Background: eIF2B is a critical translation factor and regulator of protein synthesis that is implicated in human disease.
Results: Protein-protein interactions within the five-subunit eIF2B complex are identified.
Conclusion: eIF2B complex formation requires extensive intersubunit domain interactions.
Significance: The first experimental model is proposed for eIF2Bγ and -ϵ subunit interactions.
Keywords: Protein Synthesis, Protein-Protein Interactions, Transcription Initiation Factors, Translation, Translation Regulation, eIF2, eIF2B
Abstract
In eukaryotic translation initiation, eIF2B is the guanine nucleotide exchange factor (GEF) required for reactivation of the G protein eIF2 between rounds of protein synthesis initiation. eIF2B is unusually complex with five subunits (α–ϵ) necessary for GEF activity and its control by phosphorylation of eIF2α. In addition, inherited mutations in eIF2B cause a fatal leukoencephalopathy. Here we describe experiments examining domains of eIF2Bγ and ϵ that both share sequence and predicted tertiary structure similarity with a family of phospho-hexose sugar nucleotide pyrophosphorylases. Firstly, using a genetic approach, we find no evidence to support a significant role for a potential nucleotide-binding region within the pyrophosphorylase-like domain (PLD) of eIF2Bϵ for nucleotide exchange. These findings are at odds with one mechanism for nucleotide exchange proposed previously. By using a series of constructs and a co-expression and precipitation strategy, we find that the eIF2Bϵ and -γ PLDs and a shared second domain predicted to form a left-handed β helix are all critical for interprotein interactions between eIF2B subunits necessary for eIF2B complex formation. We have identified extensive interactions between the PLDs and left-handed β helix domains that form the eIF2Bγϵ subcomplex and propose a model for domain interactions between eIF2B subunits.
Introduction
In protein synthesis, the translation initiation factor 2 (eIF2) binds GTP and initiator methionyl tRNA (tRNAiMet)2 to deliver the latter to initiating ribosomes for each translation initiation event (1). A G protein, eIF2 cycles between inactive GDP and active GTP-bound states (2). These transitions are governed by eIF5 and eIF2B. eIF5 is a dual function GTPase-accelerating protein and GDP dissociation inhibitor (3). eIF2B is a guanine nucleotide exchange factor (GEF) that promotes GDP release and GTP binding to eIF2, enabling tRNAiMet binding (4). Maintaining this eIF2 cycle is critical for continued active protein synthesis in all cells. Controlling eIF2B confers critically important regulation of the eIF2 cycle and translation initiation. In mammalian cells, four protein kinases (termed EIF2AK1–4, also known as HRI, PKR, PERK, and GCN2) each phosphorylate a serine at position 51 in the eIF2 α subunit in response to different signals. Phosphorylated eIF2 (eIF2(αP)) binds to eIF2B with higher affinity and acts as a competitive inhibitor of eIF2B, instead of its substrate (5). This reduces the activity of eIF2B and the availability of active eIF2·GTP·tRNAiMet in cells and has two divergent effects. For a majority of transcripts, eIF2(αP) causes reduced translation; however, for a subset of translationally controlled RNAs, their expression is enhanced. Many of the regulated mRNAs characterized are transcription factors important for mediating cellular reprogramming of gene expression in response to stress, for example ATF4 in mammalian cells and GCN4 in yeast (6).
The study of eIF2B has gained a new focus in the last decade as mutations within any of its five subunits have been associated with a fatal inherited brain disorder commonly called leukoencephalopathy with vanishing white matter or childhood ataxia with central nervous system hypomyelination. To date, more than 100 separate missense mutations have been described (7). Where analyzed biochemically, in assays using extracts from patient-derived cells, they typically reduce eIF2B activity by between 20 and 80% of control values (8).
eIF2B has five subunits α–ϵ, in increasing size order, encoded by separate genes (EIF2B1–5 in humans and named GCN3, GCD7, GCD1, GCD2, and GCD6, respectively, in the yeast Saccharomyces cerevisiae). Although they were initially characterized biochemically using proteins purified from mammalian sources, much of our understanding of the roles of individual subunits has come from genetic and biochemical analyses of the yeast factor. eIF2Bαβδ all share sequence similarity (see Fig. 1A) and bind together to form a subcomplex that can interact with eIF2α, with higher affinity for eIF2(αP). This was termed the regulatory subcomplex (9), and remarkably, even single amino acid substitutions in any of these subunits can disrupt this regulatory mechanism (10). Where tested, the mutants reduced affinity for eIF2(αP) (11, 12). The ϵ subunit is the catalytic subunit, and the extreme C-terminal region (ϵcat) is sufficient for nucleotide exchange in vitro (13) via interaction with two eIF2 subunits (14). Consistent findings were reported for mammalian and Drosophila eIF2Bϵ (15, 16); however, eIF2Bϵ activity is at least ∼10-fold lower than the full eIF2B complex, indicating that the other subunits act to enhance eIF2B activity as well as regulate its function. How this is achieved is not yet clear, but it is known that eIF2Bϵ binds to the γ subunit in the absence of the αβδ subunits (5) and that this γϵ catalytic subcomplex enhanced GEF activity similarly to that seen with the full five-subunit complex in assays using yeast cell lysates as a source of eIF2B.
FIGURE 1.
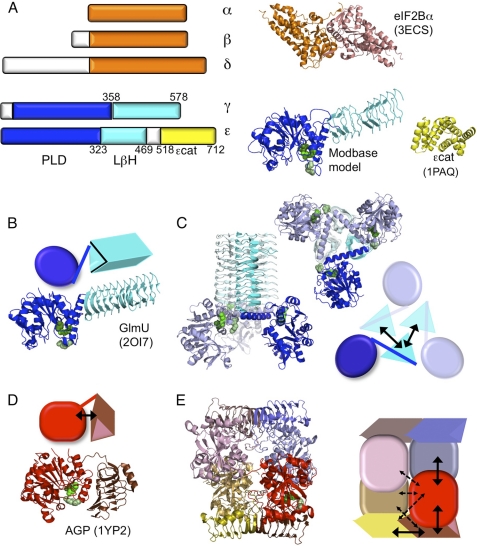
Structures and theoretical models of eIF2B subunits and homologues. A, graphic depiction of the five eIF2B subunits showing homologous regions similarly colored. Known structures for human eIF2Bα (orange, Protein Data Bank (PDB) ID = 3ECS) and yeast eIF2Bϵ catalytic domain (yellow, 1PAQ) and a predicted structure (ModBase: Database of Comparative Protein Structure Models) for yeast eIF2Bϵ PLD and LβH regions (blue and cyan, respectively) modeled on E. coli GlmU (2OI7) monomer are shown. Arg-39 (pale green), Arg-49 (green), and Asp-138 (dark green) are shown as filled spheres; see Fig. 2). B and C, structure and graphic of GlmU monomer (B) and homotrimer (C). D and E, alternative domain arrangements identified in potato AGP monomer (red) and homotetramer. Black solid and broken arrows indicate major and minor domain interactions, respectively, within or between a single monomer and adjacent partners (panels C–E).
