Background: MEF2C is essential for vascular smooth muscle development, yet the signaling pathways that regulate its function in this cell type remain largely unknown.
Results: We identify a novel regulator of MEF2C in vascular smooth muscle, called CPI-17.
Conclusion: Our data identify a genetic pathway involving CPI-17, MEF2C, and myocardin.
Significance: These findings have important ramifications during vascular development and for stem cell programming.
Keywords: Cardiovascular Disease, Cell Differentiation, Development, Vascular Biology, Vascular Smooth Muscle Cells, CPI-17, MEF2C, Muscle, Myocardin, RhoA
Abstract
Differentiation of vascular smooth muscle cells (VSMC) is a fundamental aspect of normal development and vascular disease. During contraction, VSMCs modulate calcium sensitivity through RhoA/ROCK-mediated inhibition of the myosin light chain phosphatase complex (MLCP). Previous studies have demonstrated that this signaling pathway functions in parallel to increase the expression of smooth muscle genes through the myocardin-family of co-activators. MEF2C fulfills a critical role in VSMC differentiation and regulates myocardin expression, leading us to investigate whether the RhoA/ROCK signaling cascade might regulate MEF2 activity. Depolarization-induced calcium signaling increased the expression of myocardin, which was sensitive to ROCK and p38 MAPK inhibition. We previously identified protein phosphatase 1α (PP1α), a known catalytic subunit of the MLCP in VSMCs, as a potent repressor of MEF2 activity. PP1α inhibition resulted in increased expression of myocardin, while ectopic expression of PP1α inhibited the induction of myocardin by MEF2C. Consistent with these data, shRNA-mediated suppression of a PP1α inhibitor, CPI-17, reduced myocardin expression and inhibited VSMC differentiation, suggesting a pivotal role for CPI-17 in regulating MEF2 activity. These data constitute evidence of a novel signaling cascade that links RhoA-mediated calcium sensitivity to MEF2-dependent myocardin expression in VSMCs through a mechanism involving p38 MAPK, PP1α, and CPI-17.
Introduction
During development, vascular smooth muscle cells (VSMCs)3 migrate to primitive endothelial tubes while simultaneously executing a program of differentiation to contribute to the vascular architecture (1). Upon incorporation into the vasculature, VSMCs become quiescent and primarily regulate vascular tone (2). However, unlike terminally differentiated striated muscle cell types, VSMCs retain a capacity, referred to as the activated or synthetic phenotype, to proliferate postnatally in response to vascular injury. This activated phenotype is of particular clinical interest, since it plays an important role in most stenotic vascular diseases described to date (3). The MADS-box (MCM-1, Agamous, Deficiens, Serum Response Factor) transcriptional regulators, serum response factor (SRF) and myocyte enhancer factor 2 (MEF2) play critical roles in the phenotypic modulation of VSMCs, as these transcription factors are known to regulate both immediate early genes involved in proliferation and migration, and, somewhat paradoxically, smooth muscle marker genes involved in the contractile phenotype (4–6). The cellular signals that direct SRF to these distinct sets of genes have been intensively studied, where SRF physically interacts with the myocardin family of co-activators in contractile VSMCs to induce smooth muscle marker gene expression (7). However, in response to proliferative growth factor stimulation, myocardin is displaced from SRF, in favor of an Elk-1 interaction, to target immediate early gene expression, such as c-fos (8). Recently, calcium signaling induced by depolarization has been shown to increase the expression of both SRF-dependent immediate early genes and smooth muscle marker genes (9). Interestingly, the induction of c-fos in this model was prevented by calcium/calmodulin-dependent kinase (CaMK) inhibition, and the induction of VSMC marker genes was attenuated by RhoA-associated kinase (ROCK) inhibition (9). These results suggest that distinct calcium-mediated signaling pathways regulate these seemingly opposing SRF-dependent genes.
Much less is known regarding the regulation of MEF2-dependent gene expression in VSMCs. Like SRF, MEF2 regulates the expression of immediate early genes, such as c-jun, and recent studies have suggested that c-jun expression in VSMCs is CaMK-dependent (6). However, MEF2C has also been shown to be genetically upstream of myocardin and of critical importance to VSMC differentiation (5, 10). Yet, the signaling pathways that regulate MEF2-dependent myocardin expression in VSMCs remain unknown; however, recent studies suggest that RhoA signaling may be involved (11, 12). We recently identified protein phosphatase 1α (PP1α) as a potent trans-dominant repressor of MEF2 activity (13). Interestingly, in VSMCs PP1α serves as the catalytic subunit of the myosin light chain phosphatase complex (MLCP) and is regulated by RhoA signaling to control calcium sensitivity during contraction (14). In addition, signals emanating from RhoA in VSMCs have been previously shown to activate p38 MAP kinase (MAPK) signaling (15), a known activator of MEF2 transcriptional activity in multiple cell types (16–18). In this report we document for the first time, a novel signaling pathway in VSMCs that links RhoA-mediated regulation of calcium sensitivity to MEF2-dependent expression of myocardin. This pathway involves the de-repression of MEF2 from PP1α inhibition by a two-step mechanism involving p38 MAPK and ROCK-mediated activation of the PP1α inhibitor, CPI-17 (PKC-potentiated protein phosphatase inhibitor of 17 kDa). Thus, this is the first report to identify a dominant signaling cascade that regulates myocardin expression in VSMCs, which may prove critical to our understanding of vascular development and stenotic vascular disease.
EXPERIMENTAL PROCEDURES
Plasmids
MEF2, PP1α, p38 MAPK (p38 MAPK), MKK6EE, and luciferase constructs were described previously (6, 13). The RhoA L63 and C3 transferase expression vectors were kindly provided by A. Hall, while the CPI-17 expression vector was a generous gift from A. Aitken. The activated ROCK and PKN constructs were generous gifts from M. Scheid and Y. Ono, respectively. Thr-38 mutations in CPI-17 were generated by site-directly mutagenesis. The shRNAs targeting MEF2C and CPI-17 were generated by ligating annealed oligonucleotides into the pSilencer 3.0 H1 vector, per the manufacturer's instructions, where the target sequence for the MEF2C 3′-UTR was 5′-AACAGAAATGCTGAGATACGC-3′ and the target sequence for CPI-17 was 5′-AAAGCCCAGATTGTTTCTAAG-3′.
