Abstract
Calcineurin B-like proteins (CBLs) represent a family of calcium sensor proteins that interact with a group of serine/threonine kinases designated as CBL-interacting protein kinases (CIPKs). CBL-CIPK complexes are crucially involved in relaying plant responses to many environmental signals and in regulating ion fluxes. However, the biochemical characterization of CBL-CIPK complexes has so far been hampered by low activities of recombinant CIPKs. Here, we report on an efficient wheat germ extract-based in vitro transcription/translation protocol that yields active full-length wild-type CIPK proteins. We identified a conserved serine residue within the C terminus of CBLs as being phosphorylated by their interacting CIPKs. Remarkably, our studies revealed that CIPK-dependent CBL phosphorylation is strictly dependent on CBL-CIPK interaction via the CIPK NAF domain. The phosphorylation status of CBLs does not appear to influence the stability, localization, or CIPK interaction of these calcium sensor proteins in general. However, proper phosphorylation of CBL1 is absolutely required for the in vivo activation of the AKT1 K+ channel by CBL1-CIPK23 and CBL9-CIPK23 complexes in oocytes. Moreover, we show that by combining CBL1, CIPK23, and AKT1, we can faithfully reconstitute CBL-dependent enhancement of phosphorylation of target proteins by CIPKs in vitro. In addition, we report that phosphorylation of CBL1 by CIPK23 is also required for the CBL1-dependent enhancement of CIPK23 activity toward its substrate. Together, these data identify a novel general regulatory mechanism of CBL-CIPK complexes in that CBL phosphorylation at their flexible C terminus likely provokes conformational changes that enhance specificity and activity of CBL-CIPK complexes toward their target proteins.
Keywords: Calcium-binding Proteins, Calcium Signaling, Plant, Protein Kinases, Protein Phosphorylation
Introduction
Intracellular release of Ca2+ ions belongs to the earliest events in signal perception in eukaryotes (1–3). In plants, a wide range of abiotic and biotic stimuli, including light, high and low temperature, mechanical disturbance, drought, salt and osmotic stresses, plant hormones, and pathogen elicitors induce specific and distinct spatio-temporal patterns of changes in cytosolic free Ca2+ concentration, designated as “Ca2+ signatures” (4–6). In addition, physiological processes like stomata movement, root hair elongation, and pollen tube growth are accompanied by distinct spatio-temporal changes in Ca2+ concentration (5, 7). Ca2+-binding proteins are involved in sensing and relaying these signals to downstream signaling and adaptation responses. Such calcium sensor proteins, like calcineurin B, calmodulin, and neuronal calcium sensors in animals and calcineurin B-like proteins (CBLs),3 calmodulin, and calmodulin-like proteins in plants, sense and forward local changes in Ca2+ concentration to their target proteins or to the regulation of transcriptional responses (8–14). Although, with the exception of calcium-dependent protein kinases, these Ca2+-binding proteins do not have an enzymatic activity on their own, binding of Ca2+ ions results in an increased affinity for and subsequent activation or deactivation of interacting proteins (6, 15, 16).
CBL proteins, which are most similar to calcineurin B and neuronal calcium sensor proteins, have been recognized as fulfilling crucial functions in diverse Ca2+-dependent processes in plants (17, 18). In Arabidopsis, 10 CBLs that specifically interact with distinct family members of the 26 CIPKs form a network-like signaling system for specific stimulus-response coupling (17, 19). The calcineurin B-like calcium sensor SOS3/CBL4 and the CIPK-type kinase SOS2/CIPK24 have been identified in genetic screens. They appear to be part of a calcium-regulated signaling pathway that specifically mediates salt stress signaling and adaptation by regulating the Na+/H+ antiporter SOS1 (20–24). Especially reverse genetics analyses, often in combination with electrophysiological studies, have uncovered important functions of CBLs and CIPKs in abiotic stress responses and in the regulation of plant ion homeostasis. Analyses of two CBL1 loss-of-function alleles revealed that disruption of the CBL1 gene renders plants hypersensitive to drought and salt stress and impairs the regulation of stress-responsive genes (25, 26). Alternative complex formation of the kinase CIPK1 with either CBL1 or CBL9 mediates abscisic acid (ABA)-dependent and ABA-independent signaling responses (27). Recently, several studies reported that CBL1 and CBL9 form complexes with and function synergistically in regulating their target kinase CIPK23 (28, 29). Upon activation by CBL1 or CBL9, CIPK23 phosphorylates the K+ channel AKT1 and contributes to K+ uptake under limiting K+ supply conditions. Moreover, CBL1/CBL9 and CIPK23 appear to function in regulating nitrate sensing and uptake, because biochemical and reverse genetics analyses identified CIPK23 as a critical regulator mediating the switch between low and high affinity nitrate transport modes by phosphorylating the nitrate transporter CHL1/NRT1.1 (30). Together these findings point to a central role of the CBL-CIPK network in regulating ion fluxes in plants (31).
All CBL proteins share a rather conserved core region consisting of four EF hand Ca2+-binding sites that are arranged in completely invariant spacing within the protein (9). CIPK-type kinases are composed of a conserved N-terminal kinase domain and a C-terminal regulatory domain, which are separated by a variable junction domain. Within the rather divergent regulatory domain, the conserved NAF domain has been identified as required and sufficient for mediating CBL interaction (19, 32). With regard to the biochemical mechanisms of CBL-CIPK function, analyses of the crystal structures of CBL4/SOS3 and CBL2 in complex with Ca2+ revealed how these proteins are able to sense Ca2+ (33, 34). Increases in cellular Ca2+ lead to changes in the global conformation of the CBL proteins that increase the hydrophobic character of the macromolecule. Hydrophobic interactions mediate the binding of the NAF domain of their target kinases into a hydrophobic crevice of the Ca2+ sensor proteins (35, 36). It is assumed that CBL binding to the NAF domain releases the C-terminal autoinhibitory moiety from the kinase domain thereby transforming the kinase into an active conformation (32, 37). In addition, the activation of CIPKs is further enhanced by auto-phosphorylation and by trans-phosphorylation of the activation loop within the kinase domain by unidentified kinases that together contribute to full CIPK activation (6, 32). An interesting new facet of CBL-CIPK interaction was provided by the very first report that a CBL from Pisum sativum was phosphorylated by its interacting CIPK (38). Subsequently, Lin et al. (39) reported that the kinase CIPK24/SOS2 specifically phosphorylates CBL10, but not another investigated CBL protein CBL4/SOS3, and that this phosphorylation appears to enhance the CBL10-CIPK24 interaction. Recently, it was reported that additional CBLs from Arabidopsis, like CBL1 and CBL2, can be phosphorylated by their interacting CIPKs and that CBL phosphorylation is required for activation of the plasma membrane H+-ATPase AHA2 by CBL-CIPK complexes (40). The subcellular localization of CIPKs incorporated into distinct CBL-CIPK complexes is determined by the identity of their CBL moiety (41–43). However, the further biochemical elucidation of CBL-CIPK activation/regulation and target phosphorylation has been severely hampered by the extremely low kinase activity of recombinant full-length wild-type CIPK proteins that have been expressed and purified from Escherichia coli. In the original biochemical characterization of the kinases CIPK1 and CIPK24/SOS2, it required microgram amounts of recombinant proteins to detect autophosphorylation activities of these proteins (21, 44). This constraint has been circumvented to some extent by introducing mutations in the activation loop of the kinase domain that exchange functionally conserved threonine (Thr) or serine (Ser) residues to aspartate (Asp) resulting in constitutively hyperactive enzymes, whose activity were no longer CBL-dependent (31, 45, 46). Even stronger hyperactivation of kinase activity was achieved by deleting the inhibitory C-terminal regions of CIPK24/SOS2, including the NAF domain (24, 31, 39). However, because these artificial kinases do not interact with CBLs, investigations of these recombinant proteins may be error-prone and cannot address CBL-dependent aspects of kinase regulation that bring about Ca2+ dependence and cellular targeting of CBL-CIPK complexes.
Here, we report the establishment of an efficient wheat germ extract-based in vitro transcription/translation protocol that yields active full-length wild-type CIPK proteins. In vitro ana-lyses revealed strong Mn2+ dependence of the autophosphorylation activity of all CIPKs investigated. Using these recombinant proteins, we identified phosphorylation of a conserved single Ser residue within the C terminus of CBLs by their interacting CIPKs as a general feature of CBL-CIPK complexes. This finding suggests that CBL calcium sensors not only regulate CIPK kinase activity but also simultaneously represent genuine targets of these kinases. Our evolutionary analysis suggests that this feature of CBL-CIPK complex modulation arose during the evolution of land plants. Remarkably, our studies revealed that interaction via the NAF domain in the C-terminal regulatory region of CIPKs represents a prerequisite for CIPK-mediated CBL phosphorylation. In contrast, in vitro phosphorylation of targets like SOS1 can occur independent of the NAF domain suggesting the existence of dual phosphorylation mechanisms of CIPKs. The phosphorylation status of CBLs does not appear to influence their stability and localization. However, proper phosphorylation of CBLs is absolutely required for the in vivo activation of the AKT1 K+ channel by CBL1-CIPK23 as well as CBL9-CIPK23 complexes in oocytes. Using wheat germ expressed kinase protein we for the first time succeeded to reconstitute CBL-dependent enhancement of phosphorylation of target proteins by CIPKs in vitro. In these studies, we found that proper phosphorylation of CBL1 is also required for the CBL1-dependent enhancement of CIPK23 activity toward its substrate in vitro. Together, these data suggest that CIPK-mediated phosphorylation of CBL proteins likely results in conformational changes of their flexible C terminus that enhance activity of CBL-CIPK complexes toward their target proteins.
EXPERIMENTAL PROCEDURES
Plasmid Construction
Molecular biology methods were performed according to standard procedures (47). For in vitro synthesis of N-terminally StrepII-tagged proteins, the pIVEX 1.3 WG vector (5 PRIME) was modified by replacing the original multiple cloning site region and the His6 tag coding region with a StrepII tag (WSHPQFEK)-encoding sequence and another multiple cloning site by PCR-based mutagenesis. The composition of the resulting vector, designated as pIVEX-WG-StrepII, is provided in supplemental Fig. S1. The coding sequences of CIPKs, CBLs, and SOS1-Ct (Gly441–Leu1146) were amplified by PCR and cloned into the pIVEX-WG-StrepII vector. Site-directed mutagenesis was performed to generate CBL1 S201A, CBL4 S205A, and CBL10 S237A to prevent phosphorylation and to generate CBL1 S201D, CBL4 S205D, and CBL10 S237D to mimic phosphorylation. The primers used in this work are listed in supplemental Table S1.
