Background: Maltogenic amylases that are known to date form dimers to perform hydrolysis.
Results: The structure of maltogenic amylase from Staphylothermus showed a novel domain at the N terminus associated with the active site.
Conclusion: Staphylothermus amylase has all of its substrate-binding structural components in a single monomer.
Significance: This is the first report of the newly observed domain arrangement adopted by hyperthermophilic archaic maltogenic amylase.
Keywords: Archaea, Carbohydrate-binding Protein, Carbohydrate Processing, Crystal Structure, Enzyme Mechanisms
Abstract
Staphylothermus marinus maltogenic amylase (SMMA) is a novel extreme thermophile maltogenic amylase with an optimal temperature of 100 °C, which hydrolyzes α-(1–4)-glycosyl linkages in cyclodextrins and in linear malto-oligosaccharides. This enzyme has a long N-terminal extension that is conserved among archaic hyperthermophilic amylases but is not found in other hydrolyzing enzymes from the glycoside hydrolase 13 family. The SMMA crystal structure revealed that the N-terminal extension forms an N′ domain that is similar to carbohydrate-binding module 48, with the strand-loop-strand region forming a part of the substrate binding pocket with several aromatic residues, including Phe-95, Phe-96, and Tyr-99. A structural comparison with conventional cyclodextrin-hydrolyzing enzymes revealed a striking resemblance between the SMMA N′ domain position and the dimeric N domain position in bacterial enzymes. This result suggests that extremophilic archaea that live at high temperatures may have adopted a novel domain arrangement that combines all of the substrate binding components within a monomeric subunit. The SMMA structure provides a molecular basis for the functional properties that are unique to hyperthermophile maltogenic amylases from archaea and that distinguish SMMA from moderate thermophilic or mesophilic bacterial enzymes.
Introduction
Many organisms live in physically or geochemically extreme conditions that are detrimental to most organisms. Most eukaryotic organisms cannot tolerate temperatures higher than 50 °C, due to the sensitivity of certain cellular components. The discovery of hyperthermophilic microorganisms living near, at, and above 100 °C has revolutionized scientific thought in this area, and many scientists have become interested in such systems because they have developed specific physicochemical characteristics. Due to these particular properties, enzymes from extremophiles offer great potential for basic research and for biotechnological applications. For example, most industrial starch processes require high temperatures for liquefaction and saccharification. Therefore, there is enthusiastic interest in finding new sources of thermostable amylolytic enzymes.
Recently, we reported a novel maltogenic amylase from Staphylothermus marinus (1) that was isolated from geothermal sediments from a “black smoker” on the ocean floor (2). Maltogenic amylase is an enzyme that is widely used in the starch industry. This enzyme exhibits dual activity for α-d-1,4- and α-d-1,6-glucosidic bond cleavage, which differs from the classic α-amylases in the glycoside hydrolase 13 (GH13)5 family (3). SMMA has an optimal temperature of 100 °C and a 109 °C melting temperature with enzymatic activity under acidic conditions (pH 3.5–5.0), which is a favorable property for industrial applications (2, 4). The conversion of starch at a high temperature and low pH offers several advantages, including higher substrate solubility, decreased viscosity, better bacterial decontamination, and increased reaction rates (4–8).
The GH13 family includes α-amylases and closely related subfamilies, such as maltogenic amylase (EC 3.2.1.133), neopullulanase (EC 3.2.1.135), Thermoactinomyces vulgaris amylase II (TVAII), and cyclomaltodextrinase (CDase; EC 3.2.1.54) (3). Maltogenic amylase shares catalytic characteristics with neopullulanase and CDase, which catalyze the hydrolysis of cyclodextrins (CDs), pullulan, and acarbose, and which are collectively known as CD-hydrolyzing enzymes (9). These enzyme specificities were proposed to establish the so-called neopullulanase subfamily of the α-amylase family GH13 (10), which is currently classified as the subfamily GH13_20 (11). CD-hydrolyzing enzymes possess a common domain at the N terminus (N domain) that is involved in substrate binding through a domain-swapped homodimeric structure (12–16). The narrow, deep active site generated by the N domain from another subunit in the dimer is responsible for the substrate preference, for example, for small substrates and CDs. SMMA has enzyme activity similar to the CD-hydrolyzing enzymes with broad substrate specificity.
Interestingly, the primary structure analysis of SMMA revealed that most thermostable maltogenic amylases from archaea, such as S. marinus (1), Thermofilum pendens Hrk5 (17), Thermoplasma volcanium GSS1 (18), and Pyrococcus furiosus (19), have a longer motif at the N-terminal region that is 220–250 amino acids long. However, other bacterial maltogenic amylases and CD-hydrolyzing enzymes have N-terminal regions with only 120–140 amino acids that in the CAZy (Carbohydrate-active Enzyme) server have been classified as the carbohydrate-binding module (CBM) family 34, based on the observation that this domain from the related α-amylase I (TVAI) in T. vulgaris binds carbohydrates (20, 21).
Because it is the most thermostable maltogenic amylase yet reported, we studied the SMMA structure for detailed information about its function. In this study, we show the three-dimensional structure of the enzyme, which reveals a unique domain arrangement in the active site associated with the N-terminal region that distinguishes the archaeal maltogenic amylase from classic bacterial maltogenic amylases.
EXPERIMENTAL PROCEDURES
Protein Purification
The strain Escherichia coli MC1061 (F-7, araD139, Δ(ara-leu)7696, galE15, galK16, Δ(lac)X74, rpsL,(Strr), hsdR2, (rk−mk+), mcrA, and mcrB1) harboring pSMMA6xH was cultured for 20 h in 3 liters of Luria-Bertani broth. The cells were collected by centrifugation (8000 × g, 30 min), resuspended in 300 ml of lysis buffer (50 mm Tris-HCl, pH 7.5, 300 mm NaCl, and 10 mm imidazole), and sonicated. The supernatant was collected by centrifugation (10,000 × g, 30 min, 4 °C) and heated at 70 °C for 20 min to remove the thermolabile E. coli proteins. The crude enzyme was further purified using a nickel-NTA Superflow® column, as described previously (14). The active fractions in the elution buffer were dialyzed against 50 mm Tris-HCl buffer (pH 7.5).
Crystallization and Data Collection
SMMA crystallization trials were conducted using the sitting drop method at 18 °C. We mixed 1.5 μl of a 14 mg/ml SMMA solution with an equal volume of crystallization reservoir solution containing 12% polyethylene glycol (PEG) 4000, 2% isopropyl alcohol, 0.1 m ADA, pH 6.5, and 0.1 m Li2SO4. Before data collection, rhombus-type crystals were cryocooled to 95 K using a cryoprotectant consisting of mother liquor supplemented with 25% glycerol. The crystal diffracted to a resolution of 2.28 Å, and the data were collected with a 1° rotation and a total of 340 frames.