Primary VSMC and Immortalized Cell Cultures
Primary rat aortic smooth muscle cells were isolated by enzymatic cell dispersion, as described in Hou et al. (19). Rat A10 myoblasts (ATCC; CRL-1476) were maintained in growth media consisting of 10% fetal bovine serum (FBS). Quiescence was obtained by re-feeding the cells with serum-free DMEM overnight. C3H10T1/2 mouse embryonic fibroblasts (ATCC; CCL-226) and COS7 cells were maintained in standard DMEM with 10% FBS.
shRNA Transfections
The shRNAs targeting MEF2C, CPI-17, or a nonspecific scrambled control, were transfected into A10 cells with Lipofectamine reagent (Invitrogen) according to the manufacturer's protocol. Transfected cells were enriched by puromycin selection (0.5 μg/ml) for 3 days prior to harvesting for protein extracts.
Luciferase and β-Galactosidase Assays
Transient transfections of A10, C3H10T1/2 and COS7 cells were performed by a modified calcium phosphate-DNA precipitation with pCMV-β-galactosidase serving as an internal control for transfection efficiency. Luciferase and β-galactosidase activityies were measured as described previously (6).
Immunoblot Analysis
Protein extractions were achieved using an Nonidet P-40 lysis buffer described previously (13). Protein concentrations were determined by Bradford assay, and 15 μg were resolved using SDS-PAGE and transferred to an Immobilon-P membrane (Millipore, Inc.). Immunoblotting was carried out using appropriate primary antibody in 5% powdered milk in PBS. Appropriate horseradish peroxidase-conjugated secondary antibody (Bio-Rad, 1:2000) was used in combination with chemiluminescence to visualize bands. Primary antibodies included, rabbit myocardin (Santa Cruz Biotechnology), c-Jun and c-Fos (Santa Cruz Biotechnology), p38 and p-p38 MAPK (NEB), and CPI-17 and p-CPI-17 (Santa Cruz Biotechnology), and smooth muscle α-actin (Sigma).
Co-immunoprecipitation
COS7 cells were transfected using calcium phosphate method and protein extracts were harvested, as described above. Immunoprecipitation was performed using the ExactaCruz kit (Santa Cruz Biotechnology), as per manufacturer's instructions. Elusions were analyzed by immunoblot, as described above.
Immunofluorescence
Primary VSMCs were fixed in 4% paraformaldehyde, permeabilized in ice-cold methanol, and incubated with a primary smooth muscle α-actin antibody (Sigma), CPI-17 antibody (Santa Cruz Biotechnology) with FITC- and TRITC-conjugated secondary antibodies. Cells were visualized using fluorescence microscopy.
Quantitative RT-PCR
Total RNA was isolated from primary VSMCs using a Cell-to-cDNA kit (Ambion), and quantitative PCR was performed using SYBR green (Applied Biosystems), and analyzed using the ΔΔCT method, as described previously (6).
RESULTS
Depolarization Enhances MEF2-dependent Gene Expression through Distinct Calcium-mediated Signaling Pathways in VSMCs
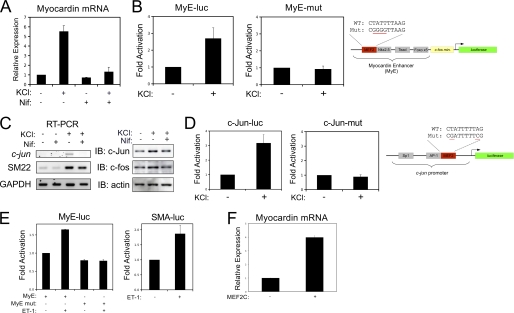
To determine the effect of depolarization-induced calcium signaling on VSMC marker gene expression, cultured VSMCs were treated with 60 mm potassium chloride (KCl) and nifedipine, an L-type calcium channel blocker. Depolarization, in both primary cultures and the A10 VSMC cell line, resulted in a nifedipine-sensitive increase in the expression of myocardin, the MEF2-dependent immediate early gene, c-jun; as well as SRF-dependent genes (Fig. 1). In addition, the induction of myocardin and c-Jun was found to be dependent on the MEF2 cis elements found within the proximal promoter regions of these genes (Fig. 1, B and D). Endothelin-1 (ET-1) has also been implicated in regulating calcium sensitivity in VSMCs during contraction through RhoA-dependent signaling, and our evidence suggests that ET-1 induces myocardin expression through the MEF2 cis element in a manner similar to depolarization (Fig. 1E) (20). Finally, to evaluate whether MEF2C could activate endogenous myocardin expression, we transfected A10 cells with MEF2C and observed an increase in myocardin expression determined by qPCR (Fig. 1F).
FIGURE 1.
Depolarization-induced expression of MEF2-target genes in VSMCs. A, primary VSMCs were depolarized with 60 mm KCl, following pretreatment with 1 μm of nifedipine (Nif), as indicated. Myocardin expression was evaluated by qPCR, corrected for GAPDH using the ΔΔCT method. B, A10 cells were transfected with a myocardin-enhancer reporter gene (MyE-luc) or with a reporter gene with the MEF2 cis element mutated (MyE-mut). Following recovery, cell were depolarized with 60 mm KCl and subjected to luciferase assay. C, quiescent A10 cells were treated with 60 mm KCl following a 15 min treatment of 5 μm Nifedipine (L-type calcium channel blocker). Immunoblotting was performed on protein extracts using c-Jun, c-Fos, and actin antibodies, and RT-PCR was performed on total RNA for c-Jun and SM22 and GAPDH. D, A10 cells were transfected with the c-jun promoter (c-Jun-luc) or with a reporter gene with the MEF2 cis element mutated (c-Jun mut). Following recovery, cells were depolarized with 60 mm KCl and subjected to luciferase assay. E, A10 cells were transfected with the myocardin enhancer and smooth muscle α-actin promoter, as indicated. Cells were treated overnight with endothelin-1 (10 nm), and extracts were subjected to luciferase assay. F, A10 cells were transfected with MEF2C (as indicated). Myocardin expression was evaluated by qPCR and corrected for GAPDH using the ΔΔCT method. Error bars indicate S.E.