Wheat Germ-based Cell-free Protein Synthesis and Protein Purification
StrepII-tagged proteins were generated using a RTS 500 wheat germ CECF kit (5 PRIME) following the manufacturer's instructions. For affinity purification, each in vitro translation reaction (1 ml) was mixed with 0.8 ml of Strep-Tactin Macroprep (IBA) and incubated for 30 min at 4 °C. StrepII-tagged proteins were eluted by gravity flow in elution buffer (100 mm Tris (pH 8.0), 150 mm NaCl, and 2.5 mm desthiobiotin) and collected in 0.4-ml fractions. Abundance of purified proteins was confirmed by Western blot analysis using Strep-Tactin horseradish peroxidase (HRP) conjugate (1:5000, IBA). Moreover, SDS-PAGE followed by Coomassie staining (R-250) was performed to estimate the concentration of each purified protein. Stained bands of the purified proteins were compared with serial dilutions of a BSA standard. In the case of StrepII-CIPK1, in vitro translation resulted in the synthesis of 210 μg of protein and yielded 12.8 μg of purified recombinant kinase protein. The concentration of the purified fraction was determined as 15 ng/μl. (supplemental Fig. S2). For storage, the eluted fractions were supplemented with 1 mg/ml BSA to ensure protein stability at low concentrations.
StrepII-tagged AKT1-Ct protein was generated by expression in and purification from E. coli. The sequence of the AKT1-Ct (His294–Ser857) with an N-terminal StrepII tag was amplified by PCR and cloned into the pET-24 vector (Novagen) with NdeI and SalI. BL21 CodonPlus(DE3)-RIL E. coli strain (Stratagene) cells transformed with the resulting vector were grown in 10-ml Luria-Bertani (LB) medium at 37 °C overnight and were subcultured until A600 reached 0.7 in 100-ml LB medium. Expression of the StrepII-AKT1-Ct was induced by adding 0.5 mm isopropyl β-d-thiogalactopyranoside at 15 °C for overnight. Cells were harvested by centrifugation, and the pellets were resuspended in a lysis buffer (100 mm Tris (pH 8.0), 150 mm NaCl, and 1 mg/ml lysozyme). After incubation on ice for 1 h, the lysate was sonicated and centrifuged at 14,000 × g for 5 min at 4 °C. The StrepII-AKT1-Ct protein was purified from the supernatant using Strep-Tactin Macroprep as described above.
In Vitro Phosphorylation Assays
Purified proteins were incubated for 30 min at 30 °C in 24-μl reactions that contained 66.7 mm Tris (pH 8.0), 100 mm NaCl, 5 mm MnSO4, 0.5 mm CaCl2, 2 mm DTT, 10 μm ATP, and 4 μCi of [γ-32P]ATP (3000 Ci/mmol). Reactions were stopped by adding 20 mm EDTA and then subjected to SDS-PAGE. SDS gels were fixed by Coomassie staining, and radioactively labeled proteins were visualized by autoradiography. All experiments were repeated at least two times with similar results. For mass spectrometric analysis, proteins were incubated as described above but with 10 μm nonradioactive ATP. The enzymatic properties of in vitro-translated and purified CIPK proteins were determined using two synthetic substrates. Syntide-2 (PLARTLSVAGLPGKK) is a substrate peptide for mammalian Ca2+/calmodulin-dependent protein kinase II as well as plant calcium-dependent protein kinases (48, 49). ALARA peptide (ALARAASAAALARRR) is a variant of a minimal substrate for several families of protein kinases (50). 150 ng of each CIPK protein were incubated with 6–200 μm of each synthetic peptide at 30 °C in 30-μl reactions that contained 66.7 mm Tris (pH 8.0), 100 mm NaCl, 5 mm MnSO4, 0.5 mm CaCl2, 2 mm DTT, 10 μm ATP, and 5 μCi of [γ-32P]ATP. After 30 min of incubation, 20 μl of each reaction was spotted onto 2 cm2 of Whatman P81 phosphocellulose paper. The papers were washed with 1% phosphoric acid five times for 5 min and then immersed into vials with 4 ml of scintillation liquid (Roth). 32P incorporation into the synthetic peptides was counted using a scintillation spectrometer (Beckman). The kinetic constants were calculated using GraphPad Prism 5 (GraphPad Software).
Mass Spectrometric Analyses
Coomassie-stained protein bands were excised from SDS gels and subjected to in-gel trypsin digestion according to established protocols (51). No reduction and alkylation steps were performed. Peptide separation was performed on an Ultimate 3000 nanoflow HPLC system (Dionex) coupled to the nanospray source of an LTQ Orbitrap XL mass spectrometer (Thermo Finnigan). The mass spectrometer was operated in positive ion mode. MS full scans (m/z 400–2000) were acquired by FT-MS in the orbitrap at a resolution of 60,000 with internal lock mass calibration on m/z 445.12003. The five most intense ions of each full scan were fragmented in the linear ion trap by collision-induced dissociation (35% normalized collision energy). Fragment ions exhibiting a neutral loss of 24.5, 32.7, 49, and 98 Da corresponding to the elimination of phosphoric acid were activated further, although the remaining MS2 ions were stored in the ion trap (multistage activation) (52). After the second activation step, MS2 and MS3 ions were simultaneously analyzed in the mass analyzer of the ion trap. For the identification of peptides, multistage activation spectra were matched against CBL peptide sequences using OMSSA 2.1.4 (53) and Sequest (54).
Yeast Two-hybrid Assays and Microscopy for Localization and BiFC Studies
The coding sequences of CIPKs, CBLs and CBL mutants, were cloned into the activation domain (AD) vector (pGAD.GH) and the DNA-binding domain (BD) vector (pGBT9.BS), respectively (19). CIPK23 and CIPK24 CDS with a deletion in the NAF domain (CIPK23 ΔNAF and CIPK24 ΔNAF) were generated as described previously (19). Construct combinations were transformed into the yeast PJ69-4A strain and selected on SD agar medium lacking Leu and Trp (SD-LW). 10-Fold serial dilutions in 2% glucose (A600 = 1 to 10−4) of the transformants were spotted on agar plates of SD medium lacking His (SD-LWH) and supplemented with 2.5 mm 3-amino-1,2,4-triazole and were incubated at 23 °C for 5–8 days.
For subcellular localization studies, the CDS of the CBLs were fused to either mVenus-, OFP (orange fluorescent protein)-, or mCherry-encoding sequences and introduced into pGPTVII vectors (55). Additionally, for BiFC analyses, CIPKs and CBLs were cloned into pSPYNE(R) and pSPYCE(M) vectors (42), respectively. Agrobacterium tumefaciens strains (GV3101/pMP90) carrying the fluorescent protein-fused constructs were infiltrated into Nicotiana benthamiana leaves according to Waadt and Kudla (56). Confocal microscopy was performed using an inverted microscope, Leica DMIRE2, equipped with a Leica TCS SP2 laser scanning device (Leica) as described in detail previously (41, 43).
Oocyte Experiments
The CDS of AKT1, CIPK23, CBL1 (WT/S201A/S201D), and CBL9 (WT/S201A/S201D) were cloned into the pGEMHE vector as described previously (57). For functional analyses, cRNAs of AKT1, the different variants of CBL1, CBL9, and CIPK23, were prepared using the mMessage mMachine T7 transcription kit (Ambion). Oocyte preparation and cRNA injection have been described elsewhere (58). AKT1 cRNA (10–20 ng) was injected in combination with 0.5 ng of cRNA of CBL variants and CIPK23. In two-electrode voltage clamp studies, Xenopus oocytes were perfused with KCl -containing solutions, based on MES/Tris buffers. The standard solution contained 10 mm MES/Tris (pH 5.6), 1 mm CaCl2, 1 mm MgCl2, and 30 mm KCl. The ionic strength was adjusted with LiCl to 100 mm. Voltage-dependent activation of AKT1 was recorded using single-pulse protocols. Starting from a holding potential (VH) of −20 mV, a series of voltage pulses were applied from +40 mV to −170 mV in steps of 10 mV.
Sequence Alignments and Comparisons
Full-length deduced amino acid sequences of 10 CBLs from Arabidopsis and 75 CBL orthologs from 1 protozoan species, 3 green algae species, and 11 land plant species were obtained from the GenBankTM data base and were aligned with the ClustalW program. GenBankTM accession numbers are listed in supplemental Table S2. The phylogenetic tree was constructed using the software Dendroscope 2.7 (59). To determine the consensus motif in the C termini of land plant CBL proteins, the extracted 21-residue amino acid sequences of all CBLs were analyzed using the on-line software WebLogo 3.0 (60).
RESULTS
Coupled in Vitro Transcription/Translation in Wheat Germ Extracts Yields Active CIPKs
Previous biochemical characterization studies of CIPKs employed bacterial expression systems to prepare recombinant CIPK proteins (21–24, 32, 37, 38, 44–46). Considering that expression in E. coli can result in aberrant phosphorylation and folding patterns of eukaryotic proteins or in missing protein modification that negatively affects recombinant protein activity (61), we sought to express CIPK proteins in eukaryotic cells. However, all our repeated attempts to overexpress full-length CIPK1 protein in either transiently or stably transformed tobacco BY-2 cell lines, transiently transformed Arabidopsis protoplasts, or stably transformed Arabidopsis plants failed due to toxicity problems or alternatively due to gene silencing that we observed after stable transformation (data not shown). We therefore chose to work with a cell-free coupled in vitro transcription/translation system on the basis of wheat germ extracts that combines the advantages of an eukaryotic expression machinery with the absence of any potential toxic expression effects due to nonrequirement of a host organism for protein expression. We generated recombinant CIPK1, CIPK23, and CIPK24 proteins by overnight incubation in wheat germ extracts and subsequent Strep-Tactin purification.
