Background: p97/VCP disease-linked mutations increase ATPase activity and destabilize the N-D1 domain interaction.
Results: Increased N-domain flexibility in p97/VCP increases ATPase activity, whereas locking down the N-domain decreases it.
Conclusion: The p97/VCP N-domain position relative to the D1 ring is linked to ATP hydrolysis ability.
Significance: p97/VCP N-domain conformational changes cause transitions between an active and inactive state.
Keywords: ATPases, Enzyme Mechanisms, Protein Conformation, Protein Domains, Protein Turnover, p97/VCP
Abstract
p97/valosin-containing protein (VCP) is a type II ATPase associated with various cellular activities that forms a homohexamer with each protomer containing an N-terminal domain (N-domain); two ATPase domains, D1 and D2; and a disordered C-terminal region. Little is known about the role of the N-domain or the C-terminal region in the p97 ATPase cycle. In the p97-associated human disease inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia, the majority of missense mutations are located at the N-domain D1 interface. Structure-based predictions suggest that such mutations affect the interaction of the N-domain with D1. Here we have tested ten major inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia-linked mutants for ATPase activity and found that all have increased activity over the wild type, with one mutant, p97A232E, having three times higher activity. Further mutagenesis of p97A232E shows that the increase in ATPase activity is mediated through D2 and requires both the N-domain and a flexible ND1 linker. A disulfide mutation that locks the N-domain to D1 in a coplanar position reversibly abrogates ATPase activity. A cryo-EM reconstruction of p97A232E suggests that the N-domains are flexible. Removal of the C-terminal region also reduces ATPase activity. Taken together, our data suggest that the conformation of the N-domain in relation to the D1-D2 hexamer is directly linked to ATP hydrolysis and that the C-terminal region is required for hexamer stability. This leads us to propose a model where the N-domain adopts either of two conformations: a flexible conformation compatible with ATP hydrolysis or a coplanar conformation that is inactive.
Introduction
Mammalian p97, a protein that is highly abundant in cells, interacts with a diverse group of adaptor proteins to perform a variety of essential cellular processes, such as endoplasmic reticulum-associated degradation (1–3), transcription factor processing (4), nuclear envelope reconstruction (5), membrane fusion (6), postmitotic Golgi reassembly (7), spindle disassembly (8), and cell cycle control (9). These activities are at least in part associated with the ubiquitin-proteasome system, in which p97 binds poly-ubiquitylated proteins and several other ubiquitin-interacting proteins to participate in the delivery of ubiquitylated substrates to the 26 S proteasome for degradation. In the endoplasmic reticulum-associated degradation pathway, which is one of the most extensively studied among p97 cellular functions, p97 plays a key role in cooperating with adaptor proteins to translocate and chaperone misfolded proteins from the endoplasmic reticulum membrane to the proteasome (10, 11) in an ATP-dependent fashion, although the exact role of the ATPase activity of p97 is not entirely clear (12, 13).
p97 is a type II ATPase associated with various cellular activities) ATPase (AAA)3 characterized by the presence of two highly conserved ATPase domains, each featuring Walker A and B motifs (14). Crystal structures of p97 show a homo-hexamer with two ring layers that encompass the D1 (amino acids 209–463 in murine p97) and D2 domains (amino acids 482–764) (15, 16). Although both D1 and D2 domains comprise the necessary motifs for ATP binding and hydrolysis, the D1 domain has less ATPase activity than D2. Its primary responsibility appears to be p97 hexamerization (17, 18). In contrast, the D2 domain confers most of the ATPase activity and is considered to be conformationally dynamic through the ATPase cycle (19, 20). p97 contains a third domain, the N-terminal domain (or N-domain, amino acids 1–192), which is connected to D1 by a flexible linker and is positioned coplanar with the D1 ring in crystal structures. The C-terminal region of p97 is not observed in the crystal structures. It is known to bind a subset of cofactors and can be phosphorylated, but its structure and significance are unknown.
The main function of the N-domain is to mediate the binding of cofactors and ubiquitylated protein substrates. Although only two conformations have been observed in p97 crystals (16, 21–26), in various solution studies, including electron microscopy and small-angle x-ray scattering, the N-domain is shown to adopt more varied conformations. Positions above, coplanar with, and below the D1 ring have all been proposed. A number of models have been proposed that link the ATP hydrolysis cycle with N-domain position, but it is unknown whether the position of the N-domain drives ATPase activity of p97 or whether it moves as a result of the ATPase cycle.
p97 is directly associated with human disease, as inherited missense mutations within the p97 gene give rise to the complex disease syndrome termed inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia (IBMPFD) (27). IBMPFD is a progressive autosomal dominant disorder with a myopathy that includes muscle fiber inclusions and rimmed vacuoles (28, 29). To date, 14 mutations at ten loci have been identified in p97 in IBMPFD patients (30–33). The majority of mutations are located on or near the interface between the N and D1 domains of p97 (34, 35). Although the pathogenesis of IBMPFD and the effect of mutations on the structure and function of p97 remain largely unknown, several of the IBMPFD-linked p97 mutations were reported to have higher ATPase activities and unusual N-domain conformations compared with wild-type proteins (36, 37) with crystal structures of several mutants showing small localized changes in the N-domain conformation (21).
To further elucidate the role of the N-domain in the p97 ATPase cycle, we have carried out a methodical analysis of the relationship between the N-domain and p97 ATPase activity. Firstly, we have analyzed ten major IBMPFD-associated mutants, with particular focus on the A232E mutation. We show that p97A232E has a significantly increased ATPase activity compared with the wild type and that this increase is due to the effect of the mutation on the N-D1 interaction. Further structure-based mutations have allowed us to probe the interaction between the N and D1 domains and investigate its effect on ATPase activity. These data lead us to propose that ATP hydrolysis by p97 critically depends on the presence of a mobile N-domain.
EXPERIMENTAL PROCEDURES
Cloning and Mutagenesis
p97 N- and C-terminal truncations were on the basis of the domain boundaries and ordered regions observed in p97 crystal structures (e.g. PDB code 1R7R). N-terminally truncated (residues 209–806) or C-terminally truncated (residues 9–764) murine p97 was amplified by PCR using gene-specific primers containing unique NdeI and NotI restriction endonuclease sites. The PCR products were then cloned using these sites into pET22b (Novagen) to generate constructs in frame with a C-terminal hexa-histidine tag. All mutations were introduced by PCR site-directed mutagenesis.
