Abstract
Herpes simplex virus 1 (HSV-1) viral glycoproteins gD (carboxyl terminus), gE, gK, and gM, the membrane protein UL20, and membrane-associated protein UL11 play important roles in cytoplasmic virion envelopment and egress from infected cells. We showed previously that a recombinant virus carrying a deletion of the carboxyl-terminal 29 amino acids of gD (gDΔct) and the entire gE gene (ΔgE) did not exhibit substantial defects in cytoplasmic virion envelopment and egress (H. C. Lee et al., J. Virol. 83:6115–6124, 2009). The recombinant virus ΔgM2, engineered not to express gM, produced a 3- to 4-fold decrease in viral titers and a 50% reduction in average plaque sizes in comparison to the HSV-1(F) parental virus. The recombinant virus containing all three mutations, gDΔct-ΔgM2-ΔgE, replicated approximately 1 log unit less efficiently than the HSV-1(F) parental virus and produced viral plaques which were on average one-third the size of those of HSV-1(F). The recombinant virus ΔUL11-ΔgM2, engineered not to express either UL11 or gM, replicated more than 1 log unit less efficiently and produced significantly smaller plaques than UL11-null or gM-null viruses alone, in agreement with the results of Leege et al. (T. Leege et al., J. Virol. 83:896-907, 2009). Analyses of particle-to-PFU ratios, relative plaque size, and kinetics of virus growth and ultrastructural visualization of glycoprotein-deficient mutant and wild-type virions indicate that gDΔct, gE, and gM function in a cooperative but not redundant manner in infectious virion morphogenesis. Overall, comparisons of single, double, and triple mutant viruses generated in the same HSV-1(F) genetic background indicated that lack of either UL20 or gK expression caused the most severe defects in cytoplasmic envelopment, egress, and infectious virus production, followed by the double deletion of UL11 and gM.
INTRODUCTION
Herpes simplex virus 1 (HSV-1) virion assembly begins in the nucleus with the construction of viral capsids, which acquire certain tegument proteins and then bud through the inner nuclear membrane, forming enveloped virions within the perinuclear space (primary envelopment). Enveloped virions fuse with the outer nuclear membranes, allowing capsid deposition in the cytoplasm of cells (10, 45, 47; reviewed in reference 42). In the cytoplasm, viral capsids are coated with tegument proteins and acquire final viral envelopes by budding into glycoprotein-enriched regions of the trans-Golgi network (TGN) membranes (secondary envelopment) (17, 51, 54, 56). This final virion morphogenesis step delivers fully enveloped virions into cytoplasmic vesicles, which are ultimately transported out of the cell (33). Secondary envelopment of cytoplasmic capsids is facilitated by interactions between tegument proteins and the cytoplasmic domains of viral glycoproteins and other membrane proteins anchored in TGN-derived membranes (17, 51, 54, 56; reviewed in references 32 and 42).
Deletion or forced retention of either gD or gH within the endoplasmic reticulum does not cause drastic defects in cytoplasmic virion envelopment and egress, although both glycoproteins are essential for viral infectivity (7, 19, 38, 55). Similarly, gB is not required for cytoplasmic envelopment and egress, inasmuch as gB-null viruses acquire envelopes and can be rendered infectious after treatment with the fusogen polyethylene glycol (8, 41). However, partial deletion of the carboxyl terminus of gB was reported to cause substantial reductions in cytoplasmic virion envelopment and egress (9), suggesting that truncated gB may cause a dominant negative effect. Recently, a gB-gD double mutant but not a gD-null virus exhibited substantial defects in late stages of virus egress, indicating that gB may cooperate with gD in facilitating virion envelopment (34). Single or simultaneous deletion of HSV-1 gE and gM genes did not cause any appreciable defects in cytoplasmic virion envelopment or infectious virus production (6). Similarly, lack of either gD or gE expression caused a mild (2- to 3-fold) reduction in enveloped virions. However, simultaneous deletion of HSV-1 gD and gE or gD, gE, and gI genes caused drastic accumulation of unenveloped capsids in the cytoplasm. Because neither of these gene deletions alone caused similar defects, it was concluded that gD and gE function in a redundant manner in cytoplasmic virion egress (16). Deletion of the UL11 gene produced mild defects in cytoplasmic virion egress (27), while deletion of either the gK or UL20 gene or specific mutations within these two genes caused drastic inhibition of cytoplasmic virion envelopment (23, 31, 41). Also, it was reported that simultaneous deletion of HSV-1 UL11 and gM caused drastic inhibition of cytoplasmic virion envelopment (37). Together, these results suggest that there are multiple cooperative relationships among viral glycoproteins and membrane proteins facilitating cytoplasmic virion envelopment.
We have reported that gK and UL20 have distinct functions in virus-induced cell fusion and cytoplasmic virion envelopment. These functions are genetically separable, since mutations in UL20 that drastically inhibit virion envelopment do not affect virus-induced cell fusion (24, 40). UL20 and gK function in virus-induced cell fusion by physically binding to gB and gH in infected cell surfaces (12, 13). Their roles in cytoplasmic envelopment and egress are not defined, but it is likely that they interact with other viral glycoproteins and tegument proteins to facilitate virion envelopment. Infectious virus production is directly dependent on the ability of viruses to assemble in the cytoplasm mature virions containing fully glycosylated glycoproteins and spread from infected to uninfected cells. Moreover, there are no known glycoprotein mutations that inhibit virion egress without affecting cytoplasmic envelopment. In the study described in this report, we determined particle-to-PFU ratios, relative plaque size (indicative of relative virus spread), and kinetics of virus growth and conducted ultrastructural visualization of glycoprotein-deficient mutant and wild-type virions in the same HSV-1(F) genetic background to gain an understanding of the relative contribution of individual viral glycoproteins in infectious virion morphogenesis and egress. The results show that UL20 and gK are the most important viral determinants for cytoplasmic virion envelopment, egress, and infectious virus production in comparison to gM, gD with deletion of the carboxyl-terminal 29 amino acids (gDΔct), gE, and UL11 alone or in various combinations.
MATERIALS AND METHODS
Cells and antibodies.