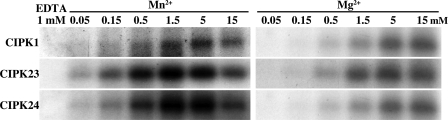
To investigate the biochemical characteristics of the in vitro-translated CIPK proteins, we first comparatively examined kinase activities of CIPK1, CIPK23, and CIPK24 in autophosphorylation assays in the presence of various concentrations of the two divalent cations, Mn2+ or Mg2+. As illustrated in Fig. 1, Mn2+ activated autophosphorylation of all analyzed CIPKs in the micro- to millimolar range. Supplementation of Mn2+ concentrations between 1.5 and 5 mm appeared to be the most effective for this reaction. In contrast, application of Mg2+ resulted in a rather weak autophosphorylation activity that was only observed at millimolar Mg2+ concentrations. We subsequently used optimal assay conditions to determine the minimal required protein amount for detecting autophosphorylation in a dilution series of various protein amounts and found that 25 ng of CIPK24 was sufficient to detect activity (supplemental Fig. S2C). The preference for Mn2+ as compared with Mg2+ was consistent with our previous study of E. coli-expressed CIPK1 (44) and studies of CIPK8 and CIPK20 (45, 46) (here designated as PKS11 and PKS18, respectively). In additional autophosphorylation analyses, we observed a similar Mn2+ dependence in autophosphorylation analyses of recombinant CIPK5, CIPK6, CIPK11, and CIPK14 proteins (Ref. 57 and data not shown). Moreover, we performed a detailed biochemical characterization of recombinant CIPK1, CIPK23, and CIPK24 to determine their Km and Vmax values in phosphorylation analyses using synthetic substrates, Syntide-2 and ALARA peptide. These experiments confirmed that the recombinant CIPK1, CIPK23, and CIPK24 proteins harbor substrate phosphorylation activity and revealed that at least toward these artificial substrates CIPK24 exhibits significantly higher Km and Vmax values than CIPK1 and CIPK23 (Table 1 and supplemental Fig. S3). Together, these results establish the cell-free transcription/translation system in wheat germ extracts as a suitable expression system for the generation of active CIPK proteins and suggest Mn2+ dependence in in vitro studies as a general characteristic of this class of kinases.
FIGURE 1.
Co-factor preference of CIPKs. Autoradiographs of in vitro (auto)phosphorylation assays with 100 ng of in vitro-translated and purified StrepII-CIPK1, -CIPK23, and -CIPK24 kinase proteins in the presence of 1 mm EDTA (left) or the indicated concentrations of MnSO4 (left panel) or MgSO4 (right panel) are shown. CIPK1, CIPK23, and CIPK24 autophosphorylation occurs more efficiently in the presence of Mn2+ when compared with Mg2+ as co-factor.
TABLE 1.
Determined kinetic constants of in vitro-translated and purified CIPK proteins
Full-length CIPK24 Phosphorylates Its Interacting Calcium Sensor CBL4
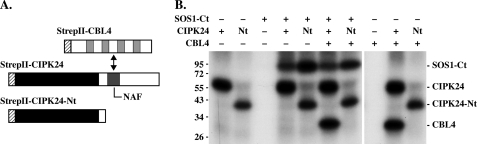
We then sought to compare the activity of a full-length CIPK24 with a C-terminally truncated CIPK24 protein version lacking amino acids Met309 to Phe446 (CIPK24-Nt, Fig. 2A). The truncated CIPK24 protein has been previously reported to exhibit similar autophosphorylation as full-length CIPK24 but enhanced kinase activity toward the ALARA peptide (32, 37). When 100 ng of both CIPK24 and CIPK24-Nt were analyzed in autophosphorylation analyses, we reproducibly observed effective autophosphorylation of both proteins that appeared to be more efficient for the full-length CIPK24 protein (Fig. 2B). We subsequently incubated both CIPK24 proteins with a C-terminal fragment of SOS1 (SOS1-Ct, amino acids Gly441–Leu1146) that represents an in vivo target of this kinase (23, 24). Both the full-length and the C-terminally truncated CIPK24 proteins phosphorylated SOS1. We next investigated the influence of the calcium sensor CBL4 on the phosphorylation of SOS1-Ct by both kinases. To allow comparability, we used protein ratios that were similar to previously published studies (100 ng of kinase and 100 ng of SOS1-Ct) (23, 24). Under these experimental conditions addition of CBL4 did not appear to enhance SOS1-Ct phosphorylation by either CIPK24 or CIPK24-Ct. However, we surprisingly observed effective phosphorylation of CBL4 by CIPK24 but not by CIPK24-Nt. Finally, we addressed a potential effect of CBL4 on the autophosphorylation activity of this kinase in the absence of SOS1-Ct by incubating CBL4 with either CIPK24 or CIPK24-Nt. The autophosphorylation capacity of both kinases was not affected by the presence of the calcium sensor (Fig. 2B). However, we again observed effective phosphorylation of CBL4 by CIPK24 but not by CIPK24-Nt. These findings indicated that CBL4 does not only constitute a regulatory moiety of CBL4-CIPK24 complexes but also represents a genuine substrate of this kinase. Moreover, this observation suggests that CIPK24 may exhibit different mechanisms of phosphorylation toward either SOS1 or CBL4 and that, at least in vitro, in contrast to the phosphorylation of SOS1, phosphorylation of CBL4 by CIPK24 strictly depends on interaction of this calcium sensor with the kinase via the NAF domain.
FIGURE 2.
Comparative kinase activity analyses of full-length and C-terminally truncated CIPK24 proteins. A, schematic representation of N-terminal StrepII-tagged CBL4, CIPK24, and CIPK24-Nt proteins. In StrepII-CIPK24-Nt, the C-terminal regulatory domain (Met309–Phe446), including the NAF domain that is necessary and sufficient for interaction with CBLs, was deleted. The StrepII tag is indicated as a dashed box, the kinase domain as a black box, and EF-hands in CBL4 and the NAF domain in CIPK24 as gray boxes. B, autoradiograph of in vitro phosphorylation assays using CIPK24 (+), CIPK24-Nt (Nt), or no kinase (−) without or in presence of SOS1-Ct and CBL4. CIPK24 and CIPK24-Nt exhibited autophosphorylation activity and also phosphorylated SOS1-Ct. However, only CIPK24 phosphorylated CBL4 indicating the importance of the C terminus with the NAF domain in CIPK24 for CBL4 phosphorylation.
Interaction-dependent Phosphorylation of CBLs by CIPKs Represents a Common Feature of CBL-CIPK Complexes
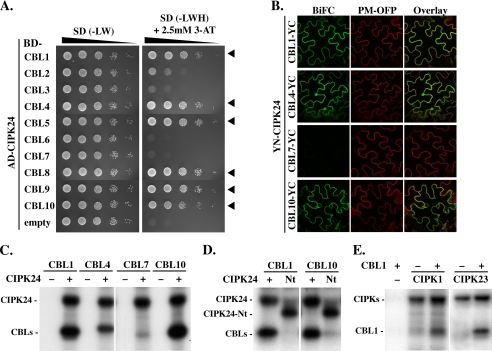
We subsequently wanted to determine whether CBL phosphorylation by CIPKs represents a common feature of CBL-CIPK complexes and especially if CBL phosphorylation is strictly dependent on the interaction with CIPKs, which results in conformational changes of both protein moieties involved (35, 36). To this end, we first investigated the interaction of CIPK24 with all 10 CBL proteins in yeast two-hybrid assays to identify the CBLs that display the ability to interact with this kinase. In these assays, we observed a distinct interaction profile of this kinase in that CIPK24 did not interact with CBL6 and CBL7 and only quite weakly interacted with the two closely related tonoplast localized proteins CBL2 and CBL3 (Fig. 3A). In contrast, we observed significant interactions with the calcium sensors CBL1, CBL4, CBL5, CBL8, CBL9, and CBL10 (Fig. 3A). BiFC analyses corroborated that CIPK24 interacted with CBL1, CBL4, and CBL10 but not with CBL7 in planta (Fig. 3B).
FIGURE 3.
CBL phosphorylation by CIPKs is a general feature of the CBL-CIPK network. A, 10-fold dilutions (A600 of 1–10−4) of the yeast PJ69-4A strain harboring an activation domain (AD)-CIPK24 and either empty DNA-binding domain (BD)-vector or different BD-CBL constructs were spotted on SD selection media without Leu and Trp (−LW; selection for positive transformants) or without Leu, Trp, and His (−LWH and 2.5 mm 3-amino-1,2,4-triazole; selection for interaction) and incubated for 8 days at 23 °C. Yeast two-hybrid assays indicate strong interaction of CIPK24 with CBL1, -4, -5, and -8–10 as indicated by arrowheads. B, BiFC analyses in transiently transformed N. benthamiana epidermal cells confirmed the interaction of YN-CIPK24 with CBL1-, CBL4-, and CBL10-YC but indicated no interaction with CBL7-YC. The plasma membrane marker (PM-OFP) (38) was co-expressed as a positive expression control. C, autoradiograph of in vitro phosphorylation assays in the absence (−) or presence (+) of CIPK24 with 50 ng of indicated CBL proteins. CIPK24 autophosphorylated and phosphorylated CBL1, CBL4, and CBL10 but not CBL7. D, only CIPK24 but not CIPK24-Nt (Nt) phosphorylated CBL1 (left) and CBL10 (right) in in vitro kinase assays. However, both kinase versions were able to autophosphorylate. E, in vitro kinase assays of CIPK1 (left) and CIPK23 (right). Both kinases phosphorylated CBL1 and exhibited autophosphorylation activity in presence (+) and in absence (−) of CBL1.
Subsequently, recombinant CBL1, CBL4, CBL7, and CBL10 proteins were generated by in vitro transcription/translation and were incubated with or without CIPK24 in phosphorylation analyses. The complete absence of any detectable radioactivity in the lanes in which CBL proteins were incubated without kinase protein in the phosphorylation reaction (− lanes in Fig. 3C) excludes nonspecific ATP binding to these calcium sensors as a potential source for radioactive signals. Remarkably, CBL1, CBL4, and CBL10 that all interacted with CIPK24 in the yeast two-hybrid and BiFC assays were phosphorylated by this kinase (Fig. 3C). Importantly, no phosphorylation of CBL7 was observed when this protein was incubated with CIPK24. Loading controls by Coomassie staining and Western blot analysis (represented in supplemental Fig. S4A) confirmed that equivalent amounts of each CBL protein were subjected to the kinase reactions. These results support the notions that our in vitro assay conditions faithfully reflect the interaction behavior of CBLs and CIPKs and that the interaction of a CBL protein with a CIPK is an essential prerequisite for phosphorylation. However, at this point of our investigations, we could not fully exclude that CBL7 does not harbor a phosphorylation site that represents a target of CIPK24.