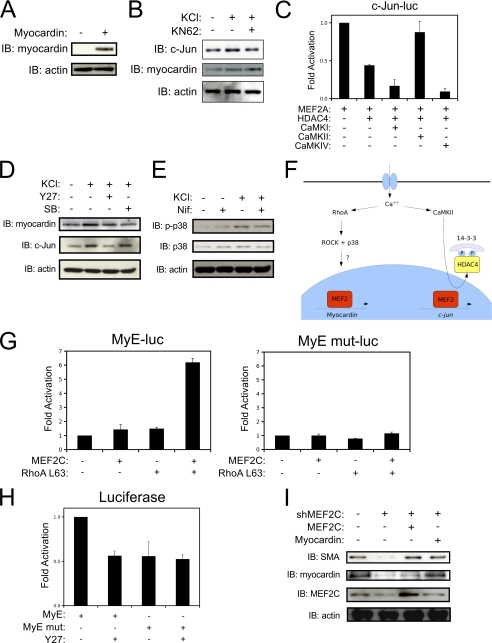
To dissect the calcium-dependent signaling pathways responsible for the induction of MEF2-dependent smooth muscle genes at the protein level, we utilized well-characterized pharmacological inhibitors in our culture model, after testing the specificity of myocardin antibodies to detect exogenous myocardin in COS7 cells (Fig. 2A). Interestingly, the depolarization-induced expression of c-Jun was attenuated by the CaMK inhibitor, KN-62; whereas, myocardin expression was not attenuated by this inhibitor (Fig. 2B). We have previously demonstrated that c-Jun expression in VSMCs is regulated by a MEF2-HDAC4 repressor complex (6). Consistent with our previous results, the c-jun promoter was repressed by ectopic expression of HDAC4; however, we now demonstrate that this repression can be counteracted by co-expression of an activated CaMKIIδ. Surprisingly, the HDAC4 repression of c-jun could not be overcome by other CaMKs, such as CaMKI or CaMKIV (Fig. 2C). In addition, depolarization resulted in a reduced nuclear content of HDAC4, suggesting that CaMKII promotes nuclear export of HDAC4 to de-repress c-jun expression (not shown).
FIGURE 2.
Distinct calcium-mediated signaling pathways regulate myocardin and c-Jun expression in VSMCs. A, COS7 cells were transfected with Myocardin-856 and subjected to immunoblotting with myocardin antibody (SC-33766, Santa Cruz Biotechnology). B, A10 cells were treated with 60 mm KCl for 2 h following 15 min pretreatment with 5 μm KN-62 (CaM kinase inhibitor) or DMSO as a vehicle control. Protein extracts were immunoblotted with c-Jun, myocardin, and actin antibodies. C, A10 cells were transfected with a c-jun reporter-gene (c-Jun-luc), MEF2A, HDAC4, and activated CaMKI, CaMKII or CaMKIV, as indicated. D, A10 cells were pretreated with either Y27632 (Y27, 5 μm) or SB203580 (SB, 5 μm), or DMSO as a vehicle control for 15 min, then depolarized for two hours. Extracts were subjected to immunoblotting as indicated. E, A10 cells were pretreated with nifedipine, depolarized, and subjected to immunoblotting. F, model of the distinct signaling pathways that regulate MEF2-dependent myocardin and c-jun expression in VSMCs. G, A10 cells were transfected with MyE or the enhancer with the MEF2 site mutated (MyE mut) along with a MEF2C, and/or an active RhoA (RhoA L63) expression vectors. Extracts were subjected to luciferase assays. H, A10 cells were transfected as described in G, treated with Y27632 (Y27, 5 μm), and harvested for luciferase assay. I, cells were transfected with a plasmid encoding a short-hairpin RNA targeting MEF2C (shMEF2C), and expression vectors for human MEF2C or myocardin, as indicated. Cultures were enriched for expression of the shRNA by puromycin selection, and extracts were subjected to immunoblotting. Error bars indicate S.E.
Intriguingly, depolarization-induced expression of myocardin was attenuated by the p38 MAPK inhibitor, SB203580; whereas, the induction of c-Jun is unaffected by this inhibitor (Fig. 2D). In addition, depolarization resulted in a nifedipine-sensitive increase in p38 MAPK activity, as indicated by an increase in phosphorylated p38 MAPK in response to KCl treatment (Fig. 2E). These results, and the work of others, indicate that distinct calcium-mediated signaling pathways regulate c-Jun and myocardin expression in VSMCs (Fig. 2F). Interestingly, the ROCK inhibitor, Y27632, could attenuate both myocardin and c-Jun expression induced by depolarization, which indicates some degree of cross-talk between these two pathways.
To further evaluate the role of RhoA/ROCK signaling in the regulation of myocardin expression, we utilized a myocardin-enhancer reporter gene that contains a MEF2 cis element (MyE). As shown in Fig. 2H, the ROCK inhibitor, Y27632, inhibited the myocardin enhancer, but not when the MEF2 cis element was mutated such that MEF2 can no longer bind (5). Additionally, the induction of this reporter-gene by MEF2C was prevented by co-expression of C3, a RhoA inhibitor (not shown). Congruently, forced expression of MEF2C and an activated RhoA (RhoA L63) cooperatively activate the myocardin enhancer, but again not when the MEF2 cis element is mutated (Fig. 2G).
To evaluate the necessity of MEF2C for myocardin expression and VSMC differentiation, we engineered a short-hairpin RNA to reduce MEF2C expression (shMEF2C). As shown in Fig. 2I, the shMEF2C reduced endogenous MEF2C expression in cultured A10 cells, which resulted in a corresponding reduction in the expression of myocardin and its down-stream VSMC target gene, smooth muscle α-actin (SMA). Furthermore, the shMEF2C-mediated reduction in myocardin expression and VSMC differentiation could be overcome by ectopic expression of human MEF2C, which is not suppressed by the shRNA, validating the specificity of the shMEF2C effect. Finally, the reduction in SMA induced by the shMEF2C could be by-passed by exogenous expression of myocardin (Fig. 2I). These results are consistent with our hypothesis that MEF2C regulates VSMC differentiation through myocardin (5), yet to our knowledge, this is the first report to evaluate this notion through both gain- and loss-of-function experiments. Collectively, these data indicate that the RhoA/ROCK signaling pathway provides an important activating stimulus for MEF2C-mediated induction of myocardin expression in VSMCs.
