Abstract
The antiviral factor CPSF6-358 interferes with the nuclear entry of human immunodeficiency virus type 1 (HIV-1). HIV-1 acquires resistance to CPSF6-358 through the N74D mutation of the capsid (CA), which alters its nuclear entry pathway. Here we show that compared to wild-type (WT) HIV-1, N74D HIV-1 is more sensitive to cyclosporine, has increased sensitivity to nevirapine, and is impaired in macrophage infection prior to reverse transcription. These phenotypes suggest a difference in the N74D reverse transcription complex that manifests early after infection and prior to interaction with the nuclear pore. Overall, our data indicate that N74D HIV-1 replication in transformed cells requires cyclophilin A but is dependent on other interactions in macrophages.
TEXT
The postentry events of human immunodeficiency virus type 1 (HIV-1) infection, particularly the interactions of viral proteins with host cell factors, have not been fully elucidated. After entry, HIV-1 reverse transcription complexes (RTCs) traffic on microtubules prior to accumulation at the nuclear membrane and passage through nuclear pores (20). It is assumed that HIV-1 utilizes host proteins in these early replication steps, but a demonstration of direct interaction and participation of cellular factors in the cytoplasmic progression of HIV-1 has been elusive. While the association of cyclophilin A (CypA) with HIV-1 RTCs aids virus replication in certain cell types (10, 16, 25), it has also been suggested that CypA can modulate intracellular immune responses (17, 18).
We previously identified a C-terminal truncation of cleavage and polyadenylation factor 6, CPSF6-358, that restricts HIV-1 infection at nuclear import as well as the isolation of the N74D capsid (CA) mutation that overcomes this restriction (15). Notably, relative to wild-type (WT) HIV-1, N74D HIV-1 differs in host nuclear transport and pore protein dependence, including the loss of NUP153 or TNPO3 utilization (15), both of which have been shown to be important for WT HIV-1 infection (5, 6, 13). These data reinforce the relevance of nuclear factors in HIV-1 infection and suggest that CA is involved in these interactions. CA is a common target for many factors that restrict retroviral infection, including CPSF6-358 (15), Fv1 (3, 4, 22), TRIM5α (28), and TRIM-Cyp (21, 23). The fact that CA modulates HIV-1 dependence on TNPO3 and nuclear pore proteins (NUPs) suggests that some CA remains associated with the RTC until nuclear pore docking, facilitating nuclear entry. Other HIV-1 CA mutants have shown impairment in reverse transcription (8) and nuclear transport (7, 30).
The ability of the N74D CA mutation to alter the requirement for nuclear import factors by HIV-1 is not understood. N74D HIV-1 differs from other CA mutants that confer CPSF6-358 resistance in that it can infect nondividing cells (15). Here we analyze the sensitivity of WT and N74D HIV-1 to cyclosporine (CsA) or reverse transcription inhibitors and their ability to infect macrophages. These data suggest that the mechanism by which the N74D RTC evades CPSF6-358 results in a loss of macrophage tropism.
N74D HIV-1 CA mutant sensitivity to CypA and TRIM5α.
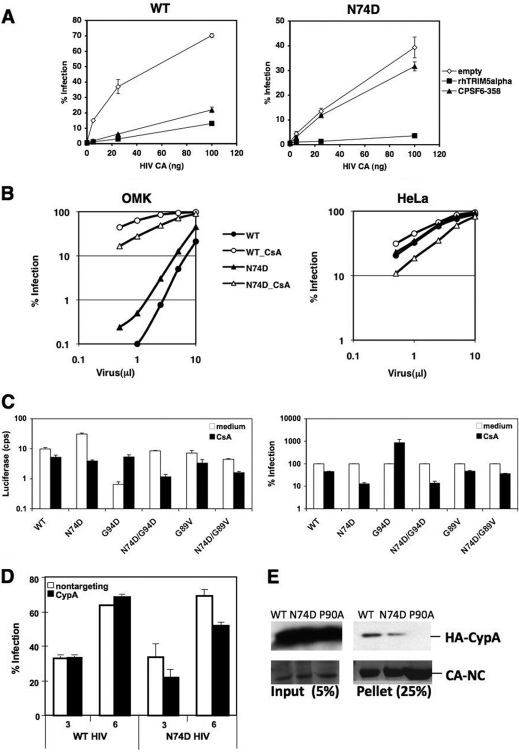
CPSF6-358 expression restricts infection by different primate lentiviruses, including WT HIV-1 (Fig. 1A), whereas N74D HIV-1 is resistant to CPSF6-358 (15). Although N74D HIV-1 has altered TNPO3/NUP requirements (15, 19), this mutant remains susceptible to restriction by rhesus TRIM5α (rhTRIM5α) (Fig. 1A), suggesting that its core early in infection is not grossly altered.