Since they were first sequenced, it was realized that eIF2Bϵ and -γ share extensive primary sequence similarity with each other (17) and with a family of enzymes called NTP-hexose sugar pyrophosphorylases (18). ADP-glucose pyrophosphorylase (AGP) binds ATP and glucose to catalyze the formation of ADP-glucose, required for starch (in plants) or glycogen synthesis (in bacteria) (19). Escherichia coli N-acetylglucosamine 1-phosphate uridyltransferase (GlmU) catalyzes a similar reaction with UTP and N-acetylglucosamine 1-phosphate necessary for peptidoglycan synthesis (20). The crystal structures of GlmU and potato tuber AGP have both been solved, and each reveals two domains, an amino-terminal globular pyrophosphorylase domain and a separate carboxyl-terminal domain sometimes referred to as “hexapeptide” repeat or “I-patch” because of a repetitive sequence motif with the loose consensus XXIGXX. This domain adopts a left-handed β-helix (LβH) structure (see Fig. 1, B and C). The structures of eIF2Bγ and the N-terminal 470 residues of eIF2Bϵ have not yet been determined, but due to sequence homology, each subunit is predicted by available modeling software tools to share similar domain architecture with these enzymes (21, 22). Both eIF2B subunits share a conserved N-terminal region, here termed pyrophosphorylase-like domain (PLD), and the LβH region. In this work, we set out to uncover the significance of these conserved domains for eIF2B GEF function and domain organization, with the aim of better understanding the relationship between the structural organization and function of this large protein complex that is critical for protein synthesis and its control. Our findings provide important structural and functional insight into the domain organization of this protein.
EXPERIMENTAL PROCEDURES
Yeast Strains, Plasmids, Genetic Methods, and Growth Conditions
All genetic procedures used standard methods (23). Yeast transformation was by the lithium acetate method (24). For complementation studies, gcd6 mutant plasmids (Table 1) were transformed into strain GP3750 (KAY16: MATα leu2-3 leu2-112 ura3-52 ino1 gcd6Δ gcn2Δ::hisG (HIS4-lacZ ura3-52) pJB5(GCD6 URA3 CEN4)) (25). Strain H1905 was used as an isogenic GCN2 control (17). Plasmid shuffling employed 5-fluororotic acid (23). To overexpress different combinations of eIF2B subunits and epitope-tagged domains, strain GP3667 (MATα trp1-Δ63 ura3-52 leu2-3 leu2-112 GAL2 gcn2Δ) (26) or a protease-deficient derivative GP4597 (GP3667 pep4::hisG) was transformed with a combination of the plasmids described in Table 1.
TABLE 1.
Plasmids used in this study
| Plasmid name | Relevant genotypea | Reference |
|---|---|---|
| pAV1265 (pJB102) | GCD6 LEU2 CEN6 | 46 |
| pAV1355 (p1871) | GCD2 GCD7 GCN3 URA3 2 μm | 47 |
| pAV1376 (p2300) | GCD6 URA3 2 μm | 9 |
| pAV1388 (p2335) | GCD6 GCD1 LEU2 2 μm | 5 |
| pAV1428 | GCD1-FLAG2-His6GCD6 URA3 2 μm | 26 |
| pAV1429 | GCD1-FLAG2-His6 TRP1 2 μm | This study |
| pAV1433 (pEG(KT)) | Pgal1-GST URA3 leu2d 2 μm | 30 |
| pAV1494 | GCN3 GCD7 GCD2 LEU2 2 μm | 26 |
| pAV1549 | GCD1-FLAG2-His6gcd6-Δ93–358 URA3 2 μm | 26 |
| pAV1578 | GCD1-FLAG2-His6gcd6-Q452* URA3 2 μm | 26 |
| pAV1535 | GCD1-FLAG2-His6gcd6-T518D* URA3 2 μm | 26 |
| pAV1694 | Pgal1-GST-gcd6 (518–712) URA3 leu2d 2 μm | 13 |
| pAV1791 | Pgal1-GST-gcd6 (161–469) URA3 leu2d 2 μm | This study |
| pAV1792 | Pgal1-GST-gcd6 (1–323) URA3 leu2d 2 μm | This study |
| pAV1793 | Pgal1-GST-gcd6 (161–323) URA3 leu2d 2 μm | This study |
| pAV1821 | Pgal1-GST-gcd6 (1–160) URA3 leu2d 2 μm | This study |
| pAV1822 | Pgal1-GST-gcd6 (324–469) URA3 leu2d 2 μm | This study |
| pAV1842 | Pgal1-GST-gcd6 (1–469) URA3 leu2d 2 μm | This study |
| pAV1857 | gcd6-D138A LEU2 CEN6 | This study |
| pAV1895 | gcd6-R39A LEU2 CEN6 | This study |
| pAV1896 | gcd6-R49A LEU2 CEN6 | This study |
| pAV2004 | Pgal10-GCD1-FLAG2-His6 TRP1 2 μm | This study |
| pAV2005 | Pgal10-gcd1(1–356)-FLAG2-His6 TRP1 2 μm | This study |
| pAV2057 | Pgal10-gcd1(357–578)-FLAG2-His6 TRP1 2 μm | This study |
| pAV2058 | Pgal10-gcd1(1–356)-FLAG2-His6 Pgal1-gcd1(357–578)-FLAG2-His6 TRP1 2 μm | This study |
a The T518D* mutation is a single base pair deletion causing a frameshift and truncated protein, with a stop nine codons downstream. The last natural amino acid is codon 517.
Strains were grown at 30 °C in Synthetic Complete (SC) medium containing glucose (2% w/v) but lacking nutrients required for plasmid selection or in Synthetic Dextrose minimal medium (SD) with necessary supplements added to complement auxotrophies (23). When inducing PGal-dependent protein expression, glucose was replaced with galactose and raffinose (2% w/v each) to make SCGal medium. For growth on solid medium, cells were typically grown to A600 = 0.1 and serially diluted before 3 μl of each dilution was spotted on the appropriate medium and grown at 30 °C for the indicated time. 3-Amino-1,2,4-triazole (3AT; Fluka) was added to 20 mm final concentration to induce a histidine starvation.
Sequence Alignments and Molecular Modeling to Define Domain Boundaries
The primary amino acid sequences of yeast eIF2Bϵ (Gcd6p residues 1–470) and -γ (Gcd1p) were aligned with equivalent sequences from other species using ClustalW (27) with manual adjustments. This allowed identification of 21 XXIGXX repeats and estimation of the extent of the LβH domains (ϵ324–469 and γ358–528). Additional confirmation for domain boundaries was obtained by structural modeling of eIF2Bϵ 1–470 using publically available websites (21, 22) that independently identified these domain structures for ϵ20–446. The N-terminal pyrophosphorylase globular domain binds nucleotides and hexose sugars. Alignments indicated that the equivalent eIF2Bϵ/γ regions are ϵ20–323 and γ38–357. We included full N termini in our constructs. As similarity is stronger within the N terminus (e.g. ϵ residues 27–93 share 25% identity/55% similarity with UDP-glucose pyrophosphorylase; see also Ref. 18), we made additional ϵ constructs (1–160 and 161–323) to split this domain and further constructs containing both regions (161–469 and 1–469; Table 1). The eIF2Bϵ catalytic domain (ϵcat, residues 518–712) has been defined previously in Refs. 13, 28, and 29.
