Background: Mutations in SIL1 can cause Marinesco-Sjögren syndrome.
Results: Deletion or mutation of the last 5–6 amino acids destabilized SIL1.
Conclusion: KELR does not represent a divergent ER retention sequence but is required for structural stability of the protein.
Significance: These new insights might be a first step toward a possible pharmacological treatment of certain types of MSS by specifically stabilizing the mutant SIL1 protein.
Keywords: Cell Biology, Chaperone Chaperonin, Endoplasmic Reticulum (ER), Exchange, Protein Folding, BiP/Hsp70, ER Protein Folding, Marinesco-Sjogren Syndrome, SIL1/BAP, Nucleotide Exchange Factor
Abstract
Marinesco-Sjögren syndrome (MSS) is an autosomal recessive, neurodegenerative, multisystem disorder characterized by severe phenotypes developing in infancy. Recently, mutations in the endoplasmic reticulum (ER)-associated co-chaperone SIL1/BAP were identified to be the major cause of MSS. SIL1 acts as a nucleotide exchange factor for BiP, the ER Hsp70 orthologue, which plays an essential role in the folding and assembly of nascent polypeptide chains in the ER. SIL1 facilitates the release of BiP from unfolded protein substrates, enabling the subsequent folding and transport of the protein. Although most mutations leading to MSS result in deletion of the majority of the protein, three separate mutations have been identified that disrupt only the last five or six amino acids of the protein, which were assumed to encode a divergent ER retention motif. This study presents an in depth analysis of two of these mutants and reveals that the phenotype in the affected individuals is not likely to be due to depletion of SIL1 from the ER via secretion. Instead, our analyses show that the mutant proteins are particularly unstable and either form large aggregates in the ER or are rapidly degraded via the proteasome. In agreement with our findings, homology modeling suggests that the very C-terminal residues of SIL1 play a role in its structural integrity rather than its localization. These new insights might be a first step toward a possible pharmacological treatment of certain types of MSS by specifically stabilizing the mutant SIL1 protein.
Introduction
The endoplasmic reticulum (ER)5 is a major site of protein biosynthesis within the cell. Proteins destined to be expressed in any of the single membrane-bound organelles, at the cell surface, or secreted are imported into the ER co-translationally as extended polypeptide chains in most eukaryotic cells. Nascent proteins are modified by N-linked glycans and folded into their mature secondary and tertiary structures and form intra- and intermolecular disulfide bonds. These processes are closely monitored by the ER quality control machinery, and once proteins mature properly, they exit the ER for residence further along the secretory pathway (1, 2). As a consequence, the ER contains large quantities of unfolded proteins and unassembled subunits. To maintain ER homeostasis and prevent the aggregation of non-native structures, this organelle contains a particularly high concentration of molecular chaperones and their co-factors. If the nascent protein is ultimately unable to achieve its proper final structure, it can be retrotranslocated to the cytosol for degradation by the 26 S proteasome, as long as the misfolded protein is maintained in a soluble state (3).
The ER possesses members of two major chaperone families; the lectins calnexin and calreticulin (4–6) and the Hsp70 orthologue BiP (7, 8). BiP has been identified as a key regulator of ER function, where it not only plays a role in protein folding and assembly (9, 10) but also maintains the permeability barrier of the ER during translocation (11), targets misfolded proteins for proteasomal degradation (12), regulates the ER localized transducers of the unfolded protein response (13, 14), and contributes to calcium stores within the ER (15). Like all Hsp70 family members, BiP has an N-terminal nucleotide binding domain and a C-terminal substrate binding domain. When ATP occupies the nucleotide binding domain, the substrate binding domain opens so that it can engage nascent proteins (16). Hydrolysis of ATP to ADP results in the lid of the substrate binding domain closing, which protects the unfolded protein from aggregation. The lid is reopened when ATP replaces ADP, thereby allowing the substrate to be released and folded. The ATPase cycle of BiP is tightly regulated by a number of co-factors, including DnaJ-like proteins that stimulate ATP hydrolysis (17, 18) and nucleotide exchange factors that release ADP and allow ATP to rebind. Two nucleotide exchange factors have been identified in the mammalian ER to date: SIL1/BAP (19) and GRP170/ORP150 (20).
A large and ever growing number of protein folding diseases are known to occur due to mutations in a single client protein (21). These mutations can lead to loss of function or the production of unstable proteins that are either aggregation-prone or rapidly degraded or proteins that fail to pass ER quality control and therefore never make it to their necessary destination. It seemed unlikely that components of the ER chaperone machinery could be targeted by mutation because it was thought that they were too essential for cell viability. For instance, BiP knock-out mice die at embryonic day 3.5 (22), and the AB5 subtilase cytotoxin exerts its lethal effects by cleaving BiP between the nucleotide binding domain and the substrate binding domain (23), thus presumably deregulating the binding and release of BiP to client proteins. Therefore, it came as no small surprise when linkage disequilibrium studies mapped MSS disease-causing mutations to the SIL1 gene (24, 25). Although affected individuals show multisystem defects, not all tissues are affected equally, and even mutations that totally disrupt the SIL1 gene are not incompatible with life in either humans or mouse models (26). To date, 17 independent mutations have been identified in the SIL1 gene (24, 25, 27–29). Many of these would be expected to result in deletion of the majority of the protein or at least those portions of the protein that are predicted to interact with BiP. In addition, three independent disease-causing mutations were identified in the SIL1 gene that disrupt only the last five or six amino acids, LLKELR (25, 28).
A large number of soluble resident ER proteins are retained in the ER by virtue of their last four amino acids, usually KDEL (30). If proteins possessing these amino acids at their C terminus escape from the ER, they engage a KDEL receptor in the intermediate compartment or cis-Golgi and are rapidly returned to the ER. A number of variations in this sequence are found on some resident ER proteins (31), arguing that the receptor is more promiscuous in its recognition than originally thought (30). This point was further demonstrated by a study in which 77 variants of the KDEL sequence were produced by altering a single amino acid at a time and examined for their ability to retain a reporter protein (31). It was found that 35 of these variants readily led to ER localization of the reporter. Targeted deletion of the “KDEL” retention sequence from resident ER chaperones can lead to their inappropriate secretion and, as a result, their depletion from this organelle (30). Thus, it was hypothesized that the KELR sequence of SIL1 represented a particularly divergent ER retention sequence (19) and furthermore that mutations that affected this sequence caused MSS due to the failure to retain SIL1 in the ER. To determine if indeed this was the case, we produced two of these mutations in the human SIL1 gene and characterized the mutant proteins. We found that deletion or mutation of the C-terminal SIL1 sequence does not lead to secretion of the protein but instead that it is critical for maintaining the stability of SIL1 so that it can function as an exchange factor for BiP. In keeping with this, modeling of the SIL1 structure suggests that its C-terminal region is structured and forms a number of stabilizing contacts with the rest of the molecule.