Structure Determination and Refinement
Diffraction data were processed and scaled using HKL2000 (22). The structure was determined using the molecular replacement method with the Phaser CCP4 suite (23) and the neopullulanase from Bacillus stearothermophilus (Protein Data Bank entry 1J0H) with the N domain omitted. The resulting model was refined through model rebuilding using CNS (24). COOT (25) was used for stereographic manual refinement and model building. The structure was validated with PROCHECK (26). Structure-based sequence alignments were generated using ClustalW (27). Molecular images, including schematics and stick figures, were produced using PyMOL (28). The detailed statistics for data collection and refinement are listed in Table 1.
TABLE 1.
Data collection and structure-solution parameters
| Parameters | Values |
|---|---|
| Crystal type | Native |
| Unit cell parameters (Å) | a = 65.39, b = 117.51, c = 199.04 |
| Resolution (Å) | 50–2.28 |
| Space group | P212121 |
| Completeness (%) | 85.6 (72.9)a |
| Rsym (%)b | 4.8 (19.8) |
| I/σ(I) | 37.1 (4.9) |
| No. of refined atoms: protein/water | 11,565/385 |
| Rfactor/Rfree (%)c | 18.7/23.7 |
| r.m.s. deviation bond length (Å) | 0.012 |
| r.m.s. deviation bond angle (degrees) | 1.116 |
| Ramachandran plot (%) | |
| Most favored region | 1293 (94.6%) |
| Additionally allowed region | 74 (5.4%) |
| Outlier region | 0 (0%) |
a Numbers in parentheses are statistics from the highest resolution shell.
b Rsym = |Iobs − Iavg|/Iobs, where Iobs is the observed individual reflection, and Iavg is the average over all symmetry equivalents.
c R factor = |Fo| − |Fc‖/|Fo|, where Fo and Fc are the observed and calculated structure factor amplitudes, respectively. Rfree was calculated using 5% of the data.
Molecular Modeling
The SMMA and β-cyclodextrin ligand complex model was constructed by overlaying the SMMA structure onto the ThMA complex structure (Protein Data Bank entry 1GVI). The substrate was optimized manually prior to energy minimization by using the steepest descent method with an 8-Å cut-off for 300 iterations using InsightII (Accerlys, San Diego, CA).
Amino Acid Sequence Alignments for Genes and Proteins
A BLAST (29) search on SMMA (GenBankTM accession number ABN69720.1) showed that the encoded protein has 23–28% identity with other GH13 proteins, including cyclomaltodextrinase from Thermococcus sp. B1001 (30), maltogenic amylase from Thermus sp. IM6501 (14), neopullulanase from B. stearothermophilus (31), and T. vulgaris α-amylase II (13).
Kinetic Parameters and Reaction Product Analysis
The copper bicinchoninate method was used to measure the concentration of reducing products to determine activity and kinetic parameters of SMMA (15, 32). For thin layer chromatography (TLC) analysis, the reaction products were spotted onto Whatman K5F silica gel plates (Whatman plc, Maidstone, UK) and developed using isopropyl alcohol/ethyl acetate/water (3:1:1, v/v/v) as the solvent system.
Analysis of Surface Tyr and Trp Residues
Proximity to the end of a secondary structure element is defined as being within 4 amino acids for helix and within 1 amino acid for strand, where the termini are assigned from the Protein Data Bank (33). Structures used in the analysis are Protein Data Bank entries 1GVI (ThMA), 1J0H (NPase), 1JI2 (TVAII), and 1EA9 (CDase). Residues in the exposed surfaces were identified using the Areaimol program (34).
RESULTS
Overall Structure
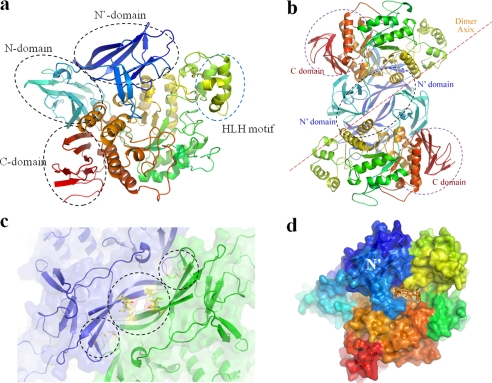
The crystal structure revealed that SMMA comprises four domains: the N, catalytic, and C domains, which are observed in most CD-hydrolyzing enzymes, and an additional novel N-terminal domain, the N′ domain, which was first observed in this study (Fig. 1a). Initially, the structure was determined and refined to a 2.28 Å resolution using molecular replacement, with the catalytic and C domains of neopullulanase from B. stearothermophilus (Protein Data Bank entry 1J0H) as the template structure. An attempt at molecular replacement with the entire three-domain region failed, which may have been due to the significantly altered orientation and geometry of the SMMA N domain (r.m.s. deviation of 2.3 Å for 463 Cα atoms). During manual model building for the N-terminal region, the electron density of the region showed two vague, separate “blobs,” which allowed the detection of the N (aa 116–219) and N′ (aa 1–115) domains of SMMA. The SMMA catalytic domain displays a conserved (β/α) 8-barrel fold with a distinct loop (aa 342–397) protruding from the barrel. Most CD-hydrolyzing enzymes have a protruding loop located near the active site, which is called the B domain and forms a portion of the substrate binding groove for subsites −2, −3, or −4. However, SMMA has a much longer insertion of aa 342–397 in this region, creating a larger domain at the entrance of the groove (Fig. 1b). In this groove, Tyr-389, which is in the middle of a helix in aa 385–391, forms an entrance gate with Tyr-257. Because SMMA and other CD-hydrolyzing enzymes from archaea have this insertion, the protruding region might serve as a signature that is unique to maltogenic amylases from archaea. Another sequence-structural feature is the presence of a glycine in the i + 4 position after the catalytic nucleophile Asp-442 (Fig. 2) because typical maltogenic amylases (i.e. members of the neopullulanase subfamily) have a glutamate in that position as a part of the four-residue signature VANE (10).
FIGURE 1.
Overall SMMA structure. a, a schematic overview of an SMMA monomer that shows the conserved N, catalytic, and C domains in CD-hydrolyzing enzymes with a novel N′ domain. The monomer is colored in a spectrum; the N terminus is in blue, and the C terminus is in red. b, the SMMA dimer structure is shown with a 2-fold axis perpendicular to the plane. c, the dimeric interface between the N′ domains is shown with the adjacent hydrophobic and charged interactions. d, a surface diagram of the monomer with a hypothetical cyclodextrin molecule (orange) is generated by superposition with the binary complex structure (Protein Data Bank entry 1GVI) to highlight the active site.
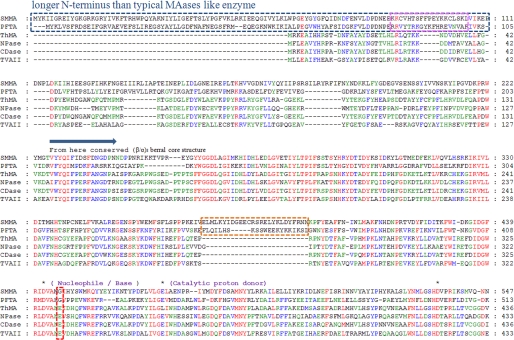
FIGURE 2.
