Abstract
Endo-β-1,4-glucanase from thermophilic Fervidobacterium nodosum Rt17-B1 (FnCel5A), a new member of glycosyl hydrolase family 5, is highly thermostable and exhibits the highest activity on carboxymethylcellulose among the reported homologues. To understand the structural basis for the thermostability and catalytic mechanism, we report here the crystal structures of FnCel5A and the complex with glucose at atomic resolution. FnCel5A exhibited a (β/α)8-barrel structure typical of clan GH-A of the glycoside hydrolase families with a large and deep catalytic pocket located in the C-terminal end of the β-strands that may permit substrate access. A comparison of the structure of FnCel5A with related structures from thermopile Clostridium thermocellum, mesophile Clostridium cellulolyticum, and psychrophile Pseudoalteromonas haloplanktis showed significant differences in intramolecular interactions (salt bridges and hydrogen bonds) that may account for the difference in their thermostabilities. The substrate complex structure in combination with a mutagenesis analysis of the catalytic residues implicates a distinctive catalytic module Glu167-His226-Glu283, which suggests that the histidine may function as an intermediate for the electron transfer network between the typical Glu-Glu catalytic module. Further investigation suggested that the aromatic residues Trp61, Trp204, Phe231, and Trp240 as well as polar residues Asn51, His127, Tyr228, and His235 in the active site not only participated in substrate binding but also provided a unique microenvironment suitable for catalysis. These results provide substantial insight into the unique characteristics of FnCel5A for catalysis and adaptation to extreme temperature.
Keywords: Carbohydrate Metabolism; Crystal Structure; Enzyme Catalysis; Enzyme Mechanisms; Enzyme Structure; Thermodynamics; Catalytic Mechanism; Endo-β-1,4-glucanase; Thermophilic Enzymes; Thermostability
Introduction
Cellulose, a main component of plant cell walls, is the most abundant biopolymer in the world and is considered to be an important alternative source of renewable energy (1–4). In the past 50 years, much effort has gone into the studies of cellulases as a potential means for obtaining sustainable, biobased products to replace depleted fossil fuels. However, the high costs of cellulase production cause difficulties in the cellulose bioconversion process (5). These challenges create a need to identify new cellulases that possess the desired properties for reducing costs. Novel cellulases from thermophiles with inherited biological stability are expected to be useful for the industrial hydrolysis of plant cellulose during processing over long periods of time and at elevated temperatures, particularly during the conversion of biomass into biofuels. The crystal structure analysis of thermophilic cellulases will provide a structural basis for understanding the catalysis and adaptation to extreme temperature.
Numerous cellulases from eukaryotes and bacteria have been investigated and exhibit different structural and catalytic properties (6–9). These enzymes hydrolyze the glycosidic bond via general acid-base catalysis with retention of configuration at the anomeric carbon. Although enzymes from different glycosidase families have little or no overall sequence homology, their catalytic domains are expected to share the same (β/α)8-barrel topology observed in some enzymes that belong to these glycosidase families. Crystal structures have been reported for members of family 1 (10, 11), family 2 (12), family 5 (13, 14), family 10 (15–18), and family 17 (19) glycosidases. The active site is located at the C-terminal end of the β-strands; this is characteristic of all known family 5 members. Two strictly conserved glutamic acids located at the C terminus of β-strands 4 and 7 have been identified as the proton donor and the nucleophile, respectively, and play an important role for catalysis (20, 21). Several aromatic and polar groups form the surface of a deep extended substrate-binding cleft that can accommodate at least four d-glucosyl subsites, two on each side of the labile glycosidic bond. For instance, the catalytic residues Glu140 and Glu280 in Clostridium thermocellum CtCelC are located at the bottom of the cleft together with residues Arg46, His90, Asn139, His198, Tyr200, and Trp313 in the structural prototype, all of which are highly conserved in family 5 glycosyl hydrolases. These amino acids interact with each other and with the substrate through a network of hydrogen bonds. Recently, His198 was reported to be essential for catalysis (22). However, whether the histidine can act as a catalytic residue requires further investigation.
Considerable efforts have been made to analyze the structural features that determine the thermal stability of proteins from thermophiles. Understanding the structural basis for the enhanced stability of proteins from thermophilic organisms relative to their mesophilic and psychrophilic counterparts is a highly relevant but complex and challenging problem. Previous comparisons of high resolution crystal structures of enzymes with the same fold and function in psychrophiles, mesophiles, and thermophiles have revealed a number of potentially stabilizing features (23). The crystal structures of cellulases from thermophilic bacterium Acidothermus cellulolyticus (AcCelE1; Protein Data Bank code 1ECE), C. thermocellum (CtCelC; Protein Data Bank code 1CEC), mesophiles Clostridium cellulolyticum (CcCel5A; Protein Data Bank code 1EDG) and Paenibacillus pabuli (PpXG5; Protein Data Bank code 2JEP), and psychrophile Pseudoalteromonas haloplanktis (PhCel5G; Protein Data Bank code 1TVP) have provided basic insight into cellulase thermostability, but many important aspects remain to be resolved.
The thermophilic bacterium Fervidobacterium nodosum Rt17-B1 belongs to the eubacterial order Thermotogales, which comprises the most extremely thermophilic eubacteria presently known and represents the deepest branch within the bacteria (24). It is capable of growing at temperature above 60 °C with an optimal growth temperature of ∼80 °C. Several genes encoding putative cellulases have been annotated in the genome, but none have been characterized. Recently, a novel cellulase gene encoding a thermostable endoglucanase from F. nodosum was cloned and expressed; it was designated as FnCel5A for being a member of glycoside hydrolase family 5, and it is the first cellulase cloned from organisms of the genus Fervidobacterium (25). The purified recombinant FnCel5A is remarkably active and stable at high temperatures. Specifically, it shows a high hydrolase activity on carboxylmethylcellulose (CMC3; 440 IU/mg), regenerated amorphous cellulose (402 IU/mg), β-d-glucan from barley (1360 IU/mg), and galactomannan (895 IU/mg) with an optimal activity temperature of 80–83 °C. Furthermore, this enzyme is highly thermostable and has a half-life of 48 h at 80 °C. The well characterized cellulases CtCelC and CcCel5A only show 65 IU/mg activity on the typical endoglucanase substrate CMC at Topt 62 °C and 117 IU/mg at Topt 50 °C, respectively. To our knowledge, FnCel5A has the highest endoglucanase activity among the reported homologues.