PP1α Regulates MEF2-dependent Gene Expression in VSMCs
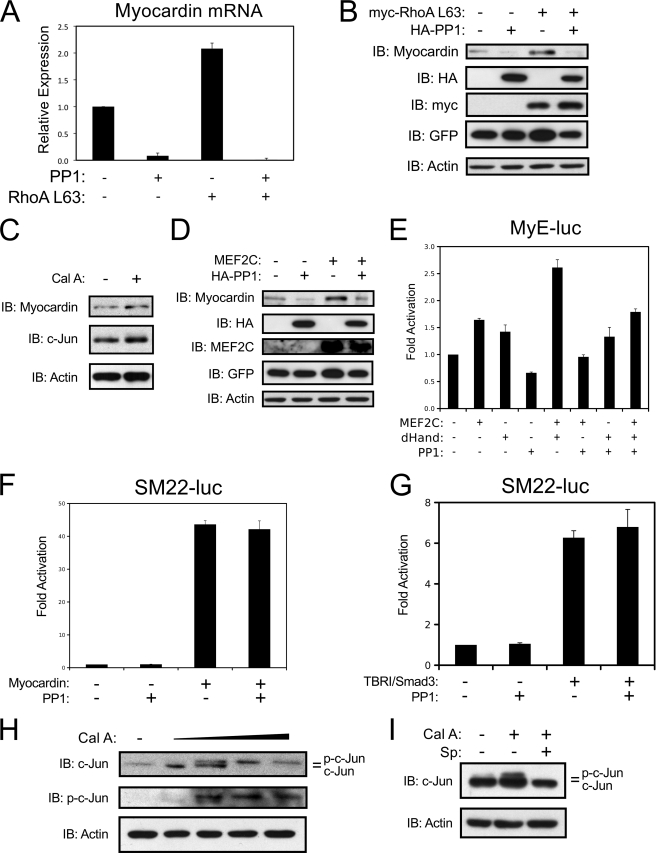
We have recently identified PP1α as a dominant repressor of MEF2 transcriptional activity (13). In smooth muscle, PP1α is the catalytic subunit of the myosin light chain phosphatase complex (MLCP) (21). Exogenous expression of PP1α inhibits endogenous myocardin expression and powerfully attenuates the induction of myocardin by ectopic expression of activated RhoA in cultured A10 cells (Fig. 3, A and B). In addition, exogenous expression of PP1α completely prevented the induction of endogenous myocardin expression by MEF2C (Fig. 3D). Furthermore, we utilized the PP1α inhibitor, calyculin A, to address the role of PP1α in MEF2-dependent gene expression in VSMCs. Calyculin A treatment increased the expression of both myocardin and c-Jun (Fig. 3C). Thus, ROCK regulation of PP1α might be an important mechanism for the attenuated expression of myocardin and c-Jun with Y27632 treatment (Fig. 2D). MEF2C has previously been shown to interact with the bHLH transcription factors of the Hand (heart and neural crest derived) family (22). In addition, genetic ablation of MEF2C, dHand (Hand2), and myocardin all result in some degree of neural crest-derived vascular defect (23–25). Therefore, we chose to evaluate whether PP1α could inhibit a functional cooperation between MEF2C and dHand. As shown in Fig. 3E, MEF2C and dHand cooperatively activated the myocardin enhancer, yet exogenous expression of PP1α attenuated this effect. To evaluate whether PP1α could block smooth muscle gene expression directly (i.e. downstream of MEF2C), we utilized a SM22-dependent reporter gene. As shown in Fig. 3, F and G, forced expression of PP1α could not overcome the induction of this promoter by myocardin or Smad3. These results indicate that PP1α regulates smooth muscle gene expression through MEF2C and not through genetically downstream transcription factors, such as SRF.
FIGURE 3.
Myocardin expression is opposed by PP1α. A, A10 cells were transfected with HA-PP1α (PP1) and activated myc-RhoA (Myc-RhoA L63) using Lipofectamine reagent and puromycin-selected overnight. Myocardin expression was evaluated by qPCR and corrected for GAPDH using the ΔΔCT method. B, A10 cells were transfected as described in A. Protein extracts were subjected to immunoblotting as indicated. C, A10 cells were treated with Calyculin A (0.5 ng/ml) or DMSO as a vehicle control for 2 h. Protein extracts were immunoblotted as indicated. D, A10 cells were transfected with HA-PP1α and MEF2C as described in A. Protein extracts were subjected to immunoblotting as indicated. E, COS7 cells were transfected with the myocardin enhancer, MEF2C, dHand, and/or PP1α (PP1) as indicated. Luciferase assays were performed on the cells extracts. F and G, A10 cells were transfected with the SM22 promoter, myocardin, an activated type I TGF-β receptor (TBRI), Smad3, or PP1α, as indicated. Extracts were harvested for luciferase. H, A10s were treated with increasing amounts of calyculin A (Cal A; 0.25 ng/ml, 0.5 ng/ml, 1 ng/ml, 2 ng/ml), and I, A10 cells were treated with 0.5 ng/ml of calyculin A and 5 μm of SP600125 for 2 h and harvested for immunoblotting. Error bars indicate S.E.
To further evaluate the role of PP1α in c-Jun expression, we performed a titration of calyculin A in A10 cells and observed an increase in phosphorylated c-Jun at higher concentrations of calyculin A (Fig. 3H). Previous studies in lung epithelial cells have shown that calyculin A can activate JNK signaling to induce c-Jun phosphorylation (26). This appears to be consistent in VSMCs, as phosphorylation of c-Jun by calyculin A treatment is attenuated by pre-treatment with the JNK inhibitor, SP600125 (Fig. 3I). In addition, our previous work has shown that PP1α helps recruit HDAC4 to MEF2 proteins, and that HDAC4 acts to repress c-Jun expression, but not myocardin expression, in VSMCs (6, 13). Thus, it appears that PP1α acts to repress c-Jun expression, in part, by inactivating JNK activity and recruiting HDAC4 to MEF2 proteins; however, myocardin expression appears to be regulated in a different manner.
