Background: Both SecA and the ribosome need to interact with the translocon during membrane protein insertion.
Results: SecA competes with ribosomes and ribosome-nascent chain complexes for binding to the translocon.
Conclusion: SecA and ribosome binding to the translocon is mutually exclusive, implying that during membrane protein insertion, both ligands bind the translocon in a sequential manner.
Significance: Insight in the mechanism of membrane protein insertion.
Keywords: Membrane Biogenesis, Membrane Proteins, Membrane Transport, Protein Translocation, Single Molecule Biophysics, SecA, SecYEG, Nascent Chain, Ribosome
Abstract
During co-translational membrane insertion of membrane proteins with large periplasmic domains, the bacterial SecYEG complex needs to interact both with the ribosome and the SecA ATPase. Although the binding sites for SecA and the ribosome overlap, it has been suggested that these ligands can interact simultaneously with SecYEG. We used surface plasmon resonance and fluorescence correlation spectroscopy to examine the interaction of SecA and ribosomes with the SecYEG complex present in membrane vesicles and the purified SecYEG complex present in a detergent-solubilized state or reconstituted into nanodiscs. Ribosome binding to the SecYEG complex is strongly stimulated when the ribosomes are charged with nascent chains of the monotopic membrane protein FtsQ. This binding is competed by an excess of SecA, indicating that binding of SecA and ribosomes to SecYEG is mutually exclusive.
Introduction
In Escherichia coli the conserved heterotrimeric membrane protein complex SecYEG (also termed translocon) mediates the transport of secretory proteins across and the insertion of membrane proteins into the cytoplasmic membrane (for review, see Ref. 1). Secretory proteins are mostly targeted post-translationally to the SecYEG complex, after which translocation is driven by the peripheral motor domain, the ATPase SecA. Membrane proteins mostly follow a co-translational targeting route in which ribosome-nascent chain complexes (RNCs)2 are directed to the SecYEG complex by signal recognition particle and its membrane-associated receptor FtsY (for review, see Ref. 2)). After docking of the ribosome on SecYEG, insertion occurs concomitantly with the elongation of the polypeptide chain on the ribosome, whereas translocation of large periplasmic loops requires the assistance of SecA (3, 4).
The initiation of protein translocation or membrane protein insertion is dependent on the high affinity interactions between the SecYEG complex and SecA or the ribosome. In recent years, structural, biochemical, and computational approaches have provided detailed insights in the interaction between SecYEG and its cytosolic binding partners. Both SecA and the ribosome interact primarily with the largest subunit of the SecYEG complex, i.e. SecY. This interaction involves multiple contact points, and the main connections are formed by the ribosomal 23S rRNA, the ribosomal protein L23, and the fourth and fifth cytoplasmic loops (C4 and C5) of SecY (5–9). Substitution of the conserved arginine residues in these SecY loops severely reduces the interaction with the ribosome (7). In addition, the C-terminus of SecY interacts with the ribosomal protein L24, and the N terminus and amphipathic helix of SecE interact with proteins L23 and L29 (7–9). Intriguingly, SecA appears to bind to the same SecY loops that are important for ribosome interaction. Indeed, a recent crystal structure of SecYEG in complex with SecA (10) demonstrated contacts between the SecY C4 and C5 loops and SecA (11, 12). Mutational analysis identified the arginine at position 357 in the C5 loop as indispensable for SecA-mediated translocation (13). However, substitution of Arg-357 does not interfere with SecA binding (11) but rather prevents the SecA-dependent initiation of protein translocation (14).
Although many substrates of the translocon only require the action of either SecA or the ribosome for translocation or membrane insertion, the biogenesis of membrane proteins with large periplasmic loops or domains, such as FtsQ and CyoA, requires both functions (15, 16). Insight in the timing and coordination of these activities is essential for the understanding of the mechanism of membrane protein insertion. It has been suggested that non-translating ribosomes and SecA do not compete for SecYEG binding and thus would be able to bind simultaneously (17). However, the overlapping binding sites (7, 8, 10) and the anticipated steric constraints upon association of these two large ligands make this difficult to envision. Here, we have monitored the binding of SecA, non-translating ribosomes and RNC complexes of the monotopic membrane protein FtsQ to the membrane-embedded and detergent-solubilized SecYEG complex using two different methods, i.e. surface plasmon resonance and fluorescence correlation spectroscopy. Our data demonstrate that SecA and the ribosome compete for binding to the SecYEG complex and that substrate loading is an important determining factor in this process.
EXPERIMENTAL PROCEDURES
Strains and Plasmids
Inner membrane vesicles (IMVs) with endogenous or overexpression levels of SecYEG were isolated from Escherichia coli SF100 transformed with the plasmids indicated in Table 1 as described previously (18). The R357E mutation and R255E,R256E double mutation were introduced into Cys-less SecY by site-directed mutagenesis using pEK20 (19) as template, yielding pNN260 (11) and pZW1, respectively. A unique cysteine at position 148 of SecY was introduced into pZW1 and pNN260 yielding pZW2 and pZW3, respectively.
TABLE 1.
Strains and plasmids used in this study
| Strains/Plasmids | Characteristics | Source |
|---|---|---|
| E. coli SF100 | F −, ΔlacX7, galE, galK, thi, rpsL, strA 4, ΔphoA(pvuII), ΔompT | (54) |
| E. coli BL21(DE3) Δtig | F −, ompT, gal, dcm, lon, hsdSB(rB− mB−) (DE3) Δtig | (24) |
| pEK20 | Cysteine-less SecYEG | (19) |
| pNN260 | Cysteine-less SecY(R357E)EG | (11) |
| pZW1 | Cysteine-less SecY(R255E, R256E) | This study |
| pEK20–148 | SecY(L148C)EG | (27) |
| pZW2 | SecY(L148C, R255E, R256E)EG | This study |
| pZW3 | SecY(L148C, R357E)EG | This study |
| pUC19Strep3FtsQSecM | RNC-FtsQ108 | (20) |
| pEK764 | RNC-FtsQ87 | (21) |
| pEK765 | RNC-FtsQ77 | This study |
| pEK767 | RNC-FtsQ108:ΔTMS | This study |
Plasmid pUC19Strep3FtsQSecM (20) was used for the isolation of FtsQ108 RNCs. For plasmids encoding FtsQ RNCs of 77 or 87 residues and FtsQ with a deletion of the first transmembrane segment (TMS), the PstI-EcoRV fragment of pUC19Strep3FtsQSecM (20) was exchanged with fragments coding for FtsQ-(3–41), FtsQ-(3–51), and FtsQ-(49–122) yielding pEK765, pEK764 (21), and pEK767, respectively. All plasmids were verified by sequence analysis.
Ribosome and RNC Complex Purification
Non-translating ribosomes were purified from E. coli MRE600 (22). Cells were cultured in LB medium at 37 °C to an A660 of 0.6 and harvested by centrifugation (9500 × g at 4 °C, 20 min). The pellet was resuspended in buffer A (20 mm Tris-HCl, pH 7.5, 100 mm NH4Cl, 10.5 mm MgOAc, 0.5 mm EDTA, 1 mm Tris(2-carboxyethyl)phosphine) and French-pressed twice at 700 p.s.i. Cell debris was removed by centrifugation (36,000 × g at 4 °C, 20 min) and the cleared lysate was laid on top of a 1.1 m sucrose cushion in buffer A with 0.5 m NH4Cl (buffer W) followed by centrifugation (72,000 × g, 4 °C, 19 h). The ribosomal pellet was washed with buffer W, and the sucrose cushion step was repeated twice to yield higher purity. The ribosome concentration was determined spectrophotometrically at a wavelength or 260 nm using an extinction coefficient of 4.2 × 107 (23).