African green monkey kidney (Vero) cells were obtained from the American Type Culture Collection (Manassas, VA). Cells were maintained in Dulbecco's modified Eagle's medium (Gibco-BRL, Grand Island, NY) supplemented with 10% fetal calf serum and antibiotics. Antibodies used include anti-HSV-1 gE monoclonal antibody (Virusys, Sykesville, MD) and Alexa Fluor 488-conjugated goat anti-mouse IgG monoclonal antibody (Invitrogen-Molecular Probes, Carlsbad, CA) for the indirect immunofluorescence assays (IFAs), as well as anti-HSV-1 gD, anti-HSV-1 gB, and anti-HSV-1 gC monoclonal antibodies (Virusys, Sykesville, MD), rabbit anti-gM antibody (a gift from Joel Baines, Cornell University, Ithaca, NY), and rabbit anti-UL11 antibody (a gift from John Wills, Pennsylvania State University, Hershey, PA) for the Western immunoblot assays.
Construction of HSV-1 mutant viruses.
Mutagenesis was accomplished in Escherichia coli using the markerless two-step Red recombination mutagenesis system and synthetic oligonucleotides (36, 53) (see Table S1 in the supplemental material) implemented on the bacterial artificial chromosome (BAC) plasmid pYEbac102 carrying the HSV-1(F) genome (52) (a kind gift from Y. Kawaguchi, University of Tokyo, Tokyo, Japan). Construction of the HSV-1 mutants gDΔct (US6), ΔgE (US8), ΔUL20, ΔgM1, ΔgE-gDΔct, and ΔgE-ΔgM1 was described previously (36). The ΔgM2 recombinant virus was constructed by altering two potential initiation codon sites (from ATG to CTG and ATG to ATT, respectively) located 57 bp apart at the beginning of the UL10 open reading frame (ORF) (5) (Fig. 1; see Table S1 in the supplemental material). The ΔUL11 virus was constructed by changing the initiation codon from ATG to CTG. The ΔgE-gDΔct recombinant virus was used as the backbone for construction of the ΔgE-gDΔct-ΔgM2 triple mutant by altering the two potential initiation codon sites in gM, as described above for ΔgM2 virus. The ΔgM2-ΔUL11 double mutant was constructed by altering the initiation codon of UL11 from ATG to CTG in the ΔgM2 virus.
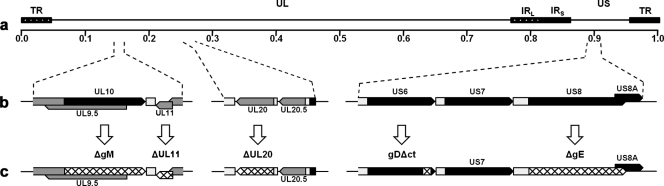
Fig 1.
Genomic map of mutated genes. (a) Prototypic arrangement of the HSV-1 genome with the unique long (UL) and unique short (US) regions flanked by the terminal repeat (TR) and internal repeat (IR; L, long; S, short) regions; (b) expanded genomic regions of the UL10, UL11, UL20, US6, and US8 ORFs; (c) effect of ATG mutagenesis on gM, gE, UL11, and UL20 gene expression (hatched regions), as well truncation of gD after gDΔct mutagenesis (hatched gD region).
Confirmation of the targeted mutations, recovery of infectious virus, and marker-rescue experiments.
HSV-1 BAC DNAs were purified from 50 ml of overnight BAC cultures with a Qiagen large-construct kit (Qiagen, Valencia, CA). Using PCR test primers designed to lie outside the target mutation site(s), all mutated DNA regions were sequenced to verify the presence of the desired mutations in BACs. Similarly, viruses recovered from infected Vero cells were sequenced to confirm the presence of the desired mutations. Viruses were recovered from cells transfected with BACs as we have described previously (36). To determine whether the ΔUL11-ΔgM2 mutant virus contained any other genomic mutations, rescue experiments were performed with approximately 1-kbp DNA fragments spanning the UL11 and gM initiation codon mutations. Vero cells were transfected with the DNA fragments, and 24 h later, transfected cells were infected with the ΔUL11-ΔgM2 virus. Virus stocks were prepared at 24 h postinfection (hpi) and plated at limiting dilution on Vero cells. Approximately 10 to 15% of viral plaques appeared to be similar to wild-type virus (data not shown). DNA sequencing of wild-type-like viral plaques confirmed the absence of the UL11 and gM mutations.
Plaque morphology of mutant viruses and relative plaque area measurements.
Visual analysis of plaque morphology of mutant viruses was performed as we have previously described (24, 27, 36, 41). Plaque area measurements and data analysis were essentially as described previously (36), except that photographs of viral plaques analyzed were taken at ×50 magnification and 30 randomly selected plaques were imaged for each of the mutant and wild-type viruses under consideration.
One-step viral growth kinetics.
Analysis of one-step growth kinetics was performed as we have described previously (20, 25). Briefly, nearly confluent Vero cell monolayers were infected with each virus at 4°C for 1 h at multiplicities of infection (MOIs) of 0.2 and 3. Thereafter, plates were incubated at 37°C in 5% CO2 and virus was allowed to penetrate for 1 h at 37°C. Any remaining extracellular virus was inactivated by low-pH treatment (pH 3.0), and cells were incubated at 37°C in 5% CO2. Supernatants and cell pellets were separated at different times postinfection and stored at −80°C.
SDS-PAGE, Western immunoblotting, and indirect immunofluorescence assay.
Subconfluent Vero cell monolayers were infected with the indicated virus at an MOI of 3. At 24 hpi, cells were collected by low-speed centrifugation, washed with phosphate-buffered saline (PBS), and processed as described previously (21, 36). The indirect immunofluorescence assay for the detection of gE expression was performed as described previously (36).
Electron microscopy.
The ultrastructural morphology of virions within infected cells was examined by transmission electron microscopy essentially as described previously (22, 23, 31, 36, 41). All infected cells processed for electron microscopy were prepared at 16 h postinfection and visualized by transmission electron microscopy.
Preparation of cytoplasmic and extracellular virions.