CPI-17 Rescues MEF2 Repression by PP1α
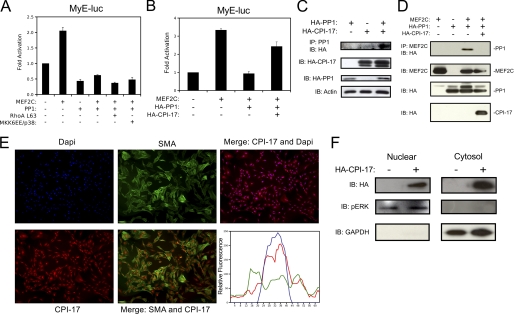
Next, we evaluated whether exogenous expression of activated p38 MAPK or RhoA might be able to overcome PP1α repression of MEF2C to induce myocardin expression. However, as shown in Fig. 4A, once repressed by PP1α, MEF2C is unresponsive to activated MKK6/p38 MAPK or RhoA signaling in COS7 cells. In VSMCs, the MLCP is regulated by a smooth muscle-enriched phosphatase inhibitor called CPI-17, which is not expressed in COS7 cells (not shown). Consistent with previously published structural data, Fig. 4C demonstrates that CPI-17 physically interacts with PP1α, evaluated by co-immunoprecipitation, which leads to inhibition of phosphatase activity (27). In addition to being potentiated by PKC, CPI-17 has also been shown to be activated by ROCK and PKN (28, 29). Therefore, we determined if exogenous expression of CPI-17 in COS7 cells could inhibit PP1α repression of MEF2 activity. As shown in Fig. 4B, CPI-17 can antagonize PP1α repression of the myocardin enhancer and restore the activation induced by MEF2C. In addition, Fig. 4D demonstrates that CPI-17 can compete away the physical interaction between MEF2C and PP1α, determined by co-immunoprecipitation. To indicate whether CPI-17 could perform a nuclear role in transcriptional regulation, we investigated the cellular localization of CPI-17 by immunofluorescence microscopy in primary VSMCs (Fig. 4E). Given that previous studies have defined a role for CPI-17 in regulating calcium sensitivity, we anticipated an abundance of CPI-17 to co-localize with the actin cytoskeleton. Surprisingly, much of the cellular CPI-17 was confined to the nuclear compartment in VSMCs, suggesting a potentially important role for CPI-17 in the nucleus. These results were confirmed biochemically using nuclear and cytosolic fractionation, which demonstrated that CPI-17 is expressed in both the nuclear and cytosolic compartments (Fig. 4F).
FIGURE 4.
PP1α-induced repression of myocardin is attenuated by CPI-17. A, COS7 cells were transfected with the myocardin enhancer (MyE), MEF2C, PP1α (PP1), activated RhoA (RhoA L63), or activated MKK6 and p38 (MKK6EE/p38), as indicated. Extracts were subjected to lucifierase assay. B, cells were transfected with the myocardin enhancer, and MEF2C, PP1, or CPI-17 as indicated, followed by luciferase assay. C, COS7 cells were transfected with HA-CPI-17 and HA-PP1α. Protein extracts were immunoprecipitated with PP1α antibody and immunoblotted, as indicated. D, COS7 cells were transfected with MEF2C, HA-PP1, or HA-CPI-17 as indicated. Extracts were immunoprecipitated with MEF2C antibody and immunoblotted for antibodies to HA. Protein extracts were immunoblotted, as indicated, to demonstrate equal loading and transfection efficiency. E, primary VSMCs were fixed, permeabilized, and subjected to immunofluorescence for CPI-17, smooth muscle actin (SMA), and the DAPI nuclear stain. Cells were visualized by standard fluorescence techniques. Relative fluorescence of a representative cell was graphed with ImageJ. F, A10 cells were transfected with HA-CPI-17 and subjected to nuclear/cytosolic fractionation. Lysates were immunoblotted as indicated. Error bars indicate S.E.
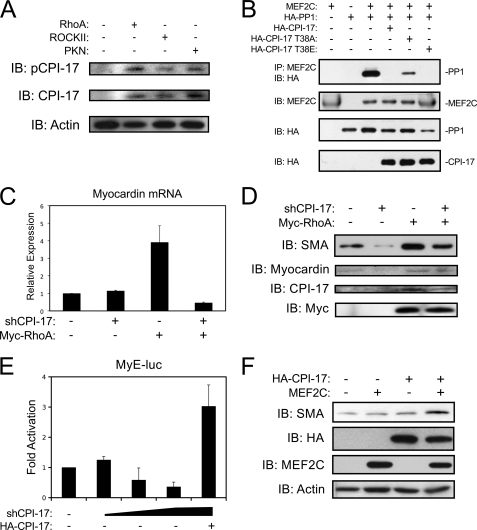
CPI-17 is activated by phosphorylation at Thr-38, and structural analysis predicts that phospho-Thr-38 serves to anchor the interaction with PP1α, resulting in maximal phosphatase inhibition (30, 31). Therefore, we utilized a Thr-38 phospho-specific antibody to evaluate the role of RhoA-dependent signaling on CPI-17 phosphorylation. As shown in Fig. 5A, expression of activated RhoA, ROCKII, and PKN, increased Thr38 phosphorylation of CPI-17 in A10 cells. Furthermore, these kinases also increased the expression of total CPI-17, which is consistent with a recent report that demonstrated CPI-17 expression was regulated in a manner similar to other smooth muscle marker genes (32). Next, we introduced both a neutralizing alanine mutation (T38A), and a phospho-mimetic glutamate mutation (T38E) at the Thr-38 residue to determine whether this site could regulate MEF2 activity. As shown in Fig. 5B, the T38E mutation was equally as effective as the wild-type CPI-17 at disrupting the MEF2C-PP1α interaction. However, as predicted, the T38A mutation was less efficient at disrupting the PP1α interaction with MEF2C indicating that phosphorylation of CPI-17 is necessary for MEF2 de-repression.