To understand the structural basis for the extreme thermostability and high catalytic efficiency, we report here the crystal structure of the recombinant FnCel5A and its complex with glucose. A comparison of the structure with its mesophilic and thermophilic counterparts in amino acid composition, intramolecular interactions, and other structural factors, which all potentially involve the thermal stability of the enzyme, was performed. In addition, mutagenesis of the residues that are related to catalysis was carried out. The structural information in combination with biochemical experiments provided an interpretation of the structural basis for the highly efficient catalysis activity of FnCel5A.
MATERIALS AND METHODS
Cloning, Expression, and Purification
The DNA fragment encoding the mature protein was amplified by PCR from F. nodosum Rt17-B1 genomic DNA using the primers listed in supplemental Table 1 and inserted into the vector pET-15b (Invitrogen). The recombinant plasmids were transformed into Escherichia coli strain BL21(DE3). Transformed cells were then cultured at 37 °C in LB medium containing 50 μg/ml ampicillin. E. coli cells were grown in LB medium at 37 °C to midexponential phase (A600 = 0.6) and for an additional 3 h after adding 1 mm isopropyl 1-thio-β-d-galactopyranoside to induce protein expression. SeMet-labeled FnCel5A was produced in E. coli B834 containing the pET15b-FnCel5A plasmid with recombinant protein expression induced by 1 mm isopropyl 1-thio-β-d-galactopyranoside and incubation at 16 °C for 20 h. Cells were harvested by centrifugation, resuspended in 20 mm Tris-HCl (pH 8.0) buffer containing 10 mm NaCl, and then homogenized by sonication. Crude bacterial extracts were subjected to heat incubation at 65 °C for 30 min and centrifuged at 20,000 × g for 30 min to remove the heat-aggregated proteins and cell debris. The supernatant was then applied onto a Ni2+-chelating affinity column (1.5 ml of Ni2+-nitrilotriacetic acid-agarose). Contaminant protein was thoroughly washed off with at least 10 bed volumes of wash buffer (20 mm Tris-HCl (pH 8.0), 10 mm NaCl, and 20 mm imidazole), and the target protein was eluted with 20 mm Tris-HCl (pH 8.0), 10 mm NaCl, and 200 mm imidazole for an approximate total of 15 ml. Resource Q anion exchange chromatography (GE Healthcare) was subsequently used with a 0–1 m NaCl gradient in 20 mm Tris-HCl (pH 8.0) buffer. The target protein was finally eluted with ∼0.2 m NaCl.
The N-terminal deletion mutant FnCel5AND was constructed in pET-11b for co-crystallization because of the great facility in substrate binding. The recombinant plasmid was transformed into E. coli strain BL21(DE3). Transformed cells were then cultured at 37 °C in LB medium containing 50 mg/ml ampicillin. When the culture density reached an A600 of 0.6–0.8, induction with 1 mm isopropyl 1-thio-β-d-galactopyranoside was performed, and cell growth continued for 4 h at 37 °C. Cells were harvested by centrifugation, resuspended in 20 mm Tris-HCl (pH 8.0) buffer containing 10 mm NaCl, and then homogenized by sonication. Crude bacterial extracts were subjected to heat incubation at 65 °C for 30 min and centrifuged at 20,000 × g for 30 min to remove heat-aggregated proteins and cell debris. The supernatant obtained by centrifugation for 30 min at 20,000 × g was then applied onto a HiTrap Q-Sepharose column. Contaminant protein was thoroughly washed off with at least 10 bed volumes of wash buffer (20 mm Tris-HCl (pH 8.0) and 10 mm NaCl), and the target protein was eluted with a linear gradient buffer (0–1 m NaCl) at a flow rate of 60 ml/h. The target fractions were collected and desalted. Resource Q anion exchange chromatography (GE Healthcare) was subsequently applied using a 0–1 m NaCl gradient in 20 mm Tris-HCl (pH 8.0) buffer. The target protein was finally eluted with ∼0.2 m NaCl. The concentrated target protein was then applied on a Superdex 200 10/300 GL column and eluted with 20 mm Tris-HCl (pH 8.0) buffer containing 10 mm NaCl.
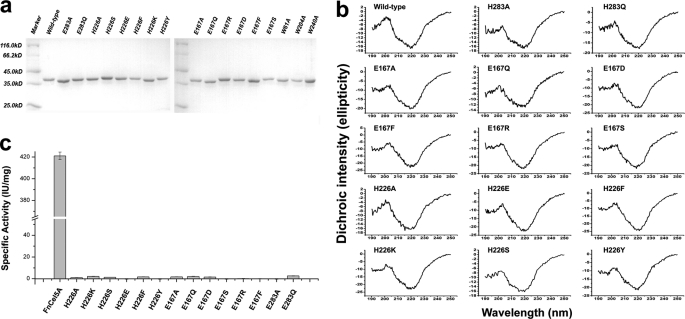
Vectors carrying the H226A, H226K, H226S, H226F, H226Y, H226E, E167A, E167Q, E167D, E167S, E167R, E167F, E283A, E283Q, W61A, W204A, and W240A mutants were generated using a PCR-based QuikChange site-directed mutagenesis kit according to the manufacturer's instructions with the plasmid pET15b-FnCel5A as the template DNA and the primers listed in supplemental Table 1. The expression and purification procedures of these mutants were the same as described for the wild type. Protein purity was checked by 12% SDS-PAGE.
Enzyme Activity Assay
Cellulase activities and enzymatic kinetics of the wild type and mutants were measured using a glucose assay kit (Bioassay Systems). The standard assay was performed by incubating 80 μg of protein and substrate CMC for 5 min at 80 °C in 50 mm phosphate buffer (pH 5.5) with 1 ml of the reaction mixture. The specific activities of the wild-type and mutant proteins were determined with a CMC concentration of 10 mg/ml. After incubation, 5 μl of reaction product was transferred into 500 μl of reagent, and the mixture was heated in a boiling water bath for 8 min. The absorbance of the specific color reaction at 630 nm is directly proportional to the concentration of reduced sugars in the products. One unit of enzyme activity was defined as the amount of enzyme required to release 1 μmol of reduced sugar from CMC in 1 min.
The Michaelis-Menten parameters kcat and Km for CMC were determined by initial rate measurements with 0.2–100 mg/ml substrate concentration ranges. The initial steady-state velocities of substrate hydrolysis were monitored at five to seven different substrate concentrations. All kinetic data were analyzed by linear regression to a Lineweaver-Burk double reciprocal plot using OriginLab Corp. software OriginPro 8.0 SR2, and the standard errors for each parameter were estimated by curve fitting.