Cytoplasmic virions were separated by glycerol shock treatment essentially as previously described by Sarmiento and Batterson (46), with the following modifications. Extracellular virions were prepared from supernatants of infected cells. Specifically, viruses were adsorbed on wells of a 12-well plate of nearly confluent Vero cell monolayers at 4°C for 1 h at an MOI of 1. Afterwards, plates were incubated at 37°C in 5% CO2 for 1 h to allow the virus to penetrate into cells. Any remaining extracellular virus was inactivated by low-pH treatment (pH 3.0), and cells were incubated at 37°C in 5% CO2 for 18 h and supernatants were collected. Infected cells were washed once with ice-cold PBS (pH 7.4), and the remaining extracellular virus was inactivated by low-pH treatment (pH 3.0). After removing the low-pH PBS, 900 μl of PBS (pH 7.4) was added to each well. To minimize nuclear disruption, the glycerol concentration was gradually increased by adding 150 μl of 90% glycerol (prewarmed to 37°C) to each well, followed by mixing and incubation at 37°C for 5 min, which was repeated three times to obtain a final concentration of 30% glycerol. The glycerol-treated samples were centrifuged at 1,200 × g for 10 min at 4°C, and supernatants were discarded. The cell pellet was suspended in 250 μl of ice-cold lysis buffer (0.01 M Tris-HCl, pH 7.4, 1.0 mM MgCl2, and 1.0 mM CaCl) by gentle mixing. The cell suspension was incubated on ice for 5 min and centrifuged at 700 × g for 10 min at 4°C to separate the nuclei and cellular debris from the cytoplasmic fraction. The supernatant (cytoplasmic fraction) was carefully transferred to new 1.5-ml tubes and used for TaqMan real-time PCR as described previously (49, 50).
Q-PCR.
Quantitative PCR (Q-PCR) was utilized to derive the number of viral genomes within cytoplasmic and extracellular samples that remained protected after DNase treatment, as described previously for Kaposi's sarcoma-associated herpesvirus (KSHV) (49, 50). Specifically, the primers and probe (6-carboxytetramethylrhodamine [TAMRA]) for the real-time PCR were designed to detect HSV-1 US6 (gD). Cytoplasmic and extracellular fractions were collected at 18 hpi, and 100 μl of each suspension was used for the extraction of viral DNA. The cytoplasmic and extracellular suspensions were treated with Turbo DNase I (Ambion, Inc.) for 1 h at 37°C. Viral DNA was extracted using a DNeasy blood and tissue kit (Qiagen, Inc.) as per the manufacturer's instructions. Equal volumes of viral DNA were used for TaqMan PCR analysis. Purified HSV-1 bacterial artificial chromosome (YE102) DNA was used to generate the standard curve. Samples were also tested, and genome numbers were determined using validated standards provided by the Path-HSV1-genesig real-time PCR detection kit for human herpesvirus 1 (herpes simplex virus 1) (PrimerDesign, Ltd., South Hampton, United Kingdom).
RESULTS
Construction and molecular analysis of recombinant viruses.
We showed previously that recombinant viruses carrying a deletion of the carboxyl terminus of gD (gDΔct) and the entire gE gene or gE and gM were able to efficiently acquire cytoplasmic envelopes (36). To compare the relative roles of UL20, gM, gE, gD, and UL11 in cytoplasmic envelopment and virion egress, we generated additional mutant viruses lacking expression of one or more of these genes (Fig. 1). All mutant viruses were produced using the two-step Red recombination mutagenesis system (53) implemented on the pYEbac102 bacterial artificial chromosome carrying the entire HSV-1(F) genome (52), as described in Materials and Methods. Further characterization of the gE and gM double-null virus produced earlier (36) suggested that a second initiation codon located 19 amino acids downstream from the first initiation codon can be utilized to produce a truncated gM (data not shown). Therefore, to ensure a complete lack of gM expression, the mutant virus ΔgM2 was created by altering both initiation codons, and both versions of the mutated gM genes were used to sequentially construct the desired set of mutations. The gM and UL11 defects were readily rescued by DNA fragments overlapping the corresponding mutations, as evidenced by the appearance of more than 10% viral plaques that were similar to wild-type plaques. Moreover, DNA sequencing of selected rescued viruses revealed the presence of wild-type gM and UL11 sequences (data not shown). The triple mutant virus gDΔct-ΔgM2-ΔgE was generated from the previously characterized gDΔct-ΔgE mutant virus (36). Overall, these results suggest that there were no spurious nucleotide changes elsewhere in the viral genomes of viruses containing the gDΔct, gE, gM, and UL11 mutations.
Recovery and plaque morphologies of infectious viruses produced by HSV-1 BAC DNAs.
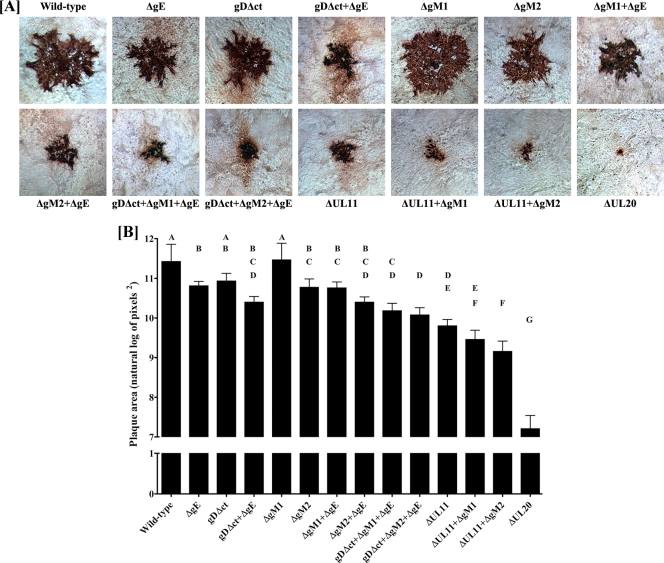
To generate virus stocks from the mutant BAC genomic constructs, individual BAC DNAs were transfected into Vero cells and initial virus stocks were recovered and characterized as detailed in Materials and Methods. The plaque morphologies of all mutant and wild-type viruses were examined in Vero cells. As expected, the HSV-1(F) wild-type virus produced the largest plaques, while the ΔUL20 mutant produced the smallest plaques, consisting of less than 5 cells (Fig. 2A). Deletion of UL11 and gM (ΔUL11-ΔgM2 virus) caused the production of viral plaques that were 4- to 5-fold larger than those of ΔUL20 virus. Deletion of either gE, the carboxyl terminus of gD (gDΔct), or gM (ΔgM2) alone did not drastically reduce the average plaque size, while simultaneous deletion of gD (gDΔct), gM (ΔgM2), and gE (ΔgE) reduced average plaque sizes to the same extent as deletion of UL11 alone (Fig. 2A). To better assess the virus plaque sizes produced by individual mutant viruses, randomly chosen viral plaques were measured and statistically analyzed as described in Materials and Methods. This analysis confirmed that the most drastic reduction in average viral plaque sizes was produced by ΔUL20, followed by the ΔUL11-ΔgM2, ΔUL11-ΔgM1, ΔUL11, and gDΔct-ΔgM2-ΔgE viruses (Fig. 2B).