To further address the NAF domain-dependent interaction requirements of CBL phosphorylation, we comparatively analyzed the phosphorylation of CBL1 and CBL10 by either full-length CIPK24 or the C-terminally truncated CIPK24-Nt (Fig. 3D). As observed before for CBL4, deletion of the NAF domain and the complete C terminus of CIPK24 did not affect the autophosphorylation of this kinase but abrogated the phosphorylation of both CBL1 and CBL10. Finally, we examined if additional CIPKs can also phosphorylate their interacting CBLs. To this end, we combined in in vitro phosphorylation assays CBL1 with either CIPK1 or CIPK23 because previous studies had established the physiological role of CBL1-CIPK1 and CBL1-CIPK23 complexes (27, 28, 30). These experiments clearly revealed phosphorylation of CBL1 by CIPK1 and CIPK23 that was only observed in the presence of recombinant kinase protein (Fig. 3E). Taken together, the results of these experiments establish the interaction-dependent phosphorylation of CBL proteins by their interacting CIPKs as a general mechanism of the CBL-CIPK network in Arabidopsis.
A Conserved Ser Residue in the C Terminus of CBL Proteins Represents the Target Site of CIPK Phosphorylation
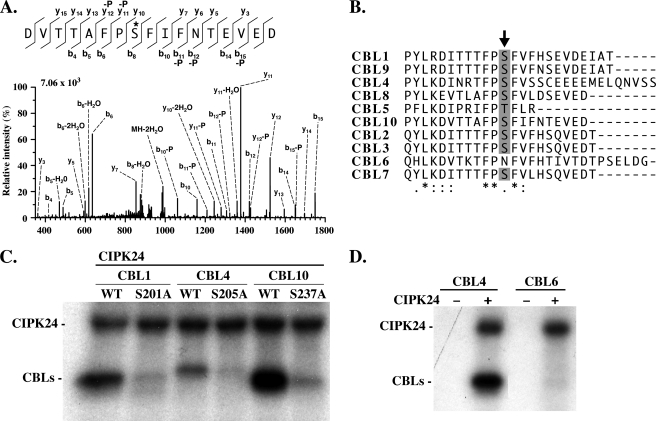
To identify the target site(s) of CIPK-mediated phosphorylation in the CBL proteins, we pursued a mass spectrometric approach. To this end, we incubated CBL1, CBL4, and CBL10 proteins with CIPK24 in the presence of 10 μm nonradioactive ATP. After SDS-PAGE, Coomassie-stained protein bands were excised from SDS gels, and after in-gel trypsin digestion, the resulting peptides were analyzed by LC-MS/MS as described under “Experimental Procedures.” Using this approach, we identified a phosphorylated peptide from the C terminus of CBL10. Peptide fragmentation revealed that Ser237 of CBL10 was phosphorylated by CIPK24 (Fig. 4A). This Ser237 in CBL10 was previously identified by scanning mutagenesis as required for phosphorylation of this calcium sensor (39). When we compared the peptide sequences of this C-terminal region of all CBL proteins from Arabidopsis, we noticed that this Ser residue is highly conserved in 8 of the 10 Arabidopsis CBLs (Fig. 4B). Only in CBL5 does Ser appear to be substituted by a phosphorylatable Thr, and in CBL6 the corresponding amino acid position is replaced by Asn. Moreover, a recent mass spectrometric analysis identified the conserved Ser in CBL2 (Ser216 in this protein) as being phosphorylated (40). To further corroborate if the conserved Ser is indeed subject to phosphorylation by CIPKs in additional CBL proteins and to address the possibility of the presence of additional phosphorylation sites, we introduced nonphosphorylatable Ala substitutions at CBL1 Ser201, CBL4 Ser205, and CBL10 Ser237. When these Ser → Ala-mutated proteins were tested side-by-side with wild-type CBL proteins in phosphorylation assays with CIPK24, we observed that the substitution prevented phosphorylation of all three CBLs (Fig. 4C). Loading controls presented in supplemental Fig. S4B confirmed that equivalent amounts of respective CBL proteins were subjected to the kinase reactions. Consequently, these experiments confirm that the Ser residues corresponding to Ser237 in CBL10 represent a conserved phosphorylation site in CBL proteins and make the existence of additional CIPK target phosphorylation sites in these calcium sensor proteins rather unlikely. Moreover, these results underscore the remarkable specificity of our phosphorylation analyses presented in Fig. 3C. Despite the presence of a phosphorylatable Ser202 in CBL7, this protein was not phosphorylated by CIPK24 due to the absence of an effective CBL-CIPK complex formation. In addition, we investigated if CBL6, which has lost the phosphorylatable residue corresponding to Ser237 in CBL10, may still be subject to phosphorylation at different residues. In phosphorylation assays that combined CBL6 with CIPK24, we did not detect any phosphorylation of CBL6 (Fig. 4D). However, this result could also reflect the inability of CBL6 to interact with CIPK24. Taken together, these data identify a single conserved residue in the CBL calcium sensor proteins of Arabidopsis as a target of CIPK phosphorylation.
FIGURE 4.
The target site of CIPK-mediated phosphorylation is conserved in Arabidopsis CBL proteins. A, LC-multistage activation spectrum showing phosphorylation of CBL10 Ser237. Fragmentation of the precursor (m/z 1006.9353, doubly charged) was performed by collision-induced dissociation (CID). The major b-type and y-type ions as well as ions corresponding to the neutral loss of phosphoric acid (-P) are indicated in the spectrum and in the corresponding amino acid sequence depicted above the spectrum. The phosphorylation site is marked by an asterisk. The +80-Da mass shift observed for b8 to b15 and y10 to y15 in addition the occurrence of corresponding neutral loss ions revealed the phosphorylation of Ser237. B, ClustalW alignment of the Arabidopsis CBL C-terminal region. The conserved phosphorylation site is indicated by an arrow. Semi-conserved, conserved, and fully conserved amino acid residues are indicated by dots, double dots, and asterisks below the alignment, respectively. C, compared with CBL wild-type (WT) proteins, mutation of the corresponding phosphorylation site in CBL1 (Ser201), CBL4 (Ser205), and CBL10 (Ser237) to Ala prevented phosphorylation by CIPK24 in in vitro kinase assays. D, CIPK24 did not phosphorylate CBL6, which lacks the conserved Ser/Thr residue in the C terminus. Autoradiograph of in vitro phosphorylation assays in the absence (−) or presence (+) of CIPK24 with 50 ng of CBL4 (left) and CBL6 proteins (right). CBL4 as the positive control was phosphorylated by CIPK24.
CIPK-dependent Phosphorylation Arose with the Evolution of Multicellular Land Plants
The CBL-CIPK signaling network represents an ancient Ca2+ signaling system that is not restricted to the plant kingdom. Excavata protozoan species like the human pathogens Trichomonas vaginalis and Naegleria gruberi harbor CBL and CIPK proteins (17). Also, evolutionary basal green algae like Chlorella sp., Ostreococcus lucimarinus, and Ostreococcus tauri encode single CBL and CIPK protein pairs in their genomes (18). With the evolution of multicellular land plants that can survive in fluctuating habitats, the complexity of this signaling system gradually increased with four CBL proteins and up to seven CIPK proteins present in the genomes of the moss Physcomytrella patens and the lycophyte Selaginella moellendorffii, respectively (18, 41). This process led to a complexity of up to 10 CBL proteins that can form an interaction network with more than 30 CIPKs in angiosperms (18).
To determine the conservation of the identified phosphorylation site in CBLs on a broader scale and to examine the evolutionary history of this mechanism, we performed a clustering analysis using the ClustalW algorithm of 85 CBL proteins, including the above mentioned species and yeast calcineurin B as outgroup, and constructed a phylogenetic tree (supplemental Fig. S5A). These investigations identified a highly conserved motif encompassing 21 amino acids in the C terminus of almost every CBL from land plant species, including the moss and the lycophyte that were analyzed (supplemental Fig. S5A). Within this motif, the central four-amino acid sequence (Phe-Pro-Ser-Phe) that contains the Ser phosphorylation site was absolutely invariant (supplemental Fig. S5B). Therefore, we designated this conserved and functionally relevant domain of CBL proteins as the “FPSF motif.” Blast searches using this domain did not identify sequences with significant amino acid sequence identity in other plant or animal proteins. Therefore, we conclude that this domain is specific for this class of calcium sensors. Only very few land plant CBLs, like SmCBL4, SbCBL8, and AtCBL5, did not appear to possess an FPSF motif. However, in these cases the proteins also lost the conserved phosphorylatable Ser residue, suggesting a secondary loss of this biochemical feature of these CBLs. The CBL proteins from the non-plant protozoan species lack the C-terminal extension that is present in CBLs from algae and plants. Remarkably, although we did not detect a conserved FPSF motif nor a phosphorylatable Ser in the CBL proteins from green algae, their CBL proteins harbor a C-terminal extension with “phospho-mimicking” Asp or Glu residues at the position corresponding to the phosphorylatable Ser in land plants (supplemental Fig. S5A). These findings strongly suggest that the phosphorylation of CBLs by their interacting CIPKs represents a biochemical mechanism that arose during the evolution of land plants. Moreover, the fact that the occurrence of CBL phosphorylation appears to be rather conserved among land plant CBL proteins is suggestive for an essential function of this biochemical modification.