FIGURE 5.
Phosphorylation of CPI-17 at threonine 38 regulates MEF2-dependent VSMC differentiation. A, A10 cells were transfected with activated RhoA, ROCKII, or PKN using Lipofectamine reagent and puromycin-selected overnight. Protein extracts were subjected to immunoblotting as indicated. B, COS7 cells were transfected, as described in 4D) with the addition of Thr-38 mutants of CPI-17. Protein extracts were immunoprecipitated and immunoblotted as previously described. C, A10 cells were transfected as described in A with a plasmid encoding a short hairpin RNA targeting CPI-17 (shCPI-17) and an expression vector for active RhoA. Following puromycin selection, myocardin expression was evaluated by qPCR, corrected for GAPDH using the ΔΔCT method. D, A10 were transfected as described in C. Following puromycin selection, extracts were subjected to immunoblotting. E, rat primary VSMCs were transfected by Lipofectamine reagent with the myocardin reporter gene (MyE-luc), increasing amounts of shCPI-17, and an expression plasmid for human CPI-17. Extracts were subjected to luciferase assay. F, 10T1/2 mouse embryonic fibroblast cells were transfected with MEF2C or HA-CPI-17 as indicate. Cells were placed in low serum conditions (5% horse serum) for 96 h and subjected to immunoblotting as indicated. Error bars indicate S.E.
Finally, to evaluate the endogenous role of CPI-17 in myocardin expression, we engineered a short-hairpin RNA to reduce CPI-17 expression (shCPI-17). Shown in Fig. 5C, shCPI-17 attenuated the induction of myocardin by activated RhoA, determined by qPCR. Furthermore, shown in Fig. 5D, the shCPI-17 reduced endogenous CPI-17 expression and attenuated its induction following exogenous expression of RhoA. Interestingly, reduced CPI-17 expression was accompanied by a corresponding decrease in myocardin and SMA protein expression, implicating CPI-17 as a critical regulator of VSMC differentiation. Titration of the shCPI-17 on the myocardin enhancer (MyE-luc) resulted in a dose-dependent decrease in myocardin expression that could be rescued with forced expression of human CPI-17, which is resistant to the shCPI-17 and validates the specificity of the shRNA. Finally, we determined if MEF2C and CPI-17 could convert a pluripotent cell-line toward a VSMC phenotype. Utilizing C3H10T1/2 mouse embryonic fibroblasts, we exogenously expressed CPI-17 and MEF2C, alone and in combination and determined the effect on SMA expression. As shown in Fig. 5F, neither CPI-17 nor MEF2C expression had an impact on basal levels of SMA expression in this cell-line. However, when combined, CPI-17 and MEF2C markedly induced SMA expression, indicating that MEF2C requires derepression by CPI-17 to activate a VSMC phenotype.
DISCUSSION
MEF2C plays an essential role in VSMC differentiation and is genetically upstream of the SRF-coactivator, myocardin (5, 10). MEF2 proteins are integrators of a number of cellular signaling pathways, and are also regulated by several interacting co-factors that either enhance or repress transcriptional activity. We document in this report that cellular signals emanating from RhoA serve to relieve MEF2C from the repressive effects of PP1α to increase myocardin expression in VSMCs (Fig. 6). Furthermore, we demonstrate, for the first time, that this genetic pathway connecting CPI-17, MEF2C, and myocardin is critical for VSMC differentiation (Figs. 1 and 5). In addition, PP1α serves to modulate c-Jun expression through an entirely different mechanism involving recruitment of HDAC4 to MEF2 proteins and phosphatase-dependent regulation of JNK signaling.
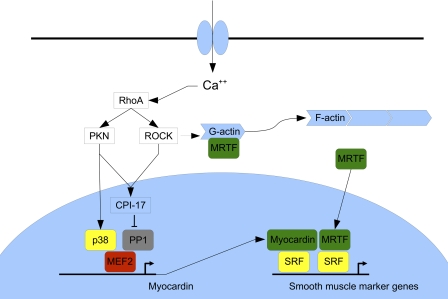
FIGURE 6.
Model of calcium-mediated induction of myocardin expression in VSMCs. This is based on the work presented in this report and previously published observations. MEF2-dependent myocardin expression is regulated by p38 MAPK and RhoA-induced derepression of PP1α by CPI-17. Myocardin activates SRF-dependent VSMC gene expression directly and by dimerizing with myocardin-related transcription factors (MRTFs) that translocate to the nucleus when G- actin polymerizes to form F-actin.
Signal-dependent Control of PP1α
The cellular distribution and substrate specificity of PP1α is regulated by physical interaction with regulatory subunits, that typically contain a conserved RVXF domain (21). In VSMCs, PP1α is targeted to the myosin light chains by physical interaction with MYPT1; however, this RVXF domain is also conserved among MADS-box proteins, such as MEF2A-D and may serve to target PP1α to nuclear MEF2 proteins (13, 14). Interestingly, SRF also contains a conserved RVXF domain within its MADS-box, yet our data suggest that PP1α cannot overcome myocardin or TGF-β induction of SRF-target genes (Fig. 3F). In addition, the phosphatase activity of PP1α is regulated through interaction with specific inhibitor proteins like Inhibitor 1 and 2 (I1 and I2), and CPI-17. The potency of these inhibitor proteins is regulated by phosphorylation and dephosphorylation by cellular kinases and phosphatases, such as PKA, calcineurin, ROCK, and PKN (14, 21, 33). Our data demonstrates that phosphorylation of CPI-17 at Thr-38 by ROCK and/or PKN regulates PP1α ability to modulate gene expression; whereas, I1 and I2 have no effect on MEF2 transcriptional activity (13). The reason for this specificity is not known; however, it may be related to the proposed cytosolic distribution of I1 and I2 compared with the relatively nuclear distribution of CPI-17, and/or the ability of CPI-17 to compete with MEF2C for PP1α binding (21).