For purification of RNCs, E. coli BL21(DE3)Δtig (24), which lacks the trigger factor gene, was transformed with either pUC19Strep3FtsQSecM, pEK764, pEK765, or pEK767 and grown in LB supplemented with 100 μg/ml ampicillin at 30 °C to A600 of 0.5. After harvesting, cells were lysed according to Evans et al. (25). In short, cells were resuspended in buffer R (50 mm Tris-HCl pH 7.5, 150 mm KCl, 10 mm MgCl2) and lysed by two freeze-thaw cycles in the presence of 400 μg/ml lysozyme. Cell debris was pelleted by centrifugation (twice at 30,000 × g, 4 °C, 30 min), and the cleared lysate was laid on a 1 m sucrose cushion prepared in buffer R. After centrifugation (112,000 × g, 4 °C, 17 h), the ribosomal pellet was dissolved in buffer R and loaded on a StrepTactin column (IBA). The column was washed with two column volumes of buffer R containing 0.5 m KCl and five column volumes of buffer R. RNCs were eluted with two column volumes of buffer R containing 2.5 mm desthiobiotin. The eluate was concentrated using Millipore Amicon Ultra-4 or Ultra-15 centrifugal tubes (cut-off 50 kDa), and the ribosome concentration was determined. The presence of stalled nascent chains was confirmed by SDS-PAGE followed by Western blotting using an antibody against the STREP-tag (IBA).
Surface Plasmon Resonance (SPR)
SPR measurements were performed on a Biacore 2000 system (GE Healthcare) as described (26). In short, IMVs containing endogenous or overexpressed levels of SecYEG (mutants) were immobilized in the separate channels of a L1 sensor chip (GE Healthcare). Buffer B (50 mm Tris-HCl, pH 7.5, 150 mm KCl, 5 mm MgCl2, 1 mm DTT, and 0.5 mg/ml BSA) containing SecA, ribosomes, or RNCs was injected into the channels at a flow rate of 20 μl/min, and SecYEG binding was probed at 25 °C. The binding surface of IMVs was regenerated by the injection of 100 mm Na2CO3, pH 10, followed by system equilibration using buffer B. Data were corrected for background binding to the IMVs containing endogenous levels of SecYEG. For SecA-ribosome competition experiments, ribosome binding was measured in the presence or absence of a saturating concentration (96 nm or as indicated) of SecA in the running buffer B. Data were fitted by nonlinear regression analysis of the response levels at equilibrium using SigmaPlot (Systat Software Inc.). For the single site (A + B ↔ AB) interaction model including a non-saturable linear component the following equation was used,
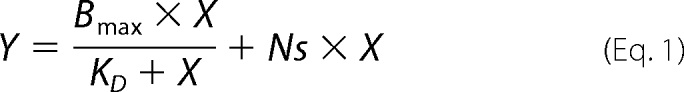 |
where Y is the SPR binding response at analyte (SecA or ribosome) concentration X, Bmax is the maximal binding response, KD is the dissociation constant, and Ns is the nonspecific binding coefficient.
For the two-site saturation model the following equation was used.
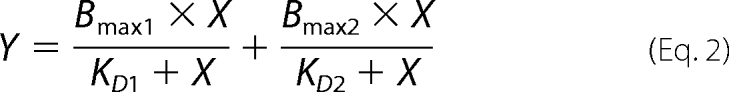 |
Fluorescence Correlation Spectroscopy (FCS)
For fluorescence microscopy, SecY(L148C)EG was overexpressed, purified, and labeled with AlexaFluor 488-C5-maleimide (Invitrogen) as described (27). Reconstitution of the fluorescently labeled SecY(L148C)EG into MSP1D1 nanodiscs (28) was carried out as described (29, 30) with the following modifications. A synthetic lipid composition consisting of 25 mol % dioleoylphosphatidylglycerol, 5 mol % cardiolipin, 30 mol % dioleoylphosphatidylethanolamine, and 40 mol % dioleoylphosphatidylcholine was used for nanodiscs formation, and lipids were destabilized by 0.5% (v/v) Triton X-100 before SecYEG reconstitution. SecYEG-containing nanodiscs were isolated using a Tricorn Superdex 200 10/300 column (GE Healthcare). The concentration of reconstituted SecYEG-AlexaFluor 488 in the collected fractions was determined based on the specific fluorophore absorbance.
FCS experiments were performed using the inverted confocal microscope LSM 710 equipped with the Confocor 3 module (Carl Zeiss GmbH). A solution of 50 nm AlexaFluor 488 dye with a known diffusion coefficient of 300 × 10−8 cm−2/s (31) was used to adjust the laser intensity at 488 nm and to calibrate the observation volume. Before the experiment SecYEG complexes were diluted to a concentration of ∼100 nm in 50 mm Tris-HCl, 100 mm NaCl, 10 mm MgCl2, and 5% (v/v) glycerol. For detergent-solubilized SecYEG the buffer was also supplied with 0.05% (w/v) n-dodecyl β-d-maltoside (DDM). Diffusion-driven fluctuations in the fluorescence intensity of SecYEG-conjugated AlexaFluor 488 were recorded over 10 s, and the measurements were repeated 10 times for each selected area. Recorded traces were averaged. For each experimental condition at least 10 measurements were done at different positions within the sample volume.
Autocorrelation curves were built and analyzed using ZEN 2010 software package (Carl Zeiss GmbH, Germany). Triplet state population of the fluorophore was below 20%, and it was omitted in the following analysis as the autocorrelation traces were fitted within 10 μs to a 10-s time range. The data were analyzed assuming free three-dimensional diffusion of SecYEG, and the auto-correlation curves were fitted according to the equation,
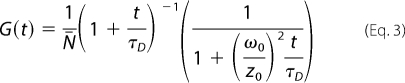 |
where G(t) is the amplitude of the auto-correlation function, N̄i is the average number of fluorescent particles in the laser focus, and τD is the diffusion time through the focus. To analyze SecYEG diffusion in the presence of ribosomes and to estimate fractions of free and ribosome-bound SecYEG, a two-component fitting model was applied,
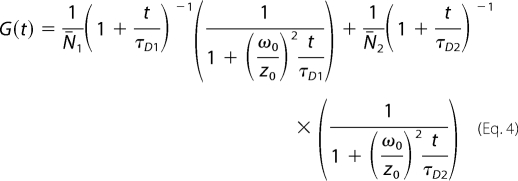 |
where N̄i and τDi are average number of fluorescent particles and diffusion time of ith species, respectively. The diffusion times of SecYEG and SecYEG-ribosome complexes were preset for the fitting procedure. For the free SecYEG the value was directly measured, whereas the diffusion time of the SecYEG-ribosome complex was approximated with that of fluorescein-labeled FtsQ108-RNC, which was experimentally determined in our set-up (∼900 μs).
SecA Co-sedimentation Assay
SecA (92 nm) was incubated with non-translating ribosomes (60 nm), FtsQ108 (50 nm), and FtsQ108ΔTMS (50 nm) in buffer B for 20 min at room temperature. Samples were loaded on a 35% (w/v) sucrose cushion and centrifuged for 2 h at 350,000 × g, 4 °C. Pellets were dissolved in SDS-PAGE sample buffer and analyzed by SDS-PAGE and silver staining.
