Background: The cis-syn cyclobutane dimer of TmCG is responsible for C-to-T mutations in skin cancer.
Results: The mC of the dimer codes as C when bypassed by the DNA damage bypass polymerase η.
Conclusion: The coding properties of the mC are consistent with a water-exposed polymerase active site.
Significance: The mC of the dimer must deaminate to T to become mutagenic.
Keywords: Cancer, DNA Damage, DNA Methylation, DNA Polymerase, Mutagenesis Mechanisms, Cyclobutane Pyrimidine Dimer, Deamination, Tautomer, Ultraviolet Light
Abstract
C-to-T mutations are a hallmark of UV light and, in humans, occur preferentially at methylated PymCG sites, which are also sites of preferential cyclobutane pyrimidine dimer (CPD) formation. In response, cells have evolved DNA damage bypass polymerases, of which polymerase η (pol η) appears to be specifically adapted to synthesize past cis-syn CPDs. Although T=T CPDs are stable, CPDs containing C or 5-methylcytosine (mC) are not and spontaneously deaminate to U or T at pH 7 and 37 °C over a period of hours or days, making their preparation and study difficult. Furthermore, there is evidence to suggest that, depending on solvent polarity, a C or an mC in a CPD can adopt three tautomeric forms, one of which could code as T. Although many in vitro studies have established that synthesis past T or U in a CPD by pol η occurs in a highly error-free manner, the only in vitro evidence that synthesis past C or mC in a CPD also occurs in an error-free manner is for an mC in the 5′-position of an mC=T CPD. Herein, we describe the preparation and characterization of an oligodeoxynucleotide containing a CPD of a TmCG site, one of the major sites of C methylation and C-to-T mutations found in the p53 gene of basal and squamous cell cancers. We also demonstrate that both yeast and human pol η synthesize past the 3′-mC CPD in a >99% error-free manner, consistent with the highly water-exposed nature of the active site.
Introduction
One of the primary causes of basal and squamous cell skin cancer is exposure to sunlight. Sequencing of the p53 gene of these skin cancers has revealed a predominance of C-to-T and CC-to-TT mutations at methylated dipyrimidine sites, PymCG (1, 2). These sites are also hotspots for cis-syn cyclobutane pyrimidine dimer (CPD)3 formation induced by the UV in sunlight (3, 4). Unlike CPDs of TT sites, C- or 5-methylcytosine (mC)-containing CPDs are unstable and deaminate to U or T (5–11). The major responses to UV damage are global genome- and transcription-coupled nucleotide excision repair (12–14) and translesion synthesis (15). CPDs do not distort DNA structure or duplex stability greatly (16–18), making them difficult to be detected and repaired by nucleotide excision repair (19). Although they block RNA polymerases and can thus be removed by transcription-coupled repair, this cannot occur for the non-transcribed strand or for either strand of inactive genes. Thus, many CPDs may go unrepaired before replication is initiated, and because they block replicative polymerases, DNA damage bypass polymerases are recruited to carry out translesion synthesis. Of these, polymerase η (pol η) is the most efficient at synthesizing past CPDs and appears to be the principal polymerase involved in CPD translesion synthesis (20–22). Early in vitro studies showed that pol η could synthesize past a T=T CPD in an essentially error-free manner, suggesting that the two Ts of the CPD were capable of directing the insertion of As. It was only by investigating DNA synthesis past N3-methyl derivatives of T=T CPDs that it was possible to demonstrate that both the 5′- and 3′-Ts of the CPD were directing the insertion of As and that the As were not being inserted by a non-templated mechanism, otherwise known as the “A rule” (23). Furthermore, studies with 7-deaza-ATP demonstrated that a Watson-Crick like intermediate was being used to direct the insertion of the A, and not a Hoogsteen-type base pair as found in the crystal structure of another Y-family polymerase (24). These conclusions were later confirmed through x-ray crystallography of intermediates in the synthesis past a T=T CPD by human pol η (25).
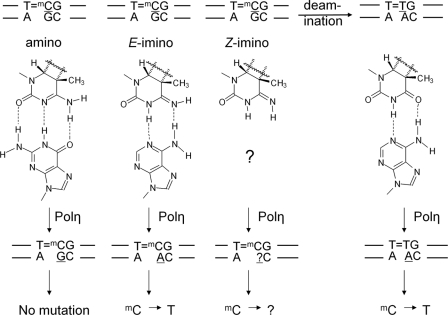
Unlike T in native DNA or in a CPD, which is known to preferentially adopt a keto tautomer and base pair with A, a C or an mC in a CPD could, in principal, adopt amino and/or imino tautomeric forms with E- or Z-stereochemisty (the imino hydrogen is trans or cis with respect to N3), some of which could thereby direct the insertion of G and/or A (Fig. 1) (26, 27). The possibility that C or mC could exist in an imino tautomeric form comes from early work demonstrating that dihydrocytosine adopts the amino tautomeric form in water but an imino tautomeric form in the much less polar solvent chloroform (28). Gas phase theoretical calculations have reproduced the preference for the imino tautomeric form in non-polar environments (29, 30) but failed to reproduce the preference for the amino form in water (30). The same type of calculations also predicted that the Cs in CPD should exist almost exclusively in the imino tautomeric state in water (27). However, evidence that Cs in CPDs exist in their amino tautomeric state comes from the highly preferential insertion of G opposite the Cs by pol η from various genetic experiments in yeast and human cells (31, 32) and from an in vitro translesion experiment with an mC=T CPD-containing template and yeast pol η (33). No work has yet been carried out on a site-specific CPD of an mCG site. Because we have previously observed that the deamination rate of an mC in a CPD is 3–4 times slower for PumCTPu than for PuTmCPu (where Pu = A or G) and depends on flanking sequence (34), it was possible that the relative proportion of tautomers may likewise depend on position and flanking sequence.
FIGURE 1.