Circular Dichroism
Far-UV CD spectra of the wild-type and mutant enzymes were recorded at 20 °C on a Jasco J-715 spectrometer in a configuration described by Jasco hardware manual P/N:0302-0265A. Secondary structure determination spectra were performed with protein concentrations of ∼0.02 mg/ml in PBS (pH 7.0). The spectra were measured in quartz Hellma 110-QS cells with a 1-cm optical path length. A sufficient signal-to-noise ratio is achieved by recording three accumulations for the far-UV CD spectra.
Crystallization and Data Collection
Crystals of Se-FnCel5A and native FnCel5A (∼20 mg/ml) were grown by the hanging drop vapor diffusion method at 291 K in 1.2 m NaH2PO4 and KH2PO4 (1:1) and 0.1 m sodium citrate (pH 5.6). Each drop contained 1 μl of protein solution and 1 μl of reservoir solution with 200 μl of reservoir solution in the well. The FnCel5A-glucose complex crystals were prepared by the hanging drop vapor diffusion method, and co-crystallization of the cellobiose and FnCel5AND (∼20 mg/ml) occurred in 23% PEG 8000, 0.2 m ammonium sulfate, and 0.1 m sodium citrate (pH 5.6). The molar ratio of proteins to polysaccharide substrates (cellobiose or CMC) was 1:1.5. All crystals were cryoprotected by the addition of 20% glycerol (v/v) to the crystallization conditions. A 1.7-Å resolution data set was collected from a SeMet-FnCel5A crystal at 100 K using an ADSC Q315 CCD detector on beamline BL5A at the Photon Factory (Japan). The diffraction data of the FnCel5A-glucose complex and native FnCel5A were collected on the home Rigaku MM-007 x-ray source. All data sets were integrated, scaled, and reduced with the HKL2000 software package (26). Crystallographic statistics for data collection are summarized in Table 2.
TABLE 2.
Structural factors potentially involved in stability of FnCel5A and homologues
| Parameters | FnCel5A | CtCelC | CcCel5A | PhCel5G |
|---|---|---|---|---|
| Fraction of buried atoms (%) | 53 | 54 | 46 | 47 |
| Accessible surface (Å2) | 4,086 | 4,127 | 4,564 | 4,398 |
| Amino acid composition (%) | ||||
| Tyrosine | 5.4 | 6.3 | 5.0 | 3.4 |
| Proline | 5.1 | 3.2 | 3.6 | 2.3 |
| Asparagine/glutamine | 7.0 | 8.4 | 14.0 | 11.6 |
| Aromatic residuesa | 16 | 14.7 | 12.6 | 10.8 |
| Charged residuesa | 28.8 | 32.4 | 21.5 | 19.6 |
| Interactions | ||||
| Salt bridges | 23 | 26 | 18 | 13 |
| Ion networks | 3 | 5 | 2 | 2 |
| Hydrogen bond | 610 | 614 | 737 | 592 |
a The charged residues include arginine, histidine, lysine, aspartic acid, and glutamic acid; the aromatic residues include tyrosine, phenylalanine, and tryptophane.
Structure Determination and Refinement
The crystal structure of FnCel5A was solved at 1.7-Å resolution by the single wavelength anomalous dispersion method from the Se-Met derivative of FnCel5A. Three of the four potential selenium atoms in one FnCel5A monomer were located by SHELEX (27), and initial phases were calculated by the program SOLVE (28). Density modification (solvent flipping) and phase extension to 1.7 Å were performed using RESOLVE (29). The initial model of Se-FnCel5A was automatically traced using the program ARP/wARP (30) to ∼90% completeness, and the refinement of the Se-FnCel5A model was performed with the program REFMAC5 (31) of the CCP4 program suite and by manual adjustment in COOT (32) in the space group P212121. After several rounds of adjustment and refinement, the Rwork and Rfree converged to 20.8 and 24.5%, respectively, in the resolution range of 50.0–1.7 Å. It is worth mentioning that the structure of the native FnCel5A was solved at 2.4-Å resolution using the molecular replacement method with the Se-FnCel5A as a searching model by using the PHASER program (33) and subsequently refined to an Rwork of 17.8% and an Rfree of 24.6%. The crystal showed space group P21212, and the model consisted of only one FnCel5A monomer. The crystal structure of the FnCel5A-glucose complex was solved at 2.2-Å resolution by the molecular replacement method with the Se-FnCel5A as a searching model. The structure was subsequently refined to an Rwork of 17.6% and an Rfree of 24.7% over the resolution range 50.0–2.2 Å. The model consisted of one FnCel5A monomer, three glucose molecules, and an additional 299 solvent molecules. Data collection and model refinement statistics are summarized in Table 2. Sequence alignment was performed using the programs ClustalW and ESPript. Structure comparison was carried out using the DALI server. All structure figures were created using the program PyMOL.
RESULTS AND DISCUSSION
Overall Topology of FnCel5A
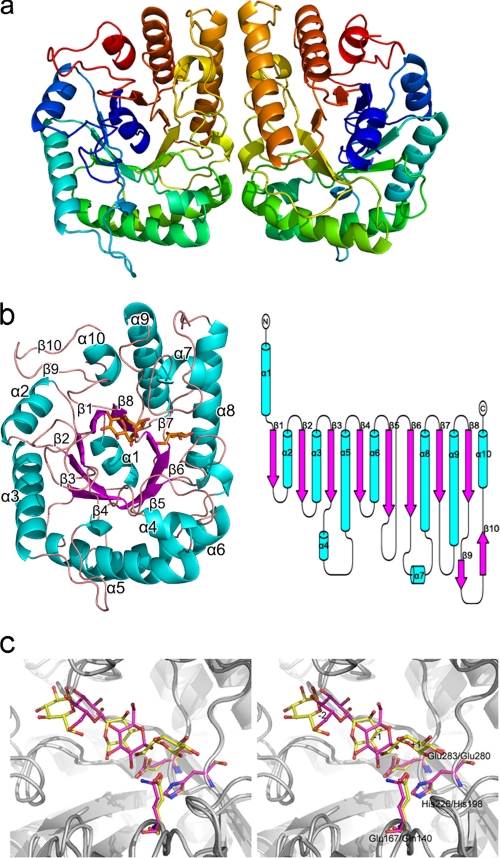
To gain a better understanding of the structural basis for the extraordinary catalytic characteristics and thermal stability of FnCel5A, the recombinant FnCel5A, Se-Met derivative, and the FnCel5A complex with glucose were crystallized at 291 K using NaH2PO4/KH2PO4 as a precipitant (34). The crystal structure of the Se-FnCel5A was determined using the single wavelength anomalous dispersion method and refined to 1.7-Å resolution with a final Rwork value of 17.8% (Rfree = 24.5%). There are two FnCel5A molecules in an asymmetric unit with a Matthews coefficient of 2.6 Å3/Da (corresponding to 48% solvent content) (Fig. 1a). Each monomer is composed of 10 β-strands and 10 α-helices arranged in a α1-β1-α2-β2-α3-β3-α4-α5-β4-α6-β5-β6-α7-α8-β7-α9-β8-β9-β10-α10 topology with dimensions of 50 × 35 × 30 Å (Fig. 1b). The first subunit contains residues 28–343, and the second contains residues 29–343 due to the lack of electron density. Subunit 2 can be superimposed onto the first subunit with a root mean square deviation (r.m.s.d.) of 0.67 Å. The two FnCel5A monomers are related by a non-crystallographic 2-fold axis to form a tight dimer with an extensive subunit interface. The main contact region concerns helix α6, α8, and the loop between α4 and α6, β5 and β6, α7, and α8 of one subunit. However, according to analytical gel filtration experiments, the purified FnCel5A in buffer conditions (20 mm Tris-HCl and 10 mm NaCl) is 34 kDa (25), suggesting a monomeric state of the FnCel5A molecule in aqueous solution. Therefore, the formation of such a crystallographic dimer was a result of crystal packing.