In Vitro Protein Translocation Assay
In vitro translocation of fluorescein-labeled proOmpA (C290S) was done as described (32). Translocated, protease-resistant proOmpA was separated by SDS-PAGE and visualized with a LAS-4000 imager (FujiFilm) using the SyBr Blue Y515 Di filter.
Miscellaneous
SDS-PAGE, Western blotting, and silver staining were performed according to standard protocols. IMVs concentrations were determined using the Bio-Rad RC DC protein assay kit using BSA as standard. ProOmpA(C290S) and SecA were purified as described (33, 34). Quantification of the SDS-PAGE bands was done using Aida/2D densitometry (Raytest).
RESULTS
Detection of SecYEG-Ribosome Interaction by Surface Plasmon Resonance
We employed SPR to follow the binding of ribosomes to the membrane-embedded SecYEG complex in real time. This method was previously shown to accurately detect the high affinity interaction between SecA and SecYEG (11, 26). IMVs containing overexpressed levels of SecYEG were immobilized on a Biacore L1 chip, and the association and dissociation of SecA (Fig. 1A) and ribosomes (Fig. 1B) was followed in time. To correct for “bulk” contributions to the SPR signal and nonspecific binding to the membrane and chip surface, all measurements were corrected for binding to IMVs bearing endogenous SecYEG levels that were immobilized in a reference channel (26) (supplemental Fig. S1A). Both SecA and ribosome injection resulted in a SecYEG-dependent SPR response (Fig. 1B).
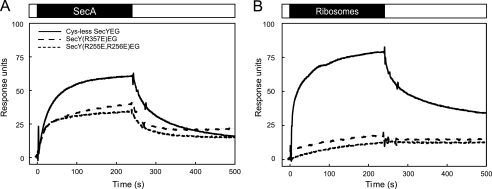
FIGURE 1.
Specific binding of SecA and ribosomes to the SecYEG complex monitored by SPR. Shown is an SPR sensogram of the binding of SecA (A) or non-translating ribosomes (B) to IMVs containing overexpressed levels of Cys-less SecYEG (solid), SecY(R357E)EG (spaced dashed), or SecY(R255E,R256E)EG (dashed). Binding was measured at 25 °C with a flow rate of 20 μl/min. Data were corrected for background binding to IMVs containing endogenous SecYEG levels. The ligand association and dissociation phases are represented above the sensograms by black and white bars, respectively. The concentration of SecA and ribosomes was 48 and 27 nm, respectively.
To validate that the observed SPR response reflected the binding of ribosomes to the SecYEG complex, we analyzed the interaction between ribosomes and two SecY mutant complexes, i.e. SecY(R255E,R256E)EG and SecY(R357E)EG. These mutations are located in the SecY cytoplasmic loops C4 and C5, respectively, and have been shown to disturb the SecYEG-ribosome interaction in detergent solution (7). Both SecYEG mutants were overexpressed to similar levels as Cys-less SecYEG (supplemental Fig. S1A). The alteration in the charge distribution in these cytosolic loops, however, resulted in a reduced mobility of SecYEG on SDS-PAGE (supplemental Fig. S1A). The SecY(R357E) mutation, which has been reported to affect the initiation of SecA-dependent protein translocation (13, 14), inhibited proOmpA translocation completely, whereas the SecY(R255E,R256E)EG mutant was normally active (supplemental Fig. S1B). Importantly, both mutants allowed for significant SecA binding (Fig. 1A), indicating that the mutations did not result in a major disturbance of the loop conformation. However, the SPR responses upon injection of ribosomes over immobilized SecY(R255E, R256E)EG or SecY(R357E)EG IMVs were dramatically reduced as compared with Cys-less SecYEG IMVs (Fig. 1B). These observations are consistent with the notion that the mutated loop residues are important for the SecYEG-ribosome interaction (7) and demonstrate that the SPR method genuinely monitors the interaction between ribosomes and the membrane-embedded SecYEG complex.
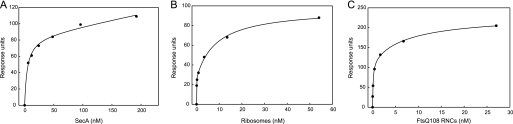
Previously, we observed that the SecA association and dissociation phases do not fit to a simple bimolecular interaction model (26, 35). Similarly, an accurate determination of the association and dissociation rates of ribosome binding could not be obtained by such simple data fitting models. Therefore, an apparent affinity of the interaction was determined by nonlinear regression analysis of the response levels at equilibrium (Fig. 2). For the SecYEG-SecA interaction, this resulted in an apparent dissociation constant (KD) of 4.5 nm when fitted according to a simple single site (A + B ↔ AB) interaction model including a non-saturable linear component (Fig. 2A, Table 2). This value is similar to those obtained previously by SPR, fluorescence microscopy, and biochemical methods (11, 26, 33, 36). In contrast, the ribosome binding responses fitted best to a two-site saturation model yielding apparent KD values of 0.03 and 8.8 nm, respectively (Fig. 2B, Table 2). Although the fit does not explain the mechanism of the interaction, the lower affinity value is in the same order as reported previously by equilibrium binding experiments, i.e. 14–17 nm with IMVs (17) and 6 nm with SecYEG proteoliposomes (37). These results demonstrate that both SecA and the ribosome interact with the SecYEG complex with nanomolar affinity.
FIGURE 2.
SecA, ribosomes, and RNCs interact with the SecYEG complex with nanomolar affinity. SecYEG binding at increasing concentrations of SecA (A), non-translating ribosomes (B), and FtsQ108 RNCs (C) was determined by SPR as described in Fig. 1. The equilibrium SPR responses were fitted by nonlinear regression analysis. The fitting for SecA was performed based on a model assuming single site with a nonspecific binding parameter. For fitting for non-translating ribosome and RNCs, a two-site saturation model was used. Binding data are summarized in Table 1.
TABLE 2.
Dissociation constants and apparent binding levels for the interaction of SecA, ribosomes, or FtsQ108 RNCs with the SecYEG complex as determined by surface plasmon resonance
R2 = coefficient of determination; KD1 and KD2 are the dissociation constants 1 and 2, respectively. Bmax1 and Bmax2 are the maximum obtained binding values associated with the two dissociation constants. Ns, nonspecific binding. RU, response units.
