Background: The transglucosidase GBD-CD2 shows a unique α-(1→2) branching specificity among GH70 family members when catalyzing dextran glucosylation from sucrose.
Results: The truncated form ΔN123-GBD-CD2 was biochemically studied and structurally characterized at 1.90 Å resolution.
Conclusion: Dextran recognition and regiospecificity clearly involves a residue in subsite +1.
Significance: This is the first three-dimensional structure of a GH70 enzyme that reveals determinants of α-(1→2) linkage specificity.
Keywords: Enzyme Mechanisms, Enzyme Structure, Glycoside Hydrolases, Oligosaccharide, Polysaccharide
Abstract
ΔN123-glucan-binding domain-catalytic domain 2 (ΔN123-GBD-CD2) is a truncated form of the bifunctional glucansucrase DSR-E from Leuconostoc mesenteroides NRRL B-1299. It was constructed by rational truncation of GBD-CD2, which harbors the second catalytic domain of DSR-E. Like GBD-CD2, this variant displays α-(1→2) branching activity when incubated with sucrose as glucosyl donor and (oligo-)dextran as acceptor, transferring glucosyl residues to the acceptor via a ping-pong bi-bi mechanism. This allows the formation of prebiotic molecules containing controlled amounts of α-(1→2) linkages. The crystal structure of the apo α-(1→2) branching sucrase ΔN123-GBD-CD2 was solved at 1.90 Å resolution. The protein adopts the unusual U-shape fold organized in five distinct domains, also found in GTF180-ΔN and GTF-SI glucansucrases of glycoside hydrolase family 70. Residues forming subsite −1, involved in binding the glucosyl residue of sucrose and catalysis, are strictly conserved in both GTF180-ΔN and ΔN123-GBD-CD2. Subsite +1 analysis revealed three residues (Ala-2249, Gly-2250, and Phe-2214) that are specific to ΔN123-GBD-CD2. Mutation of these residues to the corresponding residues found in GTF180-ΔN showed that Ala-2249 and Gly-2250 are not directly involved in substrate binding and regiospecificity. In contrast, mutant F2214N had lost its ability to branch dextran, although it was still active on sucrose alone. Furthermore, three loops belonging to domains A and B at the upper part of the catalytic gorge are also specific to ΔN123-GBD-CD2. These distinguishing features are also proposed to be involved in the correct positioning of dextran acceptor molecules allowing the formation of α-(1→2) branches.
Introduction
Glucansucrases from glycoside hydrolase family 70 (GH70)5 are transglucosidases produced by lactic acid bacteria from the genera Leuconostoc, Lactobacillus, Streptococcus, Weissella, and Oenococcus (1). They naturally catalyze the polymerization of glucosyl residues with concomitant fructose release from sucrose, a cheap agroresource. Depending on the enzyme specificity, a large variety of glucans containing all types of glucosidic bonds, namely α-(1→2), α-(1→3), α-(1→4), or α-(1→6), and varying in terms of size, structure, degree of branching, and spatial arrangement are synthesized. These enzymes are also able to transfer the glucosyl unit from sucrose onto hydroxylated acceptor molecules added in the reaction medium and stand as very attractive biocatalysts for the production of novel biopolymers, prebiotic oligosaccharides, and new glucoderivatives (2).
Sequence analysis, functional characterization, and protein engineering showed that glucansucrases are structurally and mechanistically related to GH family 13 (3). They are α-retaining enzymes and sucrose cleavage is predicted to occur through the formation of a β-d-glucosyl covalent intermediate. This reaction involves one unique active site and requires the concerted action of an aspartate and a glutamic acid, which act as the nucleophile and the acid-base catalyst, respectively (4). Secondary structure predictions suggested that the catalytic domain of GH70 glucansucrases consists of a circularly permuted (β/α)8 barrel compared with that of GH13 family enzymes (5). These predictions have been recently confirmed by the elucidation of the three-dimensional structures of the GH70 glucansucrases GTF180-ΔN and GTF-SI (6–9). GTF180-ΔN is a dextransucrase synthesizing mainly α-(1→6) glucosidic linkages, whereas GTF-SI mutansucrase is specific for α-(1→3) bond formation.
In the GH70 family, the enzyme DSR-E from Leuconostoc mesenteroides NRRL B-1299 drew our attention because it was one of the rare enzymes able to synthesize dextrans with high amounts of α-(1→2) branch linkages. Sequence analysis of this very large enzyme (313 kDa) revealed the presence of two catalytic domains, CD1 and CD2, separated by a glucan-binding domain (GBD). CD1 and CD2, which share 45% identity and 65% similarity, were both classified in family GH70 (10). Both contain the highly conserved amino acids proposed to be involved in the formation of the glucosyl enzyme intermediate. Biochemical characterization of two recombinant truncated forms (CD1-GBD and GBD-CD2) showed that CD1-GBD acts as a polymerase, producing a glucan containing 86% α-(1→6), 11% α-(1→3), and 3% α-(1→4) glucosidic bonds. The second form (GBD-CD2) was found to be exclusively responsible for the synthesis of α-(1→2) linkages (11). Indeed, this enzyme acts as a very efficient transglucosidase in the presence of sucrose and either linear α-(1→6) glucans (dextrans) or linear gluco-oligosaccharides, which are used as acceptors. Steady-state kinetic analysis of α-(1→2) branch formation revealed that the enzyme displays a ping-pong bi-bi mechanism (12). In addition, experimental conditions have been established that enable the production of new dextrans with controlled sizes and α-(1→2) linkage contents (12). The presence of α-(1→2) linkages renders these products resistant to the action of mammalian digestive enzymes and promotes the growth of beneficial bacteria of the gut microbiome (13–17).
Because of this unique specificity, GBD-CD2 holds a great potential for the production of novel functional foods. To further investigate structure-function relationships of this α-(1→2) branching sucrase, we performed rational truncations of GBD-CD2 to obtain a pure and crystallizable enzyme form. The specificity and kinetic properties of the variant ΔN123-GBD-CD2 were investigated, and the apo x-ray structure was solved at 1.90 Å resolution. Additionally, the x-ray structure of this enzyme was solved at 3.3 Å resolution in a different crystal form. These are the first three-dimensional structures of an α-(1→2) branching sucrase. When compared with the GTF180-ΔN glucansucrase and GTF-SI mutansucrase structures, the GBD-CD2 structure revealed common but also very distinctive features that are discussed with regard to the α-(1→2) branching properties.