Plasmid Constructions
PCR was used to amplify the desired fragments of GCD6 (yeast eIF2Bϵ) with added BamHI or stop-SalI tails to enable cloning in-frame with glutathione S-transferase (GST) in pEG(KT) (30), a high copy number yeast PGal-GST URA3 leu2d expression plasmid, to create GST-fused fragments. Similarly, PCR was used to clone GCD1 (yeast eIF2Bγ) fragments with added tandem C-terminal 2×FLAG and His6 tags (γ-FLAG) into pBEVY-GT, a yeast high copy number TRP1 plasmid with both PGal1 and PGal10 promoters to drive regulated expression (31). Site-directed mutagenesis of pAV1265 to introduce R39A, R49A, and D138A mutations into GCD6 employed QuikChange® (Stratagene). See Table 1 for plasmid details.
GST Purifications and FLAG Immunoprecipitations (IP)
Strains were grown in SCGal medium with 0.2% glucose to an A600 between 0.2 and 0.6 and an equivalent volume containing 10 A600 units were harvested by centrifugation at 5000 × g. Cells were washed in sterile ice-cold water with 1 mm PMSF (10 ml) and then in 1 ml of buffer IP (150 mm NaCl, 20 mm TRIS-HCl, pH 7.4, 2 mm MgCl2, 0.5% Triton X-100, 5 mm 2-mercaptoethanol) before being resuspended in 250 μl of buffer IP+PI (IP + mini EDTA-free protease inhibitor tablets (Roche Applied Science)). Cell suspensions were added to tubes containing 200 μl of 1-mm zirconia/silica beads (Thistle Scientific) and lysed by agitation using a FastPrep (MP Bio) for 3 × 25 s at 6.5 ms−1 with cooling on iced water for 5 min between cycles. Cell debris was removed from the cell extract by centrifugation at 10,000 × g for 15 min at 4 °C. The extracts were precleared with Sepharose 4B beads (GE Healthcare), and the affinity resin was blocked with 1 mg/ml BSA for 1 h in IP+PI buffer at 4 °C. For FLAG purifications, 250 μl of 1 mg ml−1 extract was incubated with 40 μl of packed volume of anti-FLAG M2 affinity resin (Sigma-Aldrich). For GST purifications, 50 μl of packed volume of anti-GST magnetic resin (Pierce) was used. Extract-resin mixes were incubated with rotation for 2 h at 4 °C. The supernatant was removed, and the beads were washed 3 × 10 min in IP+PI. The beads were resuspended in 125 μl of 1× Laemmli sample buffer. 2× Laemmli sample buffer was added to each input and supernatant. Samples were heated at 95 °C for 5 min before loading onto a 10% SDS-PAGE analysis followed by Western blotting and probing with relevant antibodies. Input and supernatant samples contained 5 μg of protein, whereas immunoprecipitated samples contained extract from an equivalent of 20 μg of input. Anti-GST, -His, and -FLAG and rabbit polyclonal antibodies to yeast eIF2B subunits were used as described previously (32). Detection used HRP-linked secondary antibodies and chemiluminescence detection (Pierce).
RESULTS
Mutations Affecting Potential Nucleotide-binding Site in eIF2Bϵ PLD Have Minimal Impact on eIF2B Activity
As outlined in the Introduction, it has been noted previously that the eIF2Bγ/ϵ subunits share sequence similarity with pyrophosphorylases such as GlmU and AGP (18). We wished to explore its significance. Multiple sequence alignments of the full-length proteins indicated that the amino-terminal half of the eIF2Bγ/ϵ PLD was most conserved (data not shown, see “Experimental Procedures”). In GlmU and AGP and other similar enzymes, this region is responsible for nucleotide binding. Single GlmU missense mutations at Arg-18 and Lys-25 dramatically impaired uridyltransferase activity (20). In addition, the same study identified Asp-105 as a residue important for nucleotide binding, being necessary for coordination of a stabilizing magnesium ion. These residues are conserved in AGP and in human and yeast eIF2Bϵ (residues Arg-39, Arg-49, and Asp-138 in Gcd6p; Figs. 1 and 2A), raising the possibility that this sequence similarity reflected a shared nucleotide-binding region. Although the minimal eIF2B catalytic domain (ϵcat) resides at the C terminus of eIF2Bϵ (Fig. 1A), the full complex has enhanced activity (5). Additionally, one proposed mechanism for eIF2B GEF activity, based upon enzyme kinetic analyses, is a sequential mechanism, whereby GTP binds eIF2B before GDP is released from eIF2 (33, 34). This differs from the more widely accepted substituted enzyme mechanism, where GDP is released from the G protein, forming a nucleotide-free state, allowing GTP to bind. Taken together, these observations prompted speculation that binding a nucleotide, or nucleotide derivative, to this region of eIF2B may be important for regulating eIF2B GEF activity. To evaluate this idea experimentally, we used site-directed mutagenesis to introduce single alanine residues at Arg-39, Arg-49, and Asp-138 in a plasmid clone of GCD6.
FIGURE 2.
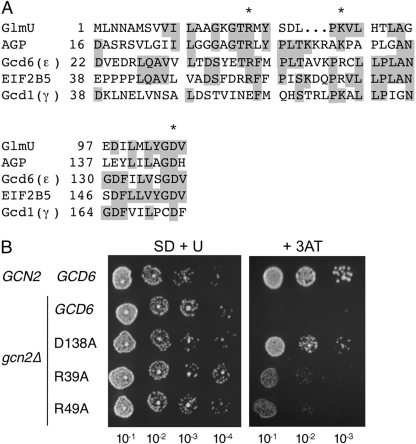
Mutations that impair pyrophosphorylase catalytic function have minimal effect on eIF2Bϵ essential function in yeast. A, segments of a sequence alignment between the pyrophosphorylase domains of E. coli GlmU, potato tuber AGP, and yeast eIF2Bγ and -ϵ subunits. * indicates the residues mutated. Identical residues are shaded. These residues are also highlighted in Fig. 1 as green spheres (N-terminal Arg, pale green; middle Lys/Arg, bright green; and conserved Asp, dark green). B, yeast growth assay for indicated single substitution mutants. Serial dilutions of cultures were spotted on minimal medium (SD + U) or amino acid starvation medium (+ 3AT).
Transformation of a gcd6Δ strain and plasmid shuffling generated strains bearing each allele as the sole source of eIF2Bϵ. Each strain grew indistinguishably from the wild-type strain on standard growth medium, indicating that no significant impairment to the essential function of eIF2B was conferred by these mutations (Fig. 2A). Western blotting confirmed no change in expression levels (data not shown). Further analyses revealed no defect at 10 or 36 °C or growth phenotypes using alternate carbon sources: raffinose, galactose, or maltose (data not shown). The transcription factor GCN4 is translationally controlled by amino acid starvation and highly sensitive to even minor alterations in eIF2B activity. Growth following amino acid starvation, or in the presence of the histidine inhibitor 3AT or the proline analog azetidine-2-carboxylic acid, requires the eIF2α kinase Gcn2p. gcn2Δ cells cannot normally grow on medium containing 3AT or azetidine-2-carboxylic acid; however, mutations that reduce eIF2B GEF activity facilitate growth (see Ref. 35). We used these assays and found that both drugs could enable growth of the D138A mutant in gcn2Δ cells, whereas 3AT also allowed weak growth of both R39A and R49A mutants (Fig. 2B and data not shown). Taken together, these results are consistent with the idea that each mutant confers only a modest reduction in eIF2B activity, not significant enough to affect growth rate in the absence of amino acid starvation. These results are not consistent with the hypothesis that these three residues are critical for eIF2B function and suggest that the sequence conservation does not imply that nucleotide or nucleotide-analog binding to this region is critical for eIF2B essential nucleotide exchange function. The implication of these results for proposed eIF2B catalytic mechanisms is discussed later.