| Analyte | Bmax1 | Bmax2 | KD1 | KD2 | Ns | R2 |
|---|---|---|---|---|---|---|
| RU | RU | nm | nm | |||
| SecA | 85 | 4.5 | 0.15 | 0.9931 | ||
| Ribosomes | 27 | 70 | 0.03 | 8.8 | 0.9978 | |
| FtsQ108RNCs | 117 | 114 | 0.12 | 7.9 | 0.9973 |
Increased Ribosome Binding in Presence of Nascent Chain
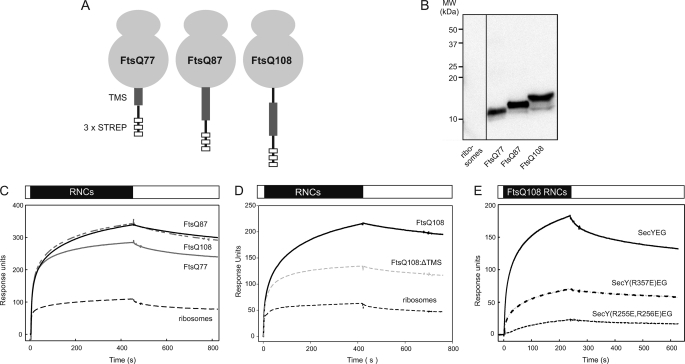
During membrane protein insertion, the SecYEG complex interacts with translating ribosomes. To determine whether the presence of a nascent chain influences the SecYEG-ribosome interaction, ribosomes carrying chimeric nascent chains of the E. coli inner membrane protein FtsQ were isolated. To halt translation, constructs were used in which the stalling motif of the E. coli secretion monitor protein SecM (38) was fused behind the first 41, 51, or 72 amino acids of FtsQ, resulting in nascent chains of 77, 87, or 108 amino acids long (FtsQ77, FtsQ87, FtsQ108 (20), respectively). At these nascent chain lengths the FtsQ TMS is expected to be either half-exposed (FtsQ77), nearly completely exposed (FtsQ87), and completely exposed (FtsQ108) from the ribosomal exit site (4) (Fig. 3A). Constructs were preceded by a triple STREP-tag to allow separation of RNCs from non-translating ribosomes (20). After isolation, the presence of the nascent chains was verified by immunoblotting against an STREP-tag antibody (Fig. 3B). Surprisingly, injection of the FtsQ87 and FtsQ108 RNCs resulted in an SPR response that was 3–5-fold higher compared with non-translating ribosomes (Fig. 3C), indicating that the SecYEG-ribosome interaction is strongly stimulated by the presence of a nascent chain. The shorter FtsQ77 RNC increased binding to a lesser extent (Fig. 3C), suggesting that the enhanced interaction of translating ribosomes with the SecYEG complex is dependent on the length of the nascent chain with an optimal interaction when the TMS of FtsQ is exposed from the ribosome tunnel. Indeed, injection of a FtsQ nascent chain of 108 residues without TMS (FtsQ108-ΔTMS) yielded a significantly reduced binding response compared with FtsQ108 exposing the TMS (Fig. 3D), suggesting that TMS contributes to the enhanced binding of RNCs to the translocon.
FIGURE 3.
The presence of a nascent chain enhances the ribosome-SecYEG interaction. A, shown is a schematic illustration of the FtsQ RNCs with various chain lengths. B, shown is Western blotting of RNCs of increasing chain length pretreated with 100 mm NaOH. To overcome poor transfer efficiency, a double amount of the FtsQ77 RNC was loaded. C, shown is binding of 27 nm ribosomes (dashed black) or RNCs with a FtsQ nascent chain length of 77 (solid gray), 87 (solid black), and 108 (dash gray) residues to immobilized IMVs containing overexpressed Cys-less SecYEG. D, binding of 13.5 nm non-translating ribosomes (black dashed), FtsQ108 RNCs (solid line), and FtsQ108-ΔTMS RNCs (gray dashed) is shown. E, binding of 27 nm FtsQ108 RNCs to immobilized IMVs containing overexpressed Cys-less SecYEG (solid line), SecY(R357E)EG (spaced dashed), and SecY(R255E,R256E)EG (dashed) is shown.
The interaction between RNCs and SecYEG was further studied using the FtsQ108 construct that has previously also been used for cross-linking and structural studies (4, 6). As observed for non-translating ribosomes, the R255E,R256E mutations in the C4 loop of SecY abrogated the interaction with FtsQ108 RNCs (Fig. 3E, dashed line). However, these RNCs showed substantial interaction with SecY(R357E)EG (Fig. 3E, spaced dashed line), which is consistent with our previous data that indicated that the R357E mutation still supports membrane protein insertion (14). These data demonstrate that translating and non-translating ribosomes interact differently with the SecYEG complex.
Interaction of Purified SecYEG Complex with SecA and Ribosomes
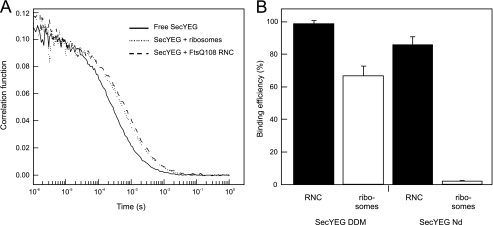
To verify the effect of a nascent chain on the SecYEG-ribosome interactions, we monitored the binding reaction between the purified components. Binding was probed using FCS, a sensitive confocal microscopy technique that allows for the analysis of molecular interactions in equilibrium at nanomolar substrate concentrations (39). In FCS, fluctuations of fluorescence resulting from fluorescently labeled proteins diffusing through the femtoliter-sized confocal excitation volume are monitored and auto-correlated over the measurement time. Temporal decay in the correlation function provides a precise estimate of the diffusional mobility of the fluorescently labeled biomolecule. Binding of the fluorescent protein to large non-labeled components can be detected as alterations of its mobility (Fig. 4A). First, the interaction between the solubilized SecYEG complex and ribosomes was determined in a solution containing 0.05% of the detergent DDM. Purified and fluorescently labeled SecY(C148)EG-AlexaFluor 488 (∼100 nm) was illuminated when diffusing through the laser focal volume, and temporary fluctuations in the fluorescence intensity were recorded and used to build an autocorrelation curve (Fig. 4A). The FCS data were analyzed assuming unrestricted three-dimensional diffusion of the labeled SecYEG in solution, and diffusion coefficient (D) of 26 ± 3 × 10−8 cm−2/s was obtained. The addition of an excess SecA (1.8 μm) decreased the diffusion coefficient of solubilized SecYEG only slightly by ∼10% to 23.9 ± 1.5 × 10−8 cm−2/s (data not shown). However, upon the addition of an excess FtsQ108 RNCs (250 nm), the mobility of SecY(L148C)EG-AlexaFluor 488 reduced substantially, resulting in a pronounced shift of the autocorrelation trace (Fig. 4A). The average diffusion time decreased ∼3-fold, and the D reduced to 10.0 ± 0.9 cm−2/s, suggesting that SecYEG was bound to RNC and diffused slowly as a part of the large complex. In agreement with the Einstein-Stokes equation, the observed reduction in the diffusion coefficient correlated with the 3-fold difference in dimensions of SecYEG protein enlarged by a belt of detergent molecules (radius R ∼ 3 nm (40, 41) and the ribosome (R ∼ 10 nm (42)). The addition of an excess (250 nm) of non-translating ribosomes to SecYEG reduced the translocon mobility to a lesser extent (Fig. 4A), suggesting that a large fraction of the SecYEG was not bound to these ribosomes and retained their high diffusional mobility. To quantify the binding efficiency, the autocorrelation traces of SecYEG were fitted assuming two SecYEG fractions in the ensemble: uncomplexed and ribosome/RNC-bound. The data suggested that more than 98% of SecYEG was bound with FtsQ108 RNCs, whereas only 67% was bound to non-translating ribosomes (Fig. 4B). As was reported previously (7), the R255E,R256E and R357E mutations in SecYEG reduced binding of non-translating ribosomes in the detergent environment (supplemental Fig. S2). However, in agreement with the SPR data presented above, the binding defect of the R357E mutant could be partially restored by the presence of a nascent chain (supplemental Fig. S2).
FIGURE 4.
The SecYEG-ribosome interaction is stimulated by the presence of a nascent chain and the lipid environment. A, fluorescence autocorrelation traces describe diffusion properties of DDM-solubilized SecYEG-AlexaFluor 488. Binding of ribosomes/RNCs reduces SecYEG mobility and causes the traces to shift toward longer diffusion times. B, when assayed in the presence of the detergent 0.05% (w/v) DDM, non-translating ribosomes bind SecYEG less efficiently than RNCs. This effect is further enhanced for SecYEG reconstituted into the lipid environment of nanodiscs.