Sequence alignment for SMMA, PFTA, ThMA, NPase, TVAII, and CDase. The sequences of six CD-hydrolyzing enzymes, two hyperthermophilic archaic maltogenic amylases, and four conventional bacterial CD-hydrolyzing enzymes were aligned using ClustalW (27). The novel SMMA N′ domain is indicated with a blue dotted line. The helix-loop-helix regions observed in the structure are indicated with orange dotted lines. The β-7-loop 7-β-8 region protruding from the N′ domain is indicated with a pink dotted line. The three conserved catalytic residues in the GH13 family are indicated with black asterisks. Red, blue, and green highlighting indicates the residues that are conserved in 100, 75, and 50% of the proteins, respectively. The accession numbers for these sequences are ABN69720.1 (SMMA), AAL82063.1 (PFTA from Pyrococcus furiosus), AAC15072.1 (ThMA from Thermus sp. strain IM6501), AAA22622.1 (NPase from B. stearothermophilus), BAA02473.1 (TVAII), and AAA92925.1 (CDase from alkalophilic Bacillus sp. I-5).
A Dali search (35) using the SMMA structure identified the neopullulanase from B. stearothermophilus (29% identity) as its closest structural homologue, with a 2.6 Å r.m.s. deviation (536 Cα atoms). The α amylase II (TVAII) from T. vulgaris R-47 (30% identity) was identified as the second closest homologue with a 2.6 Å r.m.s. deviation (537 Cα atoms). The loop regions comprising aa 236–264 and 653–689 had high B-factors (61.3 average) at the surface, and the electron density map for the loop of aa 671–677 in the C domain was too weak to build a model, which suggests that it may be flexible. SMMA forms a homodimer via an interaction between the adjacent, novel N′ domains, which have a 2-fold axis perpendicular to the arc shape of the β-strands' interface (Fig. 1c). Each monomer is primarily associated through hydrophobic interactions at the center of the region of aa 5–19 (Ile-5 and -19 from one molecule against Ile-9 from the other). This interaction is supplemented by salt bridges (Arg-181/Asp-422 and Arg-50/Glu-198) at both ends of the strands, which yield a 2140.7 Å2 interface (Fig. 1d). Most CD-hydrolyzing enzymes form dimers with the N domain intertwined. However, the SMMA dimer configuration is different from previously reported CD-hydrolyzing enzymes, in that the dimer is arranged with adjacent monomers and an interface unrelated to the active sites.
CBM48 Topology of the N′ Domain
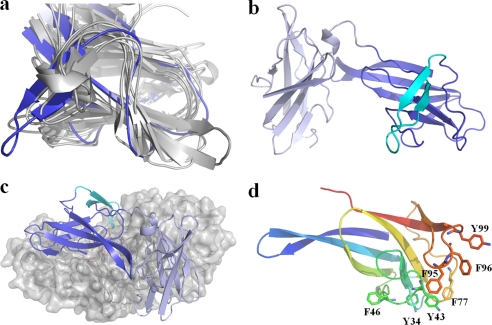
The long N-terminal region of SMMA includes two repeated motifs, the N and N′ domains, with a β-sandwich fold (6.2 Å r.m.s. deviation for 12 Cα atoms) (Fig. 3a). A structural homology search for the truncated N′ domain generated the β-subunit of the 5′-AMP-activated protein kinase (AMPK) with a 1.9 Å r.m.s. deviation over 80 residues (Z score, 10.2) using the Dali and 1.71 Å over 80 residues (Z score, 6.2) using the SSM server (36). The AMPK β subunit, which is known to bind glycogen, belongs to CBM48 (37). Currently, the CBM48 family has more than 3000 entries in the CAZy database, and 15 entries have structural information available (38). Despite the low overall sequence similarity, all of the structures superimposed well onto the SMMA N′ domain and share 6–8 β-strands from the β-sandwich fold, except for the long extended loop of aa 88–110 between the 7th and 8th β strands in SMMA (Fig. 3b). Eight of 15 structures have protruding loops that correspond to the loop of aa 88–110 in SMMA, but the SMMA β-strand loop is much longer (8.2 Å) than the other corresponding loops. The AMPK β subunit has a corresponding loop with the Leu-146 residue at the tip that interacts with cyclodextrin through several aromatic residues along the surfaces of the other β-strands to aid in the carbohydrate stacking interaction (37). In the SMMA N′ domain, Phe-95, Phe-96, and Tyr-99 are along the loop, and numerous aromatic residues, such as Tyr-43, Phe-46, and Phe-77, lie on the β-sandwich fold surfaces (Fig. 3d). Interestingly, the SMMA N′ domain is in the position of the N domain in other neopullulanase subunits, with its protruding loop being extended to the active site of the catalytic domain. With this orientation and geometry, the SMMA N′ domain forms part of the active site pocket via residues Phe-95, Phe-96, and Tyr-99, which make up the substrate binding groove.
FIGURE 3.
CBM48 topology in the N′ domain. a, superposition of the eight CBM48 structures that carry the loop protruding from the seventh and eighth β-strands to the N′ domain in SMMA. b, schematic diagram showing the antiparallel β-sandwich fold for the N and N′ domains. The loop protruding from the N′ domain is colored cyan. c, the positions of the N and N′ domains at the surface of the remaining portion of the SMMA molecule. d, aromatic residues located near the loop protruding from the N′ domain. The domain has been colored in a spectrum; the N terminus is in blue, and the C terminus is in orange.
Shaping Active Site with Novel N′ Domain
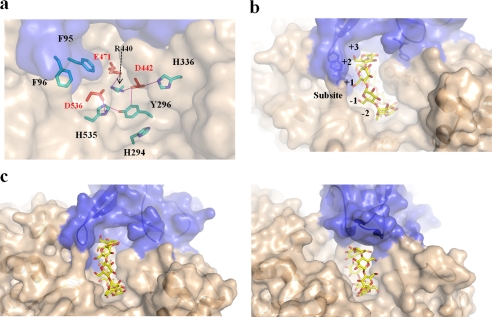
In SMMA, the substrate binding pocket was easily identified by the three highly conserved catalytic residues Asp-442, Glu-471, and Asp-536 at the bottom of the pocket (Fig. 4a). This active site pocket is located at the center of the (β/α)8 domain, is composed partly of the catalytic domain regions (residues 285–310, 397–417, 471–475, 534–546, and 568–594) and the protruding N′ domain loop, and is 14.8 Å deep, 18.33 Å long, and 9.1 Å wide (Fig. 4b). The positions and orientations of the Asp-442 nucleophile and the Glu-471 acid/base are in appropriate ranges for retaining enzymatic activity because the carboxylate group of Asp-536 is held in position by a hydrogen bond to the nitrogen atoms (Nδ1 and Nϵ2) in His-535 (Fig. 4a). Similarly, the oxygen atoms (Oϵ1 and Nϵ2) in the Asp-442 carboxylate group are hydrogen-bonded to His-336 (Nϵ2), Tyr-296, and Arg-440 (NH1), and these positions may be responsible for maintaining the ionization state of the nucleophile. The pocket contains several aromatic residues; Phe-404 and Phe-405 form subsite −1, and Tyr-296 provides the essential stacking interaction with the sugar ring at subsite −1 that is conserved in GH13 and forms a hydrogen bond with the nucleophile Asp-442.