FIGURE 1.
Overall structure of FnCel5A from F. nodosum Rt17-B1. a, ribbon representation of the two Se-FnCel5A molecules in one asymmetric unit. The coloring is from blue at the N terminus to red at the C terminus. b, ribbon and topology diagram of FnCel5A showing the (β/α)8-barrel architecture of the monomer. Helices are shown in cyan, and strands are shown in magenta. Glucose molecules are shown in orange and ball-and-stick representation. c, a stereoview of superposition between FnCel5A and CtCelC complex model. The subsite numbers are labeled from −2 to +1. Proteins are shown in ribbon. Ligands and catalytic residues of FnCel5A are shown in yellow and ball-and-stick representation compared with the magenta representation in CtCelC.
The co-crystallization was performed with FnCel5A and its natural inhibitor, cellobiose; however, the electron density map of the complex showed that we eventually captured the enzyme complex with the product glucose due to the slow hydrolysis of cellobiose. The complex structure of FnCel5A-glucose refined to 2.2-Å resolution reveals a large, deep pocket in front of the C-terminal end of the (β/α)8-barrel. Three glucose molecules are located in the pocket. The surface electrostatic potential of the substrate-binding cleft site shows highly acidic character. Structural comparison of FnCel5A-glucose and CtCelC-cellobiose (Protein Data Bank code 1CEN) showed that the spatial position of the three glucose units was similar to the CtCelC-cellobiose complex structure to some extent (Fig. 1c). Therefore, three glucose molecules were defined by +1, −1, and −2 pyranose rings in the complex structure of FnCel5A-glucose. The details of the data collection and structure refinement are summarized in Table 1.
TABLE 1.
Data collection and refinement statistics
| Structure | Se-FnCel5A | Native FnCel5A | FnCel5A-glucose complex |
|---|---|---|---|
| Data collection statistics | |||
| Cell parameters (Å) | a = 107.4, b = 82.0, c = 85.8 | a = 81.7, b = 85.2, c = 53.5 | a = 47.3, b = 47.3, c = 271.4 |
| Space group | P212121 | P21212 | P41212 |
| Wavelength used (Å) | 0.9798 | 1.5418 | 1.5418 |
| Resolution (Å) | 50.0–1.7 (1.75–1.67)a | 50.0–2.4 (2.49–2.40)a | 50.0–2.2 (2.28–2.20)a |
| No. of all reflections | 808,544 (89,834)a | 58,334 (5,841)a | 117,210 (9,629)a |
| No. of unique reflections | 122,364 (12,477)a | 12,870 (1,298)a | 16,724 (1,632)a |
| Completeness (%) | 96.8 (100)a | 84.7 (88.4)a | 98.9 (99.5)a |
| Average I/σ(I) | 30.7 (4.1)a | 9.4 (2.8)a | 19.2 (2.7)a |
| Rmergeb (%) | 7.2 (40.7)a | 13.7 (39.7)a | 9.5 (47.9)a |
| Refinement statistics | |||
| No. of reflections used ((F) > 0) | 72,739 | 12,208 | 15,844 |
| Rworkc (%) | 20.8 | 17.8 | 17.6 |
| Rfreec (%) | 24.5 | 24.6 | 24.7 |
| r.m.s.d. bond distance (Å) | 0.016 | 0.012 | 0.021 |
| r.m.s.d. bond angle (°) | 1.44 | 1.31 | 1.84 |
| Average B value (Å2) | 22.8 | 18.0 | 28.8 |
| Ramachandran plot | |||
| Most favored regions (%) | 91.2% | 88.8% | 88.7% |
| Additionally allowed regions (%) | 8.1% | 10.8% | 10.5% |
| Generously allowed regions (%) | 0.7% | 0.4% | 0.7% |
| Protein Data Bank code | 3NCO | 3RJX | 3RJY |
a Values in parentheses correspond to the highest resolution shell.
b Rmerge = ΣhklΣi|I(hkl)i − I(hkl)|/ΣhklΣiI(hkl)i where I(hkl) is the mean intensity of the observations I(hkl)i of reflection hkl.
c Rwork = Σ‖Fobs| − |Fcalc‖/Σ|Fobs| where Fobs and Fcalc are the observed and calculated structure factor amplitudes, respectively. Rfree was calculated as Rwork using a randomly selected subset of ∼10% of unique reflections not used for structure refinement.
Sequence and Structure Similarity
Although glycoside hydrolases have over 120 families and thousands of members (CAZy database), the FnCel5A enzyme is almost unique in sequence. A BLAST search in GenBankTM (non-redundant protein sequences) showed that the high sequence identity (47–53%) was shared with only eight annotated glycoside hydrolases or hypothetical protein sequences from Dictyoglomus turgidum DSM 6724 (cellulase; Gene IDs 7083331, 7083099, and 7083332), Dictyoglomus thermophilum H-6-12 (endoglucanase; Gene IDs 6945586, 6945794, and 6946331), Thermotoga maritima MSB8 (endoglucanase; Gene ID 897863), and Thermotoga sp. RQ2 (glycoside hydrolase family protein; Gene ID 6092507).