eIF2Bϵ Catalytic Domain Is Not Required for eIF2B Complex Formation
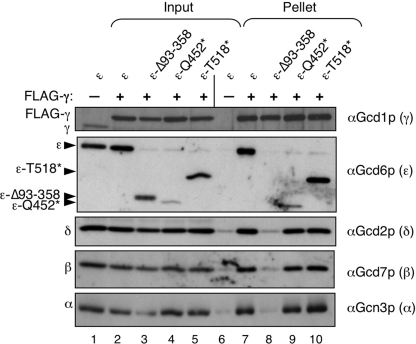
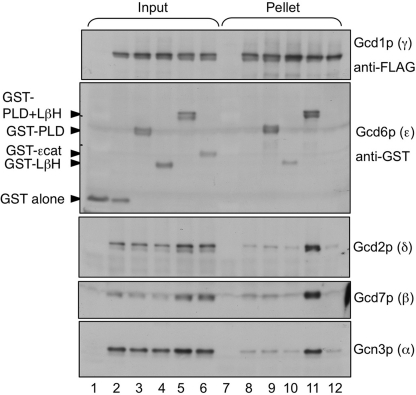
Next, we decided to examine the role of the PLD and LβH domains in eIF2B complex formation. Previously, we isolated several mutations in yeast eIF2Bϵ (GCD6) that affected its activity. Three of these truncated the protein (Thr-518*, Gln-500*, and Gln-452*) and eliminated the catalytic ϵcat domain (26). In addition, we found that an internal deletion Δ93–358 that interrupted both the PLD and the LβH domains disrupted eIF2B complex formation between eIF2Bγ and any other subunit. Our published results suggested that the eIF2Bϵ PLD and LβH regions contributed to holo-complex formation, but that ϵcat was not required. To demonstrate this directly, we co-expressed from high copy plasmids eIF2Bαβδ, γ-FLAG, and either wild-type or mutated eIF2Bϵ in yeast cells, precipitated the γ-FLAG from cell extracts, and assessed which eIF2B subunits were co-precipitated. Overexpression was necessary for our approach as GCD6 is an essential gene and the mutants used do not complement gcd6Δ (26). As shown in Fig. 3, the wild-type eIF2B complex was efficiently precipitated with γ-FLAG, but not with an untagged control (lanes 6 and 7). As reported previously, Δ93–358 disrupted interactions between γ-FLAG and all eIF2B subunits. In contrast, Gln-452* and Thr-518* efficiently formed five-subunit complexes, showing that the ϵcat domain is not required for eIF2B complex formation.
FIGURE 3.
N-terminal domains of eIF2Bϵ are necessary and sufficient for eIF2B complex formation. Immune precipitation of eIF2B complexes with FLAG-tagged eIF2Bγ from yeast whole cell extracts (strain GP3667) expressing wild-type or mutant forms of eIF2Bϵ (Gcd6p) is shown. Western blotting was used to identify the fate of each eIF2B subunit in the experiment.
eIF2Bϵ PLD and LβH Domains Independently Contribute to Interactions with eIF2Bγ, but Both Are Required for eIF2B Complex Formation
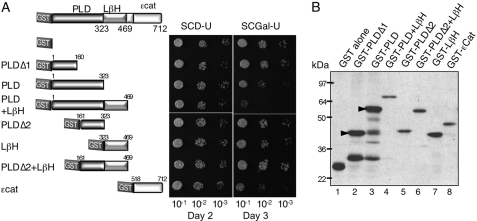
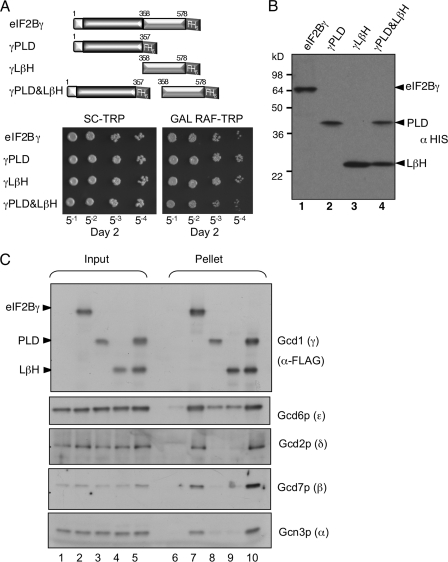
The experiment shown in Fig. 3 suggests that eIF2Bϵ PLD and LβH domains shared with eIF2Bγ, GlmU, and AGP are sufficient for eIF2B complex formation. GlmU forms a homotrimer, with extensive contacts between the LβH domains and no contact between the pyrophosphorylase domains (Fig. 1C). In contrast, the crystal structure for potato tuber AGP shows that the LβH domain is rotated around to make contacts with the pyrophosphorylase domain (Fig. 1D) and forms a homotetramer with extensive contacts between both domains and each monomer (Fig. 1E). Thus from homology modeling alone, it is not possible to infer a structure for the eIF2Bγϵ catalytic subcomplex. To examine the interactions directly, we made a series of constructs fusing the PLD, LβH, or both domains to GST (Fig. 4A) and expressed from a galactose-inducible promoter in a high copy number yeast vector. We used a combination of multiple sequence alignments and a protein structure models database (ModBase: Database of Comparative Protein Structure Models; Ref. 22) to predict the domain boundaries. In addition, we made three further constructs, two bearing approximately each half of the PLD (termed PLDΔ1 and Δ2; Fig. 4A) and one with both PLDΔ2 and LβH combined. We also used a GST-ϵcat construct made previously (13).
FIGURE 4.
PLD and LβH regions of eIF2Bϵ confer dominant slow growth when expressed in yeast. A, the indicated domains of eIF2Bϵ were fused to GST in plasmid pEG(KT) where expression is regulated by a galactose-inducible promoter. Plasmids were individually transformed into yeast strain GP4597. Cells were serially diluted and grown on the indicated media. SCD-U, SCD lacking uracil; SCGal-U, SCGal lacking uracil. B, Western blot from SCGal-U-grown cells with GST antibody reveals relative expression.
We noted that several constructs conferred a galactose-dependent slow growth to the yeast cells. The most severe slow growth phenotype was with the construct containing both the PLD and the LβH domains (Fig. 4A). Anti-GST Western blotting confirmed that all constructs were expressed; importantly, overexpression levels did not correlate with slow growth (Fig. 4B). One interpretation of the slow growth phenotype is that the excess ϵPLD+LβH protein forms an eIF2B complex lacking GEF activity by replacing endogenous eIF2Bϵ from a proportion of the eIF2B complexes, thereby slowing translation and cell growth.
To assess complex formation between these GST domains and eIF2Bγ, cells were co-transformed with a high copy GCD1-FLAG2-His6 plasmid overexpressing tagged eIF2Bγ. Reciprocal precipitations were performed to ask which GST-eIF2Bϵ domains would interact with full-length eIF2Bγ using FLAG affinity resin (Fig. 5A) or glutathione resin (Fig. 5B). These experiments showed that three fragments of eIF2Bϵ could interact strongly with eIF2Bγ. The PLD and LβH domains individually or combined together are shown in Fig. 5 (panel A, lanes 11, 14, and 12, respectively; and panel B, lanes 11, 15, and 12, respectively). These experiments show that both domains of eIF2Bϵ independently contribute to binding to eIF2Bγ. This strongly suggests that the eIF2Bγϵ interdomain interactions do not resemble those in GlmU, but possibly resemble some of those found in the AGP homotetramer (Fig. 1).