FIGURE 4.
SMMA active site. a, catalytic (red) and related subsite residues (cyan) are shown as sticks. The hydrogen bond network in the active site is shown with the additional aromatic N′ domain residues (blue). b, a surface representation of the active site pocket is shown with a hypothetical CD molecule modeled at the active site. c, a structural comparison of the active sites in SMMA (left) and NPase (right) from B. stearothermophilus (Protein Data Bank entry 1J0H); the substrate-binding grooves at the active sites are shown.
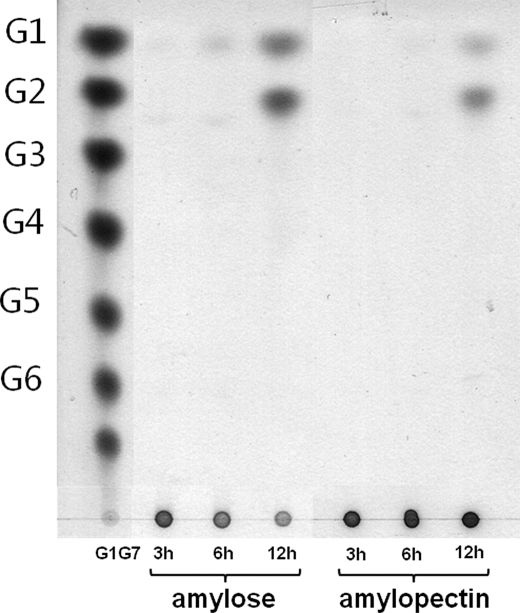
The active site pockets in bacterial CD-hydrolyzing enzymes are generated by the N domain from the other subunit and yield a groove that is slightly extended between the catalytic domain and the N domain, probably above subsites +3 and +4 (Fig. 4d). In comparison, the SMMA active site pocket is generated by the N′ domain of the same subunit without a significant groove above the subsites +3 and +4, at which the position of the putative subsite +3 is filled and blocked by Phe-405 and Phe-77 from the N′ domain (Fig. 4c). In addition, the N domain in bacterial CD-hydrolyzing enzymes protrudes above the surface and forms a structural lid for the active site pocket, whereas the SMMA N′ domain does not have the structural lid, which may explain the lack of SMMA transfer activity. Glu-332, which is known to be a key residue in ThMA transglycosylation activity, is replaced by Gly in SMMA (39). Because the active site pocket is significantly altered in SMMA, we have examined the degradation pattern with the long-chain substrates amylose and amylopectin. As shown in Fig. 5, SMMA hydrolyzes both amylose and amylopectin to produce maltose and glucose exclusively. A kinetic analysis in Table 2 showed that SMMA has lower Km value for amylose substrate (0.27) than for amylopectin (0.51), which exhibits a similar pattern to classic maltogenic amylases. However, Km values for both substrates were significantly lower than those of some maltogenic enzymes. For amylose substrate, it is 5-fold lower than that of CDase (0.27 versus 1.51) and 70-fold lower than that of ThMA (0.27 versus 19.7). For amylopectin, it is at least 2 orders of magnitude lower than CDase and ThMA (0.51 versus 55.15 and 0.51 versus 44.5, respectively), whereas the specificity constant kcat/Km is similar (1.17 versus 0.92 and 1.17 versus 3.03, respectively), which suggests that SMMA has a high affinity for polysaccharide substrates. SMMA showed a uniquely higher kcat value for γ-CD substrate than for α-CD and β-CD.
FIGURE 5.
SMMA enzymatic characteristics for amylose and amylopectin substrates. A time course analysis of SMMA with amylose and amylopectin from potato was shown. The reaction products of glucose and maltose were further analyzed (reaction conditions: SMMA 0.6 unit/mgsub, substrate 0.25%, 50 mm NaOAc, pH 5.0, 92 °C).
TABLE 2.
Kinetic parameters for some substrates of SMMA
| Enzyme | Substrate | kcat | KM | kcat/KM | Source/Reference | ||
|---|---|---|---|---|---|---|---|
| s−1 | mg ml−1 | mm | s−1ml mg−1 | s−1mm−1 | |||
| SMMA | Amylose | 0.80 | 0.27 | 2.90 | This study | ||
| Amylopectin | 0.59 | 0.51 | 1.17 | This study | |||
| α-Cyclodextrin | 1.45 | 0.43 | 3.37 | This study | |||
| β-Cyclodextrin | 5.36 | 1.48 | 3.62 | This study | |||
| γ-Cyclodextrin | 29.10 | 2.68 | 10.86 | This study | |||
| CDase I-5 | Amylose | 22.11 | 1.52 | 14.60 | Ref. 5 | ||
| Amylopectin | 50.36 | 55.15 | 0.92 | Ref. 5 | |||
| β-Cyclodextrin | 78.18 | 0.515 | 151.67 | Ref. 5 | |||
| ThMA | Amylose | 96.4 | 19.7 | 4.89 | Ref. 72 | ||
| Amylopectin | 135 | 44.5 | 3.03 | Ref. 72 | |||
| β-Cyclodextrin | 126 | 0.174 | 726.0 | Ref. 15 | |||
| TVAII | β-Cyclodextrin | 43.40 | 1.31 | 33.13 | Ref. 16 | ||
SMMA Thermostability
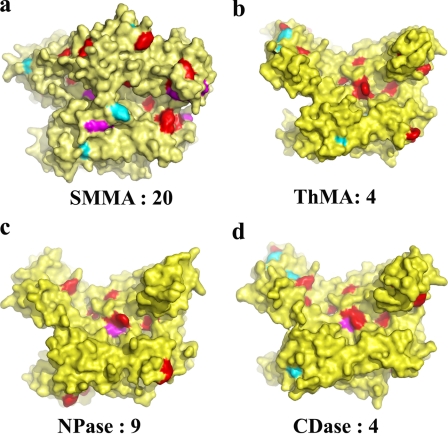
The reported SMMA melting temperature is 109 °C; thus, it is the most thermostable maltogenic amylase reported to date. Previously, the high frequency of surface hydrophobic residues with bulky side chains, such as Trp and Tyr in the α-helices and β-strand termini, were reported to be related to the thermostability of many proteins (33). Thus, we have investigated the distribution of Trp and Tyr in comparison with CDase, ThMa, neopullulanase, and TVAII. The structural analysis showed that a significantly high proportion of Tyr and Trp residues are located on the solvent-accessible surface throughout the entire molecule (Table 3). Although the total number of Tyr and Trp residues is not significantly higher than in the other enzymes, 67 (9.6%) compared with 44 (7.7%), these residues are primarily located on the surface in SMMA. Fig. 6 shows that the relative surface distribution of Tyr and Trp at the termini of the secondary structure is much wider than in those bacterial enzymes. Many extremophiles use sugars as a compatible solute to ease osmotic stress, and such solutes stabilize proteins, which may explain the abundance of aromatic residues that can bind carbohydrate sugars at the protein surface (33, 40–42). Amino acid analysis showed that SMMA has a significantly higher frequency of Ile (74, 10.6%) than do the other four enzymes (average 24, 4.0%), but it has significantly lower Gln content (9, 1.3%). Ile occurs often in thermophilic proteins, and Gln is a labile amino acid that is deaminated at high temperatures, which suggests that known structural factors involved in thermostability correlate well with the SMMA structure (43–45).