Mutagenic properties of mC and its deamination product, T, in cis-syn dimer. The major tautomers of mC in a cis-syn dimer and their base pairing properties are compared with those of the deamination product, T.
Part of the difficulty in working with C- or mC-containing CPDs in vitro or in vivo is their spontaneous deamination to U or T within hours to days (34) compared with ∼50,000 years for canonical C or mC in duplex DNA (35, 36). This lability has also impeded the development of a phosphoramidite building block for site-specific incorporation of C-containing CPDs into DNA templates by automated synthesis. At the moment, the only route to such templates involves UV irradiation of an oligodeoxynucleotide (ODN) followed by HPLC purification, as was originally developed for the preparation of a T=C CPD-containing 11-mer (37). We used this approach to prepare a 14-mer containing the CPD of an mCTA site (33). An mCTA site was chosen for our initial studies because the CPD was found to have a much lower deamination rate and to be produced in a higher yield than at the more biologically relevant TmCG site (34). Herein, we report the preparation and characterization of a CPD of a TmCG site, a known hotspot for C methylation, CPD formation, and UV light-induced C-to-T mutations, and demonstrate the highly error-free, non-mutagenic insertion of G opposite the mC in the CPD by yeast and human pol η. We also report the deamination rate of the mC in the CPD and confirm the error-free but mutagenic insertion of A opposite the resulting T.
EXPERIMENTAL PROCEDURES
Enzymes, Substrates, and Equipment
T4 polynucleotide kinase was purchased from New England Biolabs, [γ-32P]ATP from Amersham Biosciences, and dNTPs from Invitrogen. The catalytic core of yeast pol η with an N-terminal His6 tag was prepared as described previously (38). Full-length human pol η with a C-terminal His6 tag was also prepared as described previously (39). The mC-containing template ODN and primer ODNs were synthesized by Integrated DNA Technologies, Inc. Mass spectrometry was carried out on an LCQ Classic ion trap mass spectrometer (Thermo Finnigan, San Jose, CA).
Preparation and Deamination of cis-syn T=mC CPD-containing 14-mer DNA Template
100 μg of TmC 14-mer in 200 μl of 20 mm Tris (pH 8.8) on a piece of Saran Wrap was irradiated for 1 h on top of a 302-nm transilluminator (∼1.9 milliwatts/cm2) at 4 °C in a cold room. HPLC purification was carried out with 50 mm triethylamine acetate at pH 8.5 to minimize deamination to yield ∼5.2 μg of the cis-syn dimer eluting in 1 ml of ∼10% acetonitrile. This sample was immediately stored at −20 °C prior to use. A sample was also completely deaminated by adjusting the pH of the T=mC 14-mer sample to pH 6.5 by adding 0.5 m Mes buffer and heated overnight at 67 °C.
DNA Photoproduct Identification and Characterization by HPLC and Mass Spectrometry
The fractions corresponding to the major photoproduct peaks from the HPLC of the TmC 14-mer irradiation mixture, with and without heating, were analyzed by an enzyme-coupled mass spectrometry assay. In a typical assay, 1 μl of a 1 unit/μl aqueous solution of nuclease P1 was added to a 10-μl aliquot of the ODN sample, incubated at room temperature for 3 min, cooled on ice, and immediately analyzed by electrospray ionization coupled to MS/MS. MS/MS data were acquired on the selected [M − H] ions. To select both the deaminated and undeaminated components for fragmentation, the mass width for precursor selection was set at 5–6 m/z units
Single-hit Competitive Insertion Assay
In a cold room (4 °C), a 5-μl aliquot of the T=mC 14-mer template (120 nm) was added to 5 μl of 40 nm 5′-32P end-labeled primer, 20 mm Tris-HCl (pH 7.5), and 10 mm DTT and allowed to anneal for 10 min before the addition of pol η (500 nm). Each sample was incubated for an additional 7 min before initiating single-hit synthesis by the addition of 30 μl of all dNTPs (200 μm each) containing 10 mm MgCl2, 10 mm Tris-HCl (pH 7.5), and 10 mg/ml sonicated/denatured salmon sperm DNA as a polymerase trap. The synthesis reaction was quenched after 10 s by the addition of 80 μl of formamide containing 0.1% xylene cyanol, 0.2% SDS, 25 mm EDTA, and 1 μg of unlabeled primer (stop mixture). The samples were then heated at 100 °C for 10 min before loading onto a 10% polyacrylamide gel (40 cm × 0.8 mm) containing 25 mm citrate (pH 3.5), which was the same as the reservoir buffer. The gel was polymerized by adding 1.3 ml of ferrous sulfate (250 mg/100 ml), 1.3 ml of 10% ascorbate, and 300 μl of 3% hydrogen peroxide. The gel was run at 2000 V until the xylene cyanol dye marker reached 25 cm (∼3 h). For biased pool experiments, the dNTP of interest was held at 100 μm while the dGTP concentration was varied as indicated.
Multiple-hit Competitive Nucleotide Insertion Assays
In a cold room (4 °C), primer-templates were prepared by annealing 20 nm primer with 60 nm template, to which 200 μm each dNTP, 10 mm MgCl2, 5 mm DTT, 10 mm Tris-HCl (pH 7.5), and 500 nm pol η were added. After 2 min, a 10-μl aliquot was removed and added to a tube containing 300 μg of sonicated and denatured salmon sperm DNA before quenching the reaction with 80 μl of stop mixture. Aliquots from extension times that yielded almost exclusively full-length product were then assayed by electrophoresis on a 20% polyacrylamide gel containing 25 mm citrate (pH 3.5).