Fig 2.
Plaque phenotypes of wild-type and mutant viruses. (A) Confluent Vero cell monolayers were infected with each virus at an MOI of 0.001, and viral plaques were visualized at 48 hpi by immunohistochemistry as described in Materials and Methods. (B) Thirty different viral plaques were randomly selected, imaged, measured, and statistically analyzed as described in Materials and Methods. Natural log-transformed data depicted as bar graphs for each virus are shown as geometric means with 95% confidence intervals. Tukey's test was performed after one-way analysis of variance to examine pairwise differences between the means for each of the 14 viruses. Viruses that were significantly different from each other (P ≤ 0.05) are labeled with different capital letters (A, B, C, D, E, F, and G).
Replication characteristics of HSV-1 mutants.
To examine the effect of the various engineered mutations on virus replication, Vero cells were infected at an MOI of either 0.2 or 3.0 with either the wild-type or each mutant virus, and viral titers were obtained at 24 and 48 hpi (Table 1). At an MOI of 0.2, ΔUL20 replicated approximately 3 log units less efficiently at both 24 and 48 hpi than the HSV-1(F) virus. All other mutant viruses achieved titers that were approximately 1 log unit or higher than the ΔUL20 virus titer at 24 hpi and 48 hpi (Table 1). Similar results were obtained when an MOI of 3.0 was used, with the exception that all mutant viruses other than ΔUL20 virus replicated more efficiently at the high MOI than at the low MOI, with viral titers approaching those produced by the HSV-1(F) wild-type virus at 48 hpi (Table 1).
Table 1.
Comparison of wild-type and mutant virus replication
| MOI and mutant virus | Viral titera |
||
|---|---|---|---|
| 0 hpi | 24 hpi | 48 hpi | |
| MOI, 0.2 | |||
| Wild type | 6.00 × 101 ± 28.3 | 1.67 × 107 ± 9.02 × 105 | 1.63 × 107 ± 6.11 × 105 |
| ΔgE | 4.00 × 101 ± 0.00 | 8.67 × 106 ± 1.16 × 106 | 2.31 × 107 ± 1.35 × 106 |
| gDΔct | 2.00 × 101 ± 0.00 | 2.88 × 106 ± 1.68 × 105 | 1.56 × 107 ± 8.72 × 105 |
| gDΔct-ΔgE | 3.00 × 101 ± 14.1 | 5.67 × 105 ± 4.37 × 104 | 6.53 × 106 ± 1.12 × 106 |
| ΔgM2 | 2.00 × 101 ± 0.00 | 4.27 × 106 ± 7.69 × 105 | 4.80 × 106 ± 6.11 × 105 |
| ΔgM2-ΔgE | 1.00 × 101 ± 14.1 | 4.07 × 106 ± 5.21 × 105 | 9.20 × 106 ± 5.03 × 105 |
| gDΔct-ΔgM2-ΔgE | 3.00 × 101 ± 14.1 | 5.73 × 105 ± 5.93 × 104 | 2.55 × 106 ± 1.55 × 105 |
| ΔUL11 | 1.00 × 101 ± 14.1 | 4.53 × 105 ± 4.16 × 104 | 1.14 × 107 ± 1.44 × 106 |
| ΔUL11-ΔgM2 | 2.00 × 101 ± 0.00 | 1.13 × 105 ± 1.62 × 104 | 1.64 × 106 ± 8.00 × 104 |
| ΔUL20 | 4.00 × 101 ± 28.3 | 3.81 × 104 ± 6.22 × 102 | 6.13 × 104 ± 2.31 × 103 |
| MOI, 3.0 | |||
| Wild type | 4.60 × 102 ± 84.8 | 1.03 × 107 ± 8.97 × 105 | 7.61 × 106 ± 2.39 × 106 |
| ΔgE | 2.50 × 102 ± 42.4 | 1.27 × 107 ± 1.43 × 106 | 8.55 × 106 ± 3.76 × 106 |
| gDΔct | 2.40 × 102 ± 28.3 | 3.99 × 106 ± 3.15 × 105 | 4.40 × 106 ± 5.77 × 105 |
| gDΔct-ΔgE | 1.80 × 102 ± 56.6 | 3.40 × 105 ± 2.31 × 104 | 3.60 × 106 ± 6.43 × 105 |
| ΔgM2 | 1.60 × 102 ± 56.6 | 4.73 × 106 ± 5.21 × 105 | 4.73 × 106 ± 7.33 × 105 |
| ΔgM2-ΔgE | 7.00 × 101 ± 14.1 | 7.13 × 106 ± 1.10 × 106 | 7.13 × 106 ± 8.51 × 105 |
| gDΔct-ΔgM2-ΔgE | 1.50 × 102 ± 14.1 | 3.17 × 106 ± 1.89 × 105 | 2.43 × 106 ± 1.30 × 105 |
| ΔUL11 | 3.80 × 102 ± 28.3 | 1.99 × 107 ± 4.81 × 105 | 2.41 × 107 ± 1.33 × 105 |
| ΔUL11-ΔgM2 | ND | ND | ND |
| ΔUL20 | 2.70 × 102 ± 14.1 | 1.34 × 105 ± 2.34 × 103 | 1.61 × 105 ± 3.51 × 103 |
Viral titers were determined at different times after infection of Vero cells at an MOI of 0.2 or an MOI of 3.0. The experiment was performed a second time, and the titers obtained were averaged, with the standard deviation calculated for each time point. ND, not done.
Protein expression profiles of viral mutants.
To confirm that the engineered gene mutations resulted in lack of expression of the relevant protein and to investigate whether deletion of one or more viral glycoproteins affected the synthesis of other viral glycoproteins, all mutant viruses were tested for the expression of gB, gC, gD, gE, gM, and UL11. As expected, all mutant viruses containing the ΔgM2 mutation failed to express gM (Fig. 3). The ΔgM1 virus expressed a truncated gM glycoprotein migrating in a manner consistent with the deletion of 19 amino acids from the amino terminus of gM, as previously reported by others (5) (data not shown). The ΔUL11 mutation caused a lack of UL11 expression for all recombinant viruses containing this mutation. The gDΔct deletion caused the appearance of a truncated gD migrating with the expected apparent molecular mass, as we have reported previously (36). Mutant viruses unable to produce gM and gE or gM, gE, and gDΔct (DME2) appeared to express substantially smaller amounts of gB in than all other viruses tested, while all viruses synthesized equivalent levels of gC. In addition, the gDΔct-gE or gM-gE-gDΔct viruses showed substantial reductions in the relative production of the UL11 protein (Fig. 3). The expression of gE was tested by indirect immunofluorescence, since the available antibody did not react strongly enough in immunoblots. IFA results showed that gE was not expressed in cells infected with viruses specifying the ΔgE mutation, while gE expression was unaffected by any of the other gene deletions (Fig. 4).