FIGURE 5.
eIF2Bϵ PLD and LβH regions independently contribute to eIF2Bϵ-γ subunit interactions. Co-immune precipitation experiments using total cell extracts from yeast strain GP4597 co-expressing full-length eIF2Bγ-FLAG-His6 (Gcd1p) and the indicated regions of eIF2Bϵ (Gcd6p) fused to GST are shown. Co-immune precipitation used anti-FLAG M2 (A) or GST resin (B). See “Experimental Procedures” for further details.
Next, we extended our analysis to examine whether these individual eIF2Bϵ domains are sufficient for eIF2B complex formation. We transformed our cells with a plasmid co-expressing eIF2Bαβδ and used FLAG immune precipitation to assess this. In contrast to results presented in Fig. 5, but in agreement with findings shown in Fig. 3, we find that both PLD and LβH domains of eIF2Bϵ are required to stably bind eIF2Bαβδ to eIF2Bγ (Fig. 6).
FIGURE 6.
Both eIF2Bϵ PLD and LβH regions are necessary for holo-complex formation with eIF2Bαβγδ. Co-immune precipitation experiments using total cell extracts from strain GP4597 co-expressing full-length eIF2Bγ-FLAG-His6 (Gcd1p) and regions of eIF2Bϵ (Gcd6p) fused to GST and eIF2Bαβδ (Gcn3p, Gcd7p and Gcd2p) are shown. Co-immune precipitation used anti-FLAG M2 affinity resin.
eIF2Bγ PLD and LβH Domains Independently Contribute to Interactions with eIF2Bϵ, but Both Are Required for Full eIF2B Complex Formation
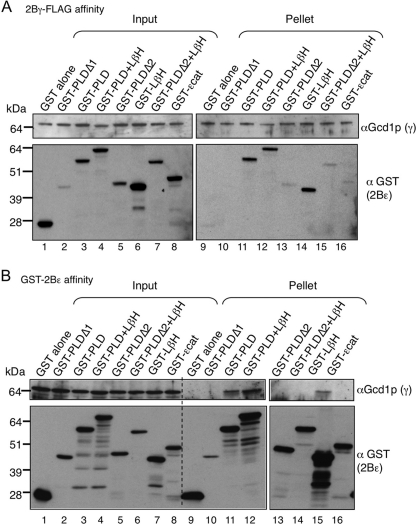
We then assessed domain interaction requirements for eIF2Bγ. Using a similar strategy to that described above, constructs expressing γ-FLAG domains from galactose-inducible promoters were made and introduced into yeast, either singly or as a pair (Fig. 7). Only the construct expressing both domains independently conferred a modest slow growth phenotype, although all fragments were expressed (Fig. 7, A and B). These cells were co-transformed with eIF2Bαβδϵ genes, and anti-FLAG immune precipitations were performed. The results were similar to those obtained with the equivalent eIF2Bϵ domains (Fig. 7C) because each γ domain could independently form a complex with the ϵ subunit, albeit at reduced levels when compared with full-length γ. When both domains were present, the αβδ subunits could also join, forming a five-subunit complex. This was true whether full-length γ was used, or remarkably, even when both separate domains were co-expressed (compare lanes 7 and 10). These results are consistent with the idea that each domain independently contributes to γϵ complex formation, but that both γ domains together are required to recruit ϵ and the αβδ subcomplex.
FIGURE 7.
Both eIF2Bγ PLD and LβH regions are necessary for holo-complex formation with eIF2Bαβδϵ. A, top, graphic of eIF2Bγ fragments fused to FLAG and His6 (FH6) cloned into pBEVY-GT (31) and transformed into yeast strain GP4597. Bottom, growth assay of 5-fold serially diluted cultures on noninducing (SC-TRP) and inducing (GAL RAF-TRP) media. B, Western blotting of the strains in panel A grown in medium containing galactose and raffinose carbon sources. C, Western blotting of co-immune precipitation experiments using total cell extracts from strain GP4597 co-expressing full-length eIF2Bϵ (Gcd6p) and regions of eIF2Bγ-FLAG2His6 (Gcd1p) and eIF2Bαβδ (Gcn3p, Gcd7p, and Gcd2p). Co-immune precipitation used anti-FLAG M2 affinity resin.
Extensive Interactions between γ and ϵ PLD and LβH Domains
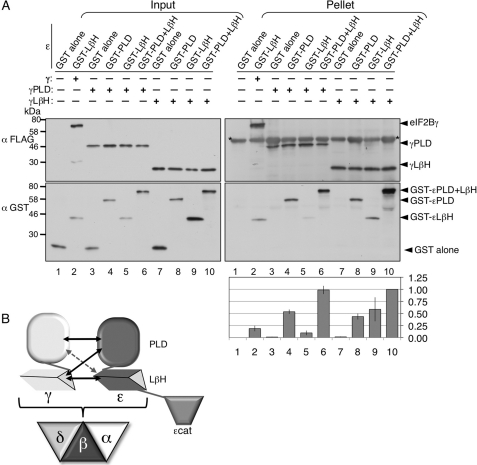
To determine which individual ϵ domains the eIF2Bγ PLD and LβH domains interact with, we co-expressed pairs of GST-ϵ and γ-FLAG domains in yeast and performed anti-FLAG IPs. As controls, we showed that GST alone would not interact with either γ domain or the FLAG resin alone (Fig. 8A, right panel, lanes 1, 3, and 7) and that both γ domains could interact with the GST-ϵPLD+LβH construct (lanes 6 and 10). The experiments showed that either γ domain could interact with both GST-ϵPLD and GST-ϵ LβH separately (lanes 4, 5, 8, and 9). Quantification of the relative interaction efficiency of GST domains with γ-FLAG was consistent with the idea that the γPLD domain interacts strongly with ϵPLD but very weakly with ϵLβH. In contrast, the γLβH construct interacted equally with both ϵPLD and ϵLβH. The level of interaction between γPLD and GST-ϵPLD+LβH appeared stronger than the sum of the strength of the individual domains (compare lanes 4, 5, and 6). Taken together, these data suggest that there are extensive interactions between the γ and ϵ structural domains that are necessary for eIF2B complex formation. These observations allow us to propose a model for domain interactions within this protein complex (Fig. 8B; see “Discussion”).
FIGURE 8.
Interdomain interactions between eIF2Bϵ and -γ. A, co-immune precipitation experiments using total cell extracts from strain GP4597 co-expressing indicated GST-eIF2Bϵ and eIF2Bγ-FLAG-His6 fragments using anti-FLAG M2 affinity resin. * indicates nonspecific cross-reaction between mouse antibodies in the pellet fractions. Quantification indicates fraction of GST-ϵ in pellets relative to γ-FLAG pulled down (not input level). The signal was normalized to the interaction shown in lane 10 ± S.E., n = 4. B, model for eIF2Bγϵ complex domain interactions based on experiments and homology to AGP (Fig. 1E) and eIF2B holo-complex formation. Arrow sizes indicate the relative strength of the individual domain interactions identified in this study.