EXPERIMENTAL PROCEDURES
Site-directed Mutagenesis of Human SIL1 cDNA
Site-directed mutagenesis of a human SIL1 cDNA clone (19) was conducted using the QuikChange® XL site-directed mutagenesis kit (Stratagene) either to delete thymidine 1366 of the SIL1 coding sequence (Δ1366) or to exchange thymidine 1370 to cytidine, altering the C-terminal protein sequence from LLKELR to LPKELR. After sequencing to verify the mutation, the wild-type and mutated SIL1 cDNAs were inserted into both the DSL (a kind gift from Dr. Daesik Lim, St. Jude Children's Research Hospital) and pSVL (Amersham Biosciences) vectors for expression in mammalian cells.
Cell Culture
293T and COS-1 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (v/v) fetal bovine serum, 2 mm l-glutamine, and a 1% (v/v) antibiotic-antimycotic solution (25 μg/ml amphotenicin B, 10,000 μg/ml streptomycin, and 10,000 units of penicillin; Cellgro, Mediatech, Inc.) at 5 and 3% CO2, respectively.
Cell Transfection
293T cells or COS-1 cells were seeded 24 h prior to transfection. In both cases, cells were transfected with a total of 2 μg of DNA. The calcium chloride method (32) was used for 293T cells. For COS-1 cells, either the FuGENE reagent (Roche Applied Science) or Genecellin (BioCellChallenge) was used, following the manufacturer's protocol. Where indicated, hamster BiP was co-expressed using a pMT vector.
Metabolic Labeling and Immunoprecipitation
Forty-eight hours post-transfection, cells were starved for 45 min in 1 ml of DMEM labeling medium (Fisher) supplemented with 10% dialyzed FBS (HyClone), 2 mm l-glutamine, 1% (v/v) antibiotic-antimycotic solution and then pulse-labeled with 100 μCi of [35S]methionine/cysteine (ICN Radiochemicals). After labeling, dishes were washed twice with 1 ml of PBS on ice and then chased for the indicated times in complete DMEM supplemented with 2 mm cold methionine and cysteine. Medium from the final time point was collected to monitor secretion of SIL1. For proteasome inhibition experiments, 10 μm MG132 was added 2 h prior to the starvation period and included throughout the metabolic labeling and chase. For lysosomal inhibition, 10 mm NH4Cl was added during the 45-min starvation period and throughout the labeling and chase. To determine glycosylation patterns of wild-type and mutant SIL1, cells were preincubated with 2.5 μg/ml tunicamycin (Sigma-Aldrich) for 30 min and labeled with [35S]methionine/cysteine for 2 h.
Preparation of Nonidet P-40-soluble and -insoluble Fractions for Immunoprecipitation
Following labeling, cells were washed twice with PBS, and each p60 dish was lysed in 1 ml of Nonidet P-40 lysis buffer (0.05 m Tris-HCl, pH 7.5, 0.15 m NaCl, 0.5% deoxycholic acid, and 0.5% Nonidet P-40) supplemented with 0.25 mm phenylmethylsulfonyl fluoride, complete EDTA free protease inhibitor tablets (Roche Applied Science), and 10 units/ml apyrase (Sigma-Aldrich). Cell lysates were collected into Eppendorf tubes and centrifuged for 15 min at 14,000 rpm at 4 °C. The supernatant (Nonidet P-40-soluble) and pellet (Nonidet P-40-insoluble) were separated and stored frozen at −80 °C until further use. Nonidet P-40-insoluble pellets were thawed, and 50 μl of SDS solubilization buffer (50 mm Tris-HCl, pH 8.0, with 2% SDS) was added. Pellets were heated to 95 °C for 20 min and regularly agitated to solubilize the Nonidet P-40-insoluble material. The 2% SDS solution was then diluted to 1 ml with Nonidet P-40 lysis buffer for immunoprecipitation. Both Nonidet P-40-soluble and -insoluble fractions were immunoprecipitated with a polyclonal anti-SIL1 antiserum (19) and Protein A-Sepharose beads and subjected to SDS-PAGE analysis. Signals were enhanced with Amplify (Amersham Biosciences) and visualized by autoradiography or quantitated by phosphorimaging, and the bands present in the chase samples were expressed as a percentage of the signal present in the pulse lane.
Immunofluorescence Staining
COS-1 cells were cultured on coverslips in p35 dishes and transfected as indicated. Forty-eight hours after transfection, slides were washed twice with cold PBS and then blocked with 5% FBS in PBS for 5 min. Slides were washed once more with PBS, fixed in an acid/alcohol (95% pure ethanol, 5% glacial acetic acid) solution at −20 °C for 30 min, rehydrated in PBS, and stored at 4 °C. Cells were stained using a polyclonal rabbit anti-SIL1 antiserum and mouse anti-ERp57 (a kind gift of Dr. A. Tsuru, Nara Institute of Science & Technology) as an ER marker. Cy3-conjugated goat anti-mouse IgG (Jackson ImmunoResearch) was used to detect ERp57 and FITC-conjugated goat anti-rabbit IgG (Southern Biotech) to detect SIL1. Slides were mounted using VectaShield (Vector Laboratories) containing 10 μg/ml DAPI and visualized by fluorescence microscopy (Olympus BX50). Photos were taken with a Sensys camera (Photometrics), and images were processed by V++ digital imaging software (Roper Scientific, Auckland, New Zealand) and Photoshop (Adobe).
Structural Modeling and Secondary Structure Prediction
A homology model of SIL1 was built with YASARA Structure (available on the World Wide Web) based on the available human HspBP1 core domain crystal structures (Protein Data Bank codes 1XQR and 1XQS). A hybrid model of different individual models, which gave the overall best Z-score in YASARA, was used for further analysis. Structural alignments were generated with the MUSTANG algorithm implemented in YASARA Structure (33). Prediction of α-helical propensity for the different peptides was performed with AGADIR (34) at pH 7.5, 0.15 m ionic strength, at 37 °C, 310 K, with an acetylated N terminus and an amidated C terminus. Data analysis was performed with IGOR Pro (available from the WaveMetrics Web site).