Fig 1.
Sensitivity of N74D HIV-1 to TRIMs and CsA. (A) HeLa cells expressing empty vector, CPSF6-358, or rhTRIM5α were infected in duplicate with various amounts of WT HIV-1 or the N74D mutant (as measured by p24 enzyme-linked immunosorbent assay [ELISA]) encoding the HSA gene in place of nef and pseudotyped with vesicular stomatitis virus G (VSV-G) and assayed for HSA expression 48 h later. (B) Owl monkey kidney (OMK) or HeLa cells incubated with or without 2.5 μM CsA were infected with different volumes of WT or N74D HIV-1 encoding red fluorescent protein (RFP) and pseudotyped with VSV-G and assayed by a fluorescence-activated cell sorter (FACS) for RFP-positive cells after 48 h. (C) HeLa cells were infected with WT HIV-1 or N74D, G94D, N74D/G94D, G89V, and N74D/G89V mutants encoding the luciferase gene and pseudotyped with VSV-G (HIV-luc/VSV-G) in the presence or absence of 2.5 μM CsA and assayed for luciferase expression 48 h later. Infectivity data shown are absolute luciferase values (left) and normalized values for infectivity in the absence of CsA (right) cps, counts per second. (D) Endogenous CypA expression in HeLa cells was knocked down by small interfering RNA (siRNA) prior to infection with the WT or N74D HIV-1 encoding RFP and pseudotyped with VSV-G. Control cells were treated with nontargeting siRNA. Cells were infected with increasing microliters of virus, and infection was measured 48 h later by flow cytometry. A Western blot of cell lysates harvested on the day of infection was performed to verify protein knockdown. (E) WT, N74D, and P90A CA-NC complexes were incubated with 293T cell extracts expressing CypA tagged with hemagglutinin (HA). Reaction mixtures were isolated after centrifugation through 50% sucrose and probed by a Western blot with anti-HA antibodies (top panels). Coomassie blue staining of total protein is shown in the bottom panels. The results are representative of two independent experiments. Error bars in panels A and C represent the standard deviations of duplicate wells of an experiment.
We examined N74D HIV-1 sensitivity to TRIM-Cyp in OMK cells (Fig. 1B). N74D HIV-1 restriction in OMK cells paralleled that of WT HIV-1 over a titration curve. However, N74D HIV-1 was less susceptible overall to restriction in OMK cells. Notably, when these cells were treated with CsA, an immunosuppressive agent that binds cellular CypA, WT HIV-1 infection increased to a greater extent than N74D HIV-1 infection. These data suggested that CypA may aid N74D HIV-1 infection. Parallel infection of HeLa cells, which do not express TRIM-Cyp, supported this interpretation. In the absence of a drug, WT and N74D HIV-1 displayed similar infection curves. In the presence of CsA, WT HIV-1 infection was only slightly decreased at low multiplicities of infection (MOIs), as has been previously reported (10, 25), but N74D HIV-1 infection decreased.