DISCUSSION
eIF2B is the GEF for eIF2 during eukaryotic protein synthesis. With five different subunits, it has a significantly greater structural complexity than GEFs for other G proteins, many of which are single subunit factors (29, 36, 37). In this study, we have focused our attention on the roles of conserved domains within the ϵ and γ subunits. Both subunits share homology with sugar nucleotide pyrophosphorylase enzymes found in all organisms. These domains are the PLD and LβH regions (Fig. 1). We first examined whether residues important for nucleotide binding in this enzyme family and conserved in eIF2Bϵ PLD were critical for eIF2B essential function. Site-directed mutagenesis and genetic assays revealed only modest defects (Fig. 2). These findings are not consistent with the idea that binding a nucleotide or a nucleotide derivative to this motif significantly influences an essential eIF2B function. These results are in contrast to mutations characterized previously within conserved residues of the PLD at ϵAsn-249 and ϵPhe-250. Both N249K and F250L severely impaired growth and eIF2B activity, with N249K identified as a lethal allele (26). These new results are of interest because previous enzyme kinetic analyses of eIF2B have proposed two competing mechanisms to account for eIF2B GEF activity: i) a standard substituted enzyme mechanism where GDP is released, allowing GTP to bind a nucleotide-free eIF2; or ii) a sequential mechanism where GTP binds before GDP is released (33, 34). The latter, more unusual mechanism would require a second nucleotide-binding site. The best candidate for this site is the PLD of eIF2Bϵ. However, the mutations to this motif that we have analyzed here, known to significantly impair nucleotide binding function in other proteins, have only minimal impact on yeast eIF2B function. Therefore a model where GTP binds directly to this motif before being transferred to eIF2 could probably be discounted (38). Similarly, ideas that nucleotide binding here may have an allosteric activation effect on eIF2B GEF function, as suggested for mammalian eIF2B, seem unlikely for the yeast protein, based on our findings (39).
Experiments examining the role of eIF2Bϵ domains for complex formation revealed that ϵcat was not required (Fig. 3). Instead the PLD and LβH regions were implicated, so we examined the roles of these domains in mediating protein-protein interactions between eIF2Bϵ and -γ in the catalytic subcomplex. We overexpressed GST-fused eIF2Bϵ domains or FLAG-His-tagged eIF2Bγ domains in yeast cells along with the full-length partner subunit. Results were similar for both sets of experiments as interactions between eIF2Bγ and -ϵ were mediated by either PLD alone or LβH alone as well as PLD+LβH domains combined (Figs. 5 and 7). We further showed that there are interactions possible between all four domains (Fig. 8A). The domains appear to form homologous interacting pairs as the PLDs appear to interact more strongly with each other than they do with the LβH domains, and similarly, the LβH domains bind more strongly to each other than to the PLDs. This domain interaction arrangement differs from that found in GlmU, where only LβH domains interact together (Fig. 1C) (20). However, these interactions are possibly broadly compatible with one of the two possible dimer pair combinations that comprise the potato tuber AGP homotetramer structure (Fig. 1E) (19). One dimer pair interacts exclusively through pyrophosphorylase interactions (e.g. Fig. 1E, the left upper and lower monomers). Our data do not favor this arrangement. A second monomer pair (as exemplified in Fig. 1E by the lower left and right pair) combine through interactions between all domains. Our data for eIF2Bγ/ϵ are broadly compatible with these domain interactions. These ideas are shown in the model in Fig. 8B, but we do acknowledge that other domain arrangements or orientations are possible.
In contrast to the requirements for γϵ complex formation, we found that only constructs comprising both pairs of PLD and LβH domains together were able to bind stably to the α, β, and δ subunits to form a five-subunit eIF2B complex (Figs. 6 and 7C). Interestingly, when we co-expressed both γPLD and γLβH as separate polypeptides together in the same strain, this was also capable of forming a five-subunit complex similar to full-length eIF2Bγ (Fig. 7C, lanes 7 and 10). Previous work has shown that the αβδ subunits can bind together to form a three-subunit subcomplex that can bind eIF2α with high affinity when it is phosphorylated (9, 11). However, how these subunits interact with each other and with the γϵ subcomplex is less clear. The eIF2Bα crystal structure formed dimers (Fig. 1A) and higher order octamer arrangements (40) and is highly similar to archaeal proteins, some of which form hexamers rather than octamers within their crystals. This hexameric arrangement has prompted the suggestion that the αβδ trimer may resemble one-half of the hexamer (41). Other experiments indicate that the α and δ subunits may be more peripheral in the complex than in the β subunit. Firstly, the α subunit is nonessential, and a β-ϵ complex forms that retains GEF function (5, 12, 42). Next, recent experiments depleting the yeast δ subunit genetically, using a “degron” strain, could also form an αβγϵ complex (43). In contrast, similar degron depletion of eIF2Bβ caused co-depletion of eIF2Bδ. A similar result was obtained previously with a missense mutation (eIF2Bβ-V341D). The mutation destabilized both eIF2Bβ and eIF2Bδ binding to the eIF2B complex (35). These results are consistent with eIF2Bβ being more central in the complex, or at least stabilizing eIF2Bδ binding. Yeast genetic evidence also suggested that eIF2Bαβγ can form an independent subcomplex (43), although this complex was not directly shown, and we have not been able to demonstrate significant levels of interaction between eIF2Bαβγ in our experiments (data not shown). Taken together, these data suggest that the α and δ subunits can be more readily excluded from the eIF2B complex and therefore likely occupy more peripheral positions in it. Experiments with mammalian eIF2B subunits also provide some support to these ideas. Purification of human subunits from 293 cells identified stable γϵ and βγδϵ complexes (44). Experiments purifying rat eIF2B from an insect cell co-expression system suggested that the N-terminal half of the ϵPLD (1–164) could bind to eIF2Bβ and that the remaining part (ϵ159–716) could bind βγδ, but not α (45). As studies have used a variety of expression and purification systems, sources of eIF2B subunits, and differing washing stringencies in their experimental systems, these elements may account for any apparently different findings. When taken together, the experiments suggest that eIF2Bβ is central to the αβδ subcomplex. We suggest that our data (Figs. 6 and 7C) are compatible with two possible modes for αβδ subcomplex binding to the γϵ subcomplex. One option is that αβδ directly bind to all four interacting domains within the γϵ subcomplex. A second possibility is that all four domains are necessary to adopt or constrain a specific γϵ conformation and that this facilitates stable binding of αβδ to only one of the other subunits, possibly eIF2Bγ.
An independent study conducted in parallel and examining domain interactions within human eIF2B has led to similar conclusions to our work (48). Using a transient transfection approach, cDNAs were co-expressed in HEK293 cells, and complexes were purified from these cells. By examining the effects of missense mutations in ϵ or γ and by co-expression of domain deletion mutants, the contribution of each domain to protein-protein interactions is evaluated. In general, the experiments nicely complement the findings reported here. They confirm our results that mutations within human eIF2Bϵ equivalent to our R39A and D138A have no significant negative impact on eIF2B activity as measured in vitro. In addition, they also find that the γϵ PLDs are important for domain interactions with each other and that both ϵPLD+LβH domains are necessary for complex formation with the αβγδ subunits. The only apparent difference is that human γ-LβH was not necessary for human αβγδϵ complex formation but was required for the yeast intersubunit interactions. This most likely reflects experimental differences in the extent of the γ-LβH domain. Yeast γ-LβH is defined here as 220 amino acids, whereas human γ-LβH domain deleted in the companion study is only 82 residues. A simple explanation for the apparent difference between experiments is that our yeast γ-LβH deletion removed an element critical for complex formation, which is retained in the human γPLD construct used.