PP1α Regulates MEF2-Target Genes
We have previously shown that PP1α regulates the transcriptional activity of MEF2 proteins through a number of mechanisms: 1) PP1α physically interacts with both the N terminus and C terminus of MEF2A, -C, and -D to inhibit transcriptional activity directly; 2) PP1α dephosphorylates Ser-408 of MEF2A; and 3) PP1α serves to recruit HDAC4 to MEF2 (13). We now document, within the cellular context of VSMCs, that these previously identified mechanisms operate in a MEF2-target gene-specific manner, where PP1α regulates myocardin expression through direct interaction with MEF2C, and regulates c-jun expression by recruiting HDAC4 to MEF2 proteins and dephosphorylation of JNK. Furthermore, we identify a nuclear role for CPI-17 in regulating VSMC gene expression. Recent analysis of CPI-17 expression in mouse embryos has revealed that in addition to its restricted expression pattern in smooth muscle postnatally, CPI-17 is also expressed transiently in the developing heart and intercostal muscles (32). Interestingly, myocardin and several smooth muscle marker genes, such as SM22, all display at least transient expression in striated and smooth muscle types during development (34–36). In light of our evidence demonstrating the critical role of CPI-17 in the regulation of myocardin expression, these data suggest a potentially larger role for CPI-17 regulating the development of all three muscle types.
In summary, we provide novel evidence that PP1α serves as critical regulator of MEF2-dependent gene expression in VSMCs, and demonstrate for the first time that RhoA-mediated signaling plays a fundamental role in inducing myocardin expression through MEF2 proteins. These findings have important ramifications to the field of vascular smooth muscle development and in the progression of vascular stenotic diseases, and uncover potentially new therapeutic targets for manipulation of VSMC differentiation in stem cell programming.
Acknowledgments
We thank Drs. Michelle Bendeck, Guangpei Hou, and Olga Ornatsky for technical assistance with the primary cultures, and Nezeka Alli and Dr. Tetsuaki Miyake for technical assistance with cloning, mutagenesis, and RNAi experiments.
This work was supported by a grant from the Heart and Stroke Foundation of Canada (HSFC) (to J. C. M.).
- VSMC
- vascular smooth muscle cells
- MLCP
- myosin light chain phosphatase
- PP1α
- protein phosphatase 1α
- SRF
- serum response factor
- MEF
- myocyte enhancer factor
- ROCK
- RhoA-associated kinase
- CaMK
- calcium/calmodulin-dependent kinase
- CPI
- PKC-potentiated protein phosphatase inhibitor
- Hand
- heart and neural crest-derived family.
REFERENCES
- 1. Drake C. J., Hungerford J. E., Little C. D. (1998) Morphogenesis of the first blood vessels. Ann. NY Acad. Sci. 857, 155–179 [DOI] [PubMed] [Google Scholar]
- 2. Owens G. K. (1995) Regulation of differentiation of vascular smooth muscle cells. Physiol. Rev. 75, 487–517 [DOI] [PubMed] [Google Scholar]
- 3. Owens G. K., Kumar M. S., Wamhoff B. R. (2004) Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol. Rev. 84, 767–801 [DOI] [PubMed] [Google Scholar]
- 4. Miano J. M. (2003) Serum response factor: toggling between disparate programs of gene expression. J. Mol. Cell. Cardiol. 35, 577–593 [DOI] [PubMed] [Google Scholar]
- 5. Creemers E. E., Sutherland L. B., McAnally J., Richardson J. A., Olson E. N. (2006) Myocardin is a direct transcriptional target of Mef2, Tead, and Foxo proteins during cardiovascular development. Development 133, 4245–4256 [DOI] [PubMed] [Google Scholar]
- 6. Gordon J. W., Pagiatakis C., Salma J., Du M., Andreucci J. J., Zhao J., Hou G., Perry R. L., Dan Q., Courtman D., Bendeck M. P., McDermott J. C. (2009) Protein kinase A-regulated assembly of a MEF2·HDAC4 repressor complex controls c-Jun expression in vascular smooth muscle cells. J. Biol. Chem. 284, 19027–19042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang D. Z., Olson E. N. (2004) Control of smooth muscle development by the myocardin family of transcriptional coactivators. Curr. Opin. Genet. Dev. 14, 558–566 [DOI] [PubMed] [Google Scholar]
- 8. Wang Z., Wang D. Z., Hockemeyer D., McAnally J., Nordheim A., Olson E. N. (2004) Myocardin and ternary complex factors compete for SRF to control smooth muscle gene expression. Nature 428, 185–189 [DOI] [PubMed] [Google Scholar]
- 9. Wamhoff B. R., Bowles D. K., McDonald O. G., Sinha S., Somlyo A. P., Somlyo A. V., Owens G. K. (2004) L-type voltage-gated Ca2+ channels modulate expression of smooth muscle differentiation marker genes via a rho kinase/myocardin/SRF-dependent mechanism. Circulat. Res. 95, 406–414 [DOI] [PubMed] [Google Scholar]
- 10. Lin Q., Lu J., Yanagisawa H., Webb R., Lyons G. E., Richardson J. A., Olson E. N. (1998) Requirement of the MADS-box transcription factor MEF2C for vascular development. Development 125, 4565–4574 [DOI] [PubMed] [Google Scholar]
- 11. Ren J., Albinsson S., Hellstrand P. (2010) Distinct effects of voltage- and store-dependent calcium influx on stretch-induced differentiation and growth in vascular smooth muscle. J. Biol. Chem. 285, 31829–31839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Martin K., Weiss S., Metharom P., Schmeckpeper J., Hynes B., O'Sullivan J., Caplice N. (2009) Thrombin stimulates smooth muscle cell differentiation from peripheral blood mononuclear cells via protease-activated receptor-1, RhoA, and myocardin. Circ. Res. 105, 214–218 [DOI] [PubMed] [Google Scholar]
- 13. Perry R. L., Yang C., Soora N., Salma J., Marback M., Naghibi L., Ilyas H., Chan J., Gordon J. W., McDermott J. C. (2009) Direct interaction between myocyte enhancer factor 2 (MEF2) and protein phosphatase 1α represses MEF2-dependent gene expression. Mol. Cell. Biol. 29, 3355–3366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Somlyo A. P., Somlyo A. V. (2003) Ca2+ sensitivity of smooth muscle and nonmuscle myosin II: modulated by G proteins, kinases, and myosin phosphatase. Physiol. Rev. 83, 1325–1358 [DOI] [PubMed] [Google Scholar]