Expression and Purification
Protein expression in Escherichia coli Rosetta (DE3) cells was induced by the addition of isopropyl-β-d-thiogalactopyranoside at a final concentration of 1 mm at 22 °C. After 16 h, the cells were harvested, and the pellets were resuspended in buffer A (500 mm KCl, 2 mm β-mercaptoethanol, 25 mm HEPES (pH 8.0)) supplemented with a mixture of protease inhibitors (Sigma). After sonication and centrifugation, the supernatant was loaded onto a 5-ml HisTrap column (GE Healthcare) pre-equilibrated in buffer A. Protein was eluted with a linear imidazole gradient in buffer A. The p97-containing fractions were pooled and concentrated and further purified by gel filtration chromatography (Superose6, GE Healthcare) pre-equilibrated in buffer B (250 mm KCl, 2 mm β-mercaptoethanol, 25 mm HEPES (pH 7.5)). For cross-linking experiments, the wild type and the R155C/N387C protein were purified by the same method using the β-mercaptoethanol free buffer.
Electron Microscopy
For the negative stain grid preparation, wild-type p97 and mutant p97R155C and p97A232E samples (2 μl of 0.05 μg/ml) were applied to glow-discharged copper grids with a continuous carbon film, followed by 2% uranyl acetate solution and then blotted and air dried. For the cryo-EM sample preparation, purified p97 and p97 mutants, at a concentration of ∼0.5 mg/ml, were used to prepare cryo-grids. 5 μl of sample was applied to a glow-discharged, holey carbon film on copper grids. The grids were blotted and flash frozen in liquid ethane using a Vitrobot (FEI). Data were collected under low-dose conditions on a Phillips CM200 electron microscope at the Imperial College Electron Microscopy Centre. The microscope was operating at 200 kV, using ×50,000 magnification and between 2–4 μm of nominal defocus. Images were collected using a Tietz camera.
Images were processed using IMAGIC-5 (38) except were specified. Micrographs were coarsened by a factor of 2–3.52 Å/pixel, and particles were picked interactively using BOXER in EMAN. The particles were then contrast transfer function-corrected and bandpass-filtered between 15–150 Å. Class averages were prepared from aligned particles using multivariate statistical analysis. For p97A232E, two initial three-dimensional models were created from side views and assuming C6 symmetry. These initial models were refined using successive rounds of competitive multireference alignment (brute force alignment written by Timothy Grant) and projection matching to separate out the heterogeneity in the data and to ascertain the major conformation present. The resultant model was calculated from 1029 particles and filtered between 20–150 Å. The electron density map has been deposited in the EM data bank, accession code EMD-2038.
ATPase Assay
ATPase assays were performed at 37 °C using an NADH oxidation coupled system (39). To start the reaction, ATP (final concentration, 7 mm) was added to the preheated reaction mixture that included 1.0 μm protein, 0.35 mm NADH, 3 mm phosphoenolpyruvate, 7 units of pyruvate kinase, 23 units of lactate dehydrogenase, 100 mm Tris-HCl (pH 8.0), and 20 mm MgCl2 (45). NADH oxidation was monitored by change of absorbance at 340 nm. A340 measurements were taken at 1-min intervals for 40 min. The ATPase activity is defined by the decrease of A340/min.
Limited Proteolysis
Purified wild-type and mutant p97 proteins were diluted to 0.2 mg/ml and divided into two batches, and 4 mm of ATPγS was added to one batch. Trypsin was added to proteins to achieve a 1:100 trypsin-to-protein ratio. The mixtures were incubated at 37 °C. 10 μl of samples were extracted in 10-min intervals, mixed with 2 μl of 6× SDS sample buffer, and boiled before loading onto a 12% (v/v) SDS-PAGE gel.
RESULTS
IBMPFD Mutants Form Hexameric Ring Structures Similar to Wild-type p97
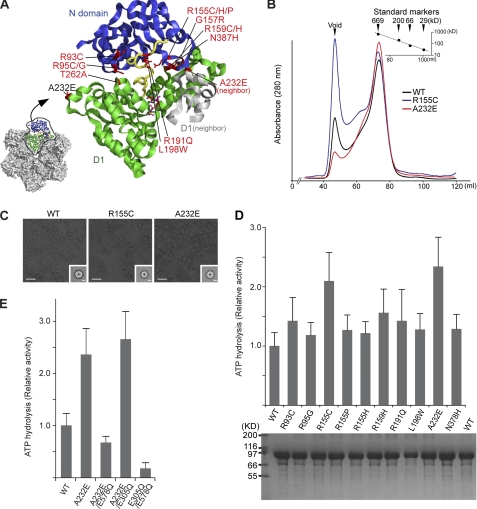
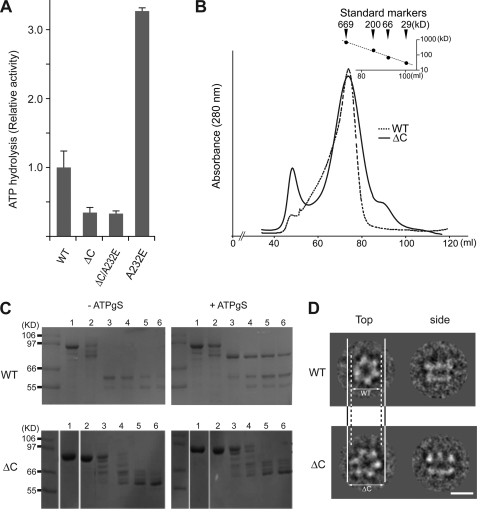
We first probed the effects of the ten major IBMPFD-linked mutations on the p97 hexamer (Fig. 1A). The IBMPFD-linked mutants (R93C, R95G, R155C, R155H, R155P, R159H, R191Q, L198W, A232E, and N387H) were expressed and purified, and the homogeneity and oligomeric states were assessed. A typical elution profile of p97 wild-type and IBMPFD mutants from a Superose6 gel filtration column is shown in Fig. 1B. All the purified mutants showed essentially the same elution profile and final purity as wild-type p97 (also see Fig. 1D, lower panel). The asymmetric shape of the peak suggests that multiple oligomeric species are present. We imaged samples from different fractions using negative stain electron microscopy. The void volume peak contains irreversibly aggregated material, and the shoulder fraction contains predominantly dodecameric p97 (dimer of hexamers) and some aggregates (supplemental Fig. S1), but the main component of the peak is hexameric ring-shaped p97 (Fig. 1C). The dodecamer and void volume fractions were found to display negligible ATPase activity showing that hexameric p97 is the only active form (supplemental Fig. S2).
FIGURE 1.