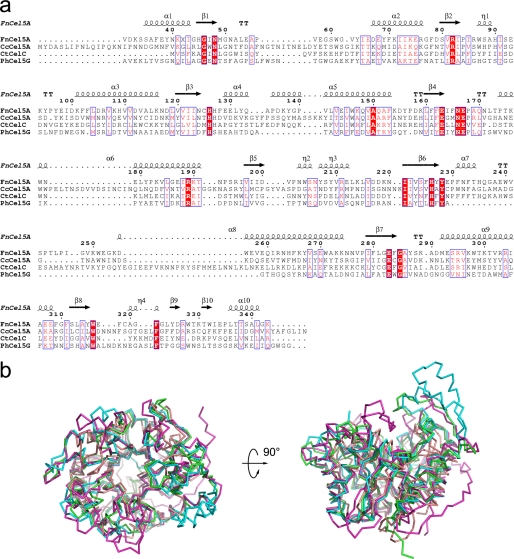
A DALI search for structural similarity revealed that the overall architecture of FnCel5A most resembled members of glycosyl hydrolase family 5: thermophilic bacterium C. thermocellum (CtCelC; Protein Data Bank code 1CEC) (22), mesophile C. cellulolyticum (CcCel5A; Protein Data Bank code 1EDG) (14), and psychrophile P. haloplanktis (PhCel5G; Protein Data Bank code 1TVP) (35). Although FnCel5A shares only 19–24% sequence identity with these homologues (Fig. 2a), all structures are arranged in a globular form and possess a (β/α)8-barrel fold that features eight β-strands surrounded by eight α-helices, which is typical of family 5 and clan GH-A enzymes. Superposition of the crystal structure of FnCel5A with those of CtCelC, CcCel5A, and PhCel5G produced the r.m.s.d. of all Cα atoms ranging from 0.88 to 1.29 Å, indicating that those four cellulases are spatially homologous (Fig. 2b). Moreover, the mutual positions of the central β-barrel structure and α-helices, which are preliminarily located at the end of the β-sheet, are similar. However, compared with the homologues, the structure-based sequence alignment also shows that FnCel5A contains residue deletion/insertion in the loops between the β-strand and α-helix of the (β/α)8-barrel fold. For instance, shorter loops can be found on the surface of the (β/α)8-barrel between strand β1 and helix α1, strand β4 and helix α6, strand β6 and strand β7, and strand β9 and helix α10 (Fig. 3).
FIGURE 2.
Structural and sequence comparison of FnCel5A with other members of family 5. a, a structure-based sequence alignment between FnCel5A and CtCelC, CcCel5A, and PhCel5G was performed using ClustalW and ESPript. Secondary structure elements are shown for FnCel5A. b, superposition of FnCel5A with three related cellulase structures from C. thermocellum (CtCelC; Protein Data Bank code 1CEC), mesophile C. cellulolyticum (CcCel5A; Protein Data Bank code 1EDG), and psychrophile P. haloplanktis (PhCel5G; Protein Data Bank code 1TVP). The r.m.s.d. between Cα atoms of the four cellulases ranged from 0.88 to 1.29 Å.
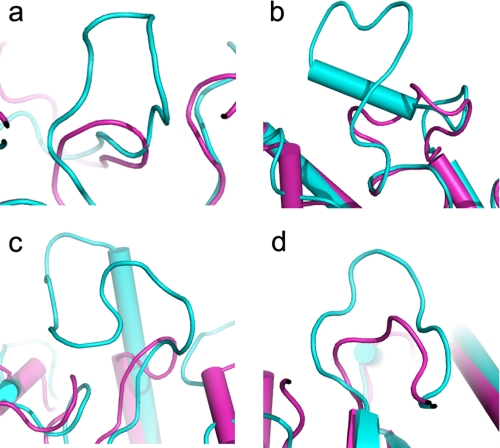
FIGURE 3.
Superposition of FnCel5A (magenta) and CcCel5A (cyan) illustrating shorter loops present in thermophilic protein. a, loop between strand β1 and helix α1; b, loop between strand β4 and helix α6; c, loop between strand β6 and strand β7; d, loop between strand β9 and helix α10.
Thermostability
Recently, more and more sequence and structural information has been reported for proteins from hyperthermophilic and thermophilic organisms. Comparative analyses indicate that hyperthermophilic proteins share a high similarity with their mesophilic homologues. Whereas no universal mechanism can explain the remarkable stability improvement of hyperthermophilic proteins, the thermostability may be due to slight alterations of protein structure that collectively contribute to these observations. Some features that may be important for thermostability are the fraction of buried atoms, accessibility to the surface area, and lengths of loops connecting the secondary structural elements (36, 37). To address the unique thermostability of this protein, the structure of FnCel5A was compared with the structures of CtCelC (Topt = 62 °C), CcCel5A (Topt = 50 °C), and PhCel5G (Topt = 20 °C). Our analysis shows that FnCel5A has a larger fraction of buried atoms (53%) compared with CcCel5A (46%). Consequently, FnCel5A shows an accessible surface area of 4086 Å2 compared with 4564 Å2 for CcCel5A, which is 12% more than FnCel5A. Another feature is the functional role of loops in protein structure and stability (38). It has been reported that molecular dynamics simulations carried out at room temperature or higher (37 °C) show that loop and turn regions undergo the largest deviations from the starting crystal structure conformations. Therefore, these are likely to be regions of the structure that unfold first during thermal denaturation (37, 39). The FnCel5A structure displays a reduction in the size of several loops compared with CcCel5A (Fig. 3). Therefore, the shortened loops in FnCel5A potentially confer an increased resistance to thermally induced unfolding.