CBL Phosphorylation Is Essential for Activity of CBL1-CIPK23 and CBL9-CIPK23 Complexes toward Their Target Protein AKT1
Having established that CIPK-mediated phosphorylation of CBL proteins represents a general feature of land plant CBL-type calcium sensors, we became especially interested in investigating its functional relevance. To this end, we generated in addition to the phosphorylation-preventing Ser → Ala mutants also Ser → Asp mutants of CBL1, CBL4, and CBL10 to mimic phosphorylation. To address whether the phosphorylation status of CBLs affects their subcellular localization, the mutated CBL Ser → Ala and Ser → Asp variants fused to mVenus were expressed in N. benthamiana leaves. To enable direct comparisons within the same cells, the wild-type versions of the respective CBLs fused to OFP/mCherry were co-expressed with the mutated CBL-mVenus variants, and protein accumulation and localization were investigated by confocal microscopy. These experiments did not reveal any discernable differences in protein accumulation (that was estimated by determining fluorescence intensity) or protein localization between the wild-type and the Ser → Ala and Ser → Asp mutants of CBL1, CBL4, and CBL10 (supplemental Fig. S6).
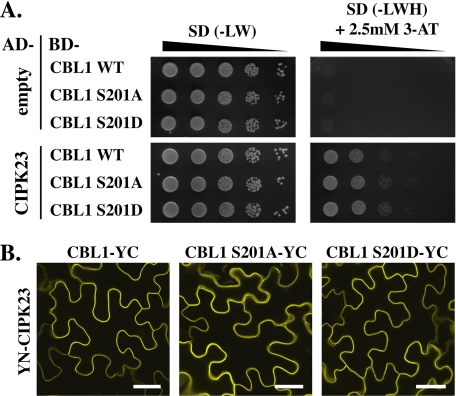
Because we intended to use the established activation of the K+ channel AKT1 by CBL1-CIPK23 complexes in oocytes as a final functional assay for the role of CBL phosphorylation, we investigated how Ser → Ala and Ser → Asp mutants of CBL1 may affect its interaction with CIPK23. However, in yeast two-hybrid analyses as well as in BiFC assays, we did not obtain any evidence that these amino acid substitutions may affect the interaction of CBL1 with CIPK23 (Fig. 5). To obtain more comprehensive insights if CBL phosphorylation may affect CBL-CIPK complex formation of other CBLs and CIPKs, we combined in further experiments the kinases CIPK1, CIPK23, and CIPK24 with wild-type versions or Ser → Ala and Ser → Asp mutants of CBL1, CBL4, and CBL10 in yeast two-hybrid assays (supplemental Fig. S7). In these experiments the interaction of CBL1 and CBL4 with CIPK24 was not affected by mutations of the Ser residue. However, in the case of CBL10-CIPK24 complex formation, the Ser → Ala mutation appeared to weaken the interaction of the calcium sensor with the kinase, although the Ser → Asp mutation had no effect on yeast growth when compared with wild-type CBL10. In contrast, the reduced growth of yeast that expressed the Ser → Asp mutant version of CBL4 together with CIPK1 was suggestive of a reduced interaction of the Ser → Asp mutant protein when compared with the growth of wild-type and Ser → Ala mutant of CBL4 with CIPK1. In summary, these data suggest that in certain cases the phosphorylation status of CBLs may affect the stability of the CBL-CIPK interaction.
FIGURE 5.
Mutations of the phosphorylation site in CBL1 did not affect its interaction with CIPK23 in yeast two-hybrid and BiFC analyses. A, dilutions (A600 of 1–10−4) of PJ69-4A yeast double transformants harboring empty activation domain (AD)-vector (top) or AD-CIPK23 (bottom) and DNA-binding domain (BD)-CBL1 WT, Ser → Ala or Ser → Asp variants were spotted on SD selection media without Leu and Trp (−LW; selection for positive transformants) or without Leu, Trp, and His (−LWH and 2.5 mm 3-amino-1,2,4-triazole (3-AT); selection for interaction). Yeast two-hybrid assays indicate no differential interaction of the CBL1 variants with CIPK23 as shown by similar growth on SD-LWH media. B, BiFC analyses in epidermal cells of infiltrated N. benthamiana leaves expressing YN-CIPK23 together with CBL1-YC or its phosphorylation site mutant variants Ser → Ala or Ser → Asp resulted in interaction and localization of all three CBL1-CIPK23 complexes at the plasma membrane. Bars, 40 μm.
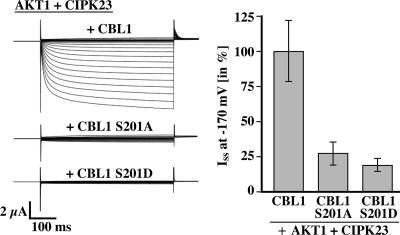
To explore the physiological function of CBL phosphorylation, we utilized an established regulatory module in which the Arabidopsis inward rectifying K+ channel AKT1 is activated by the combined function of the calcium sensors CBL1 or CBL9, which exhibits 90% amino acid sequence identity with CBL1 (62), with the kinase CIPK23 in Xenopus oocytes (28, 29, 63). Importantly, the AKT1 channel expressed alone or in combination with the kinase CIPK23 remains inactive in oocytes. However, upon co-expression of the AKT1 channel with CIPK23 and CBL1, this channel becomes activated. We used this in vivo assay system to address the importance of CBL1 phosphorylation for AKT1 activation. As expected, co-expression of wild-type CBL1 with CIPK23 and AKT1 in oocytes resulted in activation of AKT1 currents (Fig. 6). However, when we co-expressed AKT1 with CIPK23 and the CBL1 S201A mutant, we did not record significant K+ currents, suggesting that the phosphorylation of CBL1 was required for the CBL1-CIPK23 complex to regulate AKT1 activity properly. Co-expression of CBL1 S201D did not result in AKT1 activation either, suggesting that the Ser → Asp substitution is not likely sufficient to mimic the effect of phosphorylation at least in oocytes. However, considering that in all our previous analyses CIPK23 efficiently phosphorylated wild-type CBL1 and that we did not observe effects of the Ser → Asp and Ser → Ala mutants on CBL1 protein stability or localization, these data support that CIPK23-dependent phosphorylation of wild-type CBL1 is essential for the activation of AKT1 in the in vivo oocyte system. In additional control two-electrode voltage clamp studies, we replaced CIPK23 by a kinase construct (CIPK23 ΔNAF) lacking the NAF domain that is required for CBL interaction. As expected, the combination of CBL1 with CIPK23 ΔNAF and AKT1 did not yield recordable currents, further supporting the requirement of CBL1-CIPK23 interaction for efficient channel activation (supplemental Fig. S8A). In agreement with this result also in yeast two-hybrid studies deletion of the NAF domain from CIPK23 abolished interaction with CBL1 (supplemental Fig. S8B). Previous studies had shown that CBL9 together with CIPK23 can also activate AKT1 (27, 28). Because in vitro phosphorylation analysis demonstrated that CIPK23 phosphorylates CBL9 as well (supplemental Fig. S9A), we performed additional experiments in which we investigated the ability of CBL9 variants (WT/S201A/S201D) to bring about CIPK23-dependent activation of AKT1 in oocytes. Similarly as CBL1, both Ser → Ala and Ser → Asp substitution abolished activation of AKT1 by CIPK23 in oocytes (supplemental Fig. S9B).
FIGURE 6.
Mutation in CBL1 Ser201 disrupts the CBL1-CIPK23-mediated activation of the K+ channel AKT1 in Xenopus oocytes. Two-electrode voltage clamp measurements using single-pulse protocols ranging from 40 to −170 mV in 10-mV decrements starting from a holding potential of −20 mV were conducted with Xenopus oocytes expressing AKT1 with CIPK23 and CBL1, CBL1 S201A, or CBL1 S201D. The left panel shows a typical current trace. The right panel represents relative values of steady state inward K+ currents (Iss) at −170 mV (100% = the mean of Iss in AKT1 + CIPK23 + CBL1). Results represent means ± S.E. of six or more oocytes.
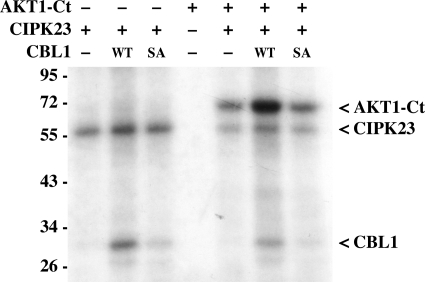
Finally, we attempted to address the impact of CIPK-dependent CBL phosphorylation on the phosphorylation activity of CBL-CIPK complexes toward their targets. To this end, we sought to use our active kinase preparations to faithfully reconstitute the CBL-dependent phosphorylation of target proteins by CIPKs in vitro, which has not been achieved before. In their native environment a single kinase can phosphorylate up to several hundred distinct target proteins, and a single kinase molecule phosphorylates a large excess of target molecules (64). However, because of the rather low activity of recombinant proteins, in many in vitro studies on CIPKs rather similar amounts of kinase protein and substrate were used (Fig. 2) (23, 24). We performed a series of AKT1-Ct phosphorylation assays in which we kept the amount of AKT1-Ct substrate constant at 800 ng and stepwise diluted the amount of CIPK23 and CBL1 added to the reaction. Consistent with our previous experiments, in all dilutions tested we observed phosphorylation of AKT1-Ct when it was incubated with CIPK23 alone. However, beginning from a 1:20 ratio of kinase to AKT1-Ct substrate, a pronounced enhancing effect of the presence of CBL1 on the phosphorylation of AKT1-Ct by CIPK23 became evident that was most pronounced at a ratio of 1:160 (supplemental Fig. S10). These results for the first time show that the presence of a CBL indeed efficiently enhances the phosphorylation of a target protein by a CIPK in vitro. We subsequently used these assay conditions to address the impact of CBL1 phosphorylation on the activity of CBL1-CIPK complexes toward AKT1-Ct (Fig. 7). Although addition of CBL1 wild-type dramatically enhanced the detectable phosphorylation of AKT1-Ct as compared with phosphorylation by CIPK23 alone, addition of CBL1 S201A resulted in a degree of phosphorylation of AKT1-Ct that was similar compared with phosphorylation by CIPK23 alone. Considering the highly controlled conditions of this in vitro analysis, which exclude variations in protein stability or amounts, this result indicates that phosphorylation of CBL1 represents an essential step in the CBL1-CIPK23 complex activation to achieve full activity toward target proteins.
FIGURE 7.