IBMPFD-linked mutants in p97. A, graphic representation of the p97 crystal structure showing the distribution of the IBMPFD-linked mutations. For clarity, only the N-domain (blue), the D1 domain (green), and the connecting linker (yellow) are shown. The neighboring subunits in the hexamer are shown in gray. B, typical purification profile from size-exclusion chromatography of wild-type p97 (black), p97R155C (blue), and p97A232E (red). The relative molecular masses (Mr) of the peaks were estimated according to protein standard markers. C, negative-stain electron micrographs and class averages for wild-type p97, p97R155C, and p97A232E with representative top-view class averages (inset). Class averages are the sum of 50 top-view images aligned and classified together by multivariate statistical analysis. Scale bar = 500 Å on the micrographs and 50 Å on the class averages. D, ATPase activities of wild-type p97 and ten IBMPFD mutants. The histogram shows the rates of ATP hydrolysis averaged from at least three independent measurements. Error bars indicate the mean ± S.D. The activities were normalized to that of wild-type. An SDS-PAGE gel of the final purified p97 proteins from size-exclusion chromatography is shown in the lower panel. 10 μl of each protein at 3 μm was loaded onto a 12% polyacrylamide gel followed by Coomassie Brilliant Blue staining. E, ATPase activities of p97A232E with D1 (E305Q) or D2 (E578Q) Walker B mutations. Note that only the Walker B mutation in D2 reduces the activity of p97A232E.
IBMPFD-linked p97 Mutants Exhibit Increased ATPase Activity in D2
We next assessed the effect of IBMPFD mutations on the ATPase activity of p97. All IBMPFD mutants showed ATPase activities similar to or higher than wild-type p97 (Fig. 1D). The A232E and R155C mutants showed the highest increase in ATPase activities, more than double the wild-type activity (Fig. 1D). A Walker B double mutant (E305Q/E578Q), deficient in ATPase activity, was used throughout as a negative control. The most active IBMPFD mutant is p97A232E, in agreement with a previous study (36, 40) and, therefore, was used for the subsequent work. To determine whether wild-type p97 and p97A232E have similar secondary structures and particle sizes, circular dichroism and dynamic light scattering were performed on purified samples. The results show that both proteins have near identical secondary structures and particle size distributions (supplemental Fig. S3), suggesting that any increased ATPase activity for p97A232E is not due to these factors. We also measured steady-state kinetic parameters for both wild-type p97 and p97A232E (54) (supplemental Table S1). Our kinetic measurements for wild-type p97 are consistent with values published previously (19, 23, 55, 56). p97A232E exhibits an increase in kcat, which is consistent with the mutant having increased ATPase activity. To establish whether this increased activity p97A232E is specifically due to increases in D1 or D2, we created two double mutants, combining A232E with Walker B hydrolysis mutants (E305Q in D1 or E578Q in D2). The activity of the p97A232E/E305Q double mutant is similar to that of p97A232E (Fig. 1E), showing that knocking out potential ATP hydrolysis activity in D1 has no effect. In contrast, the p97A232E/E578Q double mutant has a significantly reduced ATPase activity (4× reduction) compared with p97A232E (Fig. 1E). These data indicate that the increase in ATPase activity observed under steady-state conditions in p97A232E is due to increased ATP hydrolysis in D2.
Cryo-EM of p97A232E Indicates That the N-domains Are Flexible but Positioned above the D1 Ring
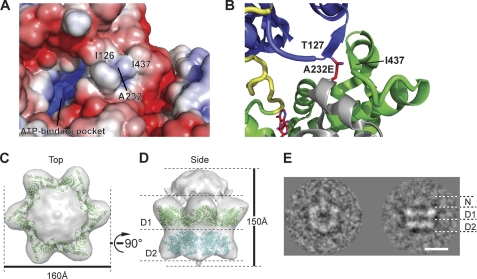
All IBMPFD mutations are clustered around the N-D1 interface of p97 (Fig. 1A). Therefore, we next investigated the implications of the A232E mutation on the position of the N-domain. A232 is located on the edge of the D1 domain, adjacent to the N-D1 domain interface of a neighboring subunit in the hexamer and surrounded by a hydrophobic environment including Ile-437 and Ile-126 (Fig. 2A). Inserting the longer glutamic acid side chain in place of alanine would disrupt the formation of a small β-sheet comprising Thr-127 in the N-domain and Ile-437 to Ala-439 in the D1 domain of the neighboring subunit (Fig. 2B). Thus, a potential structural consequence of A232E is to perturb the D1 domain and to disrupt the interaction between the N and D1 domains, which might result in an increased conformational flexibility of the N-domains. To explore this, cryo-EM single-particle analysis was carried out to create a three-dimensional reconstruction of the p97A232E hexamer. The model shows two stacked rings corresponding to the D1 and D2 domains and a cap of density above the double rings (Fig. 2, C and D). The upper ring has a large pore, indicating that the D1 ring is open in contrast to the closed ring of existing crystal structures. The D2 ring is also slightly more open in the p97A232E model than in the wild type. Model fitting of the p97 crystal structure into the EM envelope indicated a lack of corresponding density for coplanar N-domains (Fig. 2D), suggesting that the N-domains are in a different conformation to that observed in the crystal structures. It is plausible that the weak density on top of the D1 ring corresponds to the N-domains, but there is not enough density to account for their entire molecular weight. Conformational flexibility could account for this lack of observed density. Consistent with this, a class average from the p97A232E model shows similar length D1 and D2 layers with weak extra density above D1, in the same position as in the 3D reconstruction, corresponding to the N-domains (Fig. 2E). Overall, the structure is very similar to our previous reconstruction of wild-type p97 in the presence of AMPPNP (44), but the N-domains of the p97A232E model are even further removed from the coplanar position seen in the crystal structures. To test whether the p97A232E N-D1 linker A232E is more susceptible to proteolysis, we carried out limited proteolysis on wild-type p97 and p97A232E using trypsin (supplemental Fig. S4). Our results show that the N-D1 fragment from p97A232E is less stable A232E than wild-type, which is consistent with a more accessible N-D1 linker for p97A232E, although a destabilized D1 domain caused by possible rearrangements because of the A232E mutation cannot be ruled out.
FIGURE 2.
Modeling and cryo-EM structural analysis of the p97A232E IBMPFD-linked mutant. A, electrostatic surface potential of p97 around A232. Hydrophobic areas are shown in white, with positive potential in red and negative potential in blue. B, a close-up view of A232 modeled with a glutamic acid side chain. Shown are top (C) and side (D) views of the cryo-EM reconstruction of p97A232E at ∼23 Å. Overall dimensions are indicated. The density threshold was set to account for the molecular weight of p97A232E. A model of D1-D2 derived from the p97 crystal structure has been fitted into the EM envelope. The reconstruction model shows two stacked rings and a cap of density at the top. The small bottom plug is likely to be an accumulation of noise around the symmetry axis, although it may potentially be the flexible C-terminal regions. The fitted D1 and D2 domains are colored in green and cyan, respectively. E, cryo-EM class average of the top and side views of p97A232E. Scale bar = 50 Å.