Deamination Rates by Nucleotide Insertion Assay
The T=mC 14-mer template (120 nm) was adjusted to pH 7.5 with Mes buffer (0.5 m) and incubated at 37 or 50 °C. Aliquots (5 μl) were removed at various times and quickly frozen on dry ice before storing overnight at −70 °C. All subsequent steps were performed in a cold room (4 °C). To each of the 5-μl aliquots was added 5 μl of 40 nm 5′-32P end-labeled primer, 1 mm each dNTP, 20 mm Tris-HCl (pH 7.5), and 10 mm DTT, followed by annealing for 10 min before the addition of yeast pol η (500 nm). Each sample was incubated for an additional 5 min to complete multiple-hit synthesis, after which 300 μg of sonicated and denatured salmon sperm DNA was added before quenching the reaction with 80 μl of stop mixture. The samples were then heated at 95 °C for 10 min before loading onto a 25% polyacrylamide gel (40 cm × 0.8 mm) containing 25 mm citrate (pH 3.5), which was the same as the reservoir buffer, and the gel was run at 2000 V until the xylene cyanol dye marker reached 18 cm (∼4 h). The gel had been polymerized by adding 1.3 ml of ferrous sulfate (250 mg/100 ml), 1.3 ml of 10% ascorbate, and 300 μl of 3% hydrogen peroxide.
Analysis of Deamination Rate Data
The deamination rate constant was obtained from the slope of a nonlinear least squares fit of the natural log of the fraction of dGMP inserted, G/(G + A), versus time to ln(G/(G + A))0 −k*t where (G/(G + A))0 equals the fraction of dGMP inserted at time 0.
RESULTS
Template Design
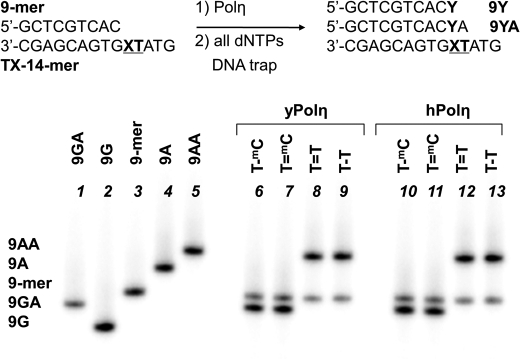
Because there is still no phosphoramidite building block available for incorporating a TmC CPD into an ODN, we resorted to preparing the TmC CPD-containing template by UV irradiation of an ODN. On the basis of prior experience with the preparation of an mC=TA CPD-containing 14-mer (33), we designed the TmC-14-mer (Fig. 2 and supplemental Fig. S1). This sequence is devoid of other dipyrimidine sites, so the T=mC CPD-containing product could be cleanly separated from other photoproducts of the TmC site and unreacted ODN by reverse phase HPLC. The sequence was long enough, however, to accommodate a 10-mer for primer extension reactions by positioning the TmC site toward the 5′-end. However, the exact sequence was modified from what was used before to accommodate the TmCG sequence and to optimize separation of the expected primer extension products by citrate-PAGE according to which nucleotides were inserted opposite the CPD by pol η (Fig. 2).
FIGURE 2.
Single-hit primer extension competition experiment opposite 14-mer templates. An equimolar mixture of all dNTPs (200 μm each) together with sonicated/denatured salmon sperm DNA was added to a preincubated mixture of the indicated 9-mer primer-14-mer template and pol η. The reactions were terminated after 10 s with EDTA and unlabeled primer, and the products were electrophoresed on a citrate (pH 3.5)-10% polyacrylamide gel. The standards corresponding to extension of the 9-mer by A or G were prepared by automated synthesis.
Preparation and Characterization of CPD-containing 14-mer Templates
The TmC-14-mer was irradiated with 302 nm light at 4 °C at pH 8.5 to suppress deamination for 1 h, after which half of the sample was allowed to completely deaminate by heating at 67 °C for 3 h at pH 6.5. The reaction mixtures were then subjected to reverse phase HPLC to separate the different photoproducts from the starting TmC-14-mer. The fractions collected by HPLC were immediately cooled on dry ice to suppress deamination prior to further analysis and use. The desired cis-syn CPD-containing product, T[c,s]mC-14-mer, was initially identified by its conversion to its deamination product, T[c,s]T-14-mer, which eluted with a longer retention time. Thus, UV irradiation of the TmC-14-mer resulted in one major product peak with a retention time of 30.5 min (supplemental Fig. S2A), which shifted to a peak with a retention time of 33.2 min after heating to effect deamination (supplemental Fig. S2B). To further confirm the assignment of the T=mC-14-mer as the cis-syn cyclobutane dimer, we analyzed the sample by nuclease P1-coupled electrospray ionization-MS. Nuclease P1 digests CPD-containing DNA to mononucleotides and a CPD-containing trinucleotide, pPy=PyN, which results in a major fragment, pPy=Pyab, corresponding to loss of the base from the 3′-nucleotide to yield an abasic site (ab) (40). MS/MS of the nuclease P1 digestion products of the HPLC fraction that eluted at 30.5 min showed an ion at m/z 953.4, corresponding to a pT=mCG photoproduct, which fragmented to a major ion at m/z 802 (supplemental Fig. S3), corresponding to pT[c,s]mCab. MS/MS of the digestion products of the fraction eluting at 33.2 min showed an ion at m/z 954.3, corresponding to pT=TG, which fragmented to an ion at m/z 803, corresponding to pT[c,s]Tab (supplemental Fig. S4).
Single-hit Competitive Nucleotide Insertion Assay Opposite CPDs by pol η
To determine the extent to which the mC in the T=mC CPD codes as C or T, we first carried out a single-hit competition assay with dATP and dGTP. In this assay, yeast pol η was first incubated with the 9-mer primer-TX 14-mer templates and then added to a solution containing equimolar dATP and dGTP and a large excess of sonicated and denatured salmon sperm DNA to trap the polymerase once it dissociated from the primer-template. To resolve the products of insertion, we used of a 25 mm citrate gel (pH 3.5), which can easily separate products according to their nucleotide composition as evidenced from the independently synthesized standards in Fig. 2. Primer extension under these single-hit conditions yielded primarily the products in which the primer was extended by two nucleotides, corresponding to the almost exclusive extension by G followed by A for the native and CPD-containing TmC templates and to extension by AA for the TT templates.