RESULTS
Generation of cDNA Encoding the MSS-associated Δ1366 Mutant
The vast majority of SIL1 mutations that cause Marinesco-Sjögren syndrome result in the deletion of large portions of the protein that would be expected to affect the predicted BiP-interacting regions encoded by exons 6 and 9. However, three equally severe disease mutations have been identified that only affect or delete the last six amino acids of SIL1 in two cases (25, 28) or introduce a proline immediately before the KELR sequence in the third (28). One of the first type of mutants arises due to deletion of the thymidine at base pair 1366 (Δ1366) of the coding sequence. The resulting frameshift causes a codon change from leucine to cysteine, which is immediately followed by a stop codon, thereby affecting only the last six amino acids of the SIL1 protein (Fig. 1). The second mutation is a base change from thymidine to adenosine at base pair 1367 (1367T→A), which alters the leucine codon to a stop codon (Fig. 1). Thus, the two mutations differ only by the presence of a C-terminal cysteine on the Δ1366 mutant, and both lack the KELR sequence, which was presumed to encode the ER retention motif. The third disease-associated change in this region is the mutation of thymidine 1370 to cytidine (1370T→C). This does not alter the C-terminal KELR sequence but replaces the leucine residue immediately preceding it with a proline (Fig. 1). The Δ1366 mutant and the 1370T→C mutants were used for this study because they represent two different classes of C-terminal SIL1 mutations: complete deletion (Δ1366) or point mutation (1370T→C) of the putative ER retention sequence or its immediate environment.
FIGURE 1.
C-terminal SIL1 mutants that are associated with Marinesco-Sjögren syndrome. Nucleotide and amino acid sequence of the C terminus of wild-type human SIL1 and three mutations found in MSS patients. The mutation site in each of the mutants is in boldface type. In the Δ1366 mutant, deletion of thymidine (asterisk) causes a frameshift, thus changing leucine to cysteine, followed by a stop codon. In the 1367T→A mutant, this same leucine becomes a stop codon. The putative ER retention motif, KELR, is indicated at the very C terminus of wild-type SIL1 and is missing from the first two mutants. The substitution in the 1370T→C mutant changes the leucine immediately preceding KELR to a proline.
SIL1 Δ1366 Is Not Secreted from Cells but Instead Is Aggregation-prone
The deletion or mutation of the last six amino acids would not be expected to directly affect the interaction of SIL1 with BiP based on the crystal structure of the HspBP1-Hsc70 complex (35) or the recently published yeast SIL1-BiP complex (36). Instead, it was assumed that these mutations affected the retention of SIL1, leading to its depletion from the ER. To determine if indeed this was the case, pulse-chase experiments were conducted on both wild-type and mutant SIL1 Δ1366. Somewhat unexpectedly, overexpression of wild-type SIL1 resulted in the secretion of a significant portion of the labeled pool of this protein (Fig. 2A). This is not unprecedented because overexpression of resident ER proteins can result in their secretion, demonstrating that ER retention and KDEL receptor interactions can be saturated (37). When we examined the Δ1366 mutant in the same way, we observed a similar loss of signal over the time course of our chase (Fig. 2A). However, there was no evidence that the mutant SIL1 was being secreted. Alternatively, it was possible that the disappearance of mutant SIL1 was due to the mutation destabilizing the protein, leading to either aggregation or degradation. To determine whether the Δ1366 mutant might be forming Nonidet P-40-insoluble aggregates, we solubilized proteins in the lysate pellet with SDS, diluted the sample, and immunoprecipitated with anti-SIL1 antiserum. Although the overexpressed wild-type protein showed almost no evidence of aggregate formation, significant quantities of mutant SIL1 were found in the Nonidet P-40-insoluble fraction (Fig. 2B), demonstrating that the last six amino acids were important for maintaining the solubility of SIL1.
FIGURE 2.
Loss of the KELR sequence does not lead to SIL1 secretion, but instead affects its stability. COS-1 cells were transfected with either wild-type or Δ1366 mutant SIL1 constructs. Forty-eight hours later, cells were pulse-labeled with [35S]methionine/cysteine for 45 min and chased for the indicated times. Culture supernatants were collected after the 6-h chase point (6m). Nonidet P-40-soluble (A) and -insoluble (B) portions of the cell lysate were prepared as described under “Experimental Procedures” and immunoprecipitated with a polyclonal anti-SIL1 antiserum and subjected to reducing SDS-PAGE analysis. The signal present in the chase time points were quantified by PhosphorImager analysis, expressed as a percentage of that at the 0 time point, and indicated below the individual lanes (A).
The Δ1366 Mutant SIL1 Is Excluded from Perinuclear/Golgi Region and Forms Large Aggregates in ER
Because no evidence of secretion could be observed for the Δ1366 mutant, the subcellular localization of both wild-type and mutant was determined. An antibody to the resident chaperone ERp57 was used to identify the ER. Although wild-type SIL1 co-localized with ERp57 and showed the same characteristic reticular staining pattern, there was a pool of SIL1 that could be readily detected in the perinuclear region that did not contain ERp57 (Fig. 3A). This pool probably represents Golgi-localized SIL1 that is being processed for secretion. Thus, although ER retention systems can be exceeded, there was no evidence that localization of ERp57 was also affected. When the Δ1366 mutant was similarly examined, we found that the signal nearly completely overlapped with that of ERp57 and was excluded from the perinuclear region (Fig. 3B). However, the retained mutant SIL1 did not exhibit the normal reticular pattern associated with ER staining and in fact appeared to affect ERp57 staining as well. The pattern most closely resembled that of large aggregate formation in keeping with the data in Fig. 2B.
FIGURE 3.
The overexpressed Δ1366 mutant aggregates in the ER, whereas the wild-type protein is observed throughout the secretory pathway. COS-1 cells growing on coverslips were transfected with wild-type SIL1 (A) or the Δ1366 mutant (B). After 48 h, cells were fixed and stained for SIL1 (green) or ERp57 (red) as described under “Experimental Procedures.” Nuclei were detected with DAPI staining. The perinuclear area is indicated with a white solid arrow in the top panel, showing cells expressing wild-type SIL1, and with a white broken arrow in the bottom panel, showing cells expressing the Δ1366 mutant.