In summary, our experiments suggest that potential nucleotide-binding elements within eIF2Bϵ have minimal impact on the rate of eIF2B nucleotide exchange. This finding is at odds with one proposed mechanism for nucleotide exchange (the sequential mechanism) but is consistent with the substituted enzyme mechanism: an issue of long controversy (33). In addition, by protein-protein interaction studies, we provide insight into the domain interactions between eIF2Bϵ and -γ necessary for eIF2B complex formation. Although further studies will be necessary to resolve precisely how the catalytic and regulatory subcomplexes bind each other, our work provides important insight toward resolving this conundrum.
Acknowledgments
We thank Clare Holtby for construction of several GST fusion plasmids used in this study, as well as Mark Ashe, Chris Grant, and members of the Pavitt laboratory for constructive comments during the course of this study and on the manuscript. We also thank Chris Proud (Southampton) for discussions and sharing experimental findings prior to publication.
This work was supported by grants from the Biotechnology and Biological Sciences Research Council.
- tRNAiMet
- initiator methionyl tRNA
- eIF2(αP)
- phosphorylated eIF2
- GEF
- guanine nucleotide factor
- PLD
- pyrophosphorylase-like domain
- LβH
- left-handed β helix
- AGP
- ADP-glucose pyrophosphorylase
- GlmU
- N-acetylglucosamine 1-phosphate uridyltransferase
- IP
- immunoprecipitation(s)
- 3AT
- 3-amino-1,2,4-triazole.
REFERENCES
- 1. Hinnebusch A. G. (2011) Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol. Mol. Biol. Rev. 75, 434–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schmitt E., Naveau M., Mechulam Y. (2010) Eukaryotic and archaeal translation initiation factor 2: a heterotrimeric tRNA carrier. FEBS Lett. 584, 405–412 [DOI] [PubMed] [Google Scholar]
- 3. Jennings M. D., Pavitt G. D. (2010) eIF5 has GDI activity necessary for translational control by eIF2 phosphorylation. Nature 465, 378–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pavitt G. D. (2005) eIF2B, a mediator of general and gene-specific translational control. Biochem. Soc. Trans. 33, 1487–1492 [DOI] [PubMed] [Google Scholar]
- 5. Pavitt G. D., Ramaiah K. V., Kimball S. R., Hinnebusch A. G. (1998) eIF2 independently binds two distinct eIF2B subcomplexes that catalyze and regulate guanine-nucleotide exchange. Genes Dev. 12, 514–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jackson R. J., Hellen C. U., Pestova T. V. (2010) The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11, 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pavitt G. D., Proud C. G. (2009) Protein synthesis and its control in neuronal cells with a focus on vanishing white matter disease. Biochem. Soc. Trans. 37, 1298–1310 [DOI] [PubMed] [Google Scholar]
- 8. Horzinski L., Huyghe A., Cardoso M. C., Gonthier C., Ouchchane L., Schiffmann R., Blanc P., Boespflug-Tanguy O., Fogli A. (2009) Eukaryotic initiation factor 2B (eIF2B) GEF activity as a diagnostic tool for EIF2B-related disorders. PLoS One 4, e8318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang W., Hinnebusch A. G. (1996) Identification of a regulatory subcomplex in the guanine nucleotide exchange factor eIF2B that mediates inhibition by phosphorylated eIF2. Mol. Cell Biol. 16, 6603–6616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pavitt G. D., Yang W., Hinnebusch A. G. (1997) Homologous segments in three subunits of the guanine nucleotide exchange factor eIF2B mediate translational regulation by phosphorylation of eIF2. Mol. Cell Biol. 17, 1298–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Krishnamoorthy T., Pavitt G. D., Zhang F., Dever T. E., Hinnebusch A. G. (2001) Tight binding of the phosphorylated α subunit of initiation factor 2 (eIF2α) to the regulatory subunits of guanine nucleotide exchange factor eIF2B is required for inhibition of translation initiation. Mol. Cell Biol. 21, 5018–5030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kimball S. R., Fabian J. R., Pavitt G. D., Hinnebusch A. G., Jefferson L. S. (1998) Regulation of guanine nucleotide exchange through phosphorylation of eukaryotic initiation factor eIF2α. Role of the α and δ subunits of eiF2b. J. Biol. Chem. 273, 12841–12845 [DOI] [PubMed] [Google Scholar]
- 13. Gomez E., Mohammad S. S., Pavitt G. D. (2002) Characterization of the minimal catalytic domain within eIF2B: the guanine-nucleotide exchange factor for translation initiation. EMBO J. 21, 5292–5301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mohammad-Qureshi S. S., Haddad R., Hemingway E. J., Richardson J. P., Pavitt G. D. (2007) Critical contacts between the eukaryotic initiation factor 2B (eIF2B) catalytic domain and both eIF2β and eIF2γ mediate guanine nucleotide exchange. Mol. Cell Biol. 27, 5225–5234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fabian J. R., Kimball S. R., Heinzinger N. K., Jefferson L. S. (1997) Subunit assembly and guanine nucleotide exchange activity of eukaryotic initiation factor-2B expressed in Sf9 cells. J. Biol. Chem. 272, 12359–12365 [DOI] [PubMed] [Google Scholar]
- 16. Williams D. D., Pavitt G. D., Proud C. G. (2001) Characterization of the initiation factor eIF2B and its regulation in Drosophila melanogaster. J. Biol. Chem. 276, 3733–3742 [DOI] [PubMed] [Google Scholar]
- 17. Bushman J. L., Asuru A. I., Matts R. L., Hinnebusch A. G. (1993) Evidence that GCD6 and GCD7, translational regulators of GCN4, are subunits of the guanine nucleotide exchange factor for eIF-2 in Saccharomyces cerevisiae. Mol. Cell Biol. 13, 1920–1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Koonin E. V. (1995) Multidomain organization of eukaryotic guanine nucleotide exchange translation initiation factor eIF-2B subunits revealed by analysis of conserved sequence motifs. Protein Sci. 4, 1608–1617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jin X., Ballicora M. A., Preiss J., Geiger J. H. (2005) Crystal structure of potato tuber ADP-glucose pyrophosphorylase. EMBO J. 24, 694–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brown K., Pompeo F., Dixon S., Mengin-Lecreulx D., Cambillau C., Bourne Y. (1999) Crystal structure of the bifunctional N-acetylglucosamine 1-phosphate uridyltransferase from Escherichia coli: a paradigm for the related pyrophosphorylase superfamily. EMBO J. 18, 4096–4107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kiefer F., Arnold K., Künzli M., Bordoli L., Schwede T. (2009) The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 37, D387–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pieper U., Webb B. M., Barkan D. T., Schneidman-Duhovny D., Schlessinger A., Braberg H., Yang Z., Meng E. C., Pettersen E. F., Huang C. C., Datta R. S., Sampathkumar P., Madhusudhan M. S., Sjölander K., Ferrin T. E., Burley S. K., Sali A. (2011) ModBase, a database of annotated comparative protein structure models, and associated resources. Nucleic Acids Res. 39, D465–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Guthrie C., Fink G. R. (eds). (1991) Guide to Yeast Genetics and Molecular Biology, Vol. 194, Academic Press Inc, San Diego, CA [Google Scholar]