In vitro phosphorylation of StrepII-AKT1-Ct by CIPK23 was enhanced by CBL1 but not by CBL1 S201A. 800 ng of StrepII-AKT1-Ct was incubated with (+) or without (−) 15 ng of CIPK23 and 30 ng of CBL1 wild-type (WT) or S201A (SA).
DISCUSSION
Recent studies have uncovered crucial functions of CBL-CIPK complexes in an increasing number of biological processes like salt tolerance, potassium transport, nitrate sensing, and stomatal regulation (6). CBL proteins determine the cellular localization of their interacting protein kinases in vivo and are essential for the activity of the resulting CBL-CIPK complexes toward their target proteins (28, 30, 41, 43). Despite the established importance of CBL-CIPK complexes in regulating the activity of ion channels and transporters like SOS1, AKT1, AKT2, and NRT1.1 (23, 24, 28–30, 57, 63), only very few target phosphorylation sites of CIPKs have been unambiguously identified. Moreover, many facets of the biochemical regulation and mutual interdependencies of CBLs and CIPKs are largely unknown. This situation was at least in part caused by the inability of many CIPKs to phosphorylate synthetic target peptides or generic target proteins like myelin basic protein (44). Therefore, the principal aims of this study were to develop methods that allow faithful analysis of recombinant CIPK activity toward their genuine target proteins and to use these recombinant proteins to unravel functional interplays of CBLs and CIPKs.
We succeeded in generating suitable amounts of active proteins for several CIPKs by using a coupled in vitro transcription/translation system based on protein extracts prepared from germinating wheat embryos. A common biochemical feature on all CIPK-type kinases that arose from our autophosphorylation analyses is their strong and rather unusual dependence on Mn2+ instead of Mg2+. We used recombinant CIPK1, CIPK23, and CIPK24 proteins to comparatively analyze their biochemical properties like Km and Vmax toward artificial synthetic target peptides. These studies suggest remarkable differences between different CIPKs with CIPK24 exhibiting the highest activity. However, we would like to interpret these results with great caution because the activity and specificity of individual CIPKs toward their genuine targets may be quite different.
An important finding of this study is the detection of phosphorylation of CBLs by CIPKs. Using mass spectrometric analysis, we identified Ser237 in the C terminus of CBL10 as the target site of phosphorylation by CIPK24. In a previous study, Lin et al. (39) applied a combination of CBL10 domain deletion and point mutation analyses in phosphorylation experiments with CIPK24 to map the phosphorylation site in this calcium sensor protein. This approach identified the very same amino acid as required for phosphorylation by CIPK24 (39). Our mass spectrometric data extend this finding by establishing that Ser237 indeed undergoes phosphorylation. A further conclusion from our work is that CIPK-mediated phosphorylation of CBLs is a general mechanistic principle of the CBL-CIPK network in land plants. This conclusion is based on our observations that CIPK24 could efficiently phosphorylate several CBLs like CBL1, CBL4, and CBL10. Moreover, other CIPKs like CIPK1 and CIPK23 could also phosphorylate their interacting calcium sensor CBL1. Recently, a study by Du et al. (40) reported that CIPK23 can phosphorylate CBL1 and CBL9 and concluded that CIPK-mediated phosphorylation of CBL proteins may constitute a general feature of this signaling network. This conclusion is in full agreement with the data reported here. Moreover, by using liquid chromatography-quadruple mass spectrometry analysis, this most recent study by Du et al. (40) also identified the conserved Ser216 in CBL2 as being phosphorylated. Together with our mass spectrometric data of CBL10, these data further strengthen the conclusion that CBL proteins in general are subject to phosphorylation by CIPKs. The growing number of experimentally verified CBL phosphorylations in interacting CBL-CIPK complexes further supports this argumentation. In agreement with Lin et al. (39) and Du et al. (40), we report CBL phosphorylation for CBL10-CIPK24, CBL1-CIPK23, and CBL9-CIPK23 complexes, and here we first report this feature of regulation for CBL1-CIPK1, CBL9-CIPK1, CBL1-CIPK24, and CBL4-CIPK24 complexes. This collection is further extended by the CBL2-CIPK11 and CBL2-CIPK14 complexes characterized recently by Du et al. (40).
Remarkably, as we report here this phosphorylation of CBLs appears to strictly depend on interaction of the CBLs with CIPKs via the NAF domain in the C terminus of these kinases. These findings are difficult to reconcile with the conclusions of a previous study that reported that CIPK24/SOS2 specifically phosphorylated CBL10 (designated as SCaBP8) but not CBL4/SOS3 (38). Although we can currently only speculate about the reasons for these different results, it appears conceivable that the use of E. coli-purified CIPK24/SOS2 in the study by Lin et al. (39) may have prevented the detection of CBL4 phosphorylation by CIPK24. Moreover, our studies show that interaction of CIPKs with CBLs is absolutely required for CBL phosphorylation. In contrast, based on in vitro phosphorylation experiments using a CIPK24 protein without the CBL interaction mediating NAF domain, the previous study concluded that the CBL10-CIPK24 interaction is not essential for phosphorylation but dramatically enhances phosphorylation. However, we noticed that deletion of the NAF domain fully abrogates interaction of CIPK24 with CBL1 and CBL4 but not with CBL10 (supplemental Fig. S11), a finding explaining residual phosphorylation of CBL10 by a CIPK24 that does not contain a NAF domain as observed by Lin et al. (39).
The occurrence of phosphorylation of CBLs by CIPKs appears not to be restricted to the model organism Arabidopsis. In our comparative sequence analysis, we identified a conserved phosphorylation domain with a central invariant FPSF motif in almost all CBL proteins of land plant species that were investigated. The C-terminal tail containing this motif and consequently the mechanism of CBL phosphorylation appears to be absent in CBL proteins from protozoan species. CBLs from single cell green algae contain this C-terminal tail but lack a recognizable FPSF motif. However, most remarkably, in CBL proteins from green algae the amino acid position corresponding to the phosphorylatable Ser in land plants is occupied by the charged amino acids Asp or Glu that are considered to represent phospho-mimicking amino acids. A most recently postulated theory provided evidence that conserved Ser phosphorylation sites in proteins like DNA topoisomerase II, enolase, and C-Raf evolved from Asp and Glu residues to conditionally restore salt bridges that were originally constitutively formed by Glu-Arg interactions (65). In this way evolution of phosphorylation sites from Glu and Asp would provide a rationale why phosphorylation can activate proteins. The conserved phosphorylation site in CBL proteins of land plants appears to provide a novel example that supports this theory. It will be most interesting to elucidate with which additional incidence in the evolution of plant signaling systems the appearance of CBL phosphorylation coincided. In this regard, it appears noteworthy that CIPK proteins can interact with the subclade A of protein phosphatases of class 2C (PP2Cs) that are involved in ABA signaling (6, 66, 67). Structural analyses of CBL-CIPK complexes revealed that interaction of CBLs with CIPKs may prevent interaction of CIPKs with PP2Cs (6, 35). Remarkably, representatives of PP2Cs of the subclade A were recently identified to have first evolved in basal land plants and appear to be absent in algae (68, 69). Because of the coincidence in the evolutionary occurrence of CBL phosphorylation and ABA-regulated PP2Cs, it is tempting to speculate that CBL phosphorylation may somehow be functionally interconnected to CIPK-PP2C interaction, an aspect that deserves further investigations in the future.
Our comparative phosphorylation analyses of full-length CIPK24 and the C-terminally truncated CIPK24-Nt protein that lacks the CBL interaction-mediating NAF domain revealed a striking difference in their ability to phosphorylate substrate proteins. Although full-length CIPK24 proteins phosphorylated both CBLs and SOS1, CIPK24-Nt phosphorylated only SOS1. This finding may point to distinct substrate phosphorylation mechanisms of CIPKs with regard to their substrate interactions. Although phosphorylation of CBL proteins strictly depends on the interaction of CIPKs with these calcium sensors via the NAF domain in the C terminus of the kinase, the phosphorylation of other substrates can occur independent of the presence of the NAF domain. In agreement with this suggestion, we noticed that isolated N termini of several CIPKs that encompass the kinase domain can interact with several substrate proteins in yeast two-hybrid assays.4 In contrast, deletion of the NAF domain abolishes interaction of CIPKs with CBL1, CBL2, and CBL4 (supplemental Figs. S8B and S11) (19). These observations suggest that CIPKs can sufficiently interact via their catalytic domain with certain substrates, whereas CBL phosphorylation in addition requires full CBL-CIPK complex formation. It appears also conceivable that interaction of CBLs with CIPKs via the NAF domain leads to conformational changes in the calcium sensor moiety that are required to expose the C terminus of CBLs for phosphorylation.
In the course of this work, we investigated several potential consequences of CBL phosphorylation for their function. We did not obtain any evidence that phosphorylation affects the stability or localization of CBL proteins. In agreement with a previous study (39), our data suggest that CBL10 phosphorylation may enhance interaction with CIPK24. In contrast, in our analyses CBL1-CIPK24 and CBL4-CIPK24 complexes that were not investigated in previous studies (39, 40) appeared not to be affected by phosphorylation. Moreover, in our yeast two-hybrid and BiFC studies, we did not observe an impact of the phosphorylation status of CBL1 on its interaction with CIPK23. This result differs from recent findings by Du et al. (40) who observed in yeast two-hybrid studies that the nonphosphorylatable CBL1 S201A mutant exhibited a clearly reduced interaction with CIPK23 compared with the wild-type CBL1, whereas the phospho-mimicking CBL1 S201D mutant appeared to have a slightly enhanced interaction with CIPK23. Differences between the two studies in terms of experimental conditions such as yeast strains and yeast expression constructs may account for the observed differences in the modulation of CBL1-CIPK23 interaction. Nevertheless, our finding that neither CBL1 S201A nor CBL1 S201D together with CIPK23 were able to bring about activation of the AKT1 K+ channel in oocytes (Fig. 6 and see below) suggests that modulation of interaction intensity is not the most physiological relevant consequence of CBL1 phosphorylation for the functionality of CBL1-CIPK23 complexes. Moreover, our additional yeast two-hybrid analyses of CBL1, CBL4, and CBL10 with several CIPKs do not support a simple model with a similar influence of CBL phosphorylation on the stability of all CBL-CIPK complexes analyzed. These differences may reflect structural differences of specific CBLs and specific CBL-CIPK complexes. In crystallization analyses of CBL4/SOS3, the C terminus encompassing the phosphorylatable FPSF motif was disordered and could not be resolved in the structure (34). In contrast, crystallization analyses of CBL2 revealed that the C-terminal region of this protein spans and covers the crevice generated by the central EF hand domains of this calcium sensor (33). Consequently, differences in the C-terminal structure of distinct CBL proteins as observed in CBL4/SOS3 and CBL2 would result in distinct effects of their phosphorylation and could in this way lead to distinct consequences that would further broaden the functional repertoire of CBL-CIPK complexes.