Overexpression of BiP Prevents Secretion of Wild-type SIL1
Because SIL1 is a co-factor for BiP, we next determined if its interaction with BiP might normally prevent it from being secreted. To examine this possibility, we co-transfected cells with SIL1 and an increasing amount of a vector encoding hamster BiP. Increasing BiP levels dramatically reduced the secretion of wild-type SIL1 from the cells (Fig. 4), arguing that its interaction with BiP may provide the mechanism for its retention in the ER and that saturation of KDEL receptor interactions is unlikely to be the reason for SIL1 secretion. When the Δ1366 mutant was examined in the same way, there was still no evidence of mutant SIL1 secretion. However, it was noted that although the signal for the wild-type SIL1 protein was more stable now that it was retained in the cells, the signal for the mutant protein disappeared at a similar or even slightly faster rate when BiP was overexpressed (Fig. 4).
FIGURE 4.
Co-expression of BiP reduces secretion of wild-type SIL1 in a dose-dependent manner but does not lead to secretion of the Δ1366 mutant. 293T cells were co-transfected with wild-type SIL1 or the Δ1366 mutant and increasing quantities of a hamster BiP construct. After 48 h, cells were pulse-labeled with [35S]methionine/cysteine for 45 min and chased for the indicated times. Culture supernatants were collected from the 6-h chase point (6m). Nonidet P-40 lysates were prepared and immunoprecipitated with a polyclonal anti-SIL1 serum and Protein A-Sepharose beads. The ratio of BiP to each of the two SIL1 vectors is indicated to the left of the panels. The signals present in the chase time points were quantified by PhosphorImager analysis, expressed as a percentage of that at the 0 time point and indicated below the individual lanes.
Overexpression of BiP Reduces Aggregation of Δ1366 Mutant but Increases Its Degradation
The failure to observe secretion of the Δ1366 mutant in the above experiment could be due to the fact that it was still aggregating even in the presence of BiP overexpression. If this was the case, we would not be able to draw any conclusions about a requirement for the KELR sequence in SIL1 retention. Thus, we examined the solubility of the mutant SIL1 when it was co-expressed with BiP and found that BiP overexpression dramatically inhibited its aggregation in a Nonidet P-40 solubility assay (data not shown). This point was further underscored by examining the localization pattern of the Δ1366 mutant in cells also overexpressing BiP. We found that BiP restored a more reticular staining pattern to the mutant protein and that it was still excluded from the perinuclear region (Fig. 5). There was no longer evidence of the large aggregate pattern seen without BiP overexpression. Because the pulse-chase experiment in Fig. 4 suggested that the signal for the Δ1366 mutant was disappearing even faster in the presence of BiP and that this was not because of enhanced aggregation (Fig. 5), we examined the possibility that the solubilized Δ1366 mutant was being recognized as an unfolded protein and targeted for degradation. To test this possibility, cells co-expressing the SIL1 mutant with BiP were subjected to pulse-chase experiments alone, in the presence of MG132, a proteasome inhibitor, or with NH4Cl, a lysosomal inhibitor. Indeed, the mutant protein was stabilized in the presence of MG132 (Fig. 6), arguing that the mutation affects the folding of the Δ1366 mutant, which leads to its degradation by the proteasome. Although in this experiment, the signal for the mutant protein was lower when cells were preincubated with MG132 to inhibit the proteasome, this decrease in expression was not consistent, and it was also observed in some instances with the wild-type protein.
FIGURE 5.
Co-expression of BiP reduces aggregation of the Δ1366 mutant but does not promote its further trafficking along the secretory pathway. COS-1 cells were transfected with the Δ1366 mutant alone (A) or with BiP (1:1.5) (B) and analyzed by immunofluorescence staining as in Fig. 3. The staining pattern of mutant SIL1 (green) and ERp57 (red), an ER marker, are shown. The perinuclear region corresponding to the Golgi is indicated with a white arrow in the bottom panel.
FIGURE 6.
The BiP-solubilized Δ1366 mutant is rapidly degraded by the proteasome. 293T cells were co-transfected with BiP and the Δ1366 mutant, and 48 h later, cells were preincubated with or without the indicated inhibitors prior to metabolic labeling. Cells were pulse-labeled with [35S]methionine/cysteine and chased for the indicated times with the appropriate inhibitors. Cell lysates were prepared with Nonidet P-40 lysis buffer, immunoprecipitated with polyclonal anti-SIL1 antiserum, and analyzed by reducing SDS-PAGE (A), or Nonidet P-40-insoluble material was solubilized with SDS, diluted in Nonidet P-40 buffer, and immunoprecipitated (B).
Structural Modeling of the Human SIL1 Protein Suggests a Stabilizing Role for Its C-terminal Residues
Our data implied that the KELR sequence of SIL1 does not represent an unusually degenerate ER retention motif but might instead play a role in the structural integrity of the protein. To address this hypothesis, we generated a structural homology model for SIL1 based on the published crystal structure of human HspBP1 (35), which is a cytosolic orthologue of SIL1 (38). Importantly, this structure corresponds to the C-terminal portion of SIL1 and allowed us to generate a model that included the KELR sequence (Fig. 7). Although a structure for the yeast SIL1 protein complexed with BiP was recently published, we were unable to verify our model on a perhaps more relevant protein because the structure of yeast SIL1 is missing too many residues at the C terminus (36).
FIGURE 7.
Structural modeling of MSS-causing mutations in SIL1 and assessment of the 1370T→C mutant. A, an overlay of human HspBP1 (orange; Protein Data Bank code 1XQR) with the modeled structure of human SIL1 (helices red, sheets cyan, loops gray) is shown. The C-terminal region of SIL1 is boxed in green, and the N and C terminus are indicated. C and D, front and side views of the C-terminal region of the human SIL1 model, respectively. In A, C, and D, the backbone of the last six residues is shown in blue, whereas their side chains and the residues they are predicted to interact with on the rest of the structure are shown in a stick representation. Hydrogen bonds and salt bridges are highlighted as dashed yellow lines. B, AGADIR prediction of the helicity of the last α-helix (Glu-443 to Arg-461) of human SIL1. Data for wild-type SIL1 are shown in black, data for Δ1366 are in blue, data for 1367T→A are in green, and data for 1370T→C are in red. E, COS-1 cells were transfected with the 1370T→C SIL1 mutant construct. Forty-eight hours later, cells were pulse-labeled with [35S]methionine/cysteine for 45 min and chased for the indicated times. Culture supernatants were collected after the 6-h chase point (6m). Nonidet P-40-soluble (top) and -insoluble (bottom) portions of the cell lysate were prepared as described under “Experimental Procedures,” immunoprecipitated with a polyclonal anti-SIL1 antiserum, and subjected to reducing SDS-PAGE analysis. F, pulse-chase experiments were performed on the 1370T→C SIL1 mutant in cells that had been preincubated with or without the indicated inhibitors prior to metabolic labeling, and the 0- and 4-h chase samples were immunoprecipitated and analyzed by SDS-PAGE. The signals were quantified by PhosphorImager analysis, and protein remaining after 4 h of chase was expressed as a percentage of that present at the end of the labeling period.