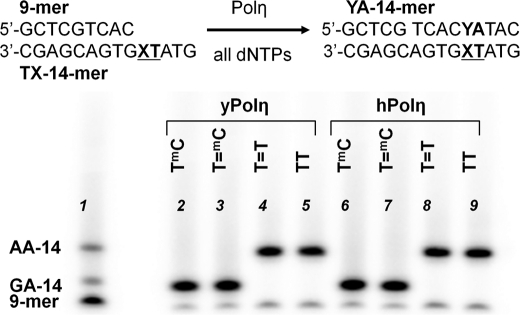
Multiple-hit Competitive Nucleotide Insertion Assay Opposite CPDs
We also examined insertion opposite the mC site in the T=mC CPD using a multiple-hit competition assay in which the full-length translesion synthesis products could also be separated according to nucleotide composition on a low pH citrate gel. In this assay, primer extension was carried out by both yeast and human pol η on the T=mC-14-mer template in the presence of equal amounts of all dNTPs. The primer extension products were then electrophoresed on a 20% Tris borate/EDTA-polyacrylamide gel to identify the full-length synthesis products in comparison with authentic 14-mer standards AA-14-mer and GA-14-mer, corresponding to insertion of dAMP or dGMP opposite the mC site, respectively. We found that both yeast and human pol η were able to fully extend the primer to the end of the T=mC and T=T-14-mer templates and that the full-length products had the same mobility on a denaturing polyacrylamide gel as the primer extension products opposite the undimerized TmC and TT-14-mer templates and both the GA-14-mer and AA-14-mer standards (supplemental Fig. S5). To determine the nucleotide insertion selectivity opposite the dipyrimidine sites, we used a 20% citrate-polyacrylamide gel to separate the full-length 14-mer synthesis products according to sequence composition (Fig. 3). As shown, the full-length synthesis products from both yeast and human pol η opposite the T=mC-14-mer were found to have the same gel mobility as those opposite the undimerized TmC-14-mer, as well as an authentic 14-mer ODN containing G (Fig. 3, lanes 1–3), indicating the preferential insertion of G opposite the mC of the CPD. Conversely, the full-length synthesis product opposite the completely deaminated T=mC-14-mer had the same mobility as that opposite the undimerized TT template and the authentic 14-mer product containing A (lanes 1, 2, and 5). These results demonstrate that the T=mC-14-mer template did not undergo any detectable deamination during its purification and subsequent handling, which would have resulted in the production of an A-containing 14-mer bypass product.
FIGURE 3.
Multiple-hit full-length primer extension experiment. The indicated 9-mer primer-14-mer templates were incubated with pol η and 200 μm each dNTP for 2 min, and the products were electrophoresed on a citrate (pH 3.5)-20% polyacrylamide denaturing gel in comparison with standards.
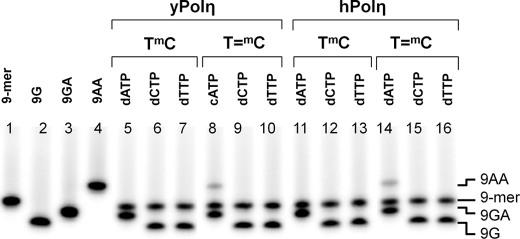
Selectivity of Nucleotide Insertion Opposite T=mCG CPD
To determine the selectivity of nucleotide insertion opposite the mC of the dimer in comparison with undamaged DNA, we carried out the single-hit primer extension experiment in the presence of a biased nucleotide pool in which the indicated nucleotide was present in a 32-fold excess over dGTP (Fig. 4). As shown, the 9-mer primer was extended only by G opposite both TmC and T=mC by both yeast and human pol η, even in the presence of 32-fold excess dCTP or dTTP. Extension of the primer by C or T would have produced bands moving slower than the 9-mer and faster than 9G, respectively (33), which was not observed. On the other hand, in the presence of 32-fold excess dATP, a small amount of a band corresponding to extension by AA opposite T=mC was observed in addition to extension by GA. This band was not observed for the undamaged TmC site, suggesting that insertion opposite the mC in a dimer proceeds with a lower fidelity of insertion than that opposite an undamaged mC.
FIGURE 4.
Single-hit primer extension biased nucleotide pool competition experiment opposite 14-mer templates. A 32:1 mixture of the indicated dNTP/dGTP (100 μm in the dNTP) together with sonicated/denatured salmon sperm DNA was added to a preincubated mixture of the indicated 9-mer primer-14-mer template and pol η. The reaction was terminated after 10 s with EDTA and unlabeled primer, and the products were electrophoresed on a citrate (pH 3.5)-10% polyacrylamide gel. The standards corresponding to extension of the 9-mer by A or G were prepared by automated synthesis.
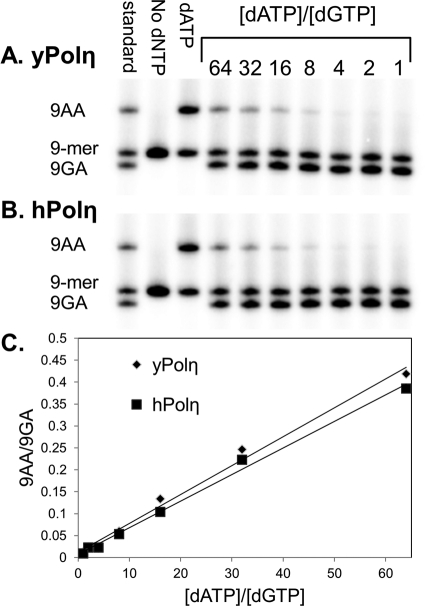
To more accurately determine the selectivity of nucleotide insertion, primer extension was carried out with different ratios of dATP to dGTP (Fig. 5). The selectivity of an A insertion relative to G could then be determined accurately from the slope of a linear fit of the ratio of the primer extension product incorporating A compared with G versus the ratio of the concentrations of dATP versus dGTP. Both yeast and human pol η showed similar selectivities for A versus G insertion of 6.6 × 10−3 and 6.1 × 10−3, respectively, corresponding to G versus A selectivities of 152 ± 7 and 165 ± 6. The non-zero intercepts of 0.012 ± 0.008 and 0.007 ± 0.006 for yeast and human pol η suggest the presence of ∼1% of the deaminated T=mC CPD and/or the E-imino tautomer.