Amino acid composition and interactions (salt bridges, ion networks, and hydrogen bonds) are also important for some thermostable enzymes. A comparison of FnCel5A and CtCelC, CcCel5A, and PhCel5G amino acid sequences showed a much lower level of identity of 19–24%. Analysis of the FnCel5A and PhCel5G amino acid sequences showed 16 and 10.8% aromatic residues, respectively. Specifically, FnCel5A showed nearly twice as many tyrosine residues compared with the PhCel5G sequence. An increase in the number of Tyr residues has been associated with the thermostability of thermophilic proteins because Tyr residues are involved with protein interior core packing through the formation of aromatic stacks or hydrogen bonds (40). FnCel5A also contains 5.1% proline residues, which is the largest proportion in the four cellulases and more than twice the number in PhCel5G. Because proline residues affect local mobility of the chain by decreasing the conformational entropy of the unfolded state, the increased rigidity of the structure of FnCel5A would be expected to increase the overall thermostability. On the other hand, the FnCel5A sequence shows half the amount of asparagine (Asn) and glutamine (Gln) residues compared with CcCel5A. The frequency of Asn and Gln, which can be classified as thermolabile due to their tendency to undergo deamidation at high temperatures and therefore may be naturally discriminated against in thermostable proteins, is substantially reduced in thermostable cellulases. Another remarkable difference is an increase in the proportion of charged residues in the thermophilic FnCel5A and CtCelC (∼30%) compared with their mesophilic and psychrophilic counterparts CcCel5A and PhCel5G (∼20%). An increase in the number of charged residues potentially enhances the occurrence of salt bridges and ion networks in thermophilic proteins (40). Specifically, a total of 23 and 26 salt bridges are formed per monomer in the thermophilic FnCel5A and CtCelC, respectively, which is almost 2-fold higher than psychrophilic PhCel5G. Most of the salt bridges are located at the surface of the (β/α)8-barrel fold, suggesting that they may contribute to the rigidity and solubility of the protein. Further investigation suggests that the thermostable FnCel5A and CtCelC contain more ion networks than their mesophilic or psychrophilic counterparts (Table 2 and 3). We found that at least two conserved ion networks are located at the active site and surface of the four homologues that might contribute to the stability of the active site and protein shell. However, ion networks formed between the peptides of the N and C termini only existed in the thermophilic enzymes FnCel5A and CtCelC. Specifically, the unique ion network Lys334-Arg76-Lys343 tightly locks the C terminus of FnCel5A because the Lys343 is the last residue. Protein termini have been defined as crucial factors for protein stability. It is also hypothesized that protein termini are the initial nucleus of protein unfolding. Therefore, we propose that the ion networks that reside between the N and C termini in FnCel5A may efficiently prevent enzyme unfolding. The above structural analysis reveals that an increased number of salt bridges and ion networks at the surface and terminus of the protein are crucial for the thermostability of members of family 5.
TABLE 3.
Ion networks of FnCel5A, CtCelC, CcCel5A, and PhCel5G
| Location | FnCel5A | CtCelC | CcCel5A | PhCel5G |
|---|---|---|---|---|
| Active site | Glu163-Arg82-Glu283 | Asp88-Arg46-Glu136-Glu280 | Glu166-Arg79-Glu307 | Asp98-Arg57-Glu131-Glu222 |
| Surface | Lys147-Glu187-Lys190 | Glu55-Arg114-Asp57 | Glu54-Arg102-Glu105 | Asp175-Arg207-Asp177 |
| Glu65-Arg125-Asp72 | ||||
| Glu107-Arg148-Glu143 | ||||
| N–C terminus | Glu334-Arg76-Lys343 | Arg327-Glu33-Lys316-Glu325 | —a | — |
a —, no ion networks in N–C terminus.
“Induced Fit” Active Site of FnCel5A
The structure of FnCel5A shows a large and deep pocket in front of the C-terminal end of the (β/α)8-barrel (Fig. 1c), which is in agreement with the observation of the active sites of all (β/α)8-barrel enzymes. The space in the pocket of FnCel5A is large enough to accommodate at least four d-glucosyl subsites, two on each side of the labile glycosidic bond. Two strictly conserved glutamic acids, Glu167 and Glu283, are located at the bottom of the cleft. The distance between the closest atoms of the catalytic side chains of Glu167 and Glu283 is 3.69 Å in the complex, which is typical of a retaining enzyme and allows for the formation of a glycosyl-enzyme intermediate situated between the two carboxyl groups (21). The scaffold for docking the substrate includes some aromatic residues and polar residues. Structure information of the complex reveals that polar residues (Asn51, His127, Trp204, Tyr228, His235, Trp240, and Trp316) are tightly bound to the substrate by forming multiple hydrogen bond interactions (Table 4). An aromatic stack effect contributed by Trp61, Trp204, Phe231, and Trp240 also enhances the binding with polysaccharide substrate. Although the structure of liganded complex is very similar to that of the native enzyme with an r.m.s.d. of 0.6 Å for all equivalent atoms, a comparison of the structure and active sites may provide insight for understanding the conformational changes due to substrate binding.
TABLE 4.
Enzyme-substrate H-bond interactions
| Substrate |
Protein |
Distance | ||
|---|---|---|---|---|
| Glucose | Atom | Residue | Atom | |
| Å | ||||
| +1 | O1 | Trp204 | O | 3.48 |
| O2 | Trp204 | O | 3.57 | |
| −1 | O1 | His127 | Nϵ2 | 3.64 |
| O3 | Tyr228 | Oη | 2.60 | |
| O4 | His235 | Nδ1 | 3.40 | |
| O4 | Trp240 | Nϵ1 | 3.20 | |
| O6 | Trp316 | Nϵ1 | 2.84 | |
| −2 | O3 | Asn51 | Oδ1 | 3.45 |
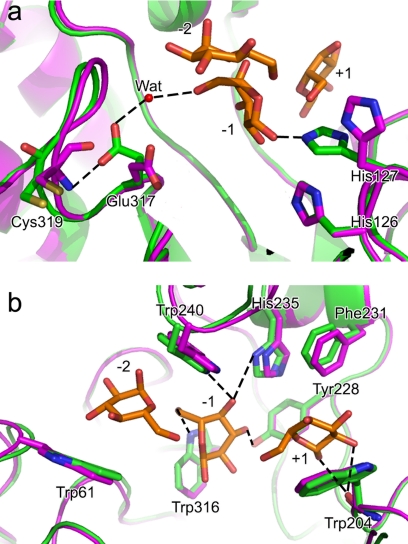
Compared with the unliganded structures, the FnCel5A-glucose complex reveals significant conformational changes of side chain rotation in the active site (Fig. 4). For instance, the imidazole side chain of His127 rotates ∼90° around the backbone axis and forms an aromatic stack with His126. Importantly, the torsion of the imidazole ring of His127 is advantageous for forming a new hydrogen bond with the −1 pyranose rings (Fig. 4). In addition, the carboxyl side chain of Glu317 in the complex rotates nearly 180° around the backbone axis. The rotation forms a steady hydrogen bond between Oϵ1 atom of Glu317 and O6 atom of the −1 pyranose ring by a water molecule. On the other hand, spatial displacement of the residues involving four loops around the active site, including Met49–Ala52, Gly59–Val63, Gln236–Pro246, and Cys319–Phe322, shows that these residues display remarkable movement. The rearrangement of these loops is related to the movements of the side chain of some adjacent residues and might influence their interactions with the neighboring residues or substrates. The Cα atoms of several substrate binding residues on the loops, such as Trp61, Trp204, Phe231, His235, and Trp240, are shifted by 0.83, 0.22, 0.39, 0.44, and 0.75 Å, respectively, with respect to the unliganded structures (Fig. 4). Their side chains are closer to −2, +1, −1, and −2 pyranose rings, which may enhance the tight binding with the substrates. Together, these results suggest that residues located in the active site show an induced fit after binding to the substrate and promote catalysis.