Because these findings suggested that the N74D mutation affected CypA interactions, we tested whether the increased susceptibility of N74D HIV-1 to CsA could be rescued by a CsA-dependent mutation, G94D (1, 10). We compared the infectivities of WT, N74D, G94D, and N74D/G94D HIV-1 in the presence or absence of CsA (Fig. 1C). As expected, compared to the WT, N74D HIV-1 infectivity was decreased and the infectivity of G94D HIV-1 was increased by CsA treatment. The presence of both the N74D and G94D mutations in HIV-1 led to a partial rescue of infectivity relative to that of N74D HIV-1 in the presence of CsA. However, N74D/G94D HIV-1 remained sensitive to CsA. The G89V mutation, which is CsA independent and does not allow CA binding to CypA (25, 29), renders HIV-1 insensitive to CsA with or without the N74D mutation. In addition, HeLa cells depleted of CypA through RNA interference are less susceptible to N74D HIV-1 infection (Fig. 1D; see also Fig. S1 in the supplemental material).
Collectively, these data indicate that the N74D mutation in CA increases HIV-1 dependence on CypA during infection. One explanation for this phenotype is that CypA binds less efficiently to N74D CA cores than to WT CA cores. To investigate this possibility, we performed an in vitro CypA-binding assay on tubes derived from WT and N74D CA-nucleocapsid (NC) (Fig. 1E). The results show that N74D tubes bind 65 to 70% less CypA than WT tubes in two independent assays, suggesting that N74D HIV-1 cores may indeed be binding CypA with less affinity and that this interaction is more easily disrupted by CsA treatment, perhaps leading to faster CA core dissociation of the mutant.
N74D HIV-1 is more susceptible to RT inhibition by nonnucleoside RT inhibitors.
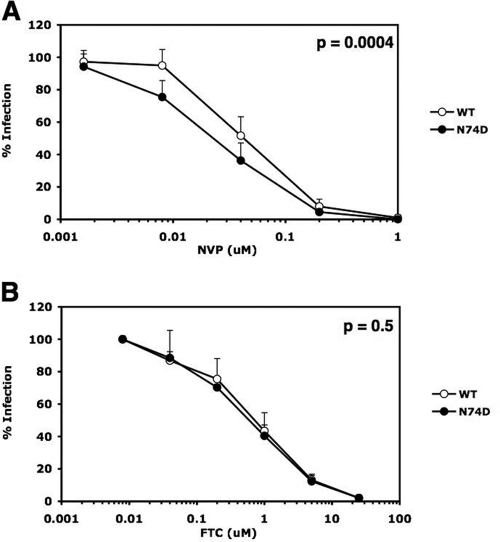
We investigated whether the mechanism of N74D CA dissociation from the RTC could accordingly affect the rate at which reverse transcriptase (RT) dissociates from the RTC. It has been suggested that reverse transcription might reciprocally influence early uncoating steps during HIV-1 infection (12). Previously, we showed that the number of functional RTs in virions influences the susceptibility of viral replication to nonnucleoside RT inhibitors (NNRTIs), which bind to RT, but not to nucleoside analogs (NRTIs), which are incorporated into viral DNA (2). Similarly, if fewer RT molecules are associated with the N74D HIV-1 RTC as a result of greater core instability, the replication of this mutant may be more sensitive to NNRTIs, but not to NRTIs, than WT HIV-1. We measured the susceptibility of infections with WT and N74D HIV-1 to inhibition by the NNRTI nevirapine (NVP) and the NRTI emtricitabine (FTC). Notably, N74D HIV-1 was significantly more sensitive to NVP than WT HIV-1 (Fig. 2A) and the viruses were equally sensitive to FTC (Fig. 2B).
Fig 2.
N74D HIV-1 is more susceptible than WT HIV-1 to the NNRTI NVP. Infections of TZM-bl cells were performed in duplicate with equal multiplicities of infection (MOIs), as determined in Ghost cells of WT and N74D HIV-luc/VSV-G in the presence or absence of NVP (A) or FTC (B). Results are the percentages of infection at each drug concentration relative to infection in the absence of the drug. Error bars represent the standard deviations from the averages of 4 and 2 independent experiments for panels A and B, respectively. Statistical P values were determined using nonlinear regression analysis to compare the curves.