FIGURE 5.
Selectivity of dGMP versus dAMP insertion opposite mC of T=mC CPD via single-hit assay. Varying ratios of dATP to dGTP (with dATP fixed at 100 μm) together with sonicated/denatured salmon sperm DNA were added to a preincubated mixture of the 9-mer primer-14-mer template and pol η. The reaction was terminated after 10 s with EDTA and unlabeled primer, and the products were electrophoresed on a citrate (pH 3.5)-10% polyacrylamide gel. The standards corresponding to extension of the 9-mer by A or G were prepared by automated synthesis.
Deamination Kinetics of T=mC CPD
The ability of pol η to faithfully insert A opposite T and G opposite mC in a CPD was then used to study the deamination kinetics of the T=mC CPD at two temperatures. The fraction of mC remaining in the CPD as a function time was determined by monitoring the fraction of G inserted opposite the mC by pol η in the full-length primer extension reaction (supplemental Fig. S6). The rate constants were then determined from the slope of linear fits of the data to a first-order rate process (supplemental Fig. S7). In this way, the deamination rate constants were determined to be 0.0014 min−1 at 37 °C and 0.0054 min−1 at 50 °C, corresponding to deamination half-lives of 8.25 and 2.1 h, respectively.
DISCUSSION
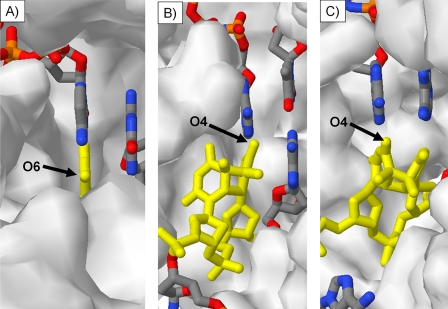
We have demonstrated that DNA synthesis by both yeast and human pol η opposite a mC in a cis-syn CPD of a TmCG site proceeds in a >99% error-free and hence non-mutagenic manner. Our original expectation was that the mC might also exist in an imino tautomeric form based on an early study showing that 5,6-dihydrocytosine could exist in either an amino or imino tautomeric form depending on the solvent (28). In the relatively non-polar solvent chloroform, dihydrocytosine was found to exist primarily in an imino tautomeric form (E and/or Z), whereas in water, it was found to exist primarily in the amino tautomeric form. If a polymerase were to surround the nascent base pair involving C or mC of a CPD and exclude water, as it does for a representative replicative polymerase, yeast pol δ (Fig. 6A) (41), one might expect an increased preference for an imino tautomer.
FIGURE 6.
Visual comparison of water accessibility of O6 of template guanine (yellow) in active site of yeast pol δ (Protein Data Bank code 3IAY) (A) and of O4 of template 3′-T (B) and template 5′-T (C) of cis-syn TT CPD (yellow) in active site of human pol η (code 3MR4).
The observation of only a dGMP insertion opposite the mC of the TmC CPD by either yeast or human pol η in the presence of equal concentrations of all four dNTPs indicates that the mC is not adopting a significant amount of the E-imino tautomer form, which would have templated the insertion of dAMP (Fig. 1). An upper limit to the amount of the E-imino tautomer comes from the non-zero intercept of ∼1% observed in the plots of dAMP versus dGMP insertion as a function of [dATP]/[dGTP], which could also more likely be attributed to the presence of the deaminated T=mC CPD. We do not have any direct way of knowing whether or not any of the Z-imino tautomer is present, as it is not Watson-Crick complementary to any of the canonical DNA bases. If it is present, it is either very poor at directing nucleotide insertion or directs the insertion of only G. Results from the single-hit primer extension experiments (Fig. 2) did not show any premature dissociation from the template compared with the undamaged template that would be indicative of significant impediment to nucleotide insertion. If the Z-imino isomer were directing insertion of G, it would likely be occurring via the equivalent of a G·T wobble base pair. Both insertion and extension of a G opposite the 3′-T of a T=T CPD by yeast pol η are known, however, to be much less efficient than for a Watson-Crick base-paired A (42, 43). Thus, the comparably efficient extension opposite the 3′-mC in the dimer and the undamaged template observed in the single-hit primer extension reaction suggest that insertion is occurring largely if not exclusively via the amino tautomer in the CPD.
The selectivity of dGMP insertion relative to dAMP by yeast and human pol η opposite the mC of the T=mC CPD is less, however, than that opposite an undamaged mC, as shown in Fig. 3, in which 32-fold excess of dATP over dGTP was used. In Fig. 3, a band for dAMP insertion opposite the mC in the CPD is clearly seen, whereas it is not seen for undamaged mC. This is consistent with experiments that determined that the selectivity for dGMP insertion relative to dAMP opposite a C in undamaged DNA was 960 for the same yeast pol η used in these experiments (44) and 909 for human pol η (45) compared with 152 and 165, respectively, which we observed for insertion opposite the mC in the T=mC CPD. The lower fidelity may have to do with the greater ability of an mC in a CPD to adopt the E-imino tautomer compared with an undamaged mC due to the loss of aromatic stabilization of the amino tautomer (27, 29, 30). Interestingly, we had observed a selectivity of only 120 for insertion opposite the mC in an mC=T CPD (33), suggesting that the 5′-mC may be more prone to tautomerization or that insertion opposite the amino tautomer is less specific opposite the 5′-mC of a CPD.