- 24. Gietz R. D., Woods R. A. (2006) Yeast transformation by the LiAc/SS Carrier DNA/PEG method. Methods Mol. Biol. 313, 107–120 [DOI] [PubMed] [Google Scholar]
- 25. Asano K., Krishnamoorthy T., Phan L., Pavitt G. D., Hinnebusch A. G. (1999) Conserved bipartite motifs in yeast eIF5 and eIF2Bϵ, GTPase-activating and GDP-GTP exchange factors in translation initiation, mediate binding to their common substrate eIF2. EMBO J. 18, 1673–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gomez E., Pavitt G. D. (2000) Identification of domains and residues within the ϵ subunit of eukaryotic translation initiation factor 2B (eIF2Bϵ) required for guanine nucleotide exchange reveals a novel activation function promoted by eIF2B complex formation. Mol. Cell Biol. 20, 3965–3976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Boesen T., Mohammad S. S., Pavitt G. D., Andersen G. R. (2004) Structure of the catalytic fragment of translation initiation factor 2B and identification of a critically important catalytic residue. J. Biol. Chem. 279, 10584–10592 [DOI] [PubMed] [Google Scholar]
- 29. Mohammad-Qureshi S. S., Jennings M. D., Pavitt G. D. (2008) Clues to the mechanism of action of eIF2B, the guanine nucleotide exchange factor for translation initiation. Biochem. Soc. Trans. 36, 658–664 [DOI] [PubMed] [Google Scholar]
- 30. Mitchell D. A., Marshall T. K., Deschenes R. J. (1993) Vectors for the inducible overexpression of glutathione S-transferase fusion proteins in yeast. Yeast 9, 715–722 [DOI] [PubMed] [Google Scholar]
- 31. Miller C. A., 3rd, Martinat M. A., Hyman L. E. (1998) Assessment of aryl hydrocarbon receptor complex interactions using pBEVY plasmids: expression vectors with bidirectional promoters for use in Saccharomyces cerevisiae. Nucleic Acids Res. 26, 3577–3583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mohammad-Qureshi S. S., Haddad R., Palmer K. S., Richardson J. P., Gomez E., Pavitt G. D. (2007) Purification of FLAG-tagged eukaryotic initiation factor 2B complexes, subcomplexes, and fragments from Saccharomyces cerevisiae. Methods Enzymol. 431, 1–13 [DOI] [PubMed] [Google Scholar]
- 33. Manchester K. L. (1997) Catalysis of guanine nucleotide exchange on eIF-2 by eIF-2B: is it a sequential or substituted enzyme mechanism? Biochem. Biophys. Res. Commun. 239, 223–227 [DOI] [PubMed] [Google Scholar]
- 34. Nika J., Yang W., Pavitt G. D., Hinnebusch A. G., Hannig E. M. (2000) Purification and kinetic analysis of eIF2B from Saccharomyces cerevisiae. J. Biol. Chem. 275, 26011–26017 [DOI] [PubMed] [Google Scholar]
- 35. Richardson J. P., Mohammad S. S., Pavitt G. D. (2004) Mutations causing childhood ataxia with central nervous system hypomyelination reduce eukaryotic initiation factor 2B complex formation and activity. Mol. Cell Biol. 24, 2352–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sprang S. R., Coleman D. E. (1998) Invasion of the nucleotide snatchers: structural insights into the mechanism of G protein GEFs. Cell 95, 155–158 [DOI] [PubMed] [Google Scholar]
- 37. Siderovski D. P., Willard F. S. (2005) The GAPs, GEFs, and GDIs of heterotrimeric G protein α subunits. Int. J. Biol. Sci. 1, 51–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Manchester K. L. (2001) Catalysis of guanine nucleotide exchange on eIF2 by eIF2B: can it be both a substituted enzyme and a sequential mechanism? Biochem. Biophys. Res. Commun. 289, 643–646 [DOI] [PubMed] [Google Scholar]
- 39. Price N., Proud C. (1994) The guanine nucleotide exchange factor, eIF-2B. Biochimie 76, 748–760 [DOI] [PubMed] [Google Scholar]
- 40. Hiyama T. B., Ito T., Imataka H., Yokoyama S. (2009) Crystal structure of the α subunit of human translation initiation factor 2B. J. Mol. Biol. 392, 937–951 [DOI] [PubMed] [Google Scholar]
- 41. Dev K., Santangelo T. J., Rothenburg S., Neculai D., Dey M., Sicheri F., Dever T. E., Reeve J. N., Hinnebusch A. G. (2009) Archaeal aIF2B interacts with eukaryotic translation initiation factors eIF2α and eIF2Bα: implications for aIF2B function and eIF2B regulation. J. Mol. Biol. 392, 701–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liu A. R., van der Lei H. D., Wang X., Wortham N. C., Tang H., van Berkel C. G., Mufunde T. A., Huang W., van der Knaap M. S., Scheper G. C., Proud C. G. (2011) Severity of vanishing white matter disease does not correlate with deficits in eIF2B activity or the integrity of eIF2B complexes. Hum. Mutat. 32, 1036–1045 [DOI] [PubMed] [Google Scholar]
- 43. Dev K., Qiu H., Dong J., Zhang F., Barthlme D., Hinnebusch A. G. (2010) The β/Gcd7 subunit of eukaryotic translation initiation factor 2B (eIF2B), a guanine nucleotide exchange factor, is crucial for binding eIF2 in vivo. Mol. Cell Biol. 30, 5218–5233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li W., Wang X., Van Der Knaap M. S., Proud C. G. (2004) Mutations linked to leukoencephalopathy with vanishing white matter impair the function of the eukaryotic initiation factor 2B complex in diverse ways. Mol. Cell Biol. 24, 3295–3306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Anthony T. G., Fabian J. R., Kimball S. R., Jefferson L. S. (2000) Identification of domains within the ϵ subunit of the translation initiation factor eIF2B that are necessary for guanine nucleotide exchange activity and eIF2B holoprotein formation. Biochim. Biophys. Acta 1492, 56–62 [DOI] [PubMed] [Google Scholar]
- 46. Bushman J. L., Foiani M., Cigan A. M., Paddon C. J., Hinnebusch A. G. (1993) Guanine nucleotide exchange factor for eukaryotic translation initiation factor 2 in Saccharomyces cerevisiae: interactions between the essential subunits GCD2, GCD6, and GCD7 and the regulatory subunit GCN3. Mol. Cell Biol. 13, 4618–4631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dever T. E., Yang W., Aström S., Byström A. S., Hinnebusch A. G. (1995) Modulation of tRNAiMet, eIF-2, and eIF-2B expression shows that GCN4 translation is inversely coupled to the level of eIF-2.GTP.Met-tRNAiMet ternary complexes. Mol. Cell Biol. 15, 6351–6363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang X., Wortham N. C., Liu R., Proud C. G. (January 11, 2012) Identification of residues that underpin interactions within the eukaryotic initiation factor (eIF2) 2B complex. J. Biol. Chem. 287, 8263–8274 [DOI] [PMC free article] [PubMed] [Google Scholar]