Increased ATPase Activity of A232E Requires Flexible N-domains
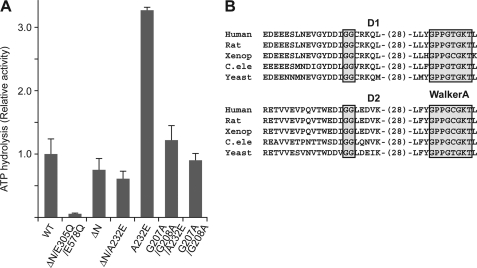
We next evaluated whether the increase in ATPase activity seen for p97A232E requires the presence and/or flexible nature of the N-domains. The ATPase activity of N-domain truncations of wild-type p97 (p97ΔN) and p97A232E were tested. p97ΔN shows similar albeit somewhat reduced ATPase activity compared with wild-type (Ref. 25, Fig. 3A). Surprisingly, unlike the increased activity of A232E in full-length p97, the p97ΔNA232E mutant shows similar activity to p97ΔN (Fig. 3A). These results suggest that N-domain plays important roles in mediating the increased ATPase activity of the A232E mutant. The significance of the increased flexible nature of p97A232E was assessed next. The N-domain is connected to D1 via a linker region containing a Gly-Gly sequence that is highly conserved among p97 homologs, is similar to conserved features seen in the D1-D2 linker, (Fig. 3B), and could be a pivoting point for potential N-domain movement. If the increase in ATPase activity of the A232E mutation required the associated increased N-domain flexibility, mutating the Gly-Gly residues to reduce the flexibility of the N-D1 linker might negate the effect of A232E on ATPase activity. Indeed, when we mutated Gly-Gly to Ala-Ala (G207A/G208A) in p97A232E, the ATPase activity was significantly reduced compared with p97A232E (Fig. 3A), whereas the linker mutation has limited effect in the context of wild-type p97. These data suggest that the elevated ATPase activity of p97A232E depends on the presence of the N-domains as well as on their increased mobility imparted by the mutation, and that wild-type p97, with its moderate ATPase activity, is less sensitive to a partially inflexible N-D1 linker.
FIGURE 3.
ATPase activity of p97A232E without a flexible N-domain. A, ATPase activities of p97A232E combined with an N-domain truncation or an N-D1 linker mutation. Measurements and presentation of results were done as for Fig. 1D. The D1 and D2 Walker B double-mutated p97ΔN was used as a negative control. B, alignment of the conserved Gly-Gly sequences in the N-D1 and D1-D2 linker regions among p97 homologs. The Gly-Gly sequences and Walker A motifs are highlighted.
Cross-linking between N and D1 Domains Inactivates p97 ATPase Activity
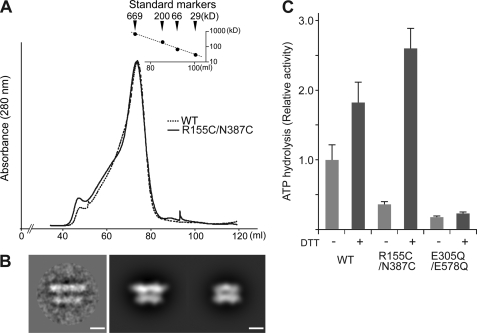
The results of the p97A232E N-D1 linker mutation suggest that the increased N-domain flexibility seen in p97A232E prevents proper regulation of ATP activity, suggesting that N-domain position and flexibility may play a role in modulating wild-type p97 ATP hydrolysis. To test this hypothesis, we created a mutant to reversibly lock the N-domain in a coplanar orientation, as observed in the p97 crystal structures. We did this by mutating two IBMPFD-related residues, Arg-155 and Asn-387, to cysteines. In the p97 crystal structures, these residues are close enough to form a disulfide bond (Fig. 1A). In non-reducing conditions, disulfide bonds would be expected to lock the N-domains in a coplanar position in the double mutant, whereas in reducing conditions the tether would be removed, allowing the N-domains to move freely. Recombinant p97R155C/N387C was purified in non-reducing conditions. The elution profile after size exclusion chromatography was identical to that of wild-type p97 (Fig. 4A), consistent with hexamer formation (B). Limited proteolysis and mass spectrometry confirmed a covalently linked peptide consistent with a disulfide bond between residues 155–387 (masses were determined by MS of 1545.6 Da and 1577.6 Da, corresponding to the fragment GDIFLVCGGMR/CMK). p97R155C/N387C was then studied by cryo-EM 2D analysis. All the class averages of p97R155C/N387C in non-reducing conditions show a D1 layer much wider than the D2 layer, indicating that the N-domains are locked in the D1 plane similar to what is observed in the crystal structures (Fig. 4B) and previous cryo-EM class averages. In ATPase assays in the absence of reducing agent, the disulfide mutant has significantly reduced activity compared with wild-type p97 (Fig. 4C). This effect could be reverted by the addition of the reducing agent DTT (Figs. 1D and 4C). This effect was corroborated by the kcat and Kmvalues for the disulfide mutant with and without DTT (supplemental Table S1). It is notable that the addition of DTT to wild-type p97 also causes a slight increase in ATPase activity. This has been observed before (41), but the reasons are unclear at present. Our data clearly show that locking the N-domain coplanar to the D1 ring correlates with an inhibition of p97 ATP hydrolysis.
FIGURE 4.
ATPase activity of the disulfide-linked p97R159C/N387C mutant. A, size-exclusion chromatographic profiles of wild-type p97 (dotted line) and the p97R159C/N387C cross-linked mutant (solid line). B, cryo-EM class average showing a side view of p97R159C/N387C in the absence of DTT (left) and back projections of the p97 crystal structure filtered to 25 Å resolution, with (center) and without (right) the N-domain. Scale bars = 50 Å. C, ATPase activity of p97R159C/N387C in the presence or absence of DTT. Measurements and representation of the histogram are the same as in Fig. 1D. Note that for p97E305Q/E578Q (D1 and D2 Walker B double mutant) there is no difference in activity with and without DTT.