EXPERIMENTAL PROCEDURES
Production of ΔN123-GBD-CD2
The gbd-cd2 gene inserted into pBAD TOPO TA vector (Invitrogen) was amplified by PCR using the forward primer CACCATGGCACAAGCAGGTCACTATATCACGAAAA and reverse primer AGCTTGAGGTAATGTTGATTTATC for ΔN123-gbd-cd2. The primers used to generate other truncated mutants are listed in supplemental Table S1. The purified PCR products were cloned into a pBAD TOPO Directional 102 vector (Invitrogen). After ligation, the N-terminal thioredoxin tag was removed by NcoI restriction endonuclease digestion (supplemental Table S1). The constructs resulted in proteins with a C-terminal V5 epitope His6 tag. The Pfu Turbo polymerase (Stratagene) and all restriction enzymes (New England Biolabs) were used according to the manufacturers' instructions. DNA sequencing of the ΔN123-gbd-cd2 gene did not reveal any mutation (MilleGen, Labège, France). Transformed Escherichia coli strain TOP10 One shot (Invitrogen) was grown in Luria-Bertani medium supplemented with 100 μg/ml ampicillin. Induction with 0.02% (w/v) l-arabinose was performed at an A600 nm of 0.5. The cells were grown for an 8 additional hours, harvested by centrifugation, resuspended in PBS buffer, pH 7.0, supplemented with EDTA free anti-protease tablets (Roche), and disrupted by sonication. The ΔN123-GBD-CD2 enzyme was recovered as inclusion bodies in the crude cell extract.
Purification of ΔN123-GBD-CD2
Inclusion bodies were recovered by three cycles of washing, with PBS buffer supplemented with 1 mm EDTA and 1% (v/v) Triton X-100, followed by centrifugation at 20,000 × g for 20 min at 4 °C. They were denaturated using 8 m urea buffer, pH 8.0. The denatured protein preparation (120 ml) was supplemented with 500 mm NaCl, 0.5% (v/v) glycerol, and 25 mm imidazole, adjusted to pH 7.4, and centrifuged at 20,000 × g for 20 min. The supernatant was injected at a flow rate of 3 ml/min onto a 20-ml nickel-nitrilotriacetic acid-Sepharose column (GE Healthcare Life Sciences), equilibrated with 8 m urea buffer, pH 8.0. Protein refolding was carried out on-column, with five column volumes of buffer (20 mm sodium phosphate, 500 mm NaCl, 1% (v/v) glycerol, pH 7.4). The protein was eluted with a gradient of imidazole and dialyzed overnight at 4 °C against 20 mm sodium phosphate, 25 mm NaCl, and 1% (v/v) glycerol, pH 7.4. The solution was then applied to Q-Sepharose resin (GE Healthcare) and eluted using a gradient of 25–500 mm NaCl in 20 mm sodium phosphate, pH 7.4, supplemented with 1% (v/v) glycerol. The eluted protein was dialyzed overnight at 4 °C against 20 mm sodium acetate, 150 mm NaCl, 2.5% (v/v) glycerol, and 1 mm CaCl2, pH 5.75. Protein purity was checked by silver-stained SDS-PAGE.
Effect of Calcium Chloride on Enzymatic Activities
See the supplemental text.
Standard Activity Determination
Standard activities were determined as described previously (12) only in the presence of sucrose.
Acceptor Reactions with ΔN123-GBD-CD2
The reaction in the presence of sucrose and maltose was performed using standard conditions for 24 h using 146 mm of maltose and 1.0 unit/ml of enzymatic activity. Branching reactions of linear dextrans were carried out using standard conditions with 292 mm sucrose and 2,470, 1,237, 463, 311, 123, 92, or 62 mm of 70-kDa dextran acceptor (dextran concentrations expressed as anhydroglucosyl unit concentrations) with 1.5 units/ml of purified enzyme. The reaction medium was analyzed by HPLC as described by Brison et al. (12) to assay glucose, fructose, and leucrose production and sucrose depletion and by determining (i) glucose production rates (hydrolysis activity), (ii) fructose production rates that reflect both α-(1→2) glucosylation and hydrolysis activity, and (iii) α-(1→2) linkage content (12). Approximately 10 mg of each purified branched dextran was purified and analyzed by 1H NMR to determine its α-(1→2) linkage content (12).
Steady-state Kinetics
The ΔΝ123-GBD-CD2 kinetic mechanism was investigated by steady-state kinetics following the methodology described by Brison et al. (12).
Crystallization and Data Collection
Freshly purified enzyme was concentrated using a centrifugal filter device (Amicon Ultra, 4 Ultracel, 50 kDa; Millipore) to 3–4 mg/ml estimated by spectroscopy at 280 nm with theoretical molar extinction coefficient and molecular weight calculated using the ExPASy ProtParam tool. The protein to reservoir volume ratio in the 2-μl hanging-drop was 1:1. The crystals were obtained from two crystallization conditions (I and II). In condition I, ΔΝ123-GBD-CD2 enzyme crystallized either as needles or plate clusters over several weeks at 285 K using 17% (w/v) PEG 3350, 0.2 m NH4I, 80 mm ammonium citrate, 2% (v/v) glycerol, pH 5.0, as precipitant. Streak seeding resulted in single or clustered plate crystals with a thickness of 10–20 μm. Crystals were cryo-protected in reservoir solution supplemented with 15% glycerol (w/v) and then cryo-cooled in a gaseous nitrogen flux at 100 K. The majority of the plate crystals usually diffracted to 5–8 Å resolution; a crystal (100 × 100 × 10 μm3), obtained by co-crystallization with the glucohexaose α-d-Glcp-(1→6)-α-d-Glcp-(1→6)-α-d-Glcp-(1→6)-α-d-Glcp-(1→6)-α-d-Glcp-(1→4)-d-Glcp, diffracted anisotropically to 3.2 Å resolution in one direction and to 3.4 Å in the other direction. This crystal was used for data collection at the European Synchrotron Radiation Facility (Grenoble, France) at 100 K on Beam Line ID14-2. The data were processed using iMOSFLM and SCALA (18–20). According to the methodology for structure determination at low resolution described by Brunger et al. (21), we fixed the resolution limit at an I/σ(I) cutoff of 1.6 (at 3.3 Å resolution).