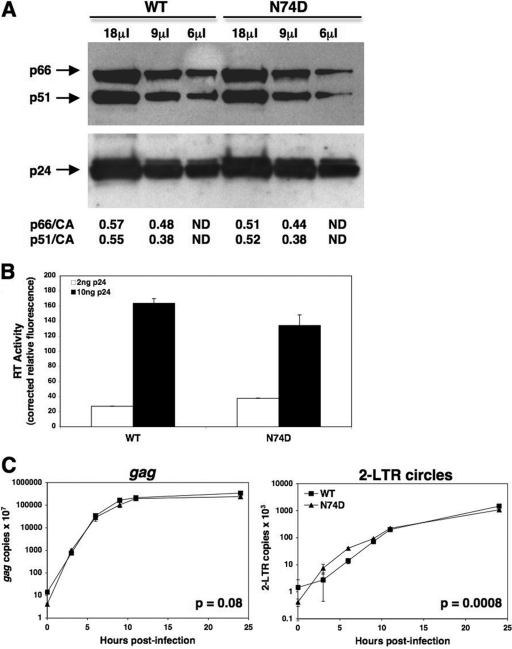
To determine whether increased NVP sensitivity of N74D HIV-1 was due to lower levels of RT packaged into virions, a Western blot analysis was performed on the WT and N74D viruses using antibodies to RT and CA (Fig. 3A). The ratios of the RT subunits (p66 and p51) to CA were similar between the two viruses, confirming that the levels of RT per particle were similar for WT and N74D HIV-1. Also, the RT activities of equal p24 levels of both WT and N74D HIV-1 were analyzed (Fig. 3B). For both 2 ng and 10 ng of p24 from each virus, we observed similar levels of RT activity. In addition, the kinetics of reverse transcription in cells by each virus were similar (Fig. 3C), with late transcripts accumulating at similar levels over an infection time course. Although 2-long-terminal-repeat (2-LTR) circles were observed more rapidly with N74D HIV-1 than with WT HIV-1, overall, these data indicate that the RT molecules from N74D HIV-1 were not diminished in function.
Fig 3.
WT and N74D HIV-luc/VSV-G package similar amounts of RT per particle and carry out similar levels of reverse transcription in vivo. (A) A Western blot was performed on WT and N74D HIV-luc/VSV-G lysates using anti-RT antibodies (a gift from N. Sluis-Cremer) to visualize p66 and p51 (upper). The blot was stripped and reprobed with anti-CA antibodies (a gift from D. Ott) to visualize p24 (lower). The p66/CA and p51/CA ratios were quantified using ImageJ software. ND, not determined. (B) RT activity was measured from WT and N74D HIV-luc/VSV-G lysates, using the EnzCheck reverse transcriptase assay (Invitrogen), and read on a fluorescent plate reader. Equal p24 levels of each virus, as measured by ELISA, were pelleted, resuspended in culture medium containing 1% Triton-X, and used in the assay. Purified WT RT protein (a gift from N. Sluis-Cremer) was used as a standard control. (C) Accumulation of gag DNA (late RT products) and 2-LTR circles was measured in TZM-bl cells infected with equal MOIs of WT and N74D HIV-luc/VSV-G. Infections were synchronized by adding virus at 4°C for 30 min and then shifting the cells to 37°C for up to 24 h. Cells were trypsinized at different times, and total DNA was isolated. qPCR was performed as previously described (15). Error bars represent standard deviations of duplicate wells of an experiment. Statistical P values were determined using nonlinear regression analysis to compare the curves.
Nondividing cells differentially restrict N74D HIV-1.