Our finding that only wild-type CBL1 in combination with CIPK23 were able to activate the K+ channel AKT1 in oocytes is in line with such a crucial function of the phosphorylation of the CBL C terminus for the activity of CBL-CIPK complexes toward their target proteins. Moreover, this result establishes the functional and physiological relevance of CBL phosphorylation. Our result that a CIPK23 lacking the CBL-interacting NAF domain (CIPK23 ΔNAF) failed to activate AKT1 in oocytes (supplemental Fig. S8) further underscores the need of CBL-dependent membrane targeting of CBL1-CIPK23 complexes. This finding is in agreement with a previous report about CBL-dependent CIPK localization (43) and with the previously described inability of a hyperactive CIPK23 mutant to activate AKT1 in oocytes in the absence of a co-expressed CBL protein (28). The inability of the phospho-mimicking S201D mutant of CBL1 to confer activation of the K+ channel AKT1 concurs with the recent finding by Du et al. (40) that a CBL2 S216D mutant together with CIPK11 was unable to mediate regulation of AHA2 H+-ATPase activity. Together, these findings suggest that Ser → Asp mutations of the phosphorylatable Ser in the C terminus of CBLs may not be fully sufficient to induce the proper conformational changes that are required for full activation of CBL-CIPK complexes. Our finding that similar to CBL1 only a wild-type protein of CBL9 but not a CBL9 S201A mutant protein together with CIPK23 can activate AKT1 in oocytes further suggests the general importance of CBL phosphorylation for the function of CBL-CIPK complexes. Considering the other results of our study that preclude significant influences of CBL phosphorylation on the localization, stability, and interactions of these calcium sensor proteins, this finding argues for an influence of CBL phosphorylation on the structure of CBL-CIPK complexes that finally affects their activity.
This conclusion is strongly supported by the results from our biochemical investigations on the influence of CBL proteins on the activity of CIPK23 toward AKT1. Here, for the very first time we succeeded to faithfully reconstitute a CBL-dependent enhancement of CIPK-mediated phosphorylation of a target protein in vitro. This important result will in the future enable similar studies for other target proteins of CIPKs and will allow to further dissect the contribution of CBL proteins to these processes. Using these in vitro assays, we observed that inclusion of a nonphosphorylatable CBL1 S201A mutant abolished the CBL-dependent enhancement of AKT1 phosphorylation by CIPK23. This result indicates the absolute requirement of CBL1 phosphorylation for enhancing the activity of CBL-CIPK complexes. Consequently, together these findings indicate that not only the targeting of CBL-CIPK complexes, which depends on the CBL moiety, but also the specific interaction of defined CBLs with specific CIPKs represent important determinants of the functional activity and specificity of CBL-CIPK complexes. In this way phosphorylation of CBL calcium sensors by their interacting CIPKs adds an additional layer to the complex array of regulatory mechanisms in this signaling network that deserves further investigations by detailed structural and biochemical analyses.
Supplementary Material
Acknowledgments
We thank Dr. Oliver Batistič and Dr. Maik Böhmer for helpful discussion during the course of this work and Sebastian Rolauffs for helping with plasmid construction.
This work was supported by Deutsche Forschungsgemeinschaft grants within the frame of the Research Unit 964 (to D. B., M. H., and J. K.).

This article contains supplemental Figs. S1–S11, Tables S1 and S2, and additional references.
M. Rehers, Hashimoto, and J. Kudla, unpublished results.
- CBL
- calcineurin B-like
- CIPK
- CBL-interacting protein kinase
- BiFC
- bimolecular fluorescence complementation
- ABA
- abscisic acid.
REFERENCES
- 1. Berridge M. J. (2003) Cardiac calcium signalling. Biochem. Soc. Trans. 31, 930–933 [DOI] [PubMed] [Google Scholar]
- 2. Clapham D. E. (2007) Calcium signaling. Cell 131, 1047–1058 [DOI] [PubMed] [Google Scholar]
- 3. Dodd A. N., Kudla J., Sanders D. (2010) The language of calcium signaling. Annu. Rev. Plant Biol. 61, 593–620 [DOI] [PubMed] [Google Scholar]
- 4. Webb A. A., McAinsh M. R., Taylor J. E., Hetherington A. M. (1996) Calcium ions as intracellular messengers in higher plants. Adv. Bot. Res. 22, 45–96 [Google Scholar]
- 5. Sanders D., Pelloux J., Brownlee C., Harper J. F. (2002) Calcium at the cross-roads of signaling. Plant Cell 14, S401–S417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kudla J., Batistic O., Hashimoto K. (2010) Calcium signals. The lead currency of plant information processing. Plant Cell 22, 541–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hetherington A. M., Brownlee C. (2004) The generation of Ca2+ signals in plants. Annu. Rev. Plant Biol. 55, 401–427 [DOI] [PubMed] [Google Scholar]
- 8. Luan S., Kudla J., Rodriguez-Concepcion M., Yalovsky S., Gruissem W. (2002) Calmodulins and calcineurin B-like proteins. Calcium sensors for specific signal response coupling in plants. Plant Cell 14, S389–S400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Batistic O., Kudla J. (2004) Integration and channeling of calcium signaling through the CBL calcium sensor/CIPK protein kinase network. Planta 219, 915–924 [DOI] [PubMed] [Google Scholar]
- 10. McCormack E., Tsai Y. C., Braam J. (2005) Handling calcium signaling: Arabidopsis CaMs and CMLs. Trends Plant Sci. 10, 383–389 [DOI] [PubMed] [Google Scholar]
- 11. DeFalco T. A., Bender K. W., Snedden W. A. (2010) Breaking the code. Ca2+ sensors in plant signaling. Biochem. J. 425, 27–40 [DOI] [PubMed] [Google Scholar]
- 12. Kim M. C., Chung W. S., Yun D. J., Cho M. J. (2009) Calcium and calmodulin-mediated regulation of gene expression in plants. Mol. Plant 2, 13–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Reddy A. S., Ali G. S., Celesnik H., Day I. S. (2011) Coping with stresses. Roles of calcium- and calcium/calmodulin-regulated gene expression. Plant Cell 23, 2010–2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hashimoto K., Kudla J. (2011) Calcium decoding mechanisms in plants. Biochimie 93, 2054–2059 [DOI] [PubMed] [Google Scholar]
- 15. Harmon A. C., Gribskov M., Harper J. F. (2000) CDPKs. A kinase for every Ca2+ signal? Trends Plant Sci. 5, 154–159 [DOI] [PubMed] [Google Scholar]
- 16. Harper J. F., Breton G., Harmon A. (2004) Decoding Ca2+ signals through plant protein kinases. Annu. Rev. Plant Biol. 55, 263–288 [DOI] [PubMed] [Google Scholar]
- 17. Batistic O., Kudla J. (2009) Plant calcineurin B-like proteins and their interacting protein kinases. Biochim. Biophys. Acta 1793, 985–992 [DOI] [PubMed] [Google Scholar]
- 18. Weinl S., Kudla J. (2009) The CBL-CIPK Ca2+-decoding signaling network. Function and perspectives. New Phytol. 184, 517–528 [DOI] [PubMed] [Google Scholar]
- 19. Albrecht V., Ritz O., Linder S., Harter K., Kudla J. (2001) The NAF domain defines a novel protein-protein interaction module conserved in Ca2+-regulated kinases. EMBO J. 20, 1051–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu J., Zhu J. K. (1998) A calcium sensor homolog required for plant salt tolerance. Science 280, 1943–1945 [DOI] [PubMed] [Google Scholar]
- 21. Halfter U., Ishitani M., Zhu J. K. (2000) The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc. Natl. Acad. Sci. U.S.A. 97, 3735–3740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Qiu Q. S., Guo Y., Dietrich M. A., Schumaker K. S., Zhu J. K. (2002) Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. U.S.A. 99, 8436–8441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Quintero F. J., Ohta M., Shi H., Zhu J. K., Pardo J. M. (2002) Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc. Natl. Acad. Sci. U.S.A. 99, 9061–9066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Quintero F. J., Martinez-Atienza J., Villalta I., Jiang X., Kim W. Y., Ali Z., Fujii H., Mendoza I., Yun D. J., Zhu J. K., Pardo J. M. (2011) Activation of the plasma membrane Na/H antiporter Salt-Overly-Sensitive 1 (SOS1) by phosphorylation of an autoinhibitory C-terminal domain. Proc. Natl. Acad. Sci. U.S.A. 108, 2611–2616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Albrecht V., Weinl S., Blazevic D., D'Angelo C., Batistic O., Kolukisaoglu U., Bock R., Schulz B., Harter K., Kudla J. (2003) The calcium sensor CBL1 integrates plant responses to abiotic stresses. Plant J. 36, 457–470 [DOI] [PubMed] [Google Scholar]
- 26. Cheong Y. H., Kim K. N., Pandey G. K., Gupta R., Grant J. J., Luan S. (2003) CBL1, a calcium sensor that differentially regulates salt, drought, and cold responses in Arabidopsis. Plant Cell 15, 1833–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. D'Angelo C., Weinl S., Batistic O., Pandey G. K., Cheong Y. H., Schültke S., Albrecht V., Ehlert B., Schulz B., Harter K., Luan S., Bock R., Kudla J. (2006) Alternative complex formation of the Ca2+-regulated protein kinase CIPK1 controls abscisic acid-dependent and -independent stress responses in Arabidopsis. Plant J. 48, 857–872 [DOI] [PubMed] [Google Scholar]
- 28. Xu J., Li H. D., Chen L. Q., Wang Y., Liu L. L., He L., Wu W. H. (2006) A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 125, 1347–1360 [DOI] [PubMed] [Google Scholar]