The C-terminal Region of p97 Is Also Important for ATPase Activity
Even though the C-terminal region is disordered in the p97 crystal structure, biochemical data suggest important functional roles for this region (42, 43). We therefore tested whether the C-terminal region is important for p97 ATPase activity. We generated and purified C-terminally truncated wild-type p97 and A232E (p97ΔC and p97ΔCA232E, respectively). Both p97ΔC and p97ΔCA232E show elution profiles on size exclusion chromatography similar that to that of wild-type p97 (Fig. 5, B and C, upper panels and lower left panel). Both p97ΔC alone and in combination with the A232E mutation showed similar and significantly reduced APTase activity, approximately half that of wild-type p97 (Fig. 5A). Limited proteolysis reveals further similarities except in the presence of ATPγS, where p97ΔC shows decreased stability compared with wild-type p97 (Fig. 5C, upper right and lower right panels). This suggests that the D2 conformational change demonstrated by Wang et al. (20) to a tighter form induced by ATP-binding did not occur in p97ΔC. 2D cryo-EM analysis of p97ΔC revealed that there is a larger open N-D1 ring structure (162 Å in diameter) compared with wild-type p97 (149 Å), as reported previously (44) (Fig. 5D). These data indicate that the C-terminal region of p97 modulates the conformational changes that take place in D2 during the ATPase cycle.
FIGURE 5.
ATPase activity of a C-terminal truncated p97. A, ATPase activities of p97ΔC and p97ΔCA232E. Measurements and representation of the histogram are the same as in Fig. 1D. B, typical purification profiles from size-exclusion chromatography of the wild type (dotted line) and ΔC (solid line) as shown in Fig. 1B. C, limited proteolysis for wild-type p97 and p97ΔC in the presence or absence of ATPγS. Reactions were analyzed by SDS-PAGE. Shown are lane 1, no trypsin; lanes 2-6, incubation with trypsin for 0, 10, 20, 30, and 40 min, respectively. D, class averages of wild-type p97 and p97ΔC cryo-EM from the top and side views with respect to the 6-fold axis. Scale bar = 100 Å. Note the wider opened D1 ring of p97ΔC compared with that of the wild type.
DISCUSSION
IBMPFD Mutants Show Elevated ATPase Activities
In this study, we evaluated the role of N-domains in the ATPase cycle of p97 by exploring p97 mutants linked to a hereditary degenerative disorder termed IBMPFD (27). We analyzed the ATPase activities of ten major IBMPFD mutants and found that some IBMPFD mutations cause increased ATPase activities under steady-state conditions compared with wild-type p97. We then correlated increased ATPase activity with the presence of the N-domains, which was a requirement for the ATP hydrolysis-increasing effect of the A232E mutation.
Under our conditions, all the IBMPFD-linked p97 mutants have increased ATPase activities. Mutations in D1 and D2 that block ATP hydrolysis show that the elevated level of ATP turnover of p97A232E is mediated by D2 and not D1. This is consistent with earlier limited-proteolysis experiments that correlated higher ATPase activity with a change in the conformation of D2 (36). Our study is the most comprehensive analysis on the ATPase activity of IBMPFD mutants and is in agreement with other studies. R95G, R155C, R155H, R155P, R191Q, and A232E have all been shown to have increased ATPase activities over wild-type p97 (34, 36, 40).
The majority of IBMPFD missense mutations cluster at the interface between the N and D1 domains (Fig. 1A). A number of previous studies suggest that the structural phenotype of IBMPFD is an altered N-domain conformation that is nucleotide-dependent (34, 36, 37). The general consensus is that IBMPFD mutations alter N-domain conformations, possibly promoting imbalances in cofactor binding (37). The available crystal structures of IBMPFD-linked mutants R155H and R95G (in the context of N-D1) bound to ATP directly show a relocation of the N domain relative to and above the D1 hexamer (21). However, it is possible that more drastic structural rearrangements occur as a consequence of the other IBMPFD mutations. The position and chemical environment of Ala-232 as observed in p97 crystal structures suggest that the A232E mutation has a destabilizing effect on the interaction between the N and D1 domains through steric hindrance and the introduction of a negative charge in a largely hydrophobic area (Fig. 2B). This prediction is corroborated by both our limited proteolysis observation and by our cryo-EM reconstruction of p97A232E that shows the N-domain on top of the D1 ring. The weak nature of the density also suggests that the N-domains are conformationally flexible, consistent with the N-D1 linker being more susceptible to limited proteolysis. Together, our data support the hypothesis that an increase in ATPase activity for p97A232E is due to the increased flexibility of the N-domain imparted by changes in the N-D1 interface caused by the mutation. Interestingly, combining the A232E mutation with a double mutation in the N-D1 linker that is predicted to decrease its flexibility reverses the increased activity observed for p97A232E alone. The removal of the N-domain in the context of the IBMPFD A232E mutant also significantly reduces ATPase activity, suggesting that the phenotype of p97A232E requires a disrupted N-domain and/or linker. We cannot exclude the possibility that the linker region itself plays a role in modulating the ATPase activity of p97A232E, as the same linker mutant had little effect on wild-type p97, but our structural data below strongly argues that the N-domain is more important.
The A232E mutant used in this study shows the highest increase in ATPase activity for IBMPFD mutants, which interestingly correlates with the most severe disease phenotype (28). All the IBMPFD-linked mutants tested show increased activity to some extent. However, we do not know if our biochemical findings for p97A232E are the same for all the IBMPFD mutants, although it is notable that all the mutants are clustered around the N-domain and D1. It is still unclear as to how these altered biochemical properties of IBMPFD mutants affect disease pathogenesis.
N-domain Conformation and p97 ATPase Activity
Several lines of evidence support a role for N-domain conformation and flexibility in p97 ATPase activity. Firstly, the binding of p47 to p97 via the N-domain causes an inhibition of p97 ATP turnover (23). Secondly, truncation of the N-domain increases the ATPase activity in VCP-like ATPase from Thermoplasma acidophilum (VAT), the archaeal p97 homologue (46). Thirdly, truncation of the N-domain in human p97, combined with a mutation in the D1 pore loop, increases p97 ATPase activity (47). Our data suggest that the presence of the N-domain is not required for ATPase activity but modulates it, either by constraining the flexibility of the molecule or by changes in its conformation with respect to the D1 ring being transmitted through the linker regions to the D2 ring. This is exemplified by our cross-linking data, where introducing a disulfide linkage between the N-domain and D1 abrogates p97 ATPase activity. Importantly, this is reversible by the addition of a reducing agent. All of our measurements have been carried out in the absence of adaptor proteins, and, thus, any N-domain conformational transitions may be more complex. Nevertheless, our cryo-EM data and normal mode analysis (44) support the hypothesis that N-domain and linker conformation relative to the D1-D2 hexamer ring is directly related to ATP turnover.