In condition II, pyramidal crystals grew over several weeks at 285 K in 15% PEG 3350 and 0.1 m NH4NO3. They were cryo-protected in 20% PEG 3350, 0.1 m NH4NO3, and 11% (v/v) glycerol. One of these crystals was used for data collection at the Soleil synchrotron (Gif-sur-Yvette, France) at 100 K on the PROXIMA1 beam line. The resulting 1.90 Å resolution data set was indexed, integrated, and scaled using XDS (22).
Structure Determination and Refinement
Details of data collection, cell parameters and processing statistics are presented in Table 1. The data set at 3.3 Å resolution was the first to be collected. CHAINSAW (23) was used to obtain a mixed homology model for domains A, B, and C using the structure of GTF180-ΔN (Protein Data Bank entry 3KLK) (8) and the ΔΝ123-GBD-CD2 sequence. This model was used as a template for molecular replacement with PHASER (24). Four loosely packed molecules were found in the asymmetric unit. Model building and refinement were done using COOT (25) and REFMAC5 (26), respectively, applying NCS restraints and resulted in an incomplete model.
TABLE 1.
Data collection and refinement statistics
The numbers in parentheses refer to statistics for the outer resolution shell.
| Triclinic crystals | Orthorhombic crystals | |
|---|---|---|
| Data collection | ||
| Wavelength (Å) | 0.933 | 0.954 |
| Cell dimensions, a, b, c (Å) and α, β, and γ (°) | 66.8, 140.0, 155.5, 85.4, 90.9, 76.9 | 68.2, 100.2, 187.2, 90.0, 90.0, 90.0 |
| Space group | P1 | P212121 |
| Molecules per asymmetric unit | 4 | 1 |
| Resolution limit (Å) | 51.6-3.3 (3.48-3.30) | 46.0-1.9 (1.95-1.90) |
| Reflections (total/unique) | 153,233/80,648 | 610,291/98,989 |
| Completeness (%) | 97.5 (97.0) | 97.2 (97.0) |
| Rmerge (%) | 20.3 (53.3) | 7.1 (45.4)c |
| I/σ(I) | 4.1 (1.6)b | 18.0 (4.4) |
| Wilson B factor (Å2) | 42.3 | 29.7 |
| Refinement statistics | ||
| Reflections (working/test) | 76,146/4,029 | 94,038/4,950 |
| Rcryst/Rfree | 0.224/0.291 | 0. 157/0.198 |
| Number of atoms | 32,344 | 9,331 |
| Protein | 8123/8106/8072/7952 | 8128 |
| Liganda | 54 | 152 |
| Ion/water | 4/33 | 2/1049 |
| B factor (Å2) | ||
| Main chain | 25.6 | 25.7 |
| Side chain | 26.5 | 27.7 |
| Ion/water/ligand | 30.0/20.0/30.0 | 30.6/34.6/54.7 |
| Stereochemical quality of the model | ||
| Root mean square deviation from bond lengths (Å) | 0.009 | 0.020 |
| Root mean square deviation from angles (°) | 1.107 | 1.835 |
| Ramachandran favored (%) | 93.1 | 97.2 |
| Ramachandran allowed (%) | 98.9 | 99.9 |
| Ramachandran disallowed (%) | 1.1 | 0.1 |
a Glycerol or polyethylene glycol molecules.
b Mean I/σ(I).
c Rmeas.
The high resolution structure of ΔΝ123-GBD-CD2 was obtained by molecular replacement using PHASER with the incomplete 3.3 Å resolution structure. Then ARP/wARP (27) was used to entirely rebuild the structure and place water molecules. The final Rwork and Rfree values for the high resolution structure were 0.157 and 0.198, respectively. The high resolution structure was in turn used to re-examine and complete the low resolution structure leading to final Rwork and Rfree values of 0.224 and 0.291, respectively. Residue numbering refers to the protein sequence of the full DSR-E dextransucrase sequence available in UniProt (accession number Q8G9Q2). The coordinates and structure factors have been deposited in the Protein Data Bank (entries 3TTO and 3TTQ).
Mutagenesis Studies
Mutants A2249W, G2250W, A2249D/G2250W, and F2214N were constructed by inverse PCR using ΔN123-gbd-cd2 as template and the primers described in supplemental Table S2. E. coli TOP10 cells (Invitrogen) were used as hosts for gene expression and mutant production. The mutants were tested in the presence of sucrose alone or with an additional maltose acceptor or 1-kDa dextran acceptor. The reaction products were analyzed by high performance anion exchange chromatography with pulsed ampero-metric detection (HPAEC-PAD) and compared with those obtained with the WT ΔN123-GBD-CD2 enzyme.
Docking Study
Docking of isomaltotriose in a model of the ΔN123-GBD-CD2 glucosyl enzyme intermediate is described in the supplemental text.
RESULTS AND DISCUSSION
Construction and Production of Truncated GBD-CD2 Enzyme
Attempts to crystallize the full-length GBD-CD2 failed. We assumed that the length and the quite hydrophobic nature of GBD (849 amino acids) did not favor crystallization. Indeed, the N-terminal part of the protein contains 41 consecutive repeat units rich in aromatic residues and homologous to the cell wall binding units (Pfam family PF01473) of C-LytA, the C-terminal choline-binding domain of Streptococcus pneumoniae autolysin A (28, 29). Therefore, the C-LytA three-dimensional structure was used to find 12 truncation sites in GBD (supplemental Fig. S1). Of the 12 constructs, the shortest active truncated form, ΔN123-GBD-CD2 (123 kDa; 1108 residues) with a glucan-binding domain reduced by 76%, was retained for further characterization. The purification procedure, which included isolation of inclusion bodies, immobilized metal ion affinity chromatography, and ion exchange chromatography yielded 14 mg of pure ΔN123-GBD-CD2 enzyme/liter of culture.
Functional Characterization of ΔN123-GBD-CD2
Using 292 mm sucrose, ΔN123-GBD-CD2 mainly catalyzed sucrose hydrolysis. Several by-products, including leucrose (α-d-Glcp-(1→5)-d-fructopyranose), kojibiose (α-d-Glcp-(1→2)-d-Glcp), and traces of maltulose (α-d-Glcp-(1→4)-d-fructofuranose), were also obtained. They result from glucosyl transfer onto the fructose and glucose units released by hydrolysis (supplemental Fig. S2A). In the presence of sucrose and maltose, the enzyme yielded the same products as with sucrose alone, indicating that maltose is not an acceptor of ΔN123-GBD-CD2 (supplemental Fig. S2B).