FIGURE 4.
View of superposition of FnCel5A (magenta) and its liganded complex form with glucoses (green) illustrating induced fit active site of FnCel5A. Conformational changes of the side chain rotation of the residues His127 and Glu317 (a) and residue movements of Trp61, Trp204, Phe231, His235, and Trp240 (b) are labeled and shown in ball-and-stick representation, respectively. Proteins are shown in ribbon. Glucose molecules are shown in orange and ball-and-stick representation. H-bonds are shown by dotted lines. Wat, water.
To elucidate the impact of the substrate binding residues on catalysis, three remarkable induced fit tryptophans were substituted by alanine scanning, and the kinetic parameters of mutants W61A, W204A, and W240A were determined (Table 5). Compared with the wild type, all of the mutants exhibited slightly lower kcat values (<30%). However, the Km values of the mutants W61A, W204A, and W240A were 1, 2.25, and 3 times higher, respectively, than the wild type, clearly suggesting that Trp61, Trp204, and Trp240 play an important role in endoglucanase activity by high substrate affinity. The results showed that the network of hydrogen bonds and aromatic interactions formed by substrate binding residues with substrate may stabilize the reactive intermediates and promote catalysis.
TABLE 5.
Kinetic parameters for wild type and mutants at 80 °C
| Enzyme | kcat | Km | kcat/Km |
|---|---|---|---|
| s−1 | mg/ml | s−1·mg−1·liter | |
| Wild type | 97.0 ± 1.4 | 20.4 ± 0.3 | 4.8 × 103 |
| W61A | 93.2 ± 2.1 | 39.5 ± 0.9 | 2.4 × 103 |
| W204A | 77.7 ± 2.8 | 45.2 ± 1.6 | 1.7 × 103 |
| W240A | 71.7 ± 1.7 | 60.2 ± 1.4 | 1.2 × 103 |
Enzymatic Mechanism
In general, cellulases most likely use a simple Glu-Glu module and acid-base catalytic mechanism similar to that proposed for lysozyme (41). Endo-β-1,4-glucanase was previously shown to catalyze the cleavage of the β-1,4 glycosidic bond with retention of the anomeric configuration, which most likely occurred through a double displacement mechanism involving a glycosyl-enzyme intermediate (42). This suggests the participation of two carboxylate residues in the Glu-Glu module acting as a nucleophile and proton donor, respectively, which have been previously identified by site-directed mutagenesis (43–46). In FnCel5A, these residues correspond to residues Glu283 and Glu167, respectively. However, there is disagreement within the field as to whether there are other residues involved in the catalysis, and a thorough understanding remains to be elucidated. In fact, studies indicate that in family β-glucosidase A, the glutamic acid (corresponding to Glu167 in FnCel5A) is not an essential nucleophile partner in catalysis (47). Substitutions involving a charged amino acid, such as His or Asp, had an even more dramatic decrease in activity. Structure analysis has revealed that His198 of CtCelC and His254 of CcCel5A, the counterparts of His226 of FnCel5A, form a hydrogen bond with the proton donor glutamic acid and therefore are hypothesized to be involved in the catalytic reaction (22). Indeed, this position of His226 is found to be completely intolerant to amino acid substitutions, such as in the case of cellulase EGZ of Erwinia chrysanthemi (48). These studies suggest that the catalytic module of endoglucanases might evolve to concise or complicated forms with the evolution of the enzyme, although they all are based on an acid-base catalytic mechanism similar to that of the ancestral lysozyme.
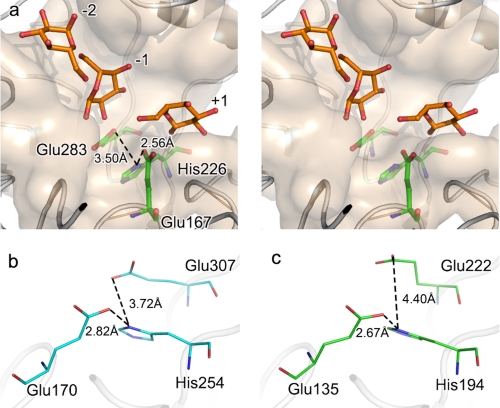
To assess the importance of the residues of the putative active center in catalysis, Glu167, Glu283, and His226 were changed by mutagenesis. A series of mutants of His226 (H226A, H226K, H226S, H226F, H226Y, and H226E), Glu167 (E167A, E167Q, and E167D, E167S, E167R, and E167F), and Glu283 (E283A and E283Q) were constructed, purified (Fig. 5a), and characterized. Conformational analysis of the recombinant proteins characterized by the far-UV CD spectra showed that the wild type and all the mutants displayed the typical pattern of (β/α)8-barrel proteins with the two characteristic minima at 222 and 208 nm of similar intensity and an almost identical signal ratio between these two minima (Fig. 5b). This indicates that the structures of the mutants have not suffered structural changes. The enzyme activity assay showed that all of the mutants completely lost their substrate hydrolysis function (Fig. 5c). We hypothesized that residue His226 may function as a catalytic residue because the structural information of FnCel5A showed that this residue resides in the active site. The Nδ1 atom of the imidazole group of His226 and the carboxyl groups of Glu167 and Glu283 are in the appropriate range for hydrogen bonding (within 3.5 Å), respectively (Fig. 6a). Considering that the catalytic residues Glu167 and Glu283 are buried in the deep interior of the binding cleft of FnCel5A and that there are several aromatic residues in close proximity, the carboxyl groups of Glu167 and Glu283 are difficult to dissociate. This might affect the nucleophile attack, protonation of the glycosidic oxygen, and deprotonation of the water during the catalytic process. The tertiary structure of FnCel5A showed that the His226 is located at the active site and is positioned within 3.50 Å of the Glu167 and Glu283 (Fig. 6a). Because of the two tautomeric forms of the neutral imidazole side chain of histidine, His226 could act as an electron donor or acceptor at an active site and commonly participate in catalysis reactions. It suggests that the interaction among Glu167-His226-Glu283 may form an electron relay network for efficient catalysis. An extensive sequence alignment (>30% sequence identity) proves that His226 of FnCel5A is highly conserved in endoglucanases of glycosyl hydrolase family 5 (supplemental Fig. 1). Based on the above structure analysis, we propose that the mechanism of FnCel5A, which includes the participation of a novel catalytic module, Glu167-His226-Glu283, may therefore be more complex than that deduced on the basis of the ancestral enzyme.