Because the above studies were performed in detergent solution, we also investigated the role of the lipids in SecA and RNC binding to SecYEG. Herein, fluorescently labeled SecY(C148)EG-AlexaFluor 488 was reconstituted into small lipid patches, known as nanodiscs (28). These provide a physiologically relevant lipid environment for the embedded membrane proteins (supplemental Fig. S3). Because nanodiscs are monodisperse in aqueous solution, they are well suited for fluorescence microscopy applications, including FCS. The diffusion coefficient of SecYEG reconstituted into nanodiscs (SecYEG-Nd) was similar to that of detergent-solubilized SecYEG (D = 27 ± 3 × 10−8 cm−2/s). In agreement with previous reports (8), no binding was observed for non-translating ribosomes. In contrast, the mobility of SecYEG-Nd decreased substantially in the presence of FtsQ108 RNCs, indicating that the presence of a nascent chain triggered ribosome binding to the membrane-embedded translocon. Fitting the autocorrelation curves with the two-component model suggested that 82% of the SecYEG formed complexes with RNCs (Fig. 4B). As observed with the native membrane-embedded and solubilized SecYEG complexes, the interaction of FtsQ108 with SecYEG-Nd was partially inhibited by the R357E mutation and more strongly blocked by the R255E,R256E mutation of SecYEG (supplemental Fig. S2). Together, these FCS results corroborate the observation made by SPR that the presence of the nascent chain strongly promotes the SecYEG-ribosome interaction the SPR data but also indicate that the interaction between SecYEG and the ribosome is influenced by the environment of the SecYEG complex (solubilized versus membrane-embedded) showing a greater interaction between empty ribosomes and SecYEG when present in detergent.
Competitive Binding of SecA and Ribosomes
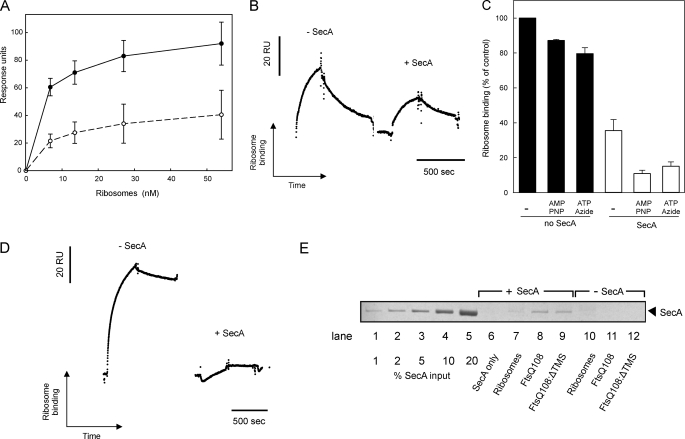
It has been proposed that SecA and ribosomes interact with the translocon non-competitively (17). This would imply that ribosomes and SecA bind to different and independent sites on SecYEG. To investigate this phenomenon, we designed an SPR competition experiment to determine whether the SecYEG complex can accommodate both SecA and the ribosome simultaneously. First, the SecYEG complexes in the immobilized IMVs were saturated with SecA using a running buffer containing excess SecA (96 nm). Subsequent injection of ribosomes in the SecA-containing running buffer resulted in a binding response that was 2–3-fold lower than in the absence of SecA (Fig. 5, A and B). Stabilizing the SecYEG-SecA interaction by the addition of AMP-PNP, which prevents the dissociation of SecA from the SecYEG complex (26, 43), or a combination of ATP and azide, which prevents the release of SecA from SecYEG (44), further decreased the ribosome binding up to 8-fold (Fig. 5C). As a control, AMP-PNP or ATP-azide alone only slightly affected the SecYEG-ribosome interaction (Fig. 5C). When the ribosomes were loaded with the FtsQ108 nascent chain, the SecYEG interaction was affected even more severely by the presence of SecA (Fig. 5D). SecA has been reported to interact with ribosomes and RNCs (45, 46). However, the reduced interaction was not caused by sequestration of ribosomes or RNCs by the SecA present in the running buffer, as co-sedimentation assays showed that under the conditions used for SPR less than 3% of the RNCs were SecA-bound (Fig. 5E). This is consistent with the observation that the SecA-ribosome interaction is of low affinity (0.9 μm) (45). The results, therefore, indicate that SecA competes with both translating and non-translating ribosomes for SecYEG binding.
FIGURE 5.
SecA competes with the ribosome for SecYEG binding. A, equilibrium SPR responses at increasing concentrations of non-translating ribosomes were measured in the presence (white) or absence (black) of SecA (96 nm) in the SPR running buffer. B, binding of non-translating ribosomes (6.75 nm) in the presence or absence of SecA (96 nm) in shown. C, normalized ribosome binding in the presence (white) or absence (black) of 96 nm SecA with or without AMP-PNP (1 mm) or ATP (1 mm) and azide (5 mm) is shown. Binding responses were normalized to the equilibrium response in the absence of SecA. D, binding of FtsQ108 RNCs (3.38 nm) in the presence or absence of a SecA (96 nm) is shown. E, co-sedimentation of SecA (92 nm) with ribosomes (60 nm), FtsQ108 (50 nm), and FtsQ108; ΔTMS (50 nm) is shown. The SecA input represents amount of SecA added to the reaction mixture. RU, response units.
Detergents Affect Interaction between SecYEG and Ligands SecA and Ribosomes
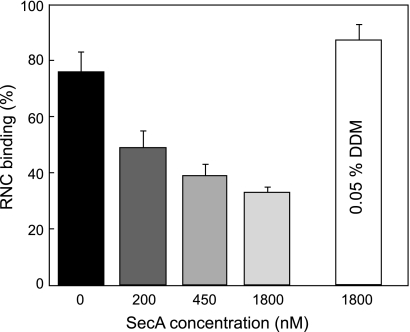
Because the diffusion coefficient of SecYEG was strongly affected by binding of a ribosome, but not by SecA (see above), the RNC- and SecA-bound SecYEG populations can be discriminated based on their diffusional mobility. SecA and FtsQ108 RNC were added to DDM-solubilized SecY(L148C)EG-AlexaFluor 488 together, and the translocon diffusion was monitored by FCS. AMP-PNP was added to stabilize the SecYEG-SecA complex. Assuming that both SecYEG-SecA and SecYEG-RNC complexes are formed within the ensemble, a two-component model can be used for analysis. Surprisingly, even in the presence of a large excess of SecA (1.8 μm versus 250 nm RNCs), 87% of the translocons were complexed with the RNCs in detergent solution (Fig. 6). To determine whether the detergent environment of the SecYEG complex caused this apparent discrepancy with the SPR results, the same experiment was performed using the nanodisc-reconstituted SecYEG complex. The SecYEG-Nd-SecA complex stabilized by AMP-PNP manifested a D of 23 ± 3 × 10−8 cm−2/s, which is similar to SecYEG-Nd alone. This allows for a discrimination between SecA and RNC FtsQ108 binding to SecYEG-Nd based on the translocon mobility only. When both SecA and FtsQ108 RNCs were added to SecY(L148C)EG-Nd in the presence of 1 mm AMP-PNP, a clear competition for SecYEG binding was observed, and the propensity to form a complex with the RNCs decreased with the SecA-RNC ratio (Fig. 6). In the presence of an elevated AMP-PNP concentration (5 mm), RNC binding was further reduced, up to 3-fold (supplemental Fig. S4). These results indicate that the interaction between SecYEG, translating ribosomes, and SecA is strongly dependent on the molecular environment; although detergents promote ribosome binding and weaken the competitive interaction with SecA, SecA and translating ribosomes bind competitively to the membrane-embedded SecYEG. Importantly, the FCS data corroborate with the SPR data and demonstrate that SecA and (non)translating ribosomes compete for binding to the membrane-embedded SecYEG complex.