It is evident from Fig. 7 that the KELR sequence of SIL1 is not predicted to be unstructured. Instead, it appears to make up the C-terminal portion of an α-helix that is predicted to comprise the last 19 residues of the protein (Fig. 7). An analysis of the helical propensity for these 19 residues with AGADIR (34) predicts an overall low helical propensity (Fig. 7B). Of note, the SIL1 deletion mutants, Δ1366 or 1367T→A, and the Leu to Pro point mutant (1370T→C) are significantly less likely to form a helix by this analysis (Fig. 7B). The overall low helical propensity of the last 19 residues of SIL1 suggests that this helix might depend on tertiary interactions in order to be formed or stabilized. Indeed, in the two deletion mutants (Δ1366 and 1367T→A), three leucine residues are missing, which, based on the structural model of SIL1, are involved in tertiary interactions with an adjacent helix, thereby stabilizing the last helix (Fig. 7, C and D). Furthermore, in the two deletion mutants, the very last residue, Arg-461, is missing, which, due to its positive charge, would be predicted to normally stabilize the helix dipole of the final helix in the wild-type SIL1 protein. Additionally, the side chain of Arg-461 is predicted to directly form stabilizing interactions with the oppositely charged side chain of Glu-428 and with the backbone carbonyl oxygen of Ala-423. The first of these three leucines is mutated to proline in the 1370T→C mutant. In addition to the well characterized helix-breaking properties of prolines, based on our modeling, this substitution would probably affect the interactions between the C-terminal helix and the rest of the molecule in this mutant. In toto, the structural model of human SIL1 predicts that the C-terminal residues affected in three different MSS mutations contribute to the stability of the C-terminal helix.
To test our hypothesis and the validity of our structural model, we generated the 1370T→C mutant of SIL1. This mutant is particularly well suited in this regard because it is predicted to interfere with the formation of a C-terminal helix but not with the putative ER retention sequence directly (Fig. 1). In pulse-chase experiments, we found that the 1370T→C mutant behaved similarly to the Δ1366 mutant in that it was not secreted and turned over rapidly (Fig. 7E). However, in contrast to the Δ1366 mutant, there was little or no formation of SDS-soluble, Nonidet P-40-insoluble aggregates (Fig. 7E). Thus, the 1370T→C mutant is not secreted but does not form aggregates like the Δ1366 mutant, arguing against the mutation disrupting an ER retention motif and suggesting degradation of this mutant in a soluble form. Indeed, treatment with MG132 inhibited turnover of the 1370T→C mutant, whereas NH4Cl did not (Fig. 7F). This suggests that the 1370T→C mutant is recognized as being a misfolded protein and degraded by the proteasome, which is in very good agreement with our hypothesis that the C-terminal SIL1 residues are important for the structural integrity of the protein rather than for its ER retention.
The C-terminal Cysteine Generates an Unusual N-Linked Glycosylation Sequence and Induces Dimerization of Δ1366 Mutant
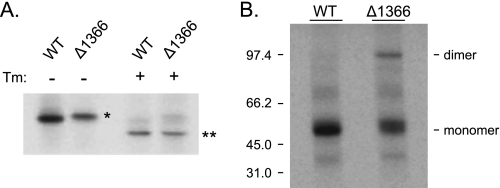
In a number of our experiments, we noticed that the Δ1366 mutant ran somewhat more slowly on SDS-polyacrylamide gels than the wild-type protein. The loss of five amino acids was unlikely to significantly affect the migration of the protein, but changes in charge can affect the binding of SDS, leading to aberrant migration. When we inspected the sequence more closely, we noted that the C-terminal sequence was now NSC, which has recently been shown to encode an N-linked glycosylation site in a few proteins (39, 40). To assess whether, in fact, this was the case, we treated cells expressing wild-type and the Δ1366 mutant with tunicamycin and performed metabolic labeling experiments. In the absence of tunicamycin, the migration of the Δ1366 mutant was slightly slower than that of the wild-type protein (Fig. 8A). Tunicamycin treatment resulted in the wild-type protein migrating faster, in keeping with the presence of two conventional N-linked glycosylation sites on SIL1. When the Δ1366 mutant was similarly examined, we found that the non-glycosylated protein now migrated the same as wild-type SIL1 (Fig. 8A), demonstrating that the altered mobility was due to the glycosylation of this unusual sequence and not to changes in SDS binding.
FIGURE 8.
The deletion of thymidine 1366 creates a novel N-linked glycosylation site and induces disulfide-mediated dimerization of the mutant. A, COS-1 cells expressing wild-type SIL1 or the Δ1366 mutant were preincubated with or without tunicamycin 30 min prior to metabolic labeling and throughout the 2-h labeling period with [35S]methionine/cysteine. Cell lysates were prepared and immunoprecipitated with anti-SIL1 antiserum and analyzed by reducing SDS-PAGE. *, mobility of the Δ1366 mutant in the absence of tunicamycin (Tm); **, its mobility after glycosylation is inhibited by tunicamycin treatment. B, COS-1 cells were co-transfected with BiP and either wild-type or mutant SIL1 and then metabolically labeled for 2 h. Cell lysates and culture supernatants were collected for immunoprecipitation. Isolated proteins were analyzed by non-reducing SDS-PAGE. The migration of monomers and dimers is indicated.
The presence of this terminal cysteine was also likely to provide an unpaired cysteine that could now cause the mutant protein to be retained in the ER by thiol-mediated retention mechanisms (41). To determine whether indeed this cysteine remained unpaired, we analyzed the wild-type and mutant protein under non-reducing conditions. As expected, the wild-type protein existed in cells predominantly as a monomer in the presence of BiP overexpression (Fig. 8B). When the Δ1366 mutant was examined under the same conditions, we found that a significant portion of the mutant existed in the form of disulfide-linked dimers, whereas the remainder migrated as a monomer (Fig. 8B). Thus, the inability of the Δ1366 mutant to be secreted even after solubilization is unlikely to be due to either thiol-mediated retention mechanisms or to disulfide-linked dimers leading to aggregation. Instead, it is likely that this mutant is retained in the ER via its association with BiP, but perhaps as an unfolded protein instead of a normal co-factor.