ΔN123-GBD-CD2 was also tested for its ability to branch dextran. 1H NMR spectra confirmed that ΔN123-GBD-CD2 catalyzes the formation of branched dextrans containing α-(1→2) glucosidic bonds (Fig. 1A). Branching amounts could be controlled by modulating the [sucrose]/[dextran] molar ratio (Fig. 1B). As shown in Fig. 1C, when the dextran concentration was increased, glucose and leucrose production was reduced. Indeed, at low molar [sucrose]/[dextran] ratios (0.12 and 0.24), the glucosyl units from sucrose are almost exclusively transferred onto dextran, similar to what has been observed for GBD-CD2 (11, 12).
FIGURE 1.
A, anomeric region of 1H NMR spectra obtained at 298 K for purified α-(1→2) branched dextrans. Black, dextran 70-kDa standard; blue, dextran α-(1→2) branched at 11%; pink, dextran α-(1→2) branched at 19%; orange, dextran α-(1→2) branched at 33%; green, dextrans α-(1→2) branched between 35 and 37%. B, percentage of α-(1→2) linkage as a function of [sucrose]/[dextran] ([Suc]/[Dex]) molar ratios used for the acceptor reactions. Empty and filled circles correspond to values obtained after 1H NMR and HPLC measurement, respectively, of the α-(1→2) linkage content in dextrans synthesized by ΔN123-GBD-CD2. Red crosses, 1H NMR results for dextrans branched by GBD-CD2 (12). C, effects of the [sucrose]/[dextran] molar ratio on α-(1→2) branched dextran yields. The reactions were carried out at 292 mm sucrose and various dextran concentrations. Main final reaction products are residual sucrose, glucose (from sucrose hydrolysis), leucrose (from fructose glucosylation), and α-(1→2) branched dextran.
ΔN123-GBD-CD2 displays Michaelis-Menten kinetics for sucrose hydrolysis. Km, SucH, and kcat,SucH values are 7.5 ± 1.0 mm and 76 s−1, respectively (Table 2). In the presence of dextran as acceptor, the initial velocities of the α-(1→2) glucosylation obey a ping-pong bi-bi mechanism. The catalytic constants kcat,T for ΔN123-GBD-CD2 and GDB-CD2 are not significantly different. In contrast, the apparent Km,DexT and Km,SucT are 1.6- and 4.9-fold higher than those observed for GBD-CD2, respectively, indicating that the deleted part of the GBD may favor binding of both sucrose and high molecular weight dextran. Deletion of the complete GBD (from Gln-1141 to Leu-1980) resulted in an almost inactive variant (11). The remaining part of GBD found in ΔN123-GBD-CD2 is thus necessary to maintain enzymatic activity.
TABLE 2.
Comparison of the apparent kinetic parameters determined for sucrose hydrolysis (subscript H) and α-(1→2) dextran branching activities (subscript T) for GBD-CD2 and ΔN123-GBD-CD2
| Apparent kinetic parameters | GBD-CD2a | ΔN123-GBD-CD2 |
|---|---|---|
| In the presence of sucrose | ||
| Vmax H (μmol·min−1·mg−1 of purified enzyme) | 34.6 ± 0.5 | 36.3 ± 0.6 |
| Km,SucH (mm) | 10.8 ± 0.8 | 7.5 ± 1.0 |
| kcat,SucH (s−1) | 109 | 76 |
| In the presence of sucrose and dextran 70 kDa | ||
| Vmax T (μmol·min−1·mg−1 of purified enzyme) | 303 ± 5 | 462 ± 45 |
| Km,SucT (mm) | 42 ± 2 | 206 ± 34 |
| Km,DexT (mm of anhydroglucosyl units) | 75 ± 3 | 125 ± 21 |
| Km,DexT (mm) | 0.174 ± 0.008 | 0.30 ± 0.05 |
| kcat T (s−1) | 970 | 947 |
| kcat T/Km,SucT (s−1·mm−1) | 23 | 4.6 |
| kcat T/Km,DexT (s−1·mm−1) | 13 | 7.6 |
a From Ref. 12.
Crystal Structures of ΔN123-GBD-CD2
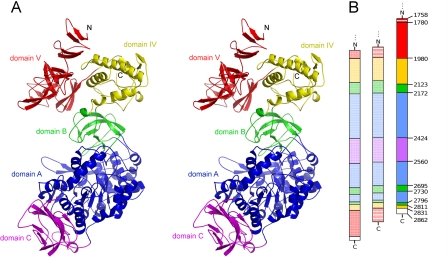
The structure of ΔN123-GBD-CD2 was solved at 1.90 Å resolution in its apo form. Refinement statistics are summarized in Table 1. The polypeptide chain (1043 residues) is organized in five distinct domains named C, A, B, IV, and V (Fig. 2A), which are not consecutively arranged along the peptide chain, similarly to the fold of other GH70 enzymes (Fig. 2B). A structure was also solved at 3.3 Å resolution in another space group. The superimposition of the two structures of ΔN123-GBD-CD2 showed that the positions of backbone atoms of domains IV and V are shifted by 1.8–7.0 Å (supplemental Fig. S3). In addition, no glucohexaose molecule was identified in the enzyme active site, although the protein was co-crystallized with this substrate.
FIGURE 2.
A, stereo view of ΔN123-GBD-CD2 domain organization. Magenta, domain C; blue, domain A, which includes the (β/α)8 barrel; green, domain B; yellow, domain IV; red, domain V. B, schematic representation of the domain arrangement along the polypeptide chains of crystallized GH70 glucansucrases, from left to right, GTF180-ΔN, GTF-SI, and ΔN123-GBD-CD2. The color code is identical to that for A. Striped red, parts of domains V of GTF-SI and ΔN123-GBD-CD2, which are not visible in the electron density map; white, purification tag.
At the interface of domains A and B, a heptacoordinated metal ion is bound that is probably a calcium ion. Indeed, the binding site is homologous to the Ca2+-binding site of GTF180-ΔN in which Asp-2164 of ΔN123-GBD-CD2 replaces Glu-979 of GTF180-ΔN (8). Distances between ligands (i.e. carbonyl group of Asp-2164, Oδ1 and Oδ2 of Asp-2170, carbonyl group of Phe-2214, Oδ1 of Asn-2693, and two water molecules) and the metal ion range from 2.32 to 2.61 Å (supplemental Fig. S4). In addition, activity measurements showed that the activity of ΔN123-GBD-CD2 decreased by 14% in the presence of the Ca2+-chelating agent EDTA. This is in agreement with calcium dependence (supplemental Table S3) (30).