The almost exclusive presence of the amino tautomeric form of the mC in the CPD during nucleotide insertion suggests that it is in a polar and/or aqueous environment (28), which is in accord with a recent crystal structure of a ternary complex of human pol η with dATP opposite the 3′-T of a T=T CPD (25). In this structure, the O4 carbonyl of the 3′-T of the T=T CPD, corresponding to the position of the N4 of the 3′-C of a Py=mC CPD, is completely exposed to water during insertion of dATP (Fig. 6B). Likewise, the O4 carbonyl of the 5′-T of the CPD, corresponding to the 5′-mC of an mC=Py CPD, is also highly exposed to water during dATP insertion (Fig. 6C). The 5′-mC of an mC=T CPD would therefore also be expected to adopt the amino tautomeric form, in accord with our previous study showing that only dGMP is inserted opposite this mC in the presence of a equal concentration of A (33). These results suggest that pol η may have evolved to bypass CPDs in an error-free manner by increasing the exposure of the N4 of a C or an mC in either or both of the 3′- and 5′-sites of a cis-syn CPD to maximize the amount of the amino tautomer.
The active site of pol η also had to evolve to accommodate the 5′-pyrimidine of the CPD when the 3′-pyrimidine of the CPD is in the templating position, otherwise the 3′-pyrimidine could not template insertion of the complementary nucleotide. Replicative polymerases achieve high selectivity for canonical bases in part by extensive contacts with the 5′-face of the templating nucleotide (Fig. 6A), which would prevent the CPD from entering the active site when insertion opposite the 3′-pyrimidine of the CPD is to take place. As a result, the polymerase would become arrested at a CPD, which would allow for the recruitment of DNA damage bypass polymerases. It has been shown, however, that in the absence of other polymerases, the replicative T7 DNA polymerase will preferentially insert A opposite the 3′-pyrimidine of all types of dipyrimidine photoproducts, irrespective of their base pairing properties, by a non-templated mechanism (46–48). If non-templated insertion were to occur opposite the 3′-C or mC of a CPD, a C-to-T mutation would result and may represent an alternate pathway for UV light-induced C-to-T mutations. Thus, the T=mC CPD could serve as a useful probe for determining if a polymerase synthesizes past CPDs by a templated or non-templated mechanism.
Although bypass of a C- or an mC-containing CPD by pol η occurs in an error-free and hence non-mutagenic manner, once the C or mC in a CPD deaminates, error-free insertion by pol η opposite the resulting U or T leads to a mutation. Thus, the mutagenicity of pol η bypass of a CPD will depend on the presence and deamination rate of a C or an mC within the CPD, which we have recently shown to be highly dependent on sequence (34), protein binding (49), and nucleosome position (50). As we have shown herein, the T=mC CPD in a single-strand ATmCG sequence context deaminates with a half-life of 8.25 h at 37 °C, which is not much different from what we previously found for a CPD with the same flanking sequence in duplex DNA at low salt concentration (7.7 h) (34). On the other hand, the methyl-CpG-binding protein MeCP2 was found to completely suppress deamination of a T=mCG CPD (49) and hence the mutagenic potential of the CPD if bypassed by pol η. In a nucleosome core particle, the deamination rate of a T=mCG CPD whose backbone faced away from the histone surface was accelerated by a factor of 9, whereas one whose backbone faced toward the surface was retarded by ∼5-fold (50). Thus, as long as pol η synthesizes past a C/mC-containing CPD prior to deamination, the bypass event will be non-mutagenic, but as soon as the CPD deaminates, the bypass will be mutagenic. Therefore, although pol η synthesizes past CPDs in an error-free manner, in the sense of faithfully inserting nucleotides that are complementary to the pyrimidines present in the CPD, if a U or T arises from deamination of a C or an mC, translesion synthesis will be mutagenic and can explain the origin of UV light-induced C-to-T mutations.
Supplementary Material
Acknowledgment
We thank Dian Su for acquiring the mass spectral data.
This work was supported, in whole or in part, by National Institutes of Health Grant CA40463 (to J.-S. T. and Z. S.).

This article contains supplemental Figs. S1–S7.
- CPD
- cyclobutane pyrimidine dimer
- mC
- 5-methylcytosine
- pol η
- polymerase η
- ODN
- oligodeoxynucleotide
- yPolη
- yeast pol η
- hPolη
- human pol η.