TABLE 3.
Relationship between thermostability and the ratio of residues Trp and Tyr
| Enzyme | Temperaturea | Total number of residues | Total number (Tyr/Trp)b | Terminal numberc | Surface numberd | Surface number/total number | Relative numbere |
|---|---|---|---|---|---|---|---|
| ºC | % | ||||||
| SMMA | 100 | 696 | 67 | 27 | 20 | 2.87 | 100 |
| ThMA | 60 | 588 | 45 | 18 | 4 | 0.68 | 23.7 |
| NPase | 50 | 588 | 48 | 15 | 9 | 1.53 | 53.3 |
| TVAII | 50 | 585 | 39 | 13 | 4 | 0.68 | 23.7 |
| CDase | 40–50 | 583 | 42 | 18 | 4 | 0.68 | 23.7 |
a The temperature indicates the optimal temperatures for maximum enzyme activity.
b The number of total Tyr and Trp residues.
c The number of Tyr and Trp residues near the end of a secondary structure element.
d The number of Tyr and Trp residues near the end of a secondary structure element and exposed at the surface.
e The relative value of the surface number to the surface number of SMMA.
FIGURE 6.
Surface distribution of aromatic residues Tyr and Trp in CD-hydrolyzing enzymes. Aromatic rings on the protein surface are colored red in SMMA (a), ThMA (b), NPas (c), and CDase (d). Residues at the termini of β-strands and α-helices are highlighted by cyan and magenta, respectively. The number of Tyr and Trp residues that are located at the termini of secondary structure elements and exposed at the surface is indicated.
DISCUSSION
Certain CD-hydrolyzing amylases have much longer extended regions in the N terminus, despite their high sequence homology with the remaining classic CD-hydrolyzing enzymes. Thus, the functional role of this region in amylase activity is especially interesting. Given the high sequence homology of this extended region among archaic enzymes, it is likely that the CD-hydrolyzing enzymes from hyperthermophilic archaea, such as Staphylothermus hellenicus (46), Thermococcus barophilus (47), Thermococcus kodakarensis (48), Thermococcus onnurineus (49), T. pendens (50), T. volcanium GSS1 (18), and P. furiosus (51), have extra domains with the CBM48 fold.
CBMs have been divided into 64 families based on amino acid sequence similarity (38). Previously, CBMs were thought to be motifs that are functionally independent from the catalytic domains located distal to the active sites, and it has been proposed that they enhance the interaction between the carbohydrate substrate and protein (52–55). For many amylolytic enzymes, CBMs were reportedly involved in starch hydrolysis by disrupting the starch granule structure, which allowed for a concentration of catalytic domains on the surface and carbohydrate starch hydrolysis by proximity (56–59). For example, CBM20 in CGTases is involved in binding and guiding linear starch chains (60). Although the evolutionary history of CBM48 has been thoroughly elucidated, indicating that it reflects the evolution of specificities, rather than the evolution of species (61), the detailed function for CBM48 is unknown. The only exception is represented by the β-subunit of AMPK (37), which showed that CBM48 is a separate domain that binds cyclodextrin to increase its binding ability for the glucosyl polymeric structures commonly found in glycogen (62). Despite the wealth of structural information on CBM48 (36, 52, 55, 63–67) (for a review, see Ref. 68)), this paper is, to the best of our knowledge, the first to demonstrate that an independent CBM domain folds to interact with the catalytic domain and participates in substrate binding at the active site. While it is adjacent to the catalytic domain, CBM48 in the N′ domain contacts the active site with the long extended loop that was previously shown to interact with cyclodextrin molecules in the AMPK β-subunit (37). The SMMA N′ domain extended loop has several aromatic residues, which is a suitable architecture for a stacking interaction with carbohydrate sugar rings. The reduced Km values toward amylose and amylopectin reported in this study may reflect the characteristic binding of CBM to carbohydrates. For CBM48 function at the active site, it is related to substrate specificity in the glycogen debranching enzymes from Deinococcus geothermalis and Deinococcus radiodurans (69). The CBM48 determines the branching pattern of glycogen at the active site, which suggests a functional role similar to the SMMA N′ domain. Thus, it would be interesting to relate the protein structure to its function (69).
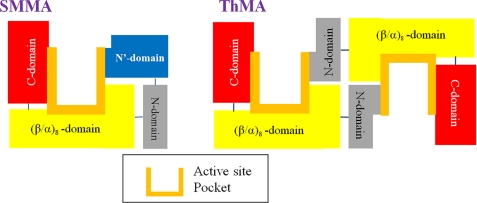
Previously, we reported that substrate binding in ThMA was influenced by the direction and arrangement of several loops located in the N domain (15). The striking similarity of the position and orientation of the SMMA N′ domain to the N domain in bacterial maltogenic amylases in this study indicates a common functional role in the substrate binding at the active site (Fig. 7). Interestingly, the SMMA N domain lacks the corresponding functional residues, such as Tyr-45 and Trp-47, which are involved in substrate recognition in ThMA (15). We suggest that the SMMA N′ domain plays the functional role of the N domain in moderate thermophile or mesophile maltogenic amylases.
FIGURE 7.
Schematic view of the SMMA active site configuration. Domains involved in the substrate binding are drawn schematically for SMMA (left) and for bacterial CD-hydrolyzing enzymes (right) to show all of the components for maltogenic activity in a single monomer.
The interface between the N′ domain and other domains in the SMMA monomeric subunit showed much stronger and tighter interactions than did the interface of the N domain in bacterial maltogenic amylases. The N′ domain is primarily associated with a 1946.0 Å2 buried interface via hydrophobic interactions, whereas the N domain in ThMA has a distribution of highly charged residues at the interface with a 1338.8-Å2 buried surface. It has been reported that high salt concentrations change the oligomeric state of bacterial maltogenic amylases (70). Although oligomerization might not be necessary for the activity of the GH13 CD-hydrolyzing enzymes, it does contribute to their high specificity (71). The charged group in the hydrophobic interface may have direct effects on thermal resilience and serve as a doorway for water molecules. High temperatures would contribute to thermal destabilization by introducing water permeation, which would result in structure perturbation. Taken together, a modified structure that positions all of the components for substrate recognition within a monomer may have been adopted to allow the archaea hyperthermophilic maltogenic enzymes to retain activity at high temperatures. The structural features of SMMA may also provide a molecular basis for engineering substrate preference and thermal stability in the starch-processing industry.
Acknowledgment
We thank the beamline staff at Photon Factory, KEK, Japan, for data collection and technical assistance.
This research was supported in part by the Marine and Extreme Genome Research Center program from the Ministry of Land, Transport, and Maritime Affairs, Republic of Korea, and the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science, and Technology (Grants 2009-0083290/2011-0017040).