Individual Domain Description
Domain C consists of eight anti-parallel β strands and includes a modified Greek key motif. The function of this domain remains unknown.
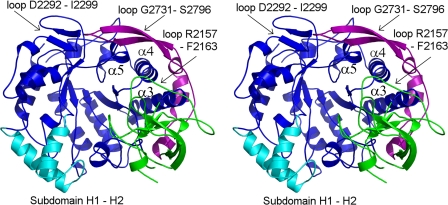
Domain A is the largest domain; it forms the catalytic core together with elements from domain B. It comprises a (β/α)8 barrel similar to that of GTF180-ΔN and GFT-SI glucansucrases, which is circularly permuted compared with that of GH13 enzymes (Fig. 3 and supplemental Fig. S5) (8, 9). Domain C is inserted between helix α8 and strand β1, and domain B connects strand β3 to helix α3. In domain A, many structural features distinguish ΔN123-GBD-CD2 from GTF180-ΔN and GTF-SI. First, helix α5, which is downstream from the putative catalytic Glu-2248, is three residues shorter than that of GTF180-ΔN and GTF-SI and adopts a different position (supplemental Fig. S5). Conversely, the loop from Gly-2731 to Ser-2796 is 25 residues longer than the equivalent loop of GTF180-ΔN (supplemental Fig. S5). Starting with two contiguous α-helices (residues Gln-2734 to Tyr-2739 and Gln-2741 to Lys-2750) followed by a β-hairpin covering helices α3, α4, and α5, it protrudes from domain B and contributes to domain A (Fig. 3). Loop Asp-2292 to Ile-2299, which connects helix α6 to strand β7 of the (β/α)8 barrel, is also four residues longer than its equivalent in GTF180-ΔN (supplemental Fig. S5). Loops Gly-2731 to Ser-2796 and Asp-2292 to Ile-2299 are both located at the upper part of the catalytic gorge, which is also delineated by a small subdomain inserted between strand β6 and helix α7 (from Gly-2324 to Asn-2368). This small subdomain comprises two helices, H1 and H2, also found in GTF-180-ΔN and GTF-SI (Fig. 3).
FIGURE 3.
Stereo view of the secondary structure elements of ΔN123-GBD-CD2 domains A and B. For domain A, blue shows the (β/α)8 barrel, cyan shows the subdomain H1-H2, and purple shows the loop Gly-2731 to Ser-2796 protruding from domain B and contributing to domain A. Domain B is green.
Domain B is folded into a five-stranded β-sheet. A comparison with domain B of GTF180-ΔΝ shows that 94 Cα of 99 Cα superimpose well (root mean square deviation, 1.2 Å). However, the loop inserted between Arg-2157 and Phe-2163, also located at the upper part of the catalytic gorge, is 11 residues shorter than the equivalent one of GTF180-ΔN (supplemental Figs. S5 and S6).
Domain IV can be superimposed with that of GTF180-ΔN with secondary structure elements conserved but slightly shifted. A DALI analysis showed no significant structural similarities of this domain to other proteins in the Protein Data Bank (31).
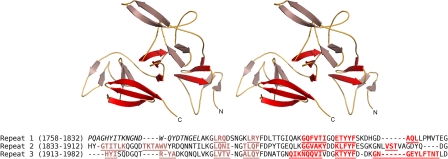
With a global V shape, domain V adopts an original fold that is similar only to domain V of GTF180-ΔN but not to any other protein domain in the Protein Data Bank (DALI analysis) (Fig. 4). Although in GTF180-ΔN both the N- and C-terminal parts of the peptide chain contribute to domain V, in ΔN123-GBD-CD2 it is only made up from residues of the N-terminal part. It comprises three subdomains exhibiting an organization in which a three-stranded β-sheet is connected to two consecutive β-hairpins. Each β-hairpin is constituted of two β-strands of 3–6 residues separated by short loops of 2–11 amino acids. The two β-hairpins and the three-stranded β-sheet of each subdomain are rotated by ∼120° with respect to each other.
FIGURE 4.
Upper panel, stereo view of the secondary structure elements of domain V (truncated glucan-binding domain) of ΔN123-GBD-CD2. The three-stranded β-sheets and β-hairpins of the three subdomains are represented in red and salmon, respectively. Lower panel, sequence alignment of the domain V. The underlined and colored residues are β-strands; N-terminal residues in italics are not visible in the electron density map.
Sucrose Specificity at Subsites −1 and +1
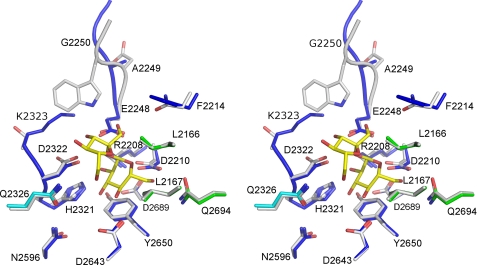
The ΔN123-GBD-CD2 active site forms a pocket into a large gorge (supplemental Fig. S7). Because ΔΝ123-GBD-CD2 and GTF180-ΔN both use sucrose as substrate, we superimposed the structure of ΔΝ123-GBD-CD2 with that of the GTF180-ΔN-sucrose complex (Protein Data Bank entry 3HZ3) to investigate the functional role of specific residues lining the active site of ΔΝ123-GBD-CD2. As shown in Fig. 5, 17 residues defining subsites −1 and +1 (32) in ΔN123-GBD-CD2 align well with the corresponding residues of GTF180-ΔN (root mean square deviation, 0.53 Å on Cα). At subsite −1, 10 conserved residues (Arg-2208, Asp-2210, Glu-2248, His-2321, Asp-2322, Asn-2596, Asp-2643, Tyr-2650, Asp-2689, and Gln-2694) correspond to residues that interact with the glucosyl moiety of sucrose in GTF180-ΔN (Fig. 5). In particular, residues Asp-2210 and Glu-2248 coincide with the nucleophile and the general acid/base catalyst of GTF180-ΔN and GH13 enzymes (8, 33, 34) and are in a position to play the same role in ΔΝ123-GBD-CD2. Likewise, Asp-2322 is equivalent to the transition state stabilizing residue Asp-1136 of GTF180-ΔN.
FIGURE 5.