FIGURE 6.
SecA competes with ribosomes for binding to SecYEG reconstituted into nanodiscs. Binding of RNCs to purified SecYEG in nanodiscs in the presence of increasing concentrations of SecA with AMP-PNP. The fraction of RNC-bound SecYEG complex was determined by FCS as described under “Results.” Detergent-solubilized SecYEG primarily interacted with RNC even in a large excess of SecA (white bar).
DISCUSSION
During the biogenesis of membrane proteins with large periplasmic domains, both the ribosome and SecA need to associate with the SecYEG complex to drive the membrane partitioning of hydrophobic segments and the translocation of hydrophilic domains, respectively. How the translocon is able to coordinate the binding of these two ligands is not well understood. Here we have used two distinct spectroscopic techniques to follow the binding of SecA and of translating and non-translating ribosomes to the SecYEG complex.
SPR and FCS readily detected the association of ribosomes with the SecYEG complex present in membrane vesicles and the purified SecYEG present in detergent solution or reconstituted into nanodiscs. Although the molecular environment influences the observed interactions, the interactions are specific and inhibited by mutations in SecY that were previously reported to result in ribosome binding defects. Interestingly, ribosome binding to the SecYEG complex was highly stimulated by the presence of a nascent chain, in particular when this nascent chain exposed a hydrophobic polypeptide sequence. The presence of a nascent chain also partially relieved the ribosome binding defect of the SecY(R357E) mutant. Taken together this suggests that translating and non-translating ribosomes interact differently with the SecYEG complex and indicates an interplay, possibly a cooperative effect, of the nascent chain in the SecYEG-ribosome interaction. A recent cryo-EM structure of an FtsQ RNC complex with the translocon indicated that the nascent chain engages in interactions between the ribosomal proteins L23 and L24 and the loops of SecY, resulting in conformational changes in the participating loops (8). These interactions and conformational changes might tighten the interaction between the RNC complex and the translocon, and this may be promoted by hydrophobic stretches in the nascent polypeptide chain.
The interaction between translating or non-translating ribosomes and the membrane-embedded SecYEG complex was competed by SecA, demonstrating that the two ligands cannot bind the translocon simultaneously. Surprisingly, even though the presence of a nascent chain enhances the ribosome-translocon interaction, SecA competed stronger with translating than with non-translating ribosomes. Because our current understanding of the exact mechanism of the translocon-RNC interaction is still limited, this observation is not readily explained. However, it is clear from the SPR data that the interaction is not a simple one-to-one binding event. Using nonlinear regression analysis of the response levels at equilibrium, ribosome binding data could only be fitted to a “two-site saturation” model according to which SecYEG partly interacted with the ribosomes with an apparent affinity in the low nanomolar scale consistent with previously reported values (17, 37) and partly with a very high affinity that was particularly prominent upon binding of RNCs. This two-site saturation model suggests that the dramatically increased binding response in the presence of a nascent chain is caused by an increase in the number of binding sites rather than an improved affinity. Possibly, RNCs recruit binding sites that were previously masked. Alternatively, binding of a RNC to a SecYEG complex causes an intrinsically higher SPR signal than binding of a non-translating ribosome, whereas the number of binding sites remains constant. This could for instance occur when the nascent chain, by interacting with SecYEG, pulls the ribosome closer to the membrane surface, which in turn would result in an increased SPR signal. In this scenario the interaction with RNCs would direct the SecYEG complex toward the high affinity state (see Table 2: 50% high affinity sites for RNCs versus 25% for non-translating ribosomes). This very tight binding of translating ribosomes to the translocon could be the result of multiple steps, so that inhibition of RNC binding by SecA could occur if the initial association is of relatively low affinity. Further studies will be required to elucidate the molecular basis of the ribosome translocon interaction.
SecA-ribosome competition only occurred when SecYEG was in its native, membrane-embedded state, as the presence of SecA did not affect the SecYEG-ribosome interaction in detergent solution. Because FCS indicated that solubilized SecYEG has a higher propensity to bind non-translating ribosomes compared with the membrane-embedded SecYEG (this study) and detergents have been shown to affect the activity and oligomeric state of SecA (47–49), it is likely that the interaction between SecA, the ribosome, and the solubilized SecYEG complex is affected by the presence of the detergent. The observation that the molecular environment is a critical factor in the interaction between the translocon and its soluble interactions partners should be considered in future studies.
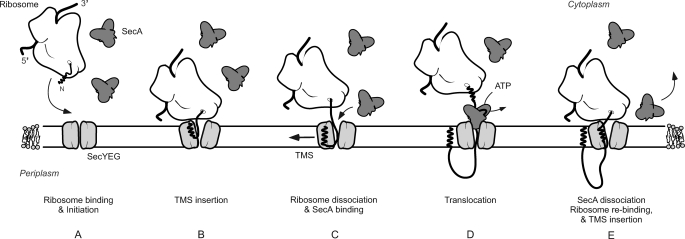
The observation that SecA and the ribosome compete for binding to the SecYEG complex even when the ribosome is charged with a nascent chain of a SecA-dependent membrane protein implies that during membrane protein insertion, SecA and the ribosome do not interact with the translocon simultaneously. Because many membrane proteins contain large periplasmic domains, the competitive interaction of SecA and the ribosome has implications for the membrane insertion mechanism. Although for single-spanning membrane proteins like FtsQ it is possible to envision a scenario where the ribosome dissociates from the SecYEG complex after membrane partitioning of the TMS to allow SecA binding, the sequence of events will be more complicated when a large periplasmic domain is followed by another TMS. For these proteins, SecA and the ribosomes might need to cycle on and off from the translocon in a sequential manner, a process for which the timing needs to be carefully orchestrated. It is plausible that the SecYEG affinity to SecA and translating ribosomes depends on the hydrophobicity of the emerging polypeptide chain. The ribosome might be released from the SecYEG complex once it encounters a large enough polar periplasmic polypeptide domain that would allow SecA to bind to this domain followed or concomitantly with SecYEG binding (Fig. 7). The micromolar SecA concentrations in the cytoplasm would ensure efficient cycling of SecA on SecYEG to translocate these polar domains (33). Likewise, SecA may dissociate from the SecYEG complex once it encounters a newly emerging TMS. In this respect, SecA has been reported to dissociate from the SecYEG complex when it encounters a potential TMS during the translocation of a secretory protein (50, 51). Early recognition of a TMS by the translating ribosome (52) may restore the high affinity interaction between SecYEG and the ribosome for the insertion of the next TMS. Future studies should be directed to unravel the interplay between SecA, RNCs, and the SecYEG complex including targeting factors such as signal recognition particle and FtsY (53).
FIGURE 7.
Scheme for the sequential binding of SecA and the translating ribosome to the SecYEG complex. A, the presence of an emerging transmembrane segment in the nascent chain stimulates binding of the ribosome to SecYEG. B, elongation of the polypeptide allows insertion of the first TMS into the membrane via the SecYEG complex. C, the ribosome dissociates from the SecYEG complex once a polar polypeptide domain emerges allowing SecA to bind. D, ATP binding and hydrolysis at SecA results in the translocation of the periplasmic polypeptide domain across the membrane. E, emergence of the next TMS from the ribosome causes the dissociation of SecA from SecYEG, allowing the ribosome to rebind to SecYEG and to insert the TMS.