Fig 3.

Western immunoblot analysis of glycoproteins specified by mutant viruses. α-gB, α-gC, α-gD, α-gM, and α-UL11 denote antibodies specific for each protein.
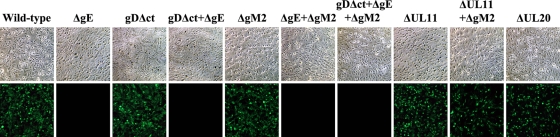
Fig 4.
Immunofluorescence detection of gE expression. Vero cells were infected at an MOI of 1 with the viruses indicated, and gE expression was detected by indirect immunofluorescence at 24 hpi. (Top row) Phase-contrast micrographs of infected cells treated with anti-gE MAb; (bottom row) fluorescent micrographs of the same infected cells. All mutant viruses containing the ATG-to-CTG mutations in gE (ΔgE) failed to react with anti-gE antibody, while viruses expressing the wild-type gE reacted with anti-gE antibody.
Ultrastructural characterization of wild-type and mutant viruses.
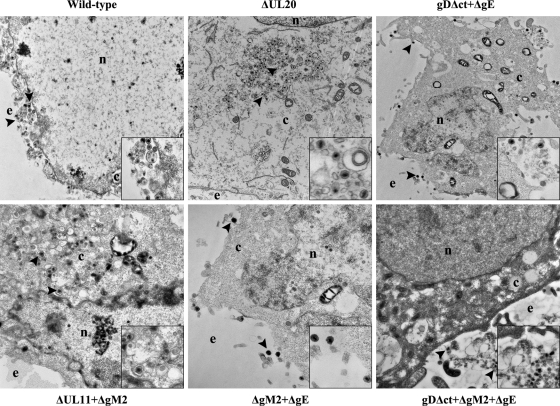
The ultrastructural phenotypes of all viruses relative to the wild-type parental virus were investigated at 16 hpi utilizing transmission electron microscopy and visually examining more than 50 individual virus-infected Vero cells. As expected, the wild-type virus did not exhibit any apparent defects in cytoplasmic virion envelopment or egress, as evidenced by the presence of fully enveloped virions intracellularly and extracellularly (Fig. 5). Ultrastructural visualization of Vero cells infected with the different mutant viruses revealed a diverse range of cytoplasmic defects in virion envelopment. The most pronounced effects were produced by the ΔUL20 and ΔUL11-ΔgM2 viruses, which produced numerous unenveloped capsids. In contrast, the DME2 virus produced fully enveloped virions that were excreted out of infected cells (Fig. 5).
Fig 5.
Ultrastructural morphology of wild-type and mutant viruses. Electron micrographs of Vero cells infected with different viruses at an MOI of 3 and processed for electron microscopy at 16 hpi are shown. Enlarged sections of each micrograph are included as insets in each panel. The nucleus (n), cytoplasm (c), and extracellular space (e) are marked. Representative virions are marked with black arrowheads.
Quantification of relative efficiency of infectious virus production and egress from infected cells.
The number of virus particles (enveloped and unenveloped capsids) can be indirectly estimated by determining the number of viral genomes obtained after DNase I treatment (see Materials and Methods). The ΔUL20 virus infection resulted in the least efficient production of infectious virus within cells, followed by infection with the ΔUL11-ΔgM2 virus. All other viruses produced infectious virions with intermediate efficiencies ranging between those of virus strains HSV-1(F), ΔUL20, and ΔUL11-ΔgM2 (Table 2). As expected, particle/PFU ratios in supernatants of infected cells were much lower than those obtained from the cytoplasmic fraction of infected cells for all viruses, with the exception of the ΔUL20 virus, which produced high numbers of noninfectious virion particles in the supernatant.
Table 2.
Determination of viral particle-to-PFU ratios
| Virus | No. of particles/PFU |
Overall defecta | |
|---|---|---|---|
| Cytoplasm | Supernatant | ||
| Wild type | 109 | 6 | + |
| ΔgE | 425 | 4 | ++ |
| gDΔct | 381 | 10 | ++ |
| gDΔct-ΔgE | 855 | 218 | +++ |
| ΔgM2 | 2,355 | 4 | ++ |
| ΔgM2-ΔgE | 417 | 9 | ++ |
| gDΔct-ΔgM2-ΔgE | 1,779 | 12 | +++ |
| ΔUL11 | 590 | 60 | +++ |
| ΔUL11-ΔgM2 | 3,460 | 70 | ++++ |
| ΔUL20 | 8,509 | 1,702b | +++++ |
Takes into account data from Q-PCR, replication kinetics, and plaque size quantification.
Cells infected with HSV-1 ΔUL20 display a more apoptotic phenotype than cells infected with wild-type virus, leading to release of more viral particles into the supernatant.
DISCUSSION
Mature virions acquire their viral envelopes by budding into cytoplasmic membranes originating from the TGN. Multiple interactions between the cytoplasmic portions of viral glycoproteins and tegument proteins facilitate cytoplasmic virion envelopment (reviewed in references 32 and 42) (Fig. 6). The fact that lack of expression of UL11, gD, gE, or gM alone does not drastically affect cytoplasmic virion envelopment has led to the hypothesis that the cytoplasmic portions of glycoproteins gD, gM, and gE and membrane-associated protein UL11 function in a redundant manner to facilitate virion envelopment (16, 32, 37, 42). To directly test this hypothesis and compare the relative importance of viral glycoproteins and membrane proteins in virion envelopment, we generated a set of mutant viruses carrying one or multiple mutations in viral glycoproteins gD, gE, and gM and in UL11 and compared them to viruses lacking expression of UL20. The results presented herein show that lack of expression of the UL20 (or gK) gene causes the most severe inhibition of cytoplasmic virion envelopment in comparison to that for all other viruses lacking expression of one or more of the gD, gE, gM, and UL11 genes.
Fig 6.