DISCUSSION
In 1987, Munro and Pelham (30) noted that a number of soluble, resident ER proteins possess the tetrapeptide, KDEL, at their C terminus and postulated that this sequence might be responsible for preventing their secretion. Indeed, by engineering this sequence at the C terminus of ovalbumin, they were able to prevent its secretion. Correspondingly, removing the sequence from BiP allowed it to be secreted, although the effects of a KDEL-less mutant on secretion were much less robust than the ability of this sequence to prevent the transport of ovalbumin (30). A number of somewhat divergent examples of this tetrapeptide have been identified on resident ER proteins since its identification, arguing that the KDEL receptor can be promiscuous to a certain extent in identifying C-terminal tetrapeptides as retention motifs. One study produced 77 variants of this sequence by changing one amino acid at a time and examined their ability to retain a reporter protein in the ER (31). They found that the −4 (lysine) and −3 (aspartic acid) residues could be quite variable and that the −1 (leucine) could be substituted with either methionine or phenylalanine and work nearly as well. However, the −2 (glutamic acid) residue was critical in the context of a KDEL sequence, and even the substitution of this residue with an aspartic acid dramatically reduced the ability of the sequence to retain the reporter. However, it is important to note that the KELR sequence has never been directly tested as a retention sequence, and inspection of SIL1 sequences from primates and rodents reveals that it is completely conserved. However it is quite different in more distal species, some of which actually possess a bona fide KDEL-type ER retention sequence (28), arguing for divergent evolution.
A recent study used immunofluorescence staining to examine the localization of the wild-type SIL1 protein and two MSS mutants, 1367T→A and 1370T→C, all of which were tagged at their extreme C terminus with an HA epitope (28). The mutant proteins showed a propensity to aggregate and co-localize with ER markers, whereas the wild-type protein could be detected in the Golgi as well as the ER, leading the authors to conclude that KELR may represent a retention sequence that was weakened during evolution (28). Our studies using untagged forms of SIL1 confirmed that the KELR sequence is not responsible for retaining it in the ER and provide a more detailed structural explanation for the behavior of all three C-terminal SIL1 mutants that have been identified in individuals with MSS as discussed below. In addition, our study provides insights into the mechanism by which wild-type SIL1 is usually retained in the ER.
Although endogenous SIL1 is retained in the ER (19), in the present study, we observed secretion of transfected SIL1 when it was expressed from a strong promoter. Our finding that overexpression of BiP suppressed the secretion of wild-type SIL1 argues that this is likely to be the normal mechanism of SIL1 retention. It is very possible that this mechanism extends to other soluble ER-resident proteins that do not have ER retention sequences, including many of the other ER-localized DnaJ-like proteins (42). In keeping with this, studies to measure the concentration of BiP and resident ER-localized DnaJ-like proteins found that the concentration of BiP was significantly greater than the combined concentration of the ERdj proteins (43). Because GRP94 possesses a KDEL tail and is one of the most abundant resident ER proteins, it is possible that this chaperone also contributes to the retention of other resident proteins that do not possess a KDEL sequence.
Our data strongly suggest that the loss of the last five amino acids of SIL1 in the Δ1366 mutant affects its folding, leading to loss of function, based on the following criteria. First, the Δ1366 mutant readily aggregates when overexpressed, whereas the wild-type SIL1 protein does not. Second, overexpression of BiP is able to solubilize the mutant protein, which is a characteristic of molecular chaperones binding to misfolded proteins. Finally, once solubilized, the Δ1366 mutant protein is degraded faster than the wild-type protein, which is another characteristic of misfolded proteins. These conclusions are further supported by the structural model we derived for human SIL1. This model suggests that, at least in the case of rodent and primate SIL1, the last few C-terminal residues are not unstructured and exposed, as would be expected for a retention sequence, although it is very possible that it is the case in species like yeast and mosquitoes that have a more conventional ER retention sequence. Instead, our model predicts that they are part of a longer α-helix and contribute to its stability through hydrophobic interactions, hydrogen bonds, and salt bridges with other portions of the protein. Deletion of these last residues or insertion of a proline as a helix breaker would thus be expected to compromise the overall ability of the last α-helix to form. This would in turn lead to exposed hydrophobic residues within this now partially unstructured final α-helix as well as in the region of SIL1 where this α-helix would normally dock. Indeed, we find the 1370T→C mutant not to be secreted, unlike overexpressed wild-type SIL1, but instead to be degraded by the proteasome. This finding strengthens our hypothesis that the C-terminal SIL1 residues are structurally important and do not represent a degenerate ER retention motif. This model also explains why overexpression of the other two C-terminal disease-associated mutations, which entirely lose their last five or six amino acids, increased their propensity to aggregate. Under normal physiological levels of synthesis, where BiP levels are in excess of the three C-terminal SIL1 mutants, it is likely that they simply turn over more rapidly, leading to their depletion from the ER and the resulting disease phenotype.
A rather unanticipated finding in the characterization of the Δ1366 mutation is that the mutation of leucine 456 to cysteine now encodes an NSC sequence that has recently been shown to serve as an N-linked glycosylation motif on a few proteins (39, 40), instead of the usual NX(S/T) motif, which is the hallmark N-glycan acceptor sequence. Although the addition of sugars usually increases the solubility of proteins, it does not appear to be sufficient to do so in this case. However, this modification is unlikely to contribute solely to the disease-associated phenotypes because the 1367T→A mutation will not be similarly modified and causes an equally severe disease.
In summary, three mutations have been identified that affect only the last few amino acids of SIL1 but that induce a disease as severe as those that affect much larger portions of the protein. This is not due to the disruption of an ER retention sequence as previously suggested but instead to changes in the structure of the remaining protein that affect its stability and/or solubility. Thus, indeed, these mutations lead to loss of SIL1 from the cell but not due to secretion. Instead, our studies indicate that normally this exchange factor is retained in the ER via its association with BiP. Our molecular modeling suggests the importance of the last few amino acids of SIL1 in stabilizing the final α-helix and probably the regions it interacts with and explains why the three C-terminal mutants cause MSS. The fact that our studies indicate that these particular mutations affect the folding of SIL1 while leaving the protein sequence almost completely intact raises the interesting possibility that these mutant proteins, and thus the patients expressing them, could benefit from the type of pharmacological stabilization being pursued for other protein folding disorders (44).