Supplementary Material
Acknowledgments
We thank Ilja Küsters, Jelger Lycklama á Nijeholt, and Marko Sustarsic for valuable discussion and technical assistance and Natalia Dudkina for cryoelectron microscopy experiments. We thank Nenad Ban (ETH, Zürich, Switzerland) and Bernd Bukau (ZMBH, Heidelbeg, Germany) for kind gifts of pUC19Strep3FtsQSecM and E. coli BL21(DE3)Δtig::kan, respectively. We thank Stephen Sligar (University of Illinois, Urbana, IL) for the plasmids expressing the MSP protein.
This work was supported by the Chemical Sciences division of the Netherlands Foundation for Scientific Research (CW-NWO).

This article contains supplemental Figs. S1–S4.
- RNC
- ribosome-bound nascent chain
- SPR
- surface plasmon resonance
- FCS
- fluorescence correlation spectroscopy
- IMV
- inner membrane vesicles
- TMS
- transmembrane segment
- DDM
- n-dodecyl β-d-maltoside
- AMP-PNP
- adenosine 5′-(β,γ-imino)triphosphate
- ND
- nanodiscs.
REFERENCES
- 1. Driessen A. J. M., Nouwen N. (2008) Protein translocation across the bacterial cytoplasmic membrane. Annu. Rev. Biochem. 77, 643–667 [DOI] [PubMed] [Google Scholar]
- 2. Luirink J., Sinning I. (2004) SRP-mediated protein targeting. Structure and function revisited. Biochim. Biophys. Acta 1694, 17–35 [DOI] [PubMed] [Google Scholar]
- 3. Andersson H., von Heijne G. (1993) Sec-dependent and sec-independent assembly of E. coli inner membrane proteins. The topological rules depend on chain length. EMBO J. 12, 683–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Urbanus M. L., Scotti P. A., Froderberg L., Saaf A., de Gier J. W., Brunner J., Samuelson J. C., Dalbey R. E., Oudega B., Luirink J. (2001) Sec-dependent membrane protein insertion. Sequential interaction of nascent FtsQ with SecY and YidC. EMBO Rep. 2, 524–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cheng Z., Jiang Y., Mandon E. C., Gilmore R. (2005) Identification of cytoplasmic residues of Sec61p involved in ribosome binding and cotranslational translocation. J. Cell Biol. 168, 67–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mitra K., Schaffitzel C., Shaikh T., Tama F., Jenni S., Brooks C. L., 3rd, Ban N., Frank J. (2005) Structure of the E. coli protein-conducting channel bound to a translating ribosome. Nature 438, 318–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ménétret J. F., Schaletzky J., Clemons W. M., Jr., Osborne A. R., Skånland S. S., Denison C., Gygi S. P., Kirkpatrick D. S., Park E., Ludtke S. J., Rapoport T. A., Akey C. W. (2007) Ribosome binding of a single copy of the SecY complex. Implications for protein translocation. Mol. Cell 28, 1083–1092 [DOI] [PubMed] [Google Scholar]
- 8. Frauenfeld J., Gumbart J., van der Sluis E. O., Funes S., Gartmann M., Beatrix B., Mielke T., Berninghausen O., Becker T., Schulten K., Beckmann R. (2011) Cryo-EM structure of the ribosome-SecYE complex in the membrane environment. Nat. Struct. Mol. Biol. 18, 614–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gumbart J., Trabuco L. G., Schreiner E., Villa E., Schulten K. (2009) Regulation of the protein-conducting channel by a bound ribosome. Structure 17, 1453–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zimmer J., Nam Y., Rapoport T. A. (2008) Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature 455, 936–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. van der Sluis E. O., Nouwen N., Koch J., de Keyzer J., van der Does C., Tampé R., Driessen A. J. M. (2006) Identification of two interaction sites in SecY that are important for the functional interaction with SecA. J. Mol. Biol. 361, 839–849 [DOI] [PubMed] [Google Scholar]
- 12. Robson A., Booth A. E., Gold V. A., Clarke A. R., Collinson I. (2007) A large conformational change couples the ATP binding site of SecA to the SecY protein channel. J. Mol. Biol. 374, 965–976 [DOI] [PubMed] [Google Scholar]
- 13. Mori H., Ito K. (2001) An essential amino acid residue in the protein translocation channel revealed by targeted random mutagenesis of SecY. Proc. Natl. Acad. Sci. U.S.A. 98, 5128–5133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. de Keyzer J., Regeling A., Driessen A. J. M. (2007) Arginine 357 of SecY is needed for SecA-dependent initiation of preprotein translocation. FEBS Lett. 581, 1859–1864 [DOI] [PubMed] [Google Scholar]
- 15. van der Laan M., Nouwen N., Driessen A. J. M. (2004) SecYEG proteoliposomes catalyze the Deltaphi-dependent membrane insertion of FtsQ. J. Biol. Chem. 279, 1659–1664 [DOI] [PubMed] [Google Scholar]
- 16. du Plessis D. J., Nouwen N., Driessen A. J. M. (2006) Subunit a of cytochrome o oxidase requires both YidC and SecYEG for membrane insertion. J. Biol. Chem. 281, 12248–12252 [DOI] [PubMed] [Google Scholar]
- 17. Zito C. R., Oliver D. (2003) Two-stage binding of SecA to the bacterial translocon regulates ribosome-translocon interaction. J. Biol. Chem. 278, 40640–40646 [DOI] [PubMed] [Google Scholar]
- 18. Kaufmann A., Manting E. H., Veenendaal A. K., Driessen A. J. M., van der Does C. (1999) Cysteine-directed cross-linking demonstrates that helix 3 of SecE is close to helix 2 of SecY and helix 3 of a neighboring SecE. Biochemistry 38, 9115–9125 [DOI] [PubMed] [Google Scholar]
- 19. van der Sluis E. O., Nouwen N., Driessen A. J. M. (2002) SecY-SecY and SecY-SecG contacts revealed by site-specific crosslinking. FEBS Lett. 527, 159–165 [DOI] [PubMed] [Google Scholar]
- 20. Schaffitzel C., Ban N. (2007) Generation of ribosome nascent chain complexes for structural and functional studies. J. Struct. Biol. 158, 463–471 [DOI] [PubMed] [Google Scholar]
- 21. Lycklama a Nijeholt J. A., Wu Z. C., Driessen A. J. M. (2011) Conformational dynamics of the plug domain of the SecYEG protein-conducting channel. J. Biol. Chem. 286, 43881–43890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Salaj-Smic E. (1978) Colicinogeny of Escherichia coli MRE 600. Antimicrob. Agents. Chemother. 14, 797–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nilsson M., Bülow L., Wahlund K. G. (1997) Use of flow field-flow fractionation for the rapid quantitation of ribosome and ribosomal subunits in Escherichia coli at different protein production conditions. Biotechnol Bioeng. 54, 461–467 [DOI] [PubMed] [Google Scholar]
- 24. Rutkowska A., Mayer M. P., Hoffmann A., Merz F., Zachmann-Brand B., Schaffitzel C., Ban N., Deuerling E., Bukau B. (2008) Dynamics of trigger factor interaction with translating ribosomes. J. Biol. Chem. 283, 4124–4132 [DOI] [PubMed] [Google Scholar]
- 25. Evans M. S., Ugrinov K. G., Frese M. A., Clark P. L. (2005) Homogeneous stalled ribosome nascent chain complexes produced in vivo or in vitro. Nat. Methods 2, 757–762 [DOI] [PubMed] [Google Scholar]