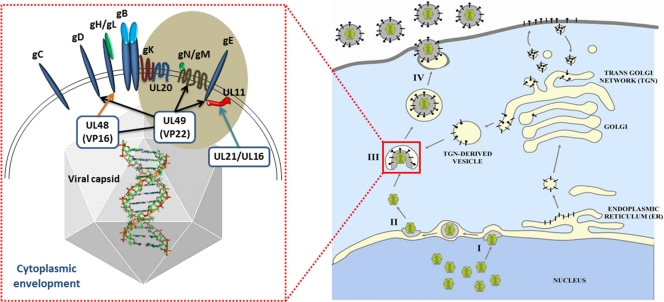
Diagrammatic description of glycoprotein-tegument protein interactions during cytoplasmic virion envelopment at TGN-derived vesicles. (Right) The different steps in virion egress from infected cells: I, budding of nuclear capsids into perinuclear spaces; II, deenvelopment at the outer nuclear membrane and release of capsids into cytoplasm; III, secondary envelopment at TGN membranes; IV, extracellular transport of enveloped virions. Glycoproteins are shown as black bars being synthesized in the endoplasmic reticulum and transported via vesicles to the Golgi apparatus and expressed on infected cell surfaces. (Left) Interactions of tegumented capsids with the carboxyl termini of glycoproteins and membrane proteins: UL49 (VP22) is shown to interact with gD, gM, and gE; UL48 (VP16) interacts with gH and VP22; UL21 and UL16 interact with UL11 (reviewed in references 32 and 42). UL20, UL11, gK, gM, and gE are shown within a brown sphere to highlight their potential cooperative relationships in infectious virion morphogenesis.
Previously, we reported that a recombinant virus lacking expression of gE (ΔgE) and expressing a truncated version of gD (gDΔct) did not exhibit any major defect in cytoplasmic virion envelopment (28). These results suggest that gE and gD do not appear to function in a redundant manner to facilitate cytoplasmic virion envelopment, despite their known interactions with tegument proteins and the membrane-associated protein UL11 (18, 30, 32, 36, 43, 48, 57) (Fig. 6). Deletion of the gM gene resulted in a reduction in the average size of viral plaques produced (5) and an increase in the accumulation of unenveloped capsids in the cytoplasm of HSV-1-infected cells (6, 39). Similar results have been reported for the alphaherpesviruses pseudorabies virus (PRV) and equine herpesvirus 1 (EHV-1) (15, 44). These defects in cytoplasmic virion envelopment are associated, at least in part, with the interactions between gM and the major tegument protein UL49 demonstrated for PRV (26) and HSV-1 (48) (Fig. 6). We show here that simultaneous deletion of the carboxyl terminus of gD and the entire gM and gE genes causes mild reductions in average plaque size, cytoplasmic virion envelopment, and infectious virus production in comparison to those for the virus carrying the gDΔct and ΔgE mutations combined, suggesting that gM does not play a significant redundant role with gD and gE in these events. Direct comparison with the ΔUL20 virus reveals that deletion of the UL20 gene is substantially more deleterious to infectious virus production than the combined effect of the gDΔct, ΔgM2, and ΔgE mutations.
Lack of gM expression caused decreased synthesis of gB, while deletion of the carboxyl-terminal 29 amino acids of gD (gDΔct) caused decreased synthesis of UL11. It was recently shown that the carboxyl terminus of gE interacts with UL11, causing their coincorporation into virion particles (48). gD and gH have been shown to bind to gB and modulate its fusogenicity (1–4, 14, 28). We have shown that gK and UL20 interact with both gB and gH (8). We have recently found that UL20 interacts with gM (V. N. Chouljenko and K. G. Kousoulas, unpublished data), suggesting that that the gK-UL20 and gM-gN complexes interact (Fig. 6). Virion tegument protein VP22 (UL49) interacts with cytoplasmic domains of gD, gE, and gM, as well as its tegument partner, VP16 (UL48) (11, 18, 26). Moreover, VP16 binds to the carboxyl terminus of gH (29). Collectively, these results suggest that there are multiple interactions among viral glycoproteins and tegument proteins and that deletion of one or more viral proteins may indirectly affect the functions and stability of other interacting proteins.
Lack of expression of both the UL11 and gM genes inhibited cytoplasmic virion envelopment in Vero cells to a lesser extent than in other cells, while a similar deletion in PRV caused drastic inhibition of infectious virus production (35, 37). Therefore, it was of interest to generate a similar mutant virus in the HSV-1(F) genetic background to directly compare it with other mutant viruses. Simultaneous deletion of the HSV-1(F) UL11 and gM genes caused a substantial reduction in cytoplasmic virion envelopment and infectious virus production which was greater than that observed in the case of the gDΔct-ΔgM2-ΔgE triple mutant virus. However, the ΔUL11 and ΔgM2 defects were markedly less severe than the ΔUL20 defect, suggesting that UL20 and its interacting partner, gK (21, 24), play more important roles that gD, gM, and gE alone or in combination. Recently, it was reported that gB may function with gD in a redundant manner to facilitate cytoplasmic virion envelopment (26). Therefore, lack of gK or UL20 may affect the binding ability of gB, gD, and gH to bind to tegument proteins. Alternatively, gK and UL20 may directly bind to tegument proteins, facilitating cytoplasmic virion envelopment. Additional experiments are needed to discern the functions of gK and UL20 in cytoplasmic virion envelopment, egress, and infectious virus production.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH, NIAID, grant AI43000 to K.G.K. D. V. Chouljenko is a previous recipient of a Louisiana Board of Regents Economic Development Graduate Assistantship. We acknowledge financial support by the LSU School of Veterinary Medicine to BIOMMED.
We thank Yulia Sokolova and Ying Xiao for technical assistance with electron microscopy and other BIOMMED staff for helpful discussions. We thank Michael T. Kearney for assistance with the statistical analysis and Nithya Jambunathan for reviewing the manuscript.