This work was supported, in whole or in part, by National Institutes of Health Grant GM54068 (to L. M. H.) and Cancer Center CORE Grant CA21765. This work was also supported by the American Lebanese Syrian Associated Charities of St. Jude Children's Research Hospital.
- ER
- endoplasmic reticulum
- MSS
- Marinesco-Sjögren syndrome.
REFERENCES
- 1. Hammond C., Helenius A. (1995) Quality control in the secretory pathway. Curr. Opin. Cell Biol. 7, 523–529 [DOI] [PubMed] [Google Scholar]
- 2. Ellgaard L., Helenius A. (2001) ER quality control. Towards an understanding at the molecular level. Curr. Opin. Cell Biol. 13, 431–437 [DOI] [PubMed] [Google Scholar]
- 3. Nishikawa S. I., Fewell S. W., Kato Y., Brodsky J. L., Endo T. (2001) Molecular chaperones in the yeast endoplasmic reticulum maintain the solubility of proteins for retrotranslocation and degradation. J. Cell Biol. 153, 1061–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Helenius A., Aebi M. (2004) Roles of N-linked glycans in the endoplasmic reticulum. Annu. Rev. Biochem. 73, 1019–1049 [DOI] [PubMed] [Google Scholar]
- 5. Moremen K. W., Molinari M. (2006) N-Linked glycan recognition and processing. The molecular basis of endoplasmic reticulum quality control. Curr. Opin. Struct. Biol. 16, 592–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Michalak M., Groenendyk J., Szabo E., Gold L. I., Opas M. (2009) Calreticulin, a multiprocess calcium-buffering chaperone of the endoplasmic reticulum. Biochem. J. 417, 651–666 [DOI] [PubMed] [Google Scholar]
- 7. Haas I. G., Wabl M. (1983) Immunoglobulin heavy chain binding protein. Nature 306, 387–389 [DOI] [PubMed] [Google Scholar]
- 8. Munro S., Pelham H. R. (1986) An Hsp70-like protein in the ER. Identity with the 78-kDa glucose-regulated protein and immunoglobulin heavy chain binding protein. Cell 46, 291–300 [DOI] [PubMed] [Google Scholar]
- 9. Bole D. G., Hendershot L. M., Kearney J. F. (1986) Posttranslational association of immunoglobulin heavy chain binding protein with nascent heavy chains in nonsecreting and secreting hybridomas. J. Cell Biol. 102, 1558–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hendershot L., Bole D., Köhler G., Kearney J. F. (1987) Assembly and secretion of heavy chains that do not associate posttranslationally with immunoglobulin heavy chain-binding protein. J. Cell Biol. 104, 761–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hamman B. D., Hendershot L. M., Johnson A. E. (1998) BiP maintains the permeability barrier of the ER membrane by sealing the lumenal end of the translocon pore before and early in translocation. Cell 92, 747–758 [DOI] [PubMed] [Google Scholar]
- 12. Skowronek M. H., Hendershot L. M., Haas I. G. (1998) The variable domain of nonassembled Ig light chains determines both their half-life and binding to the chaperone BiP. Proc. Natl. Acad. Sci. U.S.A. 95, 1574–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bertolotti A., Zhang Y., Hendershot L. M., Harding H. P., Ron D. (2000) Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat. Cell Biol. 2, 326–332 [DOI] [PubMed] [Google Scholar]
- 14. Shen J., Chen X., Hendershot L., Prywes R. (2002) ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev. Cell 3, 99–111 [DOI] [PubMed] [Google Scholar]
- 15. Lièvremont J. P., Rizzuto R., Hendershot L., Meldolesi J. (1997) BiP, a major chaperone protein of the endoplasmic reticulum lumen, plays a direct and important role in the storage of the rapidly exchanging pool of Ca2+. J. Biol. Chem. 272, 30873–30879 [DOI] [PubMed] [Google Scholar]
- 16. Liberek K., Skowyra D., Zylicz M., Johnson C., Georgopoulos C. (1991) The Escherichia coli DnaK chaperone, the 70-kDa heat shock protein eukaryotic equivalent, changes conformation upon ATP hydrolysis, thus triggering its dissociation from a bound target protein. J. Biol. Chem. 266, 14491–14496 [PubMed] [Google Scholar]
- 17. Liberek K., Marszalek J., Ang D., Georgopoulos C., Zylicz M. (1991) Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proc. Natl. Acad. Sci. U.S.A. 88, 2874–2878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cheetham M. E., Caplan A. J. (1998) Structure, function and evolution of DnaJ. Conservation and adaptation of chaperone function. Cell Stress Chaperones 3, 28–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chung K. T., Shen Y., Hendershot L. M. (2002) BAP, a mammalian BiP-associated protein, is a nucleotide exchange factor that regulates the ATPase activity of BiP. J. Biol. Chem. 277, 47557–47563 [DOI] [PubMed] [Google Scholar]
- 20. Weitzmann A., Volkmer J., Zimmermann R. (2006) The nucleotide exchange factor activity of Grp170 may explain the non-lethal phenotype of loss of Sil1 function in man and mouse. FEBS Lett. 580, 5237–5240 [DOI] [PubMed] [Google Scholar]
- 21. Herczenik E., Gebbink M. F. (2008) Molecular and cellular aspects of protein misfolding and disease. FASEB J. 22, 2115–2133 [DOI] [PubMed] [Google Scholar]
- 22. Luo S., Mao C., Lee B., Lee A. S. (2006) GRP78/BiP is required for cell proliferation and protecting the inner cell mass from apoptosis during early mouse embryonic development. Mol. Cell Biol. 26, 5688–5697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Paton A. W., Beddoe T., Thorpe C. M., Whisstock J. C., Wilce M. C., Rossjohn J., Talbot U. M., Paton J. C. (2006) AB5 subtilase cytotoxin inactivates the endoplasmic reticulum chaperone BiP. Nature 443, 548–552 [DOI] [PubMed] [Google Scholar]