- 26. de Keyzer J., van der Does C., Kloosterman T. G., Driessen A. J. M. (2003) Direct demonstration of ATP-dependent release of SecA from a translocating preprotein by surface plasmon resonance. J. Biol. Chem. 278, 29581–29586 [DOI] [PubMed] [Google Scholar]
- 27. Kedrov A., Kusters I., Krasnikov V. V., Driessen A. J. M. (2011) A single copy of SecYEG is sufficient for preprotein translocation. EMBO J. 30, 4387–4397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Denisov I. G., Grinkova Y. V., Lazarides A. A., Sligar S. G. (2004) Directed self-assembly of monodisperse phospholipid bilayer Nanodiscs with controlled size. J. Am. Chem. Soc. 126, 3477–3487 [DOI] [PubMed] [Google Scholar]
- 29. Alami M., Dalal K., Lelj-Garolla B., Sligar S. G., Duong F. (2007) Nanodiscs unravel the interaction between the SecYEG channel and its cytosolic partner SecA. EMBO J. 26, 1995–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dalal K., Duong F. (2010) Reconstitution of the SecY translocon in nanodiscs. Methods Mol. Biol. 619, 145–156 [DOI] [PubMed] [Google Scholar]
- 31. Bacia K., Schwille P. (2003) A dynamic view of cellular processes by in vivo fluorescence auto- and cross-correlation spectroscopy. Methods 29, 74–85 [DOI] [PubMed] [Google Scholar]
- 32. de Keyzer J., van Der Does C., Driessen A. J. M. (2002) Kinetic analysis of the translocation of fluorescent precursor proteins into Escherichia coli membrane vesicles. J. Biol. Chem. 277, 46059–46065 [DOI] [PubMed] [Google Scholar]
- 33. Kusters I., van den Bogaart G., Kedrov A., Krasnikov V., Fulyani F., Poolman B., Driessen A. J. M. (2011) Quaternary structure of SecA in solution and bound to SecYEG probed at the single molecule level. Structure 19, 430–439 [DOI] [PubMed] [Google Scholar]
- 34. van der Does C., de Keyzer J., van der Laan M., Driessen A. J. M. (2003) Reconstitution of purified bacterial preprotein translocase in liposomes. Methods Enzymol. 372, 86–98 [DOI] [PubMed] [Google Scholar]
- 35. de Keyzer J., van der Sluis E. O., Spelbrink R. E., Nijstad N., de Kruijff B., Nouwen N., van der Does C., Driessen A. J. M. (2005) Covalently dimerized SecA is functional in protein translocation. J. Biol. Chem. 280, 35255–35260 [DOI] [PubMed] [Google Scholar]
- 36. van der Wolk J. P., Fekkes P., Boorsma A., Huie J. L., Silhavy T. J., Driessen A. J. M. (1998) PrlA4 prevents the rejection of signal sequence-defective preproteins by stabilizing the SecA-SecY interaction during the initiation of translocation. EMBO J. 17, 3631–3639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Prinz A., Behrens C., Rapoport T. A., Hartmann E., Kalies K. U. (2000) Evolutionarily conserved binding of ribosomes to the translocation channel via the large ribosomal RNA. EMBO J. 19, 1900–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nakatogawa H., Ito K. (2002) The ribosomal exit tunnel functions as a discriminating gate. Cell 108, 629–636 [DOI] [PubMed] [Google Scholar]
- 39. Schwille P., Meyer-Almes F. J., Rigler R. (1997) Dual-color fluorescence cross-correlation spectroscopy for multicomponent diffusional analysis in solution. Biophys. J. 72, 1878–1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Van den Berg B., Clemons W. M., Jr., Collinson I., Modis Y., Hartmann E., Harrison S. C., Rapoport T. A. (2004) X-ray structure of a protein-conducting channel. Nature 427, 36–44 [DOI] [PubMed] [Google Scholar]
- 41. Kunji E. R., Harding M., Butler P. J., Akamine P. (2008) Determination of the molecular mass and dimensions of membrane proteins by size exclusion chromatography. Methods 46, 62–72 [DOI] [PubMed] [Google Scholar]
- 42. Cate J. H., Yusupov M. M., Yusupova G. Z., Earnest T. N., Noller H. F. (1999) X-ray crystal structures of 70 S ribosome functional complexes. Science 285, 2095–2104 [DOI] [PubMed] [Google Scholar]
- 43. Economou A., Wickner W. (1994) SecA promotes preprotein translocation by undergoing ATP-driven cycles of membrane insertion and deinsertion. Cell 78, 835–843 [DOI] [PubMed] [Google Scholar]
- 44. Eichler J., Rinard K., Wickner W. (1998) Endogenous SecA catalyzes preprotein translocation at SecYEG. J. Biol. Chem. 273, 21675–21681 [DOI] [PubMed] [Google Scholar]
- 45. Huber D., Rajagopalan N., Preissler S., Rocco M. A., Merz F., Kramer G., Bukau B. (2011) SecA interacts with ribosomes in order to facilitate posttranslational translocation in bacteria. Mol. Cell 41, 343–353 [DOI] [PubMed] [Google Scholar]
- 46. Karamyshev A. L., Johnson A. E. (2005) Selective SecA association with signal sequences in ribosome-bound nascent chains. A potential role for SecA in ribosome targeting to the bacterial membrane. J. Biol. Chem. 280, 37930–37940 [DOI] [PubMed] [Google Scholar]
- 47. Benach J., Chou Y. T., Fak J. J., Itkin A., Nicolae D. D., Smith P. C., Wittrock G., Floyd D. L., Golsaz C. M., Gierasch L. M., Hunt J. F. (2003) Phospholipid-induced monomerization and signal peptide-induced oligomerization of SecA. J. Biol. Chem. 278, 3628–3638 [DOI] [PubMed] [Google Scholar]
- 48. Or E., Navon A., Rapoport T. (2002) Dissociation of the dimeric SecA ATPase during protein translocation across the bacterial membrane. EMBO J. 21, 4470–4479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lill R., Dowhan W., Wickner W. (1990) The ATPase activity of SecA is regulated by acidic phospholipids, SecY, and the leader and mature domains of precursor proteins. Cell 60, 271–280 [DOI] [PubMed] [Google Scholar]
- 50. Sato K., Mori H., Yoshida M., Tagaya M., Mizushima S. (1997) In vitro analysis of the stop-transfer process during translocation across the cytoplasmic membrane of Escherichia coli. J. Biol. Chem. 272, 20082–20087 [DOI] [PubMed] [Google Scholar]
- 51. Duong F., Wickner W. (1998) Sec-dependent membrane protein biogenesis. SecYEG, preprotein hydrophobicity, and translocation kinetics control the stop-transfer function. EMBO J. 17, 696–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Woolhead C. A., McCormick P. J., Johnson A. E. (2004) Nascent membrane and secretory proteins differ in FRET-detected folding far inside the ribosome and in their exposure to ribosomal proteins. Cell 116, 725–736 [DOI] [PubMed] [Google Scholar]
- 53. Kuhn P., Weiche B., Sturm L., Sommer E., Drepper F., Warscheid B., Sourjik V., Koch H. G. (2011) The bacterial SRP receptor, SecA, and the ribosome use overlapping binding sites on the SecY translocon. Traffic 12, 563–578 [DOI] [PubMed] [Google Scholar]
- 54. Baneyx F., Georgiou G. (1990) In vivo degradation of secreted fusion proteins by the Escherichia coli outer membrane protease OmpT. J. Bacteriol. 172, 491–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.