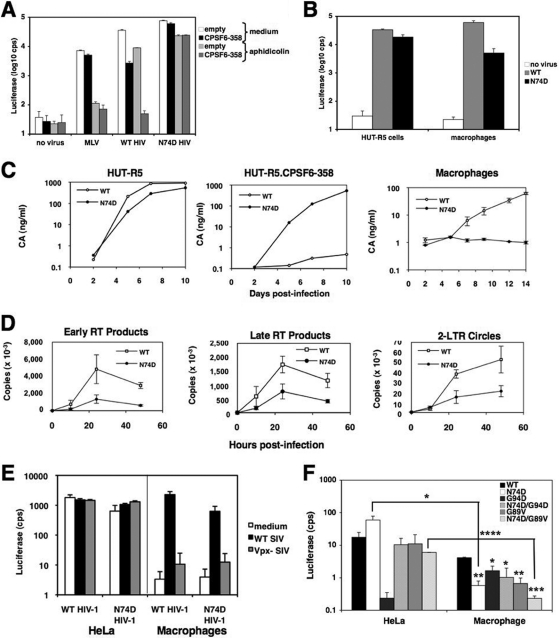
Although CPSF6-358 restricts WT HIV-1 to a greater extent in growth-arrested HeLa cells than in dividing cells, the extent of infection by N74D HIV-1 was not affected by growth arrest in the presence or absence of CPSF6-358 (Fig. 4A). The differences in the abilities of the WT and the mutant virus to infect growth-arrest cells prompted us to examine N74D HIV-1 infectivity in primary human macrophages. In contrast to the results seen in growth-arrested HeLa cells, single-cycle infection of macrophages with N74D HIV-1 was restricted relative to that of WT HIV-1 (Fig. 4B). HIV-1NL4-3/BaL exhibited sustained virus replication in macrophages, but replication of N74D HIV-1NL4-3/BaL was strongly impaired despite the fact that it replicates in a CD4+ T cell line expressing CCR5 in the presence or absence of CPSF6-358 (Fig. 4C).
Fig 4.
Infection of macrophages by the N74D mutant is inhibited prior to or at reverse transcription in a Vpx-independent manner. (A) HeLa and HeLa-CPSF6-358 cells were incubated with culture medium or with murine leukemia virus (MLV)-luc/VSV-G, WT HIV-luc/VSV-G, or N74D HIV-luc/VSV-G in the presence or absence of 2 μg/ml aphidicolin. Luciferase expression was measured 48 h later. (B) HUT-R5 cells and primary human macrophages were infected in duplicate with WT or N74D HIV-luc/VSV-G and analyzed for luciferase activity after 48 h. Representative of 4 independent experiments, using 3 donors for macrophages. (C) HUT-R5 cells, HUT-R5-CPSF6-358 cells, and macrophages were infected with WT HIV-1NL4-3/BaL (a gift from N. Landau) or N74D HIV-1NL4-3/BaL. CA was measured in the culture supernatants by p24 ELISA. Macrophages were infected in duplicate. Representative of 2 independent experiments, using 2 donors for macrophages. (D) Macrophages were infected in duplicate with WT or N74D HIV-luc/VSV-G and lysed at different time points. Early (RU5) and late (gag) RT products, as well as 2-LTR circles, were measured by qPCR. Viral DNA copies are expressed per μg of total DNA obtained from the cells. Representative of 3 independent experiments, using 2 donors. (E) HeLa cells and macrophages were incubated with culture medium or equal MOIs of WT or Vpx-negative SIV-enhanced green fluorescent protein (EGFP)/VSV-G, as determined in HeLa cells, for 4 h. Then the cells were infected with equal MOIs of WT or N74D HIV-luc/VSV-G, as determined on Ghost cells. Luciferase activity was measured 48 h postinfection. Error bars represent standard deviations of duplicates. Representative of 3 independent experiments. (F) HeLa cells or macrophages were infected with the same amounts of WT HIV-luc/VSV-G or the N74D, G94D, N74D/G94D, G89V, or N74D/G89V mutant and analyzed for luciferase activity after 48 h. Representative of 4 independent experiments. Statistical significance values of panel F were calculated by using Student's two-sided t test. Asterisks directly above bars indicate differences between WT and mutant infections in a given cell type, whereas asterisks above lines indicate differences in infectivity for the same virus between cell types. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05. Error bars in all panels represent standard deviations of duplicate wells of an experiment.