- 24. Anttonen A. K., Mahjneh I., Hämäläinen R. H., Lagier-Tourenne C., Kopra O., Waris L., Anttonen M., Joensuu T., Kalimo H., Paetau A., Tranebjaerg L., Chaigne D., Koenig M., Eeg-Olofsson O., Udd B., Somer M., Somer H., Lehesjoki A. E. (2005) The gene disrupted in Marinesco-Sjögren syndrome encodes SIL1, an HSPA5 cochaperone. Nat. Genet. 37, 1309–1311 [DOI] [PubMed] [Google Scholar]
- 25. Senderek J., Krieger M., Stendel C., Bergmann C., Moser M., Breitbach-Faller N., Rudnik-Schöneborn S., Blaschek A., Wolf N. I., Harting I., North K., Smith J., Muntoni F., Brockington M., Quijano-Roy S., Renault F., Herrmann R., Hendershot L. M., Schröder J. M., Lochmüller H., Topaloglu H., Voit T., Weis J., Ebinger F., Zerres K. (2005) Mutations in SIL1 cause Marinesco-Sjögren syndrome, a cerebellar ataxia with cataract and myopathy. Nat. Genet. 37, 1312–1314 [DOI] [PubMed] [Google Scholar]
- 26. Zhao L., Longo-Guess C., Harris B. S., Lee J. W., Ackerman S. L. (2005) Protein accumulation and neurodegeneration in the woozy mutant mouse is caused by disruption of SIL1, a cochaperone of BiP. Nat. Genet. 37, 974–979 [DOI] [PubMed] [Google Scholar]
- 27. Karim M. A., Parsian A. J., Cleves M. A., Bracey J., Elsayed M. S., Elsobky E., Parsian A. (2006) A novel mutation in BAP/SIL1 gene causes Marinesco-Sjögren syndrome in an extended pedigree. Clin. Genet. 70, 420–423 [DOI] [PubMed] [Google Scholar]
- 28. Anttonen A. K., Siintola E., Tranebjaerg L., Iwata N. K., Bijlsma E. K., Meguro H., Ichikawa Y., Goto J., Kopra O., Lehesjoki A. E. (2008) Novel SIL1 mutations and exclusion of functional candidate genes in Marinesco-Sjögren syndrome. Eur. J. Hum. Genet. 16, 961–969 [DOI] [PubMed] [Google Scholar]
- 29. Eriguchi M., Mizuta H., Kurohara K., Fujitake J., Kuroda Y. (2008) Identification of a new homozygous frameshift insertion mutation in the SIL1 gene in 3 Japanese patients with Marinesco-Sjögren syndrome. J. Neurol. Sci. 270, 197–200 [DOI] [PubMed] [Google Scholar]
- 30. Munro S., Pelham H. R. (1987) A C-terminal signal prevents secretion of luminal ER proteins. Cell 48, 899–907 [DOI] [PubMed] [Google Scholar]
- 31. Raykhel I., Alanen H., Salo K., Jurvansuu J., Nguyen V. D., Latva-Ranta M., Ruddock L. (2007) A molecular specificity code for the three mammalian KDEL receptors. J. Cell Biol. 179, 1193–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sambrook J. F., Russell D. W. (2001) Molecular Cloning: A Laboratory Manual, 3rd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 33. Konagurthu A. S., Whisstock J. C., Stuckey P. J., Lesk A. M. (2006) MUSTANG. A multiple structural alignment algorithm. Proteins 64, 559–574 [DOI] [PubMed] [Google Scholar]
- 34. Muñoz V., Serrano L. (1997) Development of the multiple sequence approximation within the AGADIR model of α-helix formation. Comparison with Zimm-Bragg and Lifson-Roig formalisms. Biopolymers 41, 495–509 [DOI] [PubMed] [Google Scholar]
- 35. Shomura Y., Dragovic Z., Chang H. C., Tzvetkov N., Young J. C., Brodsky J. L., Guerriero V., Hartl F. U., Bracher A. (2005) Regulation of Hsp70 function by HspBP1. Structural analysis reveals an alternate mechanism for Hsp70 nucleotide exchange. Mol. Cell 17, 367–379 [DOI] [PubMed] [Google Scholar]
- 36. Yan M., Li J., Sha B. (2011) Structural analysis of the Sil1-Bip complex reveals the mechanism for Sil1 to function as a nucleotide-exchange factor. Biochem. J. 438, 447–455 [DOI] [PubMed] [Google Scholar]
- 37. Bu G., Rennke S., Geuze H. J. (1997) ERD2 proteins mediate ER retention of the HNEL signal of LRP's receptor-associated protein (RAP). J. Cell Sci. 110, 65–73 [DOI] [PubMed] [Google Scholar]
- 38. Kabani M., McLellan C., Raynes D. A., Guerriero V., Brodsky J. L. (2002) HspBP1, a homologue of the yeast Fes1 and Sls1 proteins, is an Hsc70 nucleotide exchange factor. FEBS Lett. 531, 339–342 [DOI] [PubMed] [Google Scholar]
- 39. Titani K., Kumar S., Takio K., Ericsson L. H., Wade R. D., Ashida K., Walsh K. A., Chopek M. W., Sadler J. E., Fujikawa K. (1986) Amino acid sequence of human von Willebrand factor. Biochemistry 25, 3171–3184 [DOI] [PubMed] [Google Scholar]
- 40. Satomi Y., Shimonishi Y., Takao T. (2004) N-Glycosylation at Asn491 in the Asn-Xaa-Cys motif of human transferrin. FEBS Lett. 576, 51–56 [DOI] [PubMed] [Google Scholar]
- 41. Fra A. M., Fagioli C., Finazzi D., Sitia R., Alberini C. M. (1993) Quality control of ER synthesized proteins. An exposed thiol group as a three-way switch mediating assembly, retention, and degradation. EMBO J. 12, 4755–4761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Otero J. H., Lizák B., Hendershot L. M. (2010) Life and death of a BiP substrate. Semin. Cell Dev. Biol. 21, 472–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Weitzmann A., Baldes C., Dudek J., Zimmermann R. (2007) The heat shock protein 70 molecular chaperone network in the pancreatic endoplasmic reticulum. A quantitative approach. FEBS J. 274, 5175–5187 [DOI] [PubMed] [Google Scholar]
- 44. Powers E. T., Morimoto R. I., Dillin A., Kelly J. W., Balch W. E. (2009) Biological and chemical approaches to diseases of proteostasis deficiency. Annu. Rev. Biochem. 78, 959–991 [DOI] [PubMed] [Google Scholar]