The atomic coordinates and structure factors (code 4AEE) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- GH13
- glycoside hydrolase 13
- ADA
- N-(2-acetamido)-2-iminodiacetic acid
- ThMA
- maltogenic amylase from Thermus sp. strain IM6501
- NPase
- neopullulanase from B. stearothermophilus
- CDase
- cyclodextrinase from alkalophilic Bacillus sp. I-5
- TVAI and TVAII
- T. vulgaris amylase I and II, respectively
- SMMA
- S. marinus maltogenic amylase
- PFTA
- P. furiosus thermostable amylase
- CD
- cyclodextrin
- CBM
- carbohydrate-binding module
- r.m.s.
- root mean square
- aa
- amino acids
- AMPK
- 5′-AMP-activated protein kinase.
REFERENCES
- 1. Li D., Park J. T., Li X., Kim S., Lee S., Shim J. H., Park S. H., Cha J., Lee B. H., Kim J. W., Park K. H. (2010) Overexpression and characterization of an extremely thermostable maltogenic amylase, with an optimal temperature of 100 degrees C, from the hyperthermophilic archaeon Staphylothermus marinus. New Biotechnol. 27, 300–307 [DOI] [PubMed] [Google Scholar]
- 2. Fiala G., S. K., Jannasch H. W., Langworthy T. A., Madon J. (1986) Staphylothermus marinus sp. nov. represents a novel genus of extremely thermophilic submarine heterotrophic archaebacteria growing up to 98 °C. Syst. Appl. Microbiol. 8, 106–113 [Google Scholar]
- 3. MacGregor E. A., Janecek S., Svensson B. (2001) Relationship of sequence and structure to specificity in the α-amylase family of enzymes. Biochim. Biophys. Acta 1546, 1–20 [DOI] [PubMed] [Google Scholar]
- 4. Unsworth L. D., van der Oost J., Koutsopoulos S. (2007) Hyperthermophilic enzymes. Stability, activity, and implementation strategies for high temperature applications. FEBS J. 274, 4044–4056 [DOI] [PubMed] [Google Scholar]
- 5. Auh J. H., Chae H. Y., Kim Y. R., Shim K. H., Yoo S. H., Park K. H. (2006) Modification of rice starch by selective degradation of amylose using alkalophilic Bacillus cyclomaltodextrinase. J. Agric. Food Chem. 54, 2314–2319 [DOI] [PubMed] [Google Scholar]
- 6. Kawamura S., Kakuta Y., Tanaka I., Hikichi K., Kuhara S., Yamasaki N., Kimura M. (1996) Glycine 15 in the bend between two α-helices can explain the thermostability of DNA-binding protein HU from Bacillus stearothermophilus. Biochemistry 35, 1195–1200 [DOI] [PubMed] [Google Scholar]
- 7. Kim Y. W., Choi J. H., Kim J. W., Park C., Kim J. W., Cha H., Lee S. B., Oh B. H., Moon T. W., Park K. H. (2003) Directed evolution of Thermus maltogenic amylase toward enhanced thermal resistance. Appl. Environ. Microbiol. 69, 4866–4874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vieille C., Zeikus G. J. (2001) Hyperthermophilic enzymes: Sources, uses, and molecular mechanisms for thermostability. Microbiol. Mol. Biol. Rev. 65, 1–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Park K. H., Kim T. J., Cheong T. K., Kim J. W., Oh B. H., Svensson B. (2000) Structure, specificity and function of cyclomaltodextrinase, a multispecific enzyme of the α-amylase family. Biochim. Biophys. Acta 1478, 165–185 [DOI] [PubMed] [Google Scholar]
- 10. Oslancová A., Janecek S. (2002) Oligo-1,6-glucosidase and neopullulanase enzyme subfamilies from the alpha-amylase family defined by the fifth conserved sequence region. Cell Mol. Life Sci. 59, 1945–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stam M. R., Danchin E. G., Rancurel C., Coutinho P. M., Henrissat B. (2006) Dividing the large glycoside hydrolase family 13 into subfamilies. Towards improved functional annotations of α-amylase-related proteins. Protein Eng. Des. Sel. 19, 555–562 [DOI] [PubMed] [Google Scholar]
- 12. Hondoh H., Kuriki T., Matsuura Y. (2003) Three-dimensional structure and substrate binding of Bacillus stearothermophilus neopullulanase. J. Mol. Biol. 326, 177–188 [DOI] [PubMed] [Google Scholar]
- 13. Kamitori S., Abe A., Ohtaki A., Kaji A., Tonozuka T., Sakano Y. (2002) Crystal structures and structural comparison of Thermoactinomyces vulgaris R-47 α-amylase 1 (TVAI) at 1.6 Å resolution and α-amylase 2 (TVAII) at 2.3 Å resolution. J. Mol. Biol. 318, 443–453 [DOI] [PubMed] [Google Scholar]
- 14. Kim T. J., Kim M. J., Kim B. C., Kim J. C., Cheong T. K., Kim J. W., Park K. H. (1999) Modes of action of acarbose hydrolysis and transglycosylation catalyzed by a thermostable maltogenic amylase, the gene for which was cloned from a Thermus strain. Appl Environ. Microbiol. 65, 1644–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee H. S., Kim M. S., Cho H. S., Kim J. I., Kim T. J., Choi J. H., Park C., Lee H. S., Oh B. H., Park K. H. (2002) Cyclomaltodextrinase, neopullulanase, and maltogenic amylase are nearly indistinguishable from each other. J. Biol. Chem. 277, 21891–21897 [DOI] [PubMed] [Google Scholar]
- 16. Ohtaki A., Mizuno M., Yoshida H., Tonozuka T., Sakano Y., Kamitori S. (2006) Structure of a complex of Thermoactinomyces vulgaris R-47 α-amylase 2 with maltohexaose demonstrates the important role of aromatic residues at the reducing end of the substrate binding cleft. Carbohydr Res. 341, 1041–1046 [DOI] [PubMed] [Google Scholar]
- 17. Li X., Li D., Yin Y., Park K. H. (2010) Characterization of a recombinant amylolytic enzyme of hyperthermophilic archaeon Thermofilum pendens with extremely thermostable maltogenic amylase activity. Appl. Microbiol. Biotechnol. 85, 1821–1830 [DOI] [PubMed] [Google Scholar]
- 18. Kawashima T., Amano N., Koike H., Makino S., Higuchi S., Kawashima-Ohya Y., Watanabe K., Yamazaki M., Kanehori K., Kawamoto T., Nunoshiba T., Yamamoto Y., Aramaki H., Makino K., Suzuki M. (2000) Archaeal adaptation to higher temperatures revealed by genomic sequence of Thermoplasma volcanium. Proc. Natl. Acad. Sci. U.S.A. 97, 14257–14262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang S. J., Lee H. S., Park C. S., Kim Y. R., Moon T. W., Park K. H. (2004) Enzymatic analysis of an amylolytic enzyme from the hyperthermophilic archaeon Pyrococcus furiosus reveals its novel catalytic properties as both an α-amylase and a cyclodextrin-hydrolyzing enzyme. Appl. Environ. Microbiol. 70, 5988–5995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Abe A., Tonozuka T., Sakano Y., Kamitori S. (2004) Complex structures of Thermoactinomyces vulgaris R-47 α-amylase 1 with malto-oligosaccharides demonstrate the role of domain N acting as a starch-binding domain. J. Mol. Biol. 335, 811–822 [DOI] [PubMed] [Google Scholar]