Effect of the C-terminal Region on p97 ATPase Activity
The C-terminal region of p97 is highly flexible, susceptible to limited proteolysis and unstructured in the p97 full-length crystal structure (15, 16). Our data suggest that the C-terminal truncation prevents D2 from undergoing the normal conformational changes upon ATP binding. For example, p97ΔCA232E no longer shows the increased ATPase activity of D2 seen in p97A232E alone. Interestingly, this region is functionally important, as it binds adaptor proteins, including the peptide N-glycanase PNGase that is involved in endoplasmic reticulum-associated degradation substrate processing (48) and contains phosphorylation sites that modulate adaptor binding and direct subcellular localization but do not affect ATPase activity (49, 50). The C-terminal region is disordered in all p97 crystal structures solved so far, and it is difficult to propose a mechanism for how this region affects ATPase activity. It was suggested that the C-terminal region might interact directly with the N terminus of p97, a potential nuclear targeting signal, with consequences for the subcellular localization of p97 (51). Previous cryo-EM reconstructions of p97 have suggested that the C terminus may wrap around the outside of the D2 ring (52). Together with our data, this suggests that the C-terminal extension of p97 may be involved in maintaining the conformation of the D2 ring.
Model for N-domain Conformation and p97 ATPase Activity
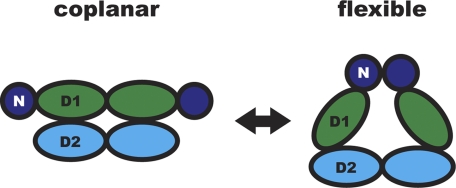
Taken together, our results support the hypothesis that the conformation of the N-domain and linker relative to the D1-D2 hexamer is directly correlated with p97 ATPase activity. Previous models for the ATPase cycle in p97 have already suggested a role for N-domains as well as negative cooperativity between the D1 and D2 rings (15, 17, 52, 53). Our biochemical and structural data obtained from IBMPFD mutants provide experimental evidence for a model where the N-domain and linker can adopt either of two conformations, namely flexible and coplanar with D1 (Fig. 6). When the N-domains are coplanar with the D1 ring, as in the N-D1 tethered mutant and in the p97 crystal structures, ATP hydrolysis is suppressed. When the N-domains are released from the D1 plane and are in a flexible state above the double rings, D2 is competent for ATP hydrolysis. The A232E mutation (and possibly other IBMPFD mutants) hinders the ability of the N-domain to assume the coplanar conformation, leading to the observed increase in ATPase activity. In wild-type p97, N-domain conformational changes could induce transitions between an active and inactive form that are part of the p97 ATP hydrolysis cycle. Such transitions could provide the necessary force for the remodeling of target proteins via adaptor interactions.
FIGURE 6.
Structural model of N-domain flexibility. p97 can adopt two conformations in the ATPase cycle. In the “flexible” conformation, the D2 domains form a compact ring and p97 hydrolyzes ATP. When N-domains are coplanar with the D1 ring, p97 is unable to hydrolyze ATP. Only two subunits of the hexamer are shown in a side view, colored as in Figs. 1A and 2D.
Supplementary Material
Acknowledgments
We thank Andreas Förster for critical reading of the manuscript.
This work was supported by the Wellcome Trust.

This article contains supplemental data, Table S1, and Figs. S1–S4.
- AAA
- ATPases associated with various cellular activities
- N-domain
- N-terminal domain
- IBMPFD
- inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia.
REFERENCES
- 1. Meyer H. H., Shorter J. G., Seemann J., Pappin D., Warren G. (2000) A complex of mammalian Ufd1 and Npl4 links the AAA-ATPase, p97, to ubiquitin and nuclear transport pathways. EMBO J. 19, 2181–2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rabinovich E., Kerem A., Fröhlich K. U., Diamant N., Bar-Nun S. (2002) AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation. Mol. Cell. Biol. 22, 626–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bays N. W., Hampton R. Y. (2002) Cdc48-Ufd1-Npl4. Stuck in the middle with Ub. Curr. Biol. 12, R366–371 [DOI] [PubMed] [Google Scholar]
- 4. Rape M., Hoppe T., Gorr I., Kalocay M., Richly H., Jentsch S. (2001) Mobilization of processed, membrane-tethered SPT23 transcription factor by CDC48(UFD1/NPL4), a ubiquitin-selective chaperone. Cell 107, 667–677 [DOI] [PubMed] [Google Scholar]
- 5. Hetzer M., Meyer H. H., Walther T. C., Bilbao-Cortes D., Warren G., Mattaj I. W. (2001) Distinct AAA-ATPase p97 complexes function in discrete steps of nuclear assembly. Nat. Cell Biol. 3, 1086–1091 [DOI] [PubMed] [Google Scholar]
- 6. Uchiyama K., Kondo H. (2005) p97/p47-Mediated biogenesis of Golgi and ER. J. Biochem. 137, 115–119 [DOI] [PubMed] [Google Scholar]
- 7. Rabouille C., Kondo H., Newman R., Hui N., Freemont P., Warren G. (1998) Syntaxin 5 is a common component of the NSF- and p97-mediated reassembly pathways of Golgi cisternae from mitotic Golgi fragments in vitro. Cell 92, 603–610 [DOI] [PubMed] [Google Scholar]
- 8. Cao K., Nakajima R., Meyer H. H., Zheng Y. (2003) The AAA-ATPase Cdc48/p97 regulates spindle disassembly at the end of mitosis. Cell 115, 355–367 [DOI] [PubMed] [Google Scholar]
- 9. Fröhlich K. U., Fries H. W., Rüdiger M., Erdmann R., Botstein D., Mecke D. (1991) Yeast cell cycle protein CDC48p shows full-length homology to the mammalian protein VCP and is a member of a protein family involved in secretion, peroxisome formation, and gene expression. J. Cell Biol. 114, 443–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jarosch E., Taxis C., Volkwein C., Bordallo J., Finley D., Wolf D. H., Sommer T. (2002) Protein dislocation from the ER requires polyubiquitination and the AAA-ATPase Cdc48. Nat. Cell Biol. 4, 134–139 [DOI] [PubMed] [Google Scholar]