Stereo view of subsites −1 and +1 of ΔN123-GBD-CD2, with sucrose from the GTF180-ΔN-sucrose complex superimposed. The catalytic residues are Asp-2210 (nucleophile), Glu-2248 (acid/base), and Asp-2322 (transition state stabilizer). Sucrose is shown with yellow carbons. Residues of the inactive GTF180-ΔN mutant (D1025N) that interact with sucrose (8) are represented in gray. The carbon atoms of their structural equivalents in ΔN123-GBD-CD2 are shown in blue (domain A), cyan (subdomain H1-H2), and green (domain B).
At subsite +1, seven residues of GTF180-ΔN have interactions with the fructosyl ring of sucrose (8). Three of them (Lue-2166, Leu-2167, and Gln-2326) are conserved between ΔΝ123-GBD-CD2 and GTF180-ΔN. The two leucine residues are involved in van der Waals interactions with the fructosyl moiety of sucrose. Residue Gln-2326 is well positioned to make a hydrogen bond with the C6 hydroxyl of the fructosyl moiety. In contrast, two residues of GTF180-ΔN subsite −1 (Asn-1029 and Trp-1065), which are H-bonded to the fructosyl ring of sucrose, have no equivalent in ΔN123-GBD-CD2. It is noteworthy that these residues are usually conserved in the GH70 family except for GBD-CD2. As shown in Fig. 5, Asn01029 is substituted by Phe-02214, and the position corresponding to Trp-1065 is occupied by Ala-2249 and Gly-2250. None of these residues can make H-bonds with the fructosyl residue. Only Lys-2323 may establish a hydrogen bond with the O4 of the fructosyl unit, suggesting a weaker sucrose binding in ΔN123-GBD-CD2. This may explain why ΔN123-GBD-CD2 has a 32-fold higher apparent Km for sucrose than GTF180-ΔN (6).
Acceptor Recognition
In GH70 enzymes, subsite +1 not only has to accommodate the fructosyl unit of the donor substrate but must also bind acceptors. We have shown that ΔN123-GBD-CD2 does not glucosylate maltose. This disaccharide is known to be the most efficient acceptor for glucansucrases (35). Analysis of the GTF180-Δ-maltose complex structure (Protein Data Bank entry 3KLL) showed that the interaction between the maltose residue and Trp-1065 is essential for maltose glucosylation, which results in the formation of panose (α-d-Glcp-(1→6)-α-d-Glcp-(1→4)-d-Glcp) (8). Trp-1065 is not conserved in ΔN123-GBD-CD2, and there is no stacking platform at this position to keep the nonreducing end residue of maltose in a correct position for glucosylation.
Mutagenesis Studies
To further investigate the possible involvement in acceptor binding of the nonconserved residues Ala-2249, Gly-2250, and Phe-2214, we constructed mutants A2249W, G2250W, A2249D/G2250W, and F2214N, in which the amino acids were replaced by the corresponding residues of GTF180-ΔN. Because the structural alignment revealed that Trp-1065 of GTF180-ΔN occupies a position in between Ala-2249 and Gly-2250, both Ala-2249 and Gly-2250 were individually replaced by a tryptophan residue. The mutants were tested in the presence of sucrose alone or with additional acceptors (maltose or 1-kDa dextran). When using only sucrose as a substrate, all of the mutants were unable to form polymer and showed reduced hydrolytic activity (Table 3). Apparently, subsite +1 is tolerant to mutations with regard to sucrose utilization.
TABLE 3.
Characterization of single and double mutants targeting nonconserved residues of subsite +1
| Relative activitya | Sucrose | Acceptor concentration | Hydrolysis yieldb | Sucrose isomer yieldc | Transfer onto acceptord | |
|---|---|---|---|---|---|---|
| % | mm | mm | % | % | % | |
| ΔN123-GBD-CD2 WT | 100 | 146 | 87 | 13 | ||
| 146 | 1-kDa dextran, 146 | 20 | 1 | 79 | ||
| 146 | Maltose, 146 | 88 | 12 | NDe | ||
| ΔN123-GBD-CD2 A2249W | 21.7 | 146 | 89 | 11 | ||
| 146 | 1-kDa dextran, 146 | 24 | 2 | 74 | ||
| 146 | Maltose, 146 | 90 | 10 | ND | ||
| ΔN123-GBD-CD2 G2250W | 1.1 | 146 | 89 | 11 | ||
| 146 | 1kDa dextran, 146 | 45 | 4 | 51 | ||
| 146 | Maltose, 146 | 90 | 10 | ND | ||
| ΔN123-GBD-CD2 A2249D-G2250W | 89.3 | 146 | 90 | 10 | ||
| 146 | 1-kDa dextran, 146 | 36 | 4 | 60 | ||
| 146 | Maltose, 146 | 90 | 10 | ND | ||
| ΔN123-GBD-CD2 F2214N | 18.6 | 146 | 100 | ND | ||
| 146 | 1-kDa dextran, 146 | 100 | ND | ND | ||
| 146 | Maltose, 146 | 100 | ND | ND |
a The relative activity was determined as the ratio of sucrose consumption (initial rate) of each mutant versus sucrose consumption (initial rate) of ΔN123-GBD-CD2.
b Hydrolysis yield = Glc (mol)/sucrose consumed (mol) ×100.
c Sucrose isomer = Glc incorporated into leucrose (mol)/sucrose consumed (mol) × 100.
d Transfer onto acceptor = 100 − hydrolysis ratio % − sucrose isomer yield %.
e ND, not detectable.
With maltose, none of the mutants showed transferase activity. In particular, the introduction of a tryptophan residue at position 2249 and/or 2250 did not promote the transfer reaction. Likely, other changes at the acceptor binding subsite +1 are necessary to generate such activity. However, when 1-kDa dextran was added as an acceptor, mutants at positions 2249 and/or 2250 were still able to catalyze the formation of α-(1→2) branches like the WT enzyme (supplemental Fig. S8). Apparently these positions are not directly involved in regiospecificity. In contrast, mutant F2214N was unable to use 1-kDa dextran as an acceptor (Table 3 and supplemental Fig. S9). Thus, this residue is critical for dextran binding and branching.