- 21. Machovic M., Janecek S. (2006) Starch-binding domains in the post-genome era. Cell Mol. Life Sci. 63, 2710–2724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Otwinowski Z., Minor M. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 23. Collaborative Computational Project, Number 4 (1994) The CCP4 suite. Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 24. Brünger A. T., Adams P. D., Rice L. M. (1998) Recent developments for the efficient crystallographic refinement of macromolecular structures. Curr. Opin. Struct. Biol. 8, 606–611 [DOI] [PubMed] [Google Scholar]
- 25. Emsley P., Cowtan K. (2004) Coot. Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 26. Laskowski R. A., Moss D. S., Thornton J. M. (1993) Main-chain bond lengths and bond angles in protein structures. J. Mol. Biol. 231, 1049–1067 [DOI] [PubMed] [Google Scholar]
- 27. Thompson J. D., Higgins D. G., Gibson T. J. (1994) ClustalW: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. DeLano W. L. (2010) The PyMOL Molecular Graphics System, version 1.3r1, Schrodinger, LLC, New York [Google Scholar]
- 29. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 30. Hashimoto Y., Yamamoto T., Fujiwara S., Takagi M., Imanaka T. (2001) Extracellular synthesis, specific recognition, and intracellular degradation of cyclomaltodextrins by the hyperthermophilic archaeon Thermococcus sp. strain B1001. J. Bacteriol. 183, 5050–5057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Imanaka T., Kuriki T. (1989) Pattern of action of Bacillus stearothermophilus neopullulanase on pullulan. J. Bacteriol. 171, 369–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fox J. D., Robyt J. F. (1991) Miniaturization of three carbohydrate analyses using a microsample plate reader. Anal. Biochem. 195, 93–96 [DOI] [PubMed] [Google Scholar]
- 33. Greaves R. B., Warwicker J. (2009) Stability and solubility of proteins from extremophiles. Biochem. Biophys. Res. Commun. 380, 581–585 [DOI] [PubMed] [Google Scholar]
- 34. Lee B., Richards F. M. (1971) The interpretation of protein structures. Estimation of static accessibility. J. Mol. Biol. 55, 379–400 [DOI] [PubMed] [Google Scholar]
- 35. Holm L., Kääriäinen S., Rosenström P., Schenkel A. (2008) Searching protein structure databases with DaliLite version 3. Bioinformatics 24, 2780–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Krissinel E., Henrick K. (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 [DOI] [PubMed] [Google Scholar]
- 37. Polekhina G., Gupta A., van Denderen B. J., Feil S. C., Kemp B. E., Stapleton D., Parker M. W. (2005) Structural basis for glycogen recognition by AMP-activated protein kinase. Structure 13, 1453–1462 [DOI] [PubMed] [Google Scholar]
- 38. Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009) The Carbohydrate-active enzymes database (CAZy). An expert resource for glycogenomics. Nucleic Acids Res. 37, D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kim T. J., Park C. S., Cho H. Y., Cha S. S., Kim J. S., Lee S. B., Moon T. W., Kim J. W., Oh B. H., Park K. H. (2000) Role of the glutamate 332 residue in the transglycosylation activity of Thermus maltogenic amylase. Biochemistry 39, 6773–6780 [DOI] [PubMed] [Google Scholar]
- 40. Greaves R. B., Warwicker J. (2007) Mechanisms for stabilization and the maintenance of solubility in proteins from thermophiles. BMC Struct. Biol. 7, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cambillau C., Claverie J. M. (2000) Structural and genomic correlates of hyperthermostability. J. Biol. Chem. 275, 32383–32386 [DOI] [PubMed] [Google Scholar]
- 42. Shoseyov O., Shani Z., Levy I. (2006) Carbohydrate binding modules. Biochemical properties and novel applications. Microbiol. Mol. Biol. Rev. 70, 283–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Volkov I. Iu., Lunina N. A., Velikodvorskaia G. A. (2004) Prospects for practical application of substrate-binding modules of glycosyl hydrolases (a review). Prikl. Biokhim. Mikrobiol. 40, 499–504 [PubMed] [Google Scholar]
- 44. Georis J., de Lemos Esteves F., Lamotte-Brasseur J., Bougnet V., Devreese B., Giannotta F., Granier B., Frère J. M. (2000) An additional aromatic interaction improves the thermostability and thermophilicity of a mesophilic family 11 xylanase. Structural basis and molecular study. Protein Sci. 9, 466–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. You C., Huang Q., Xue H., Xu Y., Lu H. (2010) Potential hydrophobic interaction between two cysteines in interior hydrophobic region improves thermostability of a family 11 xylanase from Neocallimastix patriciarum. Biotechnol. Bioeng. 105, 861–870 [DOI] [PubMed] [Google Scholar]
- 46. Arab H., Völker H., Thomm M. (2000) Thermococcus aegaeicus sp. nov. and Staphylothermus hellenicus sp. nov., two novel hyperthermophilic archaea isolated from geothermally heated vents off Palaeochori Bay, Milos, Greece. Int. J. Syst. Evol. Microbiol. 50, 2101–2108 [DOI] [PubMed] [Google Scholar]
- 47. Marteinsson V. T., Birrien J. L., Reysenbach A. L., Vernet M., Marie D., Gambacorta A., Messner P., Sleytr U. B., Prieur D. (1999) Thermococcus barophilus sp. nov., a new barophilic and hyperthermophilic archaeon isolated under high hydrostatic pressure from a deep-sea hydrothermal vent. Int. J. Syst. Bacteriol. 49, 351–359 [DOI] [PubMed] [Google Scholar]
- 48. Fukui T., Atomi H., Kanai T., Matsumi R., Fujiwara S., Imanaka T. (2005) Complete genome sequence of the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1 and comparison with Pyrococcus genomes. Genome Res. 15, 352–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lee H. S., Kang S. G., Bae S. S., Lim J. K., Cho Y., Kim Y. J., Jeon J. H., Cha S. S., Kwon K. K., Kim H. T., Park C. J., Lee H. W., Kim S. I., Chun J., Colwell R. R., Kim S. J., Lee J. H. (2008) The complete genome sequence of Thermococcus onnurineus NA1 reveals a mixed heterotrophic and carboxydotrophic metabolism. J. Bacteriol. 190, 7491–7499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Anderson I., Rodriguez J., Susanti D., Porat I., Reich C., Ulrich L. E., Elkins J. G., Mavromatis K., Lykidis A., Kim E., Thompson L. S., Nolan M., Land M., Copeland A., Lapidus A., Lucas S., Detter C., Zhulin I. B., Olsen G. J., Whitman W., Mukhopadhyay B., Bristow J., Kyrpides N. (2008) Genome sequence of Thermofilum pendens reveals an exceptional loss of biosynthetic pathways without genome reduction. J. Bacteriol. 190, 2957–2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Robb F. T., Maeder D. L., Brown J. R., DiRuggiero J., Stump M. D., Yeh R. K., Weiss R. B., Dunn D. M. (2001) Genomic sequence of hyperthermophile, Pyrococcus furiosus. Implications for physiology and enzymology. Methods Enzymol. 330, 134–157 [DOI] [PubMed] [Google Scholar]
