support and lively discussions and Alain Lamarre for his helpful comments on the manuscript. We are particularly grateful to Bevan Sawatsky and Margarita Legaspi for help with the animal studies.
This work was supported by grants NIP-79937, and PAN-310641, 83146 from the Canadian Institutes of Health Research and grant 9488 from the Canadian Foundation for Innovation to V.V. and the Armand-Frappier Foundation and FRSQ scholarships to I.M.
Footnotes
Published ahead of print 8 February 2012
REFERENCES
- 1. Chen CJ, et al. 2010. Differential localization and function of PB1-F2 derived from different strains of influenza A virus. J. Virol. 84:10051–10062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chen W, et al. 2001. A novel influenza A virus mitochondrial protein that induces cell death. Nat. Med. 7:1306–1312 [DOI] [PubMed] [Google Scholar]
- 3. Conenello GM, et al. 2011. A single N66S mutation in the PB1-F2 protein of influenza A virus increases virulence by inhibiting the early interferon response in vivo. J. Virol. 85:652–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Conenello GM, Zamarin D, Perrone LA, Tumpey T, Palese P. 2007. A single mutation in the PB1-F2 of H5N1 (HK/97) and 1918 influenza A viruses contributes to increased virulence. PLoS Pathog. 3:1414–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dudek SE, et al. 2011. The influenza virus PB1-F2 protein has interferon antagonistic activity. Biol. Chem. 392:1135–1144 [DOI] [PubMed] [Google Scholar]
- 6. Eccles R. 2005. Understanding the symptoms of the common cold and influenza. Lancet Infect. Dis. 5:718–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Esper FP, Spahlinger T, Zhou L. 2011. Rate and influence of respiratory virus co-infection on pandemic (H1N1) influenza disease. J. Infect. 63:260–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Estenssoro E, et al. 2010. Pandemic 2009 influenza A in Argentina: a study of 337 patients on mechanical ventilation. Am. J. Respir. Crit. Care Med. 182:41–48 [DOI] [PubMed] [Google Scholar]
- 9. Hai R, et al. 2010. PB1-F2 expression by the 2009 pandemic H1N1 influenza virus has minimal impact on virulence in animal models. J. Virol. 84:4442–4450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG. 2000. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc. Natl. Acad. Sci. U. S. A. 97:6108–6113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kash JC, et al. 2011. Lethal synergism of 2009 pandemic H1N1 influenza virus and Streptococcus pneumoniae coinfection is associated with loss of murine lung repair responses. mBio 2:e00172–e00111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Khanna M, Kumar P, Choudhary K, Kumar B, Vijayan VK. 2008. Emerging influenza virus: a global threat. J. Biosci. 33:475–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kosik I, Krejnusova I, Bystricka M, Polakova K, Russ G. 2011. N-terminal region of the PB1-F2 protein is responsible for increased expression of influenza A viral protein PB1. Acta Virol. 55:45–53 [DOI] [PubMed] [Google Scholar]
- 14. Krumbholz A, et al. 2011. Current knowledge on PB1-F2 of influenza A viruses. Med. Microbiol. Immunol. 200:69–75 [DOI] [PubMed] [Google Scholar]
- 15. Le Goffic R, et al. 2010. Influenza A virus protein PB1-F2 exacerbates IFN-beta expression of human respiratory epithelial cells. J. Immunol. 185:4812–4823 [DOI] [PubMed] [Google Scholar]
- 16. Le Goffic R, et al. 2011. Transcriptomic analysis of host immune and cell death responses associated with the influenza A virus PB1-F2 protein. PLoS Pathog. 7:e1002202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mazur I, et al. 2008. The proapoptotic influenza A virus protein PB1-F2 regulates viral polymerase activity by interaction with the PB1 protein. Cell. Microbiol. 10:1140–1152 [DOI] [PubMed] [Google Scholar]
- 18. McAuley JL, et al. 2010. PB1-F2 proteins from H5N1 and 20 century pandemic influenza viruses cause immunopathology. PLoS Pathog. 6:e1001014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McAuley JL, et al. 2007. Expression of the 1918 influenza A virus PB1-F2 enhances the pathogenesis of viral and secondary bacterial pneumonia. Cell Host Microbe 2:240–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McAuley JL, Zhang K, McCullers JA. 2010. The effects of influenza A virus PB1-F2 protein on polymerase activity are strain specific and do not impact pathogenesis. J. Virol. 84:558–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Meunier I, von Messling V. 2011. NS1-mediated delay of type I interferon induction contributes to influenza A virulence in ferrets. J. Gen. Virol. 92:1635–1644 [DOI] [PubMed] [Google Scholar]
- 22. Nakata M, Itou T, Sakai T. 2008. Molecular cloning and phylogenetic analysis of inflammatory cytokines of the ferret (Mustela putorius furo). J. Vet. Med. Sci. 70:543–550 [DOI] [PubMed] [Google Scholar]
- 23. Perrone LA, Plowden JK, Garcia-Sastre A, Katz JM, Tumpey TM. 2008. H5N1 and 1918 pandemic influenza virus infection results in early and excessive infiltration of macrophages and neutrophils in the lungs of mice. PLoS Pathog. 4:e1000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rudd PA, Bastien-Hamel LE, von Messling V. 2010. Acute canine distemper encephalitis is associated with rapid neuronal loss and local immune activation. J. Gen. Virol. 91:980–989 [DOI] [PubMed] [Google Scholar]
- 25. Schmolke M, et al. 2011. Differential contribution of PB1-F2 to the virulence of highly pathogenic H5N1 influenza A virus in mammalian and avian species. PLoS Pathog. 7:e1002186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Siminski JT, Kavanagh TJ, Chi E, Raghu G. 1992. Long-term maintenance of mature pulmonary parenchyma cultured in serum-free conditions. Am. J. Physiol. 262:L105–L110 [DOI] [PubMed] [Google Scholar]
- 27. Svitek N, Rudd PA, Obojes K, Pillet S, von Messling V. 2008. Severe seasonal influenza in ferrets correlates with reduced interferon and increased IL-6 induction. Virology 376:53–59 [DOI] [PubMed] [Google Scholar]
- 28. Svitek N, von Messling V. 2007. Early cytokine mRNA expression profiles predict mOrbillivirus disease outcome in ferrets. Virology 362:404–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tang JW, Shetty N, Lam TT, Hon KL. 2010. Emerging, novel, and known influenza virus infections in humans. Infect. Dis. Clin. North Am. 24:603–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Varga ZT, et al. 2011. The influenza virus protein PB1-F2 inhibits the induction of type I interferon at the level of the MAVS adaptor protein. PLoS Pathog. 7:e1002067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Watanabe T, et al. 2009. Viral RNA polymerase complex promotes optimal growth of 1918 virus in the lower respiratory tract of ferrets. Proc. Natl. Acad. Sci. U. S. A. 106:588–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wise HM, et al. 2009. A complicated message: identification of a novel PB1-related protein translated from influenza A virus segment 2 mRNA. J. Virol. 83:8021–8031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zamarin D, Garcia-Sastre A, Xiao X, Wang R, Palese P. 2005. Influenza virus PB1-F2 protein induces cell death through mitochondrial ANT3 and VDAC1. PLoS Pathog. 1:e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zamarin D, Ortigoza MB, Palese P. 2006. Influenza A virus PB1-F2 protein contributes to viral pathogenesis in mice. J. Virol. 80:7976–7983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zell R, et al. 2007. Prevalence of PB1-F2 of influenza A viruses. J. Gen. Virol. 88:536–546 [DOI] [PubMed] [Google Scholar]