- 11. Lilley B. N., Ploegh H. L. (2005) Multiprotein complexes that link dislocation, ubiquitination, and extraction of misfolded proteins from the endoplasmic reticulum membrane. Proc. Natl. Acad. Sci. U.S.A. 102, 14296–14301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kobayashi T., Tanaka K., Inoue K., Kakizuka A. (2002) Functional ATPase activity of p97/valosin-containing protein (VCP) is required for the quality control of endoplasmic reticulum in neuronally differentiated mammalian PC12 cells. J. Biol. Chem. 277, 47358–47365 [DOI] [PubMed] [Google Scholar]
- 13. Beskow A., Grimberg K. B., Bott L. C., Salomons F. A., Dantuma N. P., Young P. (2009) A conserved unfoldase activity for the p97 AAA-ATPase in proteasomal degradation. J. Mol. Biol. 394, 732–746 [DOI] [PubMed] [Google Scholar]
- 14. Ogura T., Wilkinson A. J. (2001) AAA+ superfamily ATPases. Common structure, diverse function. Genes Cells 6, 575–597 [DOI] [PubMed] [Google Scholar]
- 15. DeLaBarre B., Brunger A. T. (2003) Complete structure of p97/valosin-containing protein reveals communication between nucleotide domains. Nat. Struct. Biol. 10, 856–863 [DOI] [PubMed] [Google Scholar]
- 16. Huyton T., Pye V. E., Briggs L. C., Flynn T. C., Beuron F., Kondo H., Ma J., Zhang X., Freemont P. S. (2003) The crystal structure of murine p97/VCP at 3.6A. J. Struct. Biol. 144, 337–348 [DOI] [PubMed] [Google Scholar]
- 17. Zhang X., Shaw A., Bates P. A., Newman R. H., Gowen B., Orlova E., Gorman M. A., Kondo H., Dokurno P., Lally J., Leonard G., Meyer H., van Heel M., Freemont P. S. (2000) Structure of the AAA ATPase p97. Mol. Cell 6, 1473–1484 [DOI] [PubMed] [Google Scholar]
- 18. Wang Q., Song C., Li C. C. (2003) Hexamerization of p97-VCP is promoted by ATP binding to the D1 domain and required for ATPase and biological activities. Biochem. Biophys. Res. Commun. 300, 253–260 [DOI] [PubMed] [Google Scholar]
- 19. Song C., Wang Q., Li C. C. (2003) ATPase activity of p97-valosin-containing protein (VCP). D2 mediates the major enzyme activity, and D1 contributes to the heat-induced activity. J. Biol. Chem. 278, 3648–3655 [DOI] [PubMed] [Google Scholar]
- 20. Wang Q., Song C., Yang X., Li C. C. (2003) D1 ring is stable and nucleotide-independent, whereas D2 ring undergoes major conformational changes during the ATPase cycle of p97-VCP. J. Biol. Chem. 278, 32784–32793 [DOI] [PubMed] [Google Scholar]
- 21. Tang W. K., Li D., Li C. C., Esser L., Dai R., Guo L., Xia D. (2010) A novel ATP-dependent conformation in p97 N-D1 fragment revealed by crystal structures of disease-related mutants. EMBO J. 29, 2217–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dai R. M., Chen E., Longo D. L., Gorbea C. M., Li C. C. (1998) Involvement of valosin-containing protein, an ATPase co-purified with IκBα and 26 S proteasome, in ubiquitin-proteasome-mediated degradation of IκBα. J. Biol. Chem. 273, 3562–3573 [DOI] [PubMed] [Google Scholar]
- 23. Meyer H. H., Kondo H., Warren G. (1998) The p47 co-factor regulates the ATPase activity of the membrane fusion protein, p97. FEBS Lett. 437, 255–257 [DOI] [PubMed] [Google Scholar]
- 24. Dai R. M., Li C. C. (2001) Valosin-containing protein is a multi-ubiquitin chain-targeting factor required in ubiquitin-proteasome degradation. Nat. Cell Biol. 3, 740–744 [DOI] [PubMed] [Google Scholar]
- 25. Ye Y., Meyer H. H., Rapoport T. A. (2003) Function of the p97-Ufd1-Npl4 complex in retrotranslocation from the ER to the cytosol. Dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains. J. Cell Biol. 162, 71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dreveny I., Kondo H., Uchiyama K., Shaw A., Zhang X., Freemont P. (2004) Structural basis of the interaction between the AAA ATPase p97/VCP and its adaptor protein p47. EMBO J. 23, 1030–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guinto J. B., Ritson G. P., Taylor J. P., Forman M. S. (2007) Valosin-containing protein and the pathogenesis of frontotemporal dementia associated with inclusion body myopathy. Acta Neuropathol. 114, 55–61 [DOI] [PubMed] [Google Scholar]
- 28. Watts G. D., Wymer J., Kovach M. J., Mehta S. G., Mumm S., Darvish D., Pestronk A., Whyte M. P., Kimonis V. E. (2004) Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat. Genet. 36, 377–381 [DOI] [PubMed] [Google Scholar]
- 29. Haubenberger D., Bittner R. E., Rauch-Shorny S., Zimprich F., Mannhalter C., Wagner L., Mineva I., Vass K., Auff E., Zimprich A. (2005) Inclusion body myopathy and Paget disease is linked to a novel mutation in the VCP gene. Neurology 65, 1304–1305 [DOI] [PubMed] [Google Scholar]
- 30. Watts G. D., Thomasova D., Ramdeen S. K., Fulchiero E. C., Mehta S. G., Drachman D. A., Weihl C. C., Jamrozik Z., Kwiecinski H., Kaminska A., Kimonis V. E. (2007) Novel VCP mutations in inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia. Clin. Genet. 72, 420–426 [DOI] [PubMed] [Google Scholar]
- 31. Kimonis V. E., Fulchiero E., Vesa J., Watts G. (2008) VCP disease associated with myopathy, Paget disease of bone and frontotemporal dementia. Review of a unique disorder. Biochim. Biophys. Acta 1782, 744–748 [DOI] [PubMed] [Google Scholar]
- 32. Weihl C. C., Pestronk A., Kimonis V. E. (2009) Valosin-containing protein disease. Inclusion body myopathy with Paget's disease of the bone and fronto-temporal dementia. Neuromuscul. Disord. 19, 308–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Djamshidian A., Schaefer J., Haubenberger D., Stogmann E., Zimprich F., Auff E., Zimprich A. (2009) A novel mutation in the VCP gene (G157R) in a German family with inclusion-body myopathy with Paget disease of bone and frontotemporal dementia. Muscle Nerve 39, 389–391 [DOI] [PubMed] [Google Scholar]