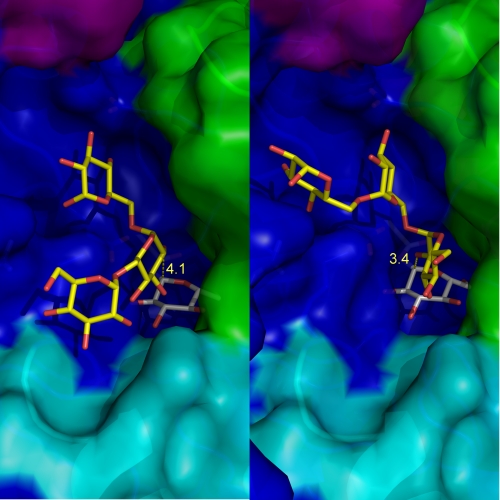
To catalyze glucosyl transfer onto dextrans, ΔN123-GBD-CD2 must first bind sucrose and then catalyze the formation of the covalent β-d-glucosyl enzyme complex. Fructose must be released from subsite +1 to allow the acceptor to enter. The dextran glucosyl residue that will be branched must bind in subsite +1 with its C2 hydroxyl group properly oriented toward the C1 of the β-d-glucosyl-enzyme intermediate. In accordance with the ping-pong bi-bi mechanism of ΔN123-GBD-CD2, the product has to leave the active site after branching to allow the binding of a new sucrose molecule. How acceptor molecules are accommodated in the catalytic gorge in a position to be α-(1→2) glucosylated remains to be elucidated. Despite numerous attempts to crystallize complexes of ΔN123-GBD-CD2 with isomaltotriose, linear, or α-(1→2) branched gluco-oligosaccharides, we were not successful. Docking of isomaltotriose was thus attempted in a model of the glucosyl-enzyme intermediate of ΔΝ123-GBD-CD2. Two main groups of docked isomaltotriose were obtained with a glucosyl unit of the acceptor chain appropriately oriented to allow the formation of an α-(1→2) linkage. In the first group, the central glucosyl unit of isomaltotriose is in a position to be α-(1→2) glucosylated (Fig. 6A), with a shortest distance of 4.10 Å between the O2 atom of the acceptor glucosyl unit and the C1 atom of the glucosyl enzyme intermediate. In the second group, the glucosyl unit at the nonreducing extremity would be glucosylated with a distance between O2 and C1 of 3.45 Å (Fig. 6B). These models reveal that α-(1→2) branching occurring at either the nonreducing end or at a residue of the dextran backbone is feasible. Moreover, structural analysis of ΔN123-GBD-CD2 has identified residues that line its catalytic gorge and that could be involved in dextran binding or enzyme regiospecificity (Fig. 7). Indeed, one of these residues, Phe-2214, was shown by site-directed mutagenesis to be crucial for dextran accommodation. The roles of the other residues, as well as residues in the three loops specific for GBD-CD2, will now be explored to further deepen our understanding of the mechanism and specificity of the enzyme.
FIGURE 6.
Isomaltotriose docking in the catalytic groove of the modeled glucosyl-enzyme intermediate of ΔN123-GBD-CD2. Two binding modes were found that allow glucosylation through α-(1→2) linkage formation onto the central glucosyl unit (A) or the nonreducing end extremity (B). The covalently linked glucosyl unit in subsite −1 is represented in gray, and isomaltotriose is in yellow. The distances between O2 atom of isomaltotriose and C1 atom of the glucosyl enzyme intermediate (in Å) are shown in yellow. Domain coloring is as in Fig. 3.
FIGURE 7.
Stereo view of the catalytic gorges of GTF180-ΔN and ΔN123-GBD-CD2. For clarity, only different residues or residues adopting different conformations are shown. Backbone atoms of ΔN123-GBD-CD2 are depicted in light blue. Residues from domain A of ΔN123-GBD-CD2 are depicted in blue, cyan, and purple (see Fig. 3). Residues from domain B of ΔN123-GBD-CD2 are shown in green. Gray residues belong to GTF180-ΔN. Sucrose from GTF180-ΔN-sucrose complex (Protein Data Bank entry 3HZ3) in subsites −1 and +1 is represented as yellow carbons.
Supplementary Material
Acknowledgments
We thank the staff of Beam Line ID14-2 at the ESRF (Grenoble, France) and Pierre Legrand from the PROXIMA1 beam line at the SOLEIL synchrotron (Gif-sur-Yvette, France) for data collection facilities and assistance. We greatly thank Nelly Monties, Pierre Escalier, Sandra Pizzut-Serin, and Virginie Rivière for technical assistance.
This work was supported by the region Midi-Pyrénées (France) and the European Molecular Biology Organization.

This article contains supplemental text, Tables S1–S3, and Figs. S1–S9.
The atomic coordinates and structure factors (codes 3TTO and 3TTQ) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- GH
- glycoside hydrolase
- GBD
- glucan-binding domain
- GBD-CD2
- GBD-catalytic domain 2
- Glcp
- glucopyranose.