- 29. Li L., Kim B. G., Cheong Y. H., Pandey G. K., Luan S. (2006) A Ca2+ signaling pathway regulates a K+ channel for low-K response in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 103, 12625–12630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ho C. H., Lin S. H., Hu H. C., Tsay Y. F. (2009) CHL1 functions as a nitrate sensor in plants. Cell 138, 1184–1194 [DOI] [PubMed] [Google Scholar]
- 31. Hedrich R., Kudla J. (2006) Calcium signaling networks channel plant K+ uptake. Cell 125, 1221–1223 [DOI] [PubMed] [Google Scholar]
- 32. Guo Y., Halfter U., Ishitani M., Zhu J. K. (2001) Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. Plant Cell 13, 1383–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nagae M., Nozawa A., Koizumi N., Sano H., Hashimoto H., Sato M., Shimizu T. (2003) Crystallization and preliminary x-ray characterization of a novel calcium-binding protein AtCBL2 from Arabidopsis thaliana. Acta Crystallogr. D Biol. Crystallogr. 59, 1079–1080 [DOI] [PubMed] [Google Scholar]
- 34. Sánchez-Barrena M. J., Martínez-Ripoll M., Zhu J. K., Albert A. (2005) The structure of the Arabidopsis thaliana SOS3. Molecular mechanism of sensing calcium for salt stress response. J. Mol. Biol. 345, 1253–1264 [DOI] [PubMed] [Google Scholar]
- 35. Sánchez-Barrena M. J., Fujii H., Angulo I., Martínez-Ripoll M., Zhu J. K., Albert A. (2007) The structure of the C-terminal domain of the protein kinase AtSOS2 bound to the calcium sensor AtSOS3. Mol. Cell 26, 427–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Akaboshi M., Hashimoto H., Ishida H., Saijo S., Koizumi N., Sato M., Shimizu T. (2008) The crystal structure of plant-specific calcium-binding protein AtCBL2 in complex with the regulatory domain of AtCIPK14. J. Mol. Biol. 377, 246–257 [DOI] [PubMed] [Google Scholar]
- 37. Gong D., Guo Y., Jagendorf A. T., Zhu J. K. (2002) Biochemical characterization of the Arabidopsis protein kinase SOS2 that functions in salt tolerance. Plant Physiol. 130, 256–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mahajan S., Sopory S. K., Tuteja N. (2006) Cloning and characterization of CBL-CIPK signaling components from a legume (Pisum sativum). FEBS J. 273, 907–925 [DOI] [PubMed] [Google Scholar]
- 39. Lin H., Yang Y., Quan R., Mendoza I., Wu Y., Du W., Zhao S., Schumaker K. S., Pardo J. M., Guo Y. (2009) Phosphorylation of SOS3-like calcium-binding protein8 by SOS2 protein kinase stabilizes their protein complex and regulates salt tolerance in Arabidopsis. Plant Cell 21, 1607–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Du W., Lin H., Chen S., Wu Y., Zhang J., Fuglsang A. T., Palmgren M. G., Wu W., Guo Y. (2011) Phosphorylation of SOS3-like calcium-binding proteins by their interacting SOS2-like protein kinases is a common regulatory mechanism in Arabidopsis. Plant Physiol. 156, 2235–2243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Batistic O., Sorek N., Schültke S., Yalovsky S., Kudla J. (2008) Dual fatty acyl modification determines the localization and plasma membrane targeting of CBL/CIPK Ca2+ signaling complexes in Arabidopsis. Plant Cell 20, 1346–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Waadt R., Schmidt L. K., Lohse M., Hashimoto K., Bock R., Kudla J. (2008) Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL-CIPK complexes in planta. Plant J. 56, 505–516 [DOI] [PubMed] [Google Scholar]
- 43. Batistic O., Waadt R., Steinhorst L., Held K., Kudla J. (2010) CBL-mediated targeting of CIPKs facilitates the decoding of calcium signals emanating from distinct cellular stores. Plant J. 61, 211–222 [DOI] [PubMed] [Google Scholar]
- 44. Shi J., Kim K. N., Ritz O., Albrecht V., Gupta R., Harter K., Luan S., Kudla J. (1999) Novel protein kinases associated with calcineurin B-like calcium sensors in Arabidopsis. Plant Cell 11, 2393–2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gong D., Gong Z., Guo Y., Chen X., Zhu J. K. (2002) Biochemical and functional characterization of PKS11, a novel Arabidopsis protein kinase. J. Biol. Chem. 277, 28340–28350 [DOI] [PubMed] [Google Scholar]
- 46. Gong D., Zhang C., Chen X., Gong Z., Zhu J. K. (2002) Constitutive activation and transgenic evaluation of the function of an Arabidopsis PKS protein kinase. J. Biol. Chem. 277, 42088–42096 [DOI] [PubMed] [Google Scholar]
- 47. Sambrook J., Russell D. W. (2001) Molecular Cloning: A Laboratory Manual, 3rd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 48. Hashimoto Y., Soderling T. R. (1987) Calcium·calmodulin-dependent protein kinase II and calcium·phospholipid-dependent protein kinase activities in rat tissues assayed with a synthetic peptide. Arch. Biochem. Biophys. 252, 418–425 [DOI] [PubMed] [Google Scholar]
- 49. Harmon A. C., Yoo B. C., McCaffery C. (1994) Pseudosubstrate inhibition of CDPK, a protein kinase with a calmodulin-like domain. Biochemistry 33, 7278–7287 [DOI] [PubMed] [Google Scholar]
- 50. Dale S., Wilson W. A., Edelman A. M., Hardie D. G. (1995) Similar substrate recognition motifs for mammalian AMP-activated protein kinase, higher plant HMG-CoA reductase kinase-A, yeast SNF1, and mammalian calmodulin-dependent protein kinase I. FEBS Lett. 361, 191–195 [DOI] [PubMed] [Google Scholar]
- 51. Shevchenko A., Tomas H., Havlis J., Olsen J., Mann M. (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860 [DOI] [PubMed] [Google Scholar]
- 52. Schroeder M. J., Shabanowitz J., Schwartz J. C., Hunt D. F., Coon J. J. (2004) A neutral loss activation method for improved phosphopeptide sequence analysis by quadrupole ion trap mass spectrometry. Anal. Chem. 76, 3590–3598 [DOI] [PubMed] [Google Scholar]
- 53. Geer L. Y., Markey S. P., Kowalak J. A., Wagner L., Xu M., Maynard D. M., Yang X., Shi W., Bryant S. H. (2004) Open mass spectrometry search algorithm. J. Proteome Res. 3, 958–964 [DOI] [PubMed] [Google Scholar]
- 54. Eng J. K., McCormack A. L., Yates J. R. (1994) An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989 [DOI] [PubMed] [Google Scholar]
- 55. Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., Kudla J. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40, 428–438 [DOI] [PubMed] [Google Scholar]
- 56. Waadt R., Kudla J. (2008) In planta visualization of protein interactions using bimolecular fluorescence complementation (BiFC). CSH Protoc., doi: 10.1101/pdb.prot4995 [DOI] [PubMed] [Google Scholar]
- 57. Held K., Pascaud F., Eckert C., Gajdanowicz P., Hashimoto K., Corratgé-Faillie C., Offenborn J. N., Lacombe B., Dreyer I., Thibaud J. B., Kudla J. (2011) Calcium-dependent modulation and plasma membrane targeting of the AKT2 potassium channel by the CBL4/CIPK6 calcium sensor/protein kinase complex. Cell Res. 21, 1116–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Becker D., Dreyer I., Hoth S., Reid J. D., Busch H., Lehnen M., Palme K., Hedrich R. (1996) Changes in voltage activation, Cs+ sensitivity, and ion permeability in H5 mutants of the plant K+ channel KAT1. Proc. Natl. Acad. Sci. U.S.A. 93, 8123–8128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Huson D. H., Richter D. C., Rausch C., Dezulian T., Franz M., Rupp R. (2007) Dendroscope: an interactive viewer for large phylogenetic trees. BMC Bioinformatics doi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. (2004) WebLogo. A sequence logo generator. Genome Res. 14, 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Netzer W. J., Hartl F. U. (1997) Recombination of protein domains facilitated by co-translational folding in eukaryotes. Nature 388, 343–349 [DOI] [PubMed] [Google Scholar]
- 62. Kolukisaoglu U., Weinl S., Blazevic D., Batistic O., Kudla J. (2004) Calcium sensors and their interacting protein kinases. Genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol. 134, 43–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Geiger D., Becker D., Vosloh D., Gambale F., Palme K., Rehers M., Anschuetz U., Dreyer I., Kudla J., Hedrich R. (2009) Heteromeric AtKC1·AKT1 channels in Arabidopsis roots facilitate growth under K+-limiting conditions. J. Biol. Chem. 284, 21288–21295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ubersax J. A., Ferrell J. E., Jr. (2007) Mechanisms of specificity in protein phosphorylation. Nat. Rev. Mol. Cell Biol. 8, 530–541 [DOI] [PubMed] [Google Scholar]
- 65. Pearlman S. M., Serber Z., Ferrell J. E., Jr. (2011) A mechanism for the evolution of phosphorylation sites. Cell 147, 934–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ohta M., Guo Y., Halfter U., Zhu J. K. (2003) A novel domain in the protein kinase SOS2 mediates interaction with the protein phosphatase 2C ABI2. Proc. Natl. Acad. Sci. U.S.A. 100, 11771–11776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Cutler S. R., Rodriguez P. L., Finkelstein R. R., Abrams S. R. (2010) Abscisic acid. Emergence of a core signaling network. Annu. Rev. Plant Biol. 61, 651–679 [DOI] [PubMed] [Google Scholar]
- 68. Hauser F., Waadt R., Schroeder J. I. (2011) Evolution of abscisic acid synthesis and signaling mechanisms. Curr. Biol. 21, R346–R355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Tougane K., Komatsu K., Bhyan S. B., Sakata Y., Ishizaki K., Yamato K. T., Kohchi T., Takezawa D. (2010) Evolutionarily conserved regulatory mechanisms of abscisic acid signaling in land plants. Characterization of abscisic acid insensitive1-like type 2C protein phosphatase in the liverwort Marchantia polymorpha. Plant Physiol. 152, 1529–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.