- 15. Deaton R. A., Su C., Valencia T. G., Grant S. R. (2005) Transforming growth factor-β1-induced expression of smooth muscle marker genes involves activation of PKN and p38 MAPK. J. Biol. Chem. 280, 31172–31181 [DOI] [PubMed] [Google Scholar]
- 16. Han J., Jiang Y., Li Z., Kravchenko V. V., Ulevitch R. J. (1997) Activation of the transcription factor MEF2C by the MAP kinase p38 in inflammation. Nature 386, 296–299 [DOI] [PubMed] [Google Scholar]
- 17. Ornatsky O. I., Cox D. M., Tangirala P., Andreucci J. J., Quinn Z. A., Wrana J. L., Prywes R., Yu Y. T., McDermott J. C. (1999) Post-translational control of the MEF2A transcriptional regulatory protein. Nucleic Acids Res. 27, 2646–2654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhao M., New L., Kravchenko V. V., Kato Y., Gram H., di Padova F., Olson E. N., Ulevitch R. J., Han J. (1999) Regulation of the MEF2 family of transcription factors by p38. Mol. Cell. Biol. 19, 21–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hou G., Vogel W., Bendeck M. P. (2001) The discoidin domain receptor tyrosine kinase DDR1 in arterial wound repair. J. Clin. Investig. 107, 727–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gong M. C., Iizuka K., Nixon G., Browne J. P., Hall A., Eccleston J. F., Sugai M., Kobayashi S., Somlyo A. V., Somlyo A. P. (1996) Role of guanine nucleotide-binding proteins-Ras-family or trimeric proteins or both-in Ca2+ sensitization of smooth muscle. Proc. Natl. Acad. Sci. U.S.A. 93, 1340–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cohen P. T. (2002) Protein phosphatase 1-targeted in many directions. J. Cell Science 115, 241–256 [DOI] [PubMed] [Google Scholar]
- 22. Morin S., Pozzulo G., Robitaille L., Cross J., Nemer M. (2005) MEF2-dependent recruitment of the HAND1 transcription factor results in synergistic activation of target promoters. J. Biol. Chem. 280, 32272–32278 [DOI] [PubMed] [Google Scholar]
- 23. Verzi M. P., Agarwal P., Brown C., McCulley D. J., Schwarz J. J., Black B. L. (2007) The transcription factor MEF2C is required for craniofacial development. Dev. Cell 12, 645–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yamagishi H., Olson E. N., Srivastava D. (2000) The basic helix-loop-helix transcription factor, dHAND, is required for vascular development. J. Clin. Investig. 105, 261–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang J., Cheng L., Li J., Chen M., Zhou D., Lu M. M., Proweller A., Epstein J. A., Parmacek M. S. (2008) Myocardin regulates expression of contractile genes in smooth muscle cells and is required for closure of the ductus arteriosus in mice. J. Clin. Investig. 118, 515–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chu S., Ferro T. J. (2006) Identification of a hydrogen peroxide-induced PP1-JNK1-Sp1 signaling pathway for gene regulation. Am. J. Physiol. Lung Cell. Mol. Physiol. 291, L983–L992 [DOI] [PubMed] [Google Scholar]
- 27. Eto M., Kitazawa T., Brautigan D. L. (2004) Phosphoprotein inhibitor CPI-17 specificity depends on allosteric regulation of protein phosphatase-1 by regulatory subunits. Proc. Natl. Acad. Sci. U. S. A. 101, 8888–8893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hamaguchi T., Ito M., Feng J., Seko T., Koyama M., Machida H., Takase K., Amano M., Kaibuchi K., Hartshorne D. J., Nakano T. (2000) Phosphorylation of CPI-17, an inhibitor of myosin phosphatase, by protein kinase N. Biochem. Biophys. Res. Commun. 274, 825–830 [DOI] [PubMed] [Google Scholar]
- 29. Koyama M., Ito M., Feng J., Seko T., Shiraki K., Takase K., Hartshorne D. J., Nakano T. (2000) Phosphorylation of CPI-17, an inhibitory phosphoprotein of smooth muscle myosin phosphatase, by Rho-kinase. FEBS Lett. 475, 197–200 [DOI] [PubMed] [Google Scholar]
- 30. Eto M., Ohmori T., Suzuki M., Furuya K., Morita F. (1995) A novel protein phosphatase-1 inhibitory protein potentiated by protein kinase C. Isolation from porcine aorta media and characterization. J. Biochem. 118, 1104–1107 [DOI] [PubMed] [Google Scholar]
- 31. Eto M., Kitazawa T., Matsuzawa F., Aikawa S., Kirkbride J. A., Isozumi N., Nishimura Y., Brautigan D. L., Ohki S. Y. (2007) Phosphorylation-induced conformational switching of CPI-17 produces a potent myosin phosphatase inhibitor. Structure 15, 1591–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim J., Young G., Jin L., Somlyo A., Eto M. (2009) Expression of CPI-17 in smooth muscle during embryonic development and in neointimal lesion formation. Histochem. Cell Biol. 132, 191–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Eto M. (2009) Regulation of cellular protein phosphatase-1 (PP1) by phosphorylation of the CPI-17 family, C-kinase-activated PP1 inhibitors. J. Biol. Chem. 284, 35273–35277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Du K. L., Ip H. S., Li J., Chen M., Dandre F., Yu W., Lu M. M., Owens G. K., Parmacek M. S. (2003) Myocardin is a critical serum response factor cofactor in the transcriptional program regulating smooth muscle cell differentiation. Mol. Cell. Biol. 23, 2425–2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang D., Chang P. S., Wang Z., Sutherland L., Richardson J. A., Small E., Krieg P. A., Olson E. N. (2001) Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell 105, 851–862 [DOI] [PubMed] [Google Scholar]
- 36. Long X., Creemers E. E., Wang D. Z., Olson E. N., Miano J. M. (2007) Myocardin is a bifunctional switch for smooth versus skeletal muscle differentiation. Proc. Natl. Acad. Sci. U.S. A. 104, 16570–16575 [DOI] [PMC free article] [PubMed] [Google Scholar]