To understand the failure of N74D HIV-1 to replicate in macrophages, we measured the progress of reverse transcription and the levels of 2-LTR circles by quantitative PCR (qPCR). There were lower levels of early and late reverse transcripts and 2-LTR circles in cells infected with N74D HIV-1 than in cells infected with WT HIV-1 (Fig. 4D). Infection of HIV-1 is impaired in macrophages but can be relieved by expression of the simian immunodeficiency virus (SIV) protein Vpx (9, 24, 27). Vpx has recently been shown to degrade the restriction factor SAMHD1 (11, 14). Therefore, we infected macrophages with WT and N74D HIV-1 in the presence of WT or Vpx-negative SIV particles. The infectivity of WT and N74D HIV-1 was increased in the presence of WT SIV but not in the presence of Vpx-negative SIV (Fig. 4E). There was a corresponding increase in viral DNA (vDNA) only when Vpx was present (data not shown). However, the infectivity of N74D HIV-1 was not rescued to the same extent as WT HIV-1.
To see if CA mutations that affect CypA binding would alter N74D HIV-1 infection of macrophages, different CA mutants were used to infect HeLa cells and macrophages (Fig. 4F). N74D HIV-1 and G94D HIV-1 displayed distinct patterns of infection. While N74D HIV-1 was more impaired in macrophage infection than in HeLa cell infection, the infectivity of G94D HIV-1 was increased in macrophages relative to that in HeLa cells. These findings are consistent with a report of a HeLa-specific restriction factor that impairs infection by this mutant (26). The addition of the N74D mutation to G94D rescued infection of HeLa cells to levels similar to those of WT or N74D HIV-1. To understand whether the impaired infection of N74D HIV-1 was CypA dependent, we tested the CypA-independent N74D/G89V double mutant HIV-1. Relative to G89V HIV-1, infectivity of the double mutant was the most impaired in macrophages of all the CA mutants, suggesting that the N74D defect is independent of CypA use or levels.
The N74D mutation in HIV-1 CA creates a virus with several notable characteristics. While this mutant replicates well in CD4+ T cell lines, is insensitive to CPSF6-358, and is not dependent on TNPO3/NUPs (15, 19), it is inefficient in macrophage infection due to a restriction prior to, or during initiation of, reverse transcription. The loss of macrophage tropism may, in part, explain why Asn74 is well conserved among primate lentiviruses (15).
Mutations in HIV-1 CA, or other retroviral CAs, can overcome CPSF6-358 restriction (15), supporting the argument that CA has a role in the nuclear import of preintegration complexes. One such CA mutant, Q63A/Q67A HIV-1, supports normal levels of reverse transcription but exhibits diminished 2-LTR circle formation in dividing HeLa-derived cells (7). A relationship between the initiation of reverse transcription and a CA association with the HIV-1 RTC has also been observed (12). Here we find additional evidence of an interaction between CA and RT, with N74D HIV-1 having modestly increased sensitivity to NVP but not FTC compared to that of WT HIV-1. Because this was not due to different levels of RT molecules packaged into WT and N74D virions, we hypothesize that N74D HIV-1 sensitivity to NVP may reflect a reduced association of RT molecules with the RTC after infection. Alternatively, NVP access to WT virion cores may be subtly different from its access to N74D virion cores. Both interpretations imply an effect on core integrity by CA mutation. While the initiation of reverse transcription may precipitate changes in this core structure (12), altered interactions of the core exterior with host cytoplasmic proteins, such as CypA, may also facilitate CA dissociation (16).
In contrast to HIV-1 CA mutations, such as A92E and G94D (10, 25), N74D CA increases HIV-1 dependence on CypA. CsA treatment of cells results in an early block to N74D HIV-1 infection (see Fig. S2 in the supplemental material). The effect of CsA on N74D HIV-1 is mediated through CypA, as N74D/G89V HIV-1, which has a CA that cannot bind CypA, is insensitive to the drug. In addition, N74D HIV-1 infection is reduced in HeLa cells depleted of CypA through RNA interference. The role of CypA in HIV-1 infection has been proven to be complex (17). The increased dependence of N74D HIV-1 on CypA provides a new tool to assess this interaction in vivo. We hypothesize that N74D HIV-1 has an increased dependence on CypA for uncoating, which leads to the loss of RT molecules more quickly than in WT HIV-1.