Footnotes
Published ahead of print 8 February 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Atanasiu D, et al. 2007. Bimolecular complementation reveals that glycoproteins gB and gH/gL of herpes simplex virus interact with each other during cell fusion. Proc. Natl. Acad. Sci. U. S. A. 104:18718–18723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Avitabile E, Forghieri C, Campadelli-Fiume G. 2007. Complexes between herpes simplex virus glycoproteins gD, gB, and gH detected in cells by complementation of split enhanced green fluorescent protein. J. Virol. 81:11532–11537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Avitabile E, Forghieri C, Campadelli-Fiume G. 2009. Cross talk among the glycoproteins involved in herpes simplex virus entry and fusion: the interaction between gB and gH/gL does not necessarily require gD. J. Virol. 83:10752–10760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Avitabile E, Lombardi G, Campadelli-Fiume G. 2003. Herpes simplex virus glycoprotein K, but not its syncytial allele, inhibits cell-cell fusion mediated by the four fusogenic glycoproteins, gD, gB, gH, and gL. J. Virol. 77:6836–6844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baines JD, Roizman B. 1993. The UL10 gene of herpes simplex virus 1 encodes a novel viral glycoprotein, gM, which is present in the virion and in the plasma membrane of infected cells. J. Virol. 67:1441–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Browne H, Bell S, Minson T. 2004. Analysis of the requirement for glycoprotein M in herpes simplex virus type 1 morphogenesis. J. Virol. 78:1039–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Browne H, Bell S, Minson T, Wilson DW. 1996. An endoplasmic reticulum-retained herpes simplex virus glycoprotein H is absent from secreted virions: evidence for reenvelopment during egress. J. Virol. 70:4311–4316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cai WH, Gu B, Person S. 1988. Role of glycoprotein B of herpes simplex virus type 1 in viral entry and cell fusion. J. Virol. 62:2596–2604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Calistri A, et al. 2007. Intracellular trafficking and maturation of herpes simplex virus type 1 gB and virus egress require functional biogenesis of multivesicular bodies. J. Virol. 81:11468–11478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Campadelli-Fiume G, Farabegoli F, Di Gaeta S, Roizman B. 1991. Origin of unenveloped capsids in the cytoplasm of cells infected with herpes simplex virus 1. J. Virol. 65:1589–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chi JH, Harley CA, Mukhopadhyay A, Wilson DW. 2005. The cytoplasmic tail of herpes simplex virus envelope glycoprotein D binds to the tegument protein VP22 and to capsids. J. Gen. Virol. 86:253–261 [DOI] [PubMed] [Google Scholar]
- 12. Chouljenko VN, Iyer AV, Chowdhury S, Chouljenko DV, Kousoulas KG. 2009. The amino terminus of herpes simplex virus type 1 glycoprotein K (gK) modulates gB-mediated virus-induced cell fusion and virion egress. J. Virol. 83:12301–12313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chouljenko VN, Iyer AV, Chowdhury S, Kim J, Kousoulas KG. 2010. The herpes simplex virus type 1 UL20 protein and the amino terminus of glycoprotein K (gK) physically interact with gB. J. Virol. 84:8596–8606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Connolly SA, Jackson JO, Jardetzky TS, Longnecker R. 2011. Fusing structure and function: a structural view of the herpesvirus entry machinery. Nat. Rev. Microbiol. 9:369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dijkstra JM, Mettenleiter TC, Klupp BG. 1997. Intracellular processing of pseudorabies virus glycoprotein M (gM): gM of strain Bartha lacks N-glycosylation. Virology 237:113–122 [DOI] [PubMed] [Google Scholar]
- 16. Farnsworth A, Goldsmith K, Johnson DC. 2003. Herpes simplex virus glycoproteins gD and gE/gI serve essential but redundant functions during acquisition of the virion envelope in the cytoplasm. J. Virol. 77:8481–8494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Farnsworth A, Johnson DC. 2006. Herpes simplex virus gE/gI must accumulate in the trans-Golgi network at early times and then redistribute to cell junctions to promote cell-cell spread. J. Virol. 80:3167–3179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Farnsworth A, Wisner TW, Johnson DC. 2007. Cytoplasmic residues of herpes simplex virus glycoprotein gE required for secondary envelopment and binding of tegument proteins VP22 and UL11 to gE and gD. J. Virol. 81:319–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Forrester A, et al. 1992. Construction and properties of a mutant of herpes simplex virus type 1 with glycoprotein H coding sequences deleted. J. Virol. 66:341–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Foster TP, Alvarez X, Kousoulas KG. 2003. Plasma membrane topology of syncytial domains of herpes simplex virus type 1 glycoprotein K (gK): the UL20 protein enables cell surface localization of gK but not gK-mediated cell-to-cell fusion. J. Virol. 77:499–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Foster TP, Chouljenko VN, Kousoulas KG. 2008. Functional and physical interactions of the herpes simplex virus type 1 UL20 membrane protein with glycoprotein K. J. Virol. 82:6310–6323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Foster TP, Kousoulas KG. 1999. Genetic analysis of the role of herpes simplex virus type 1 glycoprotein K in infectious virus production and egress. J. Virol. 73:8457–8468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Foster TP, Melancon JM, Baines JD, Kousoulas KG. 2004. The herpes simplex virus type 1 UL20 protein modulates membrane fusion events during cytoplasmic virion morphogenesis and virus-induced cell fusion. J. Virol. 78:5347–5357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Foster TP, Melancon JM, Olivier TL, Kousoulas KG. 2004. Herpes simplex virus type 1 glycoprotein K and the UL20 protein are interdependent for intracellular trafficking and trans-Golgi network localization. J. Virol. 78:13262–13277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Foster TP, Rybachuk GV, Kousoulas KG. 2001. Glycoprotein K specified by herpes simplex virus type 1 is expressed on virions as a Golgi complex-dependent glycosylated species and functions in virion entry. J. Virol. 75:12431–12438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fuchs W, et al. 2002. Physical interaction between envelope glycoproteins E and M of pseudorabies virus and the major tegument protein UL49. J. Virol. 76:8208–8217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fulmer PA, Melancon JM, Baines JD, Kousoulas KG. 2007. UL20 protein functions precede and are required for the UL11 functions of herpes simplex virus type 1 cytoplasmic virion envelopment. J. Virol. 81:3097–3108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gianni T, Amasio M, Campadelli-Fiume G. 2009. Herpes simplex virus gD forms distinct complexes with fusion executors gB and gH/gL in part through the C-terminal profusion domain. J. Biol. Chem. 284:17370–17382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gross ST, Harley CA, Wilson DW. 2003. The cytoplasmic tail of herpes simplex virus glycoprotein H binds to the tegument protein VP16 in vitro and in vivo. Virology 317:1–12 [DOI] [PubMed] [Google Scholar]