REFERENCES
- 1. Henrissat B., Bairoch A. (1996) Updating the sequence-based classification of glycosyl hydrolases. Biochem. J. 316, 695–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Monsan P., Remaud-Siméon M., André I. (2010) Transglucosidases as efficient tools for oligosaccharide and glucoconjugate synthesis. Curr. Opin. Microbiol. 13, 293–300 [DOI] [PubMed] [Google Scholar]
- 3. MacGregor E. A., Janecek S., Svensson B. (2001) Relationship of sequence and structure to specificity in the α-amylase family of enzymes. Biochim. Biophys. Acta 1546, 1–20 [DOI] [PubMed] [Google Scholar]
- 4. Moulis C., Joucla G., Harrison D., Fabre E., Potocki-Veronese G., Monsan P., Remaud-Simeon M. (2006) Understanding the polymerization mechanism of glycoside-hydrolase family 70 glucansucrases. J. Biol. Chem. 281, 31254–31267 [DOI] [PubMed] [Google Scholar]
- 5. MacGregor E. A., Jespersen H. M., Svensson B. (1996) A circularly permuted α-amylase-type α/β-barrel structure in glucan-synthesizing glucosyltransferases. FEBS Lett. 378, 263–266 [DOI] [PubMed] [Google Scholar]
- 6. Pijning T., Vujičić-Žagar A., Kralj S., Eeuwema W., Dijkhuizen L., Dijkstra B. W. (2008) Biochemical and crystallographic characterization of a glucansucrase from Lactobacillus reutri 80. Biocatal. Biotransfor. 26, 12–17 [Google Scholar]
- 7. Ito K., Ito S., Shimamura T., Kawarasaki Y., Abe K., Misaka T., Kobayashi T., Iwata S. (2010) Crystallization and preliminary X-ray analysis of a glucansucrase from the dental caries pathogen Streptococcus mutans. Acta Crystallogr. F 66, 1086–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vujicic-Zagar A., Pijning T., Kralj S., López C. A., Eeuwema W., Dijkhuizen L., Dijkstra B. W. (2010) Crystal structure of a 117 kDa glucansucrase fragment provides insight into evolution and product specificity of GH70 enzymes. Proc. Natl. Acad. Sci. U.S.A. 107, 21406–21411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ito K., Ito S., Shimamura T., Weyand S., Kawarasaki Y., Misaka T., Abe K., Kobayashi T., Cameron A. D., Iwata S. (2011) Crystal structure of glucansucrase from the dental caries pathogen Streptococcus mutans. J. Mol. Biol. 408, 177–186 [DOI] [PubMed] [Google Scholar]
- 10. Bozonnet S., Dols-Laffargue M., Fabre E., Pizzut S., Remaud-Simeon M., Monsan P., Willemot R. M. (2002) Molecular characterization of DSR-E, an α-1,2 linkage-synthesizing dextransucrase with two catalytic domains. J. Bacteriol. 184, 5753–5761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fabre E., Bozonnet S., Arcache A., Willemot R. M., Vignon M., Monsan P., Remaud-Simeon M. (2005) Role of the two catalytic domains of DSR-E dextransucrase and their involvement in the formation of highly α-1,2 branched dextran. J. Bacteriol. 187, 296–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brison Y., Fabre E., Moulis C., Portais J. C., Monsan P., Remaud-Siméon M. (2010) Synthesis of dextrans with controlled amounts of α-1,2 linkages using the transglucosidase GBD-CD2. Appl. Microbiol. Biotechnol. 86, 545–554 [DOI] [PubMed] [Google Scholar]
- 13. Valette P., Pelenc V., Djouzi Z., Andrieux C., Paul F., Monsan P., Szylit O. (1993) Bioavailability of new synthesized glucooligosaccharides in the intestinal tract of gnotobiotic rats. J. Sci. Food Agr. 62, 121–127 [Google Scholar]
- 14. Djouzi Z., Andrieux C., Pelenc V., Somarriba S., Popot F., Paul F., Monsan P., Szylit O. (1995) Degradation and fermentation of α-gluco-oligosaccharides by bacterial strains from human colon: in vitro and in vivo studies in gnotobiotic rats. J. Appl. Microbiol. 79, 117–127 [DOI] [PubMed] [Google Scholar]
- 15. Djouzi Z., Andrieux C. (1997) Compared effects of three oligosaccharides on metabolism of intestinal microflora in rats inoculated with a human faecal flora. Br. J. Nutr. 78, 313–324 [DOI] [PubMed] [Google Scholar]
- 16. Flickinger E. A., Wolf B. W., Garleb K. A., Chow J., Leyer G. J., Johns P. W., Fahey G. C., Jr. (2000) Glucose-based oligosaccharides exhibit different in vitro fermentation patterns and affect in vivo apparent nutrient digestibility and microbial populations in dogs. J. Nutr. 130, 1267–1273 [DOI] [PubMed] [Google Scholar]
- 17. Sarbini S. R., Kolida S., Naeye T., Einerhand A., Brison Y., Remaud-Simeon M., Monsan P., Gibson G. R., Rastall R. A. (2011) In vitro fermentation of linear and α-1,2-branched dextrans by the human fecal microbiota. Appl. Environ. Microbiol. 77, 5307–5315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Battye T. G., Kontogiannis L., Johnson O., Powell H. R., Leslie A. G. (2011) iMOSFLM. A new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 67, 271–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Evans P. (2006) Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 20. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brunger A. T., DeLaBarre B., Davies J. M., Weis W. I. (2009) X-ray structure determination at low resolution. Acta Crystallogr. D Biol. Crystallogr. 65, 128–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stein N. (2008) CHAINSAW: a program for mutating pdb files used as templates in molecular replacement. J. Appl. Cryst. 41, 641–643 [Google Scholar]
- 24. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 27. Langer G., Cohen S. X., Lamzin V. S., Perrakis A. (2008) Automated macromolecular model building for x-ray crystallography using ARP/wARP version 7. Nat. Protoc. 3, 1171–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fernández-Tornero C., López R., García E., Giménez-Gallego G., Romero A. (2001) A novel solenoid fold in the cell wall anchoring domain of the pneumococcal virulence factor LytA. Nat. Struct. Biol. 8, 1020–1024 [DOI] [PubMed] [Google Scholar]
- 29. Shah D. S., Joucla G., Remaud-Simeon M., Russell R. R. (2004) Conserved repeat motifs and glucan binding by glucansucrases of oral streptococci and Leuconostoc mesenteroides. J. Bacteriol. 186, 8301–8308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Robyt J. F., Walseth T. F. (1979) Production, purification, and properties of dextransucrase from Leuconostoc mesenteroides NRRL B-512F. Carbohydr. Res. 68, 95–111 [DOI] [PubMed] [Google Scholar]
- 31. Holm L., Kääriäinen S., Rosenström P., Schenkel A. (2008) Searching protein structure databases with DaliLite v.3. Bioinformatics 24, 2780–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Davies G. J., Wilson K. S., Henrissat B. (1997) Nomenclature for sugar-binding subsites in glycosyl hydrolases. Biochem. J. 321, 557–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jensen M. H., Mirza O., Albenne C., Remaud-Simeon M., Monsan P., Gajhede M., Skov L. K. (2004) Crystal structure of the covalent intermediate of amylosucrase from Neisseria polysaccharea. Biochemistry 43, 3104–3110 [DOI] [PubMed] [Google Scholar]
- 34. Uitdehaag J. C., Mosi R., Kalk K. H., van der Veen B. A., Dijkhuizen L., Withers S. G., Dijkstra B. W. (1999) X-ray structures along the reaction pathway of cyclodextrin glycosyltransferase elucidate catalysis in the α-amylase family. Nat. Struct. Biol. 6, 432–436 [DOI] [PubMed] [Google Scholar]
- 35. Remaud-Siméon M., Willemot R. M., Sarçabal P., Potocki de Montalk G., Monsan P. (2000) Glucanfucrases: molecular engineering and oligosaccharide synthesis. J. Mol. Catal. B Enzym. 10, 117–128 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.