- 34. Weihl C. C., Dalal S., Pestronk A., Hanson P. I. (2006) Inclusion body myopathy-associated mutations in p97/VCP impair endoplasmic reticulum-associated degradation. Hum. Mol. Genet. 15, 189–199 [DOI] [PubMed] [Google Scholar]
- 35. Hübbers C. U., Clemen C. S., Kesper K., Böddrich A., Hofmann A., Kämäräinen O., Tolksdorf K., Stumpf M., Reichelt J., Roth U., Krause S., Watts G., Kimonis V., Wattjes M. P., Reimann J., Thal D. R., Biermann K., Evert B. O., Lochmüller H., Wanker E. E., Schoser B. G., Noegel A. A., Schröder R. (2007) Pathological consequences of VCP mutations on human striated muscle. Brain 130, 381–393 [DOI] [PubMed] [Google Scholar]
- 36. Halawani D., LeBlanc A. C., Rouiller I., Michnick S. W., Servant M. J., Latterich M. (2009) Hereditary inclusion body myopathy-linked p97/VCP mutations in the NH2 domain and the D1 ring modulate p97/VCP ATPase activity and D2 ring conformation. Mol. Cell Biol. 29, 4484–4494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fernández-Sáiz V., Buchberger A. (2010) Imbalances in p97 co-factor interactions in human proteinopathy. EMBO Rep. 11, 479–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. van Heel M., Gowen B., Matadeen R., Orlova E. V., Finn R., Pape T., Cohen D., Stark H., Schmidt R., Schatz M., Patwardhan A. (2000) Single-particle electron cryo-microscopy. Towards atomic resolution. Q. Rev. Biophys. 33, 307–369 [DOI] [PubMed] [Google Scholar]
- 39. Nørby J. G. (1988) Coupled assay of Na+, K+-ATPase activity. Methods Enzymol. 156, 116–119 [DOI] [PubMed] [Google Scholar]
- 40. Manno A., Noguchi M., Fukushi J., Motohashi Y., Kakizuka A. (2010) Enhanced ATPase activities as a primary defect of mutant valosin-containing proteins that cause inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia. Genes Cells 15, 911–922 [DOI] [PubMed] [Google Scholar]
- 41. Noguchi M., Takata T., Kimura Y., Manno A., Murakami K., Koike M., Ohizumi H., Hori S., Kakizuka A. (2005) ATPase activity of p97/valosin-containing protein is regulated by oxidative modification of the evolutionally conserved cysteine 522 residue in Walker A motif. J. Biol. Chem. 280, 41332–41341 [DOI] [PubMed] [Google Scholar]
- 42. Zhao G., Zhou X., Wang L., Li G., Schindelin H., Lennarz W. (2007) Studies on peptide:N-glycanase-p97 interaction suggest that p97 phosphorylation modulates endoplasmic reticulum-associated degradation. Proc. Natl. Acad. Sci. U.S.A. 104, 8785–8790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Böhm S., Lamberti G., Fernández-Sáiz V., Stapf C., Buchberger A. (2011) Cellular functions of Ufd2 and Ufd3 in proteasomal protein degradation depend on Cdc48 binding. Mol. Cell. Biol. 31, 1528–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Beuron F., Dreveny I., Yuan X., Pye V. E., McKeown C., Briggs L. C., Cliff M. J., Kaneko Y., Wallis R., Isaacson R. L., Ladbury J. E., Matthews S. J., Kondo H., Zhang X., Freemont P. (2006) Conformational changes in the AAA ATPase p97-p47 adaptor complex. EMBO J. 25, 1967–1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tanner J. A., Abowath A., Miller A. D. (2002) Isothermal titration calorimetry reveals a zinc ion as an atomic switch in the diadenosine polyphosphates. J. Biol. Chem. 277, 3073–3078 [DOI] [PubMed] [Google Scholar]
- 46. Gerega A., Rockel B., Peters J., Tamura T., Baumeister W., Zwickl P. (2005) VAT, the thermoplasma homolog of mammalian p97/VCP, is an N domain-regulated protein unfoldase. J. Biol. Chem. 280, 42856–42862 [DOI] [PubMed] [Google Scholar]
- 47. Rothballer A., Tzvetkov N., Zwickl P. (2007) Mutations in p97/VCP induce unfolding activity. FEBS Lett. 581, 1197–1201 [DOI] [PubMed] [Google Scholar]
- 48. Yeung H. O., Kloppsteck P., Niwa H., Isaacson R. L., Matthews S., Zhang X., Freemont P. S. (2008) Insights into adaptor binding to the AAA protein p97. Biochem. Soc. Trans. 36, 62–67 [DOI] [PubMed] [Google Scholar]
- 49. Ewens C. A., Kloppsteck P., Förster A., Zhang X., Freemont P. S. (2010) Structural and functional implications of phosphorylation and acetylation in the regulation of the AAA+ protein p97. Biochem. Cell Biol. 88, 41–48 [DOI] [PubMed] [Google Scholar]
- 50. Egerton M., Samelson L. E. (1994) Biochemical characterization of valosin-containing protein, a protein tyrosine kinase substrate in hematopoietic cells. J. Biol. Chem. 269, 11435–11441 [PubMed] [Google Scholar]
- 51. Madeo F., Schlauer J., Zischka H., Mecke D., Fröhlich K. (1998) Tyrosine phosphorylation regulates cell cycle-dependent nuclear localization of Cdc48p. Mol. Biol. Cell 9, 131–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rouiller I., DeLaBarre B., May A. P., Weis W. I., Brunger A. T., Milligan R. A., Wilson-Kubalek E. M. (2002) Conformational changes of the multifunction p97 AAA ATPase during its ATPase cycle. Nat. Struct. Biol. 9, 950–957 [DOI] [PubMed] [Google Scholar]
- 53. Pye V., Dreveny I., Briggs L., Sands C., Beuron F., Zhang X., Freemont P. (2006) Going through the motions: the ATPase cycle of p97. J. Struct. Biol. 156, 12–28 [DOI] [PubMed] [Google Scholar]
- 54. Cogan E. B., Birrell G. B., Griffith O. H. (1999) A robotics-based automated assay for inorganic and organic phosphates. Anal. Biochem. 271, 29–35 [DOI] [PubMed] [Google Scholar]
- 55. DeLaBarre B., Christianson J. C., Kopito R. R., Brunger A. T. (2006) Central pore residues mediate the p97/VCP activity required for ERAD. Mol. Cell 22, 451–462 [DOI] [PubMed] [Google Scholar]
- 56. Zalk R., Shoshan-Barmatz V. (2003) ATP-binding sites in brain p97/VCP (valosin-containing protein), a multifunctional AAA ATPase. Biochem. J. 374, 473–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.