REFERENCES
- 1. Brash D. E., Rudolph J. A., Simon J. A., Lin A., McKenna G. J., Baden H. P., Halperin A. J., Pontén J. (1991) A role for sunlight in skin cancer: UV-induced p53 mutations in squamous cell carcinoma. Proc. Natl. Acad. Sci. U.S.A. 88, 10124–10128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ziegler A., Leffell D. J., Kunala S., Sharma H. W., Gailani M., Simon J. A., Halperin A. J., Baden H. P., Shapiro P. E., Bale A. E. (1993) Mutation hotspots due to sunlight in the p53 gene of non-melanoma skin cancers. Proc. Natl. Acad. Sci. U.S.A. 90, 4216–4220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. You Y. H., Li C., Pfeifer G. P. (1999) Involvement of 5-methylcytosine in sunlight-induced mutagenesis. J. Mol. Biol. 293, 493–503 [DOI] [PubMed] [Google Scholar]
- 4. You Y. H., Szabó P. E., Pfeifer G. P. (2000) Cyclobutane pyrimidine dimers form preferentially at the major p53 mutational hotspot in UVB-induced mouse skin tumors. Carcinogenesis 21, 2113–2117 [DOI] [PubMed] [Google Scholar]
- 5. Setlow R. B., Carrier W. L., Bollum F. J. (1965) Pyrimidine dimers in UV-irradiated poly dI:dC. Proc. Natl. Acad. Sci. U.S.A. 53, 1111–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu F. T., Yang N. C. (1978) Photochemistry of cytosine derivatives. 1. Photochemistry of thymidylyl-(3′ → 5′)-deoxycytidine. Biochemistry 17, 4865–4876 [DOI] [PubMed] [Google Scholar]
- 7. Fix D., Bockrath R. (1981) Thermal resistance to photoreactivation of specific mutations potentiated in E. coli B/r ung by ultraviolet light. Mol. Gen. Genet. 182, 7–11 [DOI] [PubMed] [Google Scholar]
- 8. Lemaire D. G., Ruzsicska B. P. (1993) Kinetic analysis of the deamination reactions of cyclobutane dimers of thymidylyl-3′,5′-2′-deoxycytidine and 2′-deoxycytidylyl-3′,5′-thymidine. Biochemistry 32, 2525–2533 [DOI] [PubMed] [Google Scholar]
- 9. Barak Y., Cohen-Fix O., Livneh Z. (1995) Deamination of cytosine-containing pyrimidine photodimers in UV-irradiated DNA. Significance for UV light mutagenesis. J. Biol. Chem. 270, 24174–24179 [DOI] [PubMed] [Google Scholar]
- 10. Peng W., Shaw B. R. (1996) Accelerated deamination of cytosine residues in UV-induced cyclobutane pyrimidine dimers leads to CC → TT transitions. Biochemistry 35, 10172–10181 [DOI] [PubMed] [Google Scholar]
- 11. Tu Y., Dammann R., Pfeifer G. P. (1998) Sequence- and time-dependent deamination of cytosine bases in UVB-induced cyclobutane pyrimidine dimers in vivo. J. Mol. Biol. 284, 297–311 [DOI] [PubMed] [Google Scholar]
- 12. Nouspikel T. (2009) DNA repair in mammalian cells: nucleotide excision repair: variations on versatility. Cell. Mol. Life Sci. 66, 994–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tornaletti S. (2009) DNA repair in mammalian cells: transcription-coupled DNA repair: directing your effort where it's most needed. Cell. Mol. Life Sci. 66, 1010–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lagerwerf S., Vrouwe M. G., Overmeer R. M., Fousteri M. I., Mullenders L. H. (2011) DNA damage response and transcription. DNA Repair 10, 743–750 [DOI] [PubMed] [Google Scholar]
- 15. Waters L. S., Minesinger B. K., Wiltrout M. E., D'Souza S., Woodruff R. V., Walker G. C. (2009) Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol. Mol. Biol. Rev. 73, 134–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang C. I., Taylor J. S. (1991) Site-specific effect of thymine dimer formation on dAn·dTn tract bending and its biological implications. Proc. Natl. Acad. Sci. U.S.A. 88, 9072–9076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jing Y., Kao J. F., Taylor J. S. (1998) Thermodynamic and base pairing studies of matched and mismatched DNA dodecamer duplexes containing cis-syn, (6-4), and Dewar photoproducts of TT. Nucleic Acids Res. 26, 3845–3853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Park H., Zhang K., Ren Y., Nadji S., Sinha N., Taylor J. S., Kang C. (2002) Crystal structure of a DNA decamer containing a cis-syn thymine dimer. Proc. Natl. Acad. Sci. U.S.A. 99, 15965–15970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huang J. C., Hsu D. S., Kazantsev A., Sancar A. (1994) Substrate spectrum of human excinuclease: repair of abasic sites, methylated bases, mismatches, and bulky adducts. Proc. Natl. Acad. Sci. U.S.A. 91, 12213–12217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Johnson R. E., Prakash S., Prakash L. (1999) Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, pol η. Science 283, 1001–1004 [DOI] [PubMed] [Google Scholar]
- 21. Masutani C., Kusumoto R., Yamada A., Dohmae N., Yokoi M., Yuasa M., Araki M., Iwai S., Takio K., Hanaoka F. (1999) The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature 399, 700–704 [DOI] [PubMed] [Google Scholar]
- 22. Prakash S., Johnson R. E., Prakash L. (2005) Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 74, 317–353 [DOI] [PubMed] [Google Scholar]
- 23. Sun L., Zhang K., Zhou L., Hohler P., Kool E. T., Yuan F., Wang Z., Taylor J. S. (2003) Yeast pol η holds a cis-syn thymine dimer loosely in the active site during elongation opposite the 3′-T of the dimer, but tightly opposite the 5′-T. Biochemistry 42, 9431–9437 [DOI] [PubMed] [Google Scholar]
- 24. Hwang H., Taylor J. S. (2005) Evidence for Watson-Crick and not Hoogsteen or wobble base pairing in the selection of nucleotides for insertion opposite pyrimidines and a thymine dimer by yeast DNA pol η. Biochemistry 44, 4850–4860 [DOI] [PubMed] [Google Scholar]
- 25. Biertümpfel C., Zhao Y., Kondo Y., Ramón-Maiques S., Gregory M., Lee J. Y., Masutani C., Lehmann A. R., Hanaoka F., Yang W. (2010) Structure and mechanism of human DNA polymerase η. Nature 465, 1044–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jiang N., Taylor J. S. (1993) In vivo evidence that UV-induced C → T mutations at dipyrimidine sites could result from the replicative bypass of cis-syn cyclobutane dimers or their deamination products. Biochemistry 32, 472–481 [DOI] [PubMed] [Google Scholar]