- 52. Feese M. D., Kato Y., Tamada T., Kato M., Komeda T., Miura Y., Hirose M., Hondo K., Kobayashi K., Kuroki R. (2000) Crystal structure of glycosyltrehalose trehalohydrolase from the hyperthermophilic archaeum Sulfolobus solfataricus. J. Mol. Biol. 301, 451–464 [DOI] [PubMed] [Google Scholar]
- 53. Koay A., Rimmer K. A., Mertens H. D., Gooley P. R., Stapleton D. (2007) Oligosaccharide recognition and binding to the carbohydrate binding module of AMP-activated protein kinase. FEBS Lett. 581, 5055–5059 [DOI] [PubMed] [Google Scholar]
- 54. Machovic M., Svensson B., MacGregor E. A., Janecek S. (2005) A new clan of CBM families based on bioinformatics of starch-binding domains from families CBM20 and CBM21. FEBS J. 272, 5497–5513 [DOI] [PubMed] [Google Scholar]
- 55. Vester-Christensen M. B., Abou Hachem M., Svensson B., Henriksen A. (2010) Crystal structure of an essential enzyme in seed starch degradation. Barley limit dextrinase in complex with cyclodextrins. J. Mol. Biol. 403, 739–750 [DOI] [PubMed] [Google Scholar]
- 56. Bolam D. N., Ciruela A., McQueen-Mason S., Simpson P., Williamson M. P., Rixon J. E., Boraston A., Hazlewood G. P., Gilbert H. J. (1998) Pseudomonas cellulose-binding domains mediate their effects by increasing enzyme substrate proximity. Biochem. J. 331, 775–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Boraston A. B., Bolam D. N., Gilbert H. J., Davies G. J. (2004) Carbohydrate-binding modules. Fine-tuning polysaccharide recognition. Biochem. J. 382, 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Giardina T., Gunning A. P., Juge N., Faulds C. B., Furniss C. S., Svensson B., Morris V. J., Williamson G. (2001) Both binding sites of the starch-binding domain of Aspergillus niger glucoamylase are essential for inducing a conformational change in amylose. J. Mol. Biol. 313, 1149–1159 [DOI] [PubMed] [Google Scholar]
- 59. Southall S. M., Simpson P. J., Gilbert H. J., Williamson G., Williamson M. P. (1999) The starch-binding domain from glucoamylase disrupts the structure of starch. FEBS Lett. 447, 58–60 [DOI] [PubMed] [Google Scholar]
- 60. Penninga D., van der Veen B. A., Knegtel R. M., van Hijum S. A., Rozeboom H. J., Kalk K. H., Dijkstra B. W., Dijkhuizen L. (1996) The raw starch binding domain of cyclodextrin glycosyltransferase from Bacillus circulans strain 251. J. Biol. Chem. 271, 32777–32784 [DOI] [PubMed] [Google Scholar]
- 61. Machovic M., Janecek S. (2008) Domain evolution in the GH13 pullulanase subfamily with focus on the carbohydrate-binding module family 48. Biologia 63, 1057–1068 [Google Scholar]
- 62. Koay A., Woodcroft B., Petrie E. J., Yue H., Emanuelle S., Bieri M., Bailey M. F., Hargreaves M., Park J. T., Park K. H., Ralph S., Neumann D., Stapleton D., Gooley P. R. (2010) AMPK β subunits display isoform-specific affinities for carbohydrates. FEBS Lett. 584, 3499–3503 [DOI] [PubMed] [Google Scholar]
- 63. Katsuya Y., Mezaki Y., Kubota M., Matsuura Y. (1998) Three-dimensional structure of Pseudomonas isoamylase at 2.2 Å resolution. J. Mol. Biol. 281, 885–897 [DOI] [PubMed] [Google Scholar]
- 64. Abad M. C., Binderup K., Rios-Steiner J., Arni R. K., Preiss J., Geiger J. H. (2002) The x-ray crystallographic structure of Escherichia coli branching enzyme. J. Biol. Chem. 277, 42164–42170 [DOI] [PubMed] [Google Scholar]
- 65. Mikami B., Iwamoto H., Malle D., Yoon H. J., Demirkan-Sarikaya E., Mezaki Y., Katsuya Y. (2006) Crystal structure of pullulanase. Evidence for parallel binding of oligosaccharides in the active site. J. Mol. Biol. 359, 690–707 [DOI] [PubMed] [Google Scholar]
- 66. Woo E. J., Lee S., Cha H., Park J. T., Yoon S. M., Song H. N., Park K. H. (2008) Structural insight into the bifunctional mechanism of the glycogen-debranching enzyme TreX from the archaeon Sulfolobus solfataricus. J. Biol. Chem. 283, 28641–28648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Vander Kooi C. W., Taylor A. O., Pace R. M., Meekins D. A., Guo H. F., Kim Y., Gentry M. S. (2010) Structural basis for the glucan phosphatase activity of Starch Excess4. Proc. Natl. Acad. Sci. U.S.A. 107, 15379–15384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Janeček Š., Svensson B., MacGregor E. A. (2011) Structural and evolutionary aspects of two families of non-catalytic domains present in starch and glycogen binding proteins from microbes, plants, and animals. Enzyme Microb. Technol. 49, 429–440 [DOI] [PubMed] [Google Scholar]
- 69. Palomo M., Kralj S., van der Maarel M. J., Dijkhuizen L. (2009) The unique branching patterns of Deinococcus glycogen branching enzymes are determined by their N-terminal domains. Appl. Environ. Microbiol. 75, 1355–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kim T. J., Nguyen V. D., Lee H. S., Kim M. J., Cho H. Y., Kim Y. W., Moon T. W., Park C. S., Kim J. W., Oh B. H., Lee S. B., Svensson B., Park K. H. (2001) Modulation of the multisubstrate specificity of Thermus maltogenic amylase by truncation of the N-terminal domain and by a salt-induced shift of the monomer/dimer equilibrium. Biochemistry 40, 14182–14190 [DOI] [PubMed] [Google Scholar]
- 71. Turner P., Nilsson C., Svensson D., Holst O., Gorton L., Nordberg Karlsson E. (2005) Monomeric and dimeric cyclomaltodextrinases reveal different modes of substrate degradation. Biologia 60, 79–87 [Google Scholar]
- 72. Park S. H., Kang H. K., Shim J. H., Woo E. J., Hong J. S., Kim J. W., Oh B. H., Lee B. H., Cha H., Park K. H. (2007) Modulation of substrate preference of Thermus maltogenic amylase by mutation of the residues at the interface of a dimer. Biosci. Biotechnol. Biochem. 71, 1564–1567 [DOI] [PubMed] [Google Scholar]