- 30. Han J, Chadha P, Meckes DG, Jr, Baird NL, Wills JW. 2011. Interaction and interdependent packaging of tegument protein UL11 and glycoprotein E of herpes simplex virus. J. Virol. 85:9437–9446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jayachandra S, Baghian A, Kousoulas KG. 1997. Herpes simplex virus type 1 glycoprotein K is not essential for infectious virus production in actively replicating cells but is required for efficient envelopment and translocation of infectious virions from the cytoplasm to the extracellular space. J. Virol. 71:5012–5024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Johnson DC, Baines JD. 2011. Herpesviruses remodel host membranes for virus egress. Nat. Rev. Microbiol. 9:382–394 [DOI] [PubMed] [Google Scholar]
- 33. Johnson DC, Huber MT. 2002. Directed egress of animal viruses promotes cell-to-cell spread. J. Virol. 76:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Johnson DC, Wisner TW, Wright CC. 2011. Herpes simplex virus glycoproteins gB and gD function in a redundant fashion to promote secondary envelopment. J. Virol. 85:4910–4926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kopp M, Granzow H, Fuchs W, Klupp B, Mettenleiter TC. 2004. Simultaneous deletion of pseudorabies virus tegument protein UL11 and glycoprotein M severely impairs secondary envelopment. J. Virol. 78:3024–3034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lee HC, Chouljenko VN, Chouljenko DV, Boudreaux MJ, Kousoulas KG. 2009. The herpes simplex virus type 1 glycoprotein D (gD) cytoplasmic terminus and full-length gE are not essential and do not function in a redundant manner for cytoplasmic virion envelopment and egress. J. Virol. 83:6115–6124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Leege T, et al. 2009. Effects of simultaneous deletion of pUL11 and glycoprotein M on virion maturation of herpes simplex virus type 1. J. Virol. 83:896–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ligas MW, Johnson DC. 1988. A herpes simplex virus mutant in which glycoprotein D sequences are replaced by beta-galactosidase sequences binds to but is unable to penetrate into cells. J. Virol. 62:1486–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. MacLean CA, Robertson LM, Jamieson FE. 1993. Characterization of the UL10 gene product of herpes simplex virus type 1 and investigation of its role in vivo. J. Gen. Virol. 74(Pt 6):975–983 [DOI] [PubMed] [Google Scholar]
- 40. Melancon JM, Fulmer PA, Kousoulas KG. 2007. The herpes simplex virus UL20 protein functions in glycoprotein K (gK) intracellular transport and virus-induced cell fusion are independent of UL20 functions in cytoplasmic virion envelopment. Virol. J. 4:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Melancon JM, Luna RE, Foster TP, Kousoulas KG. 2005. Herpes simplex virus type 1 gK is required for gB-mediated virus-induced cell fusion, while neither gB and gK nor gB and UL20p function redundantly in virion de-envelopment. J. Virol. 79:299–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mettenleiter TC, Klupp BG, Granzow H. 2009. Herpesvirus assembly: an update. Virus Res. 143:222–234 [DOI] [PubMed] [Google Scholar]
- 43. O'Regan KJ, Bucks MA, Murphy MA, Wills JW, Courtney RJ. 2007. A conserved region of the herpes simplex virus type 1 tegument protein VP22 facilitates interaction with the cytoplasmic tail of glycoprotein E (gE). Virology 358:192–200 [DOI] [PubMed] [Google Scholar]
- 44. Osterrieder N, et al. 1996. The equine herpesvirus 1 glycoprotein gp21/22a, the herpes simplex virus type 1 gM homolog, is involved in virus penetration and cell-to-cell spread of virions. J. Virol. 70:4110–4115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Reynolds AE, Wills EG, Roller RJ, Ryckman BJ, Baines JD. 2002. Ultrastructural localization of the herpes simplex virus type 1 UL31, UL34, and US3 proteins suggests specific roles in primary envelopment and egress of nucleocapsids. J. Virol. 76:8939–8952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sarmiento M, Batterson WW. 1992. Glycerol shock treatment facilitates purification of herpes simplex virus. J. Virol. Methods 36:151–157 [DOI] [PubMed] [Google Scholar]
- 47. Skepper JN, Whiteley A, Browne H, Minson A. 2001. Herpes simplex virus nucleocapsids mature to progeny virions by an envelopment → deenvelopment → reenvelopment pathway. J. Virol. 75:5697–5702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Stylianou J, Maringer K, Cook R, Bernard E, Elliott G. 2009. Virion incorporation of the herpes simplex virus type 1 tegument protein VP22 occurs via glycoprotein E-specific recruitment to the late secretory pathway. J. Virol. 83:5204–5218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Subramanian R, D'Auvergne O, Kong H, Kousoulas KG. 2008. The cytoplasmic terminus of Kaposi's sarcoma-associated herpesvirus glycoprotein B is not essential for virion egress and infectivity. J. Virol. 82:7144–7154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Subramanian R, Sehgal I, D'Auvergne O, Kousoulas KG. 2010. Kaposi's sarcoma-associated herpesvirus glycoproteins B and K8.1 regulate virion egress and synthesis of vascular endothelial growth factor and viral interleukin-6 in BCBL-1 cells. J. Virol. 84:1704–1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sugimoto K, et al. 2008. Simultaneous tracking of capsid, tegument, and envelope protein localization in living cells infected with triply fluorescent herpes simplex virus 1. J. Virol. 82:5198–5211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tanaka M, Kagawa H, Yamanashi Y, Sata T, Kawaguchi Y. 2003. Construction of an excisable bacterial artificial chromosome containing a full-length infectious clone of herpes simplex virus type 1: viruses reconstituted from the clone exhibit wild-type properties in vitro and in vivo. J. Virol. 77:1382–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tischer BK, von Einem J, Kaufer B, Osterrieder N. 2006. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 40:191–197 [DOI] [PubMed] [Google Scholar]
- 54. Turcotte S, Letellier J, Lippe R. 2005. Herpes simplex virus type 1 capsids transit by the trans-Golgi network, where viral glycoproteins accumulate independently of capsid egress. J. Virol. 79:8847–8860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Whiteley A, Bruun B, Minson T, Browne H. 1999. Effects of targeting herpes simplex virus type 1 gD to the endoplasmic reticulum and trans-Golgi network. J. Virol. 73:9515–9520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wisner TW, Johnson DC. 2004. Redistribution of cellular and herpes simplex virus proteins from the trans-Golgi network to cell junctions without enveloped capsids. J. Virol. 78:11519–11535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yeh PC, et al. 2011. Direct and specific binding of the UL16 tegument protein of herpes simplex virus to the cytoplasmic tail of glycoprotein E. J. Virol. 85:9425–9436 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.