- 27. Danilo V. I., Les A., Alderfer J. L. (2001) A theoretical study of the cis-syn pyrimidine dimers in the gas phase and water cluster and a tautomer bypass mechanism for the origin of UV-induced mutations. J. Biomol. Struct. Dyn. 19, 179–191 [DOI] [PubMed] [Google Scholar]
- 28. Brown D. M., Hewlins M. J. (1968) Dihydrocytosine and related compounds. J. Chem. Soc. C 2050–2055 [Google Scholar]
- 29. Dupuy-Mamelle N., Pullman B. (1967) No. 61.-Recherches theoriques sur la structure electronique des purine et pyrimidines bilogiques. IV. Les pyrimidines saturees sur la liaison 5–6. J. Chim. Phys. Phys. Chim. Biol. 64, 708–712 [Google Scholar]
- 30. Danilov V. I., Stewart J. J. P., Les A., Alderfer J. L. (2000) A theoretical study of pyrimidine photohydrates and a proposed mechanism for the mutagenic effect of ultraviolet light. Chem. Phys. Lett. 328, 75–82 [Google Scholar]
- 31. Yu S. L., Johnson R. E., Prakash S., Prakash L. (2001) Requirement of DNA polymerase η for error-free bypass of UV-induced CC and TC photoproducts. Mol. Cell. Biol. 21, 185–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yoon J. H., Prakash L., Prakash S. (2009) Highly error-free role of DNA polymerase η in the replicative bypass of UV-induced pyrimidine dimers in mouse and human cells. Proc. Natl. Acad. Sci. U.S.A. 106, 18219–18224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vu B., Cannistraro V. J., Sun L., Taylor J. S. (2006) DNA synthesis past a 5-methyl-C-containing cis-syn cyclobutane pyrimidine dimer by yeast pol η is highly non-mutagenic. Biochemistry 45, 9327–9335 [DOI] [PubMed] [Google Scholar]
- 34. Cannistraro V. J., Taylor J. S. (2009) Acceleration of 5-methylcytosine deamination in cyclobutane dimers by G and its implications for UV-induced C-to-T mutation hotspots. J. Mol. Biol. 392, 1145–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Frederico L. A., Kunkel T. A., Shaw B. R. (1990) A sensitive genetic assay for the detection of cytosine deamination: determination of rate constants and the activation energy. Biochemistry 29, 2532–2537 [DOI] [PubMed] [Google Scholar]
- 36. Shen J. C., Rideout W. M., 3rd, Jones P. A. (1994) The rate of hydrolytic deamination of 5-methylcytosine in double-stranded DNA. Nucleic Acids Res. 22, 972–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Horsfall M. J., Borden A., Lawrence C. W. (1997) Mutagenic properties of the T-C cyclobutane dimer. J. Bacteriol. 179, 2835–2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cannistraro V. J., Taylor J. S. (2004) DNA-thumb interactions and processivity of T7 DNA polymerase in comparison to yeast polymerase η. J. Biol. Chem. 279, 18288–18295 [DOI] [PubMed] [Google Scholar]
- 39. Sherrer S. M., Fiala K. A., Fowler J. D., Newmister S. A., Pryor J. M., Suo Z. (2011) Quantitative analysis of the efficiency and mutagenic spectra of abasic lesion bypass catalyzed by human Y-family DNA polymerases. Nucleic Acids Res. 39, 609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang Y., Taylor J. S., Gross M. L. (1999) Nuclease P1 digestion combined with tandem mass spectrometry for the structure determination of DNA photoproducts. Chem. Res. Toxicol. 12, 1077–1082 [DOI] [PubMed] [Google Scholar]
- 41. Swan M. K., Johnson R. E., Prakash L., Prakash S., Aggarwal A. K. (2009) Structural basis of high fidelity DNA synthesis by yeast DNA polymerase δ. Nat. Struct. Mol. Biol. 16, 979–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Washington M. T., Johnson R. E., Prakash S., Prakash L. (2000) Accuracy of thymine-thymine dimer bypass by Saccharomyces cerevisiae DNA polymerase η. Proc. Natl. Acad. Sci. U.S.A. 97, 3094–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Washington M. T., Johnson R. E., Prakash L., Prakash S. (2001) Accuracy of lesion bypass by yeast and human DNA polymerase η. Proc. Natl. Acad. Sci. U.S.A. 98, 8355–8360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Brown J. A., Zhang L., Sherrer S. M., Taylor J. S., Burgers P. M., Suo Z. (2010) Pre-steady-state kinetic analysis of truncated and full-length Saccharomyces cerevisiae DNA polymerase eta. J. Nucleic Acids doi: 10.4061/2010/871939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Johnson R. E., Washington M. T., Prakash S., Prakash L. (2000) Fidelity of human DNA polymerase η. J. Biol. Chem. 275, 7447–7450 [DOI] [PubMed] [Google Scholar]
- 46. Smith C. A., Baeten J., Taylor J. S. (1998) The ability of a variety of polymerases to synthesize past site-specific cis-syn, trans-syn-II, (6-4), and Dewar photoproducts of thymidylyl-(3′ → 5′)-thymidine. J. Biol. Chem. 273, 21933–21940 [DOI] [PubMed] [Google Scholar]
- 47. Sun L., Wang M., Kool E. T., Taylor J. S. (2000) Pyrene nucleotide as a mechanistic probe: evidence for a transient abasic site-like intermediate in the bypass of dipyrimidine photoproducts by T7 DNA polymerase. Biochemistry 39, 14603–14610 [DOI] [PubMed] [Google Scholar]
- 48. Taylor J. S. (2002) New structural and mechanistic insight into the A rule and the instructional and non-instructional behavior of DNA photoproducts and other lesions. Mutat. Res. 510, 55–70 [DOI] [PubMed] [Google Scholar]
- 49. Cannistraro V. J., Taylor J. S. (2010) Methyl CpG-binding protein 2 (MeCP2) enhances photodimer formation at methyl-CpG sites but suppresses dimer deamination. Nucleic Acids Res. 38, 6943–6955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Song Q., Cannistraro V. J., Taylor J. S. (2011) Rotational position of a 5-methylcytosine-containing cyclobutane pyrimidine dimer in a nucleosome greatly affects its deamination rate. J. Biol. Chem. 286, 6329–6335 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.