It is conceivable that altered N74D CA dissociation from the RTC aids in CPSF6-358 resistance. CPSF6-358 binds HIV-1 CA, which is diminished when the N74D mutation is present (15). However, N74D CA binding to CPSF6-358 is not abrogated and remains a few fold lower than that of WT CA in pulldown assays. Given the potency of the CPSF6-358 restriction of WT HIV-1, especially in nondividing cells, a difference in N74D CA association with the RTC may also contribute to the effectiveness of resistance.
There are parallels in N74D HIV-1 infection of HeLa cells and macrophages. In HeLa cells, the mutant virus can be blocked with CsA prior to or at initiation of reverse transcription, and in macrophages, N74D HIV-1 is also blocked early. Is this a result of diminished CypA levels or differential CypA localization in macrophages? While diminished CypA interactions with CA may underlie some of the macrophage restriction, experiments with CA double mutants, in particular N74D/G89V, indicate that the N74D mutation may preclude interaction with another necessary cofactor in these cells, such as CPSF6 or TNPO3. One interpretation is that the pathway of nuclear entry used by N74D HIV-1, if it is able to progress past the early reverse transcription block, remains suboptimal in macrophages.
In conclusion, our findings demonstrate that the CA N74D mutation, in addition to altering HIV-1 nuclear entry pathways, has effects on the RTC in the cytoplasm. N74D HIV-1 has an increased dependence on CypA in transformed cells that contrasts with an early, CypA-independent replication defect by N74D HIV-1 that is observed in macrophages. We hypothesize that CypA availability in transformed cells masks the N74D HIV-1 replication defect and that the N74D mutation prevents interaction with cofactors or a trafficking pathway that WT HIV-1 utilizes in macrophage infection.
ACKNOWLEDGMENTS
We thank Ned Landau, David Ott, and Nicolas Sluis-Cremer for valuable reagents used in the experiments and Tamera Franks and Christopher Kline for technical support. We thank Paul Bieniasz for suggesting examination of the N74D/G94D HIV-1 double mutant.
This work was supported by a contribution from the Pittsburgh Center for HIV Protein Interactions and was supported by the National Cancer Institute's intramural Center for Cancer Research, which supports the HIV Drug Resistance Program (S.H.H. and V.N.K.), grant number 1 UL1 RR024153 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH) and NIH Roadmap for Medical Research (Z.A.), and NIH grants GM082251 (Z.A.), AI078839 (D.U.), and AI052014 (A.E.).
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services or NCRR, and mention of trade names, commercial products, or organizations does not imply endorsement by the U.S. Government.
Footnotes
Published ahead of print 1 February 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Aberham C, Weber S, Phares W. 1996. Spontaneous mutations in the human immunodeficiency virus type 1 gag gene that affect viral replication in the presence of cyclosporins. J. Virol. 70:3536–3544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ambrose Z, Julias JG, Boyer PL, Kewalramani VN, Hughes SH. 2006. The level of reverse transcriptase (RT) in human immunodeficiency virus type 1 particles affects susceptibility to nonnucleoside RT inhibitors but not to lamivudine. J. Virol. 80:2578–2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boone LR, Innes CL, Glover PL, Linney E. 1989. Development and characterization of an Fv-1-sensitive retrovirus-packaging system: single-hit titration kinetics observed in restrictive cells. J. Virol. 63:2592–2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bowerman B, Brown PO, Bishop JM, Varmus HE. 1989. A nucleoprotein complex mediates the integration of retroviral DNA. Genes Dev. 3:469–478 [DOI] [PubMed] [Google Scholar]
- 5. Brass AL, et al. 2008. Identification of host proteins required for HIV infection through a functional genomic screen. Science 319:921–926 [DOI] [PubMed] [Google Scholar]
- 6. Christ F, et al. 2008. Transportin-SR2 imports HIV into the nucleus. Curr. Biol. 18:1192–1202 [DOI] [PubMed] [Google Scholar]
- 7. Dismuke DJ, Aiken C. 2006. Evidence for a functional link between uncoating of the human immunodeficiency virus type 1 core and nuclear import of the viral preintegration complex. J. Virol. 80:3712–3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Forshey BM, Aiken C. 2003. Disassembly of human immunodeficiency virus type 1 cores in vitro reveals association of Nef with the subviral ribonucleoprotein complex. J. Virol. 77:4409–4414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Goujon C, et al. 2006. With a little help from a friend: increasing HIV transduction of monocyte-derived dendritic cells with virion-like particles of SIV(MAC). Gene Ther. 13:991–994 [DOI] [PubMed] [Google Scholar]
- 10. Hatziioannou T, Perez-Caballero D, Cowan S, Bieniasz PD. 2005. Cyclophilin interactions with incoming human immunodeficiency virus type 1 capsids with opposing effects on infectivity in human cells. J. Virol. 79:176–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hrecka K, et al. 2011. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474:658–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hulme AE, Perez O, Hope TJ. 2011. Complementary assays reveal a relationship between HIV-1 uncoating and reverse transcription. Proc. Natl. Acad. Sci. U. S. A. 108:9975–9980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Konig R, et al. 2008. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell 135:49–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Laguette N, et al. 2011. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474:654–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee K, et al. 2010. Flexible use of nuclear import pathways by HIV-1. Cell Host Microbe 7:221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li Y, Kar AK, Sodroski J. 2009. Target cell type-dependent modulation of human immunodeficiency virus type 1 capsid disassembly by cyclophilin A. J. Virol. 83:10951–10962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Luban J. 2007. Cyclophilin A, TRIM5, and resistance to human immunodeficiency virus type 1 infection. J. Virol. 81:1054–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Manel N, et al. 2010. A cryptic sensor for HIV-1 activates antiviral innate immunity in dendritic cells. Nature 467:214–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Matreyek KA, Engelman A. 2011. The requirement for nucleoporin NUP153 during human immunodeficiency virus type 1 infection is determined by the viral capsid. J. Virol. 85:7818–7827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McDonald D, et al. 2002. Visualization of the intracellular behavior of HIV in living cells. J. Cell Biol. 159:441–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nisole S, Lynch C, Stoye JP, Yap MW. 2004. A Trim5-cyclophilin A fusion protein found in owl monkey kidney cells can restrict HIV-1. Proc. Natl. Acad. Sci. U. S. A. 101:13324–13328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rommelaere J, Donis-Keller H, Hopkins N. 1979. RNA sequencing provides evidence for allelism of determinants of the N-, B- or NB-tropism of murine leukemia viruses. Cell 16:43–50 [DOI] [PubMed] [Google Scholar]
- 23. Sayah DM, Sokolskaja E, Berthoux L, Luban J. 2004. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature 430:569–573 [DOI] [PubMed] [Google Scholar]
- 24. Sharova N, et al. 2008. Primate lentiviral Vpx commandeers DDB1 to counteract a macrophage restriction. PLoS Pathog. 4:e1000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sokolskaja E, Sayah DM, Luban J. 2004. Target cell cyclophilin A modulates human immunodeficiency virus type 1 infectivity. J. Virol. 78:12800–12808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Song C, Aiken C. 2007. Analysis of human cell heterokaryons demonstrates that target cell restriction of cyclosporine-resistant human immunodeficiency virus type 1 mutants is genetically dominant. J. Virol. 81:11946–11956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Srivastava S, et al. 2008. Lentiviral Vpx accessory factor targets VprBP/DCAF1 substrate adaptor for cullin 4 E3 ubiquitin ligase to enable macrophage infection. PLoS Pathog. 4:e1000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stremlau M, et al. 2004. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 427:848–853 [DOI] [PubMed] [Google Scholar]
- 29. Towers GJ, et al. 2003. Cyclophilin A modulates the sensitivity of HIV-1 to host restriction factors. Nat. Med. 9:1138–1143 [DOI] [PubMed] [Google Scholar]
- 30. Yamashita M, Perez O, Hope TJ, Emerman M. 2007. Evidence for direct involvement of the capsid protein in HIV infection of nondividing cells. PLoS Pathog. 3:1502–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]