FIGURE 5.
Biochemical assays of FnCel5A and its mutants. a, SDS-PAGE analysis of the purity for the wild type and all mutants. b, far-UV CD profiles of the wild-type and mutant enzymes. The mean residual ellipticity is expressed in degrees cm2/dmol. A sufficient signal-to-noise ratio is achieved by recording three accumulations. c, histogram showing the specific activity of the wild type and mutants in enzyme activity assays. Data are presented as the mean of at least three experiments. A reaction mixture without enzyme was used as the negative control. Error bars indicated the standard deviation (S.D.) calculated from at least three independent experiments.
FIGURE 6.
Stereoscopic view of FnCel5A catalytic trial (a) and comparison of distance between histidine and catalytic glutamic acid in CcCel5A (b) and PhCel5G (c). The substrate binding subset is shown in pink surface, proteins are shown in ribbon, and the catalytic glutamic acid residues and conserved histidine residue are shown in ball-and-stick representation. Glucose molecules are shown in orange and ball-and-stick representation.
Although the Glu-His-Glu catalytic motif is highly conserved in sequences of glycosyl hydrolase family 5 members, the real spatial conformation of the three residues plays the decisive role in catalysis. The mesophilic and psychrophilic counterparts of FnCel5A exhibit similar interactions (Fig. 6, b and c), which suggests that some members of the glycosidase family have evolved an electron relay network for efficient catalysis. Although the distance between His226 and the proton donor Glu167 in FnCel5A is similar to the counterparts in PhCel5G and CcCel5A, the distance between the residues equivalent to His226 and nucleophile Glu283 in FnCel5A displayed a remarkable increase in CcCel5A and PhCel5G. When the diversity of the activity is considered (FnCel5A > CcCel5A > PhCel5G), these data imply that a more refined conformation of the catalytic module in FnCel5A may provide the higher catalysis efficiency compared with its counterparts. We believe that the proposed derivative novel catalytic module Glu-His-Glu may be a general mechanism in family 5 glycoside hydrolases or extend to other close phylogenetic glycoside hydrolases due to the convergent or divergent evolution of enzymes. However, the role of the histidine in the specificity of the glycosidase superfamily will require further investigation using bioinformatics techniques.
Conclusions
In this study, we report the crystal structure of FnCel5A and its complex formation with glucose at the atomic level. Although the structure of FnCel5A shares similarity with glycoside hydrolase family 5 cellulases, some special structural features of FnCel5A allow us to understand its high thermostability and the unique catalytic mechanism of action. The increased intramolecular interactions, particularly the salt bridges and ion networks based on the amino acid composition, contribute to the high thermostability. The complex model defines possible substrate binding sites within the catalytic cleft and suggests an induced fit binding model. Importantly, this complex structure in combination with mutagenesis studies of the catalytic residues suggests a novel catalytic triad, Glu167-His226-Glu283, for endo-β-1,4-glucanase that evolved from the conservative Glu-Glu catalytic module and is the first reported in glycosyl hydrolase family 5. Additional structural analyses and biochemical assays for other members of the glycoside hydrolase family will be carried out to reveal the generality of this catalytic mechanism. In addition, our results will help elucidate the novel catalytic and thermostable mechanism of glycoside hydrolase family 5 and thus provide insight for the design of more efficient and thermostable biocatalysts.
Protein Data Bank Accession Codes
The coordinates have been deposited at The Research Collaboratory for Structural Bioinformatics Protein Data Bank under the Protein Data Bank accession codes 3NCO for Se-FnCel5A, 3RJX for the native FnCel5A, and 3RJY for the FnCel5A-glucose complex.
Supplementary Material
This work was supported by the National Basic Research Program of China (973 Program), Natural Science Foundation of China, and Science and Technology Commission of the Shanghai Municipality.

This article contains supplemental Fig. 1 and Table 1.
The atomic coordinates and structure factors (codes 3NCO, 3RJX, and 3RJY) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- CMC
- carboxylmethylcellulose
- Topt
- optimal temperature
- r.m.s.d.
- root mean square deviation.
REFERENCES
- 1. Ragauskas A. J., Williams C. K., Davison B. H., Britovsek G., Cairney J., Eckert C. A., Frederick W. J., Jr., Hallett J. P., Leak D. J., Liotta C. L., Mielenz J. R., Murphy R., Templer R., Tschaplinski T. (2006) The path forward for biofuels and biomaterials. Science 311, 484–489 [DOI] [PubMed] [Google Scholar]
- 2. Percival Zhang Y. H., Himmel M. E., Mielenz J. R. (2006) Outlook for cellulase improvement: screening and selection strategies. Biotechnol. Adv. 24, 452–481 [DOI] [PubMed] [Google Scholar]
- 3. Chang M. C. (2007) Harnessing energy from plant biomass. Curr. Opin. Chem. Biol. 11, 677–684 [DOI] [PubMed] [Google Scholar]
- 4. Rubin E. M. (2008) Genomics of cellulosic biofuels. Nature 454, 841–845 [DOI] [PubMed] [Google Scholar]
- 5. Kim J. M., Kong I. S., Yu J. H. (1987) Molecular cloning of an endoglucanase gene from an alkalophilic Bacillus sp. and its expression in Escherichia coli. Appl. Environ. Microbiol. 53, 2656–2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Henrissat B. (1991) A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280, 309–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Henrissat B., Bairoch A. (1993) New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 293, 781–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Henrissat B., Callebaut I., Fabrega S., Lehn P., Mornon J. P., Davies G. (1995) Conserved catalytic machinery and the prediction of a common fold for several families of glycosyl hydrolases. Proc. Natl. Acad. Sci. U.S.A. 92, 7090–7094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jenkins J., Lo Leggio L., Harris G., Pickersgill R. (1995) β-Glucosidase, β-galactosidase, family A cellulases, family F xylanases and two barley glycanases form a superfamily of enzymes with 8-fold β/α architecture and with two conserved glutamates near the carboxy-terminal ends of β-strands four and seven. FEBS Lett. 362, 281–285 [DOI] [PubMed] [Google Scholar]
- 10. Wiesmann C., Beste G., Hengstenberg W., Schulz G. E. (1995) The three-dimensional structure of 6-phospho-β-galactosidase from Lactococcus lactis. Structure 3, 961–968 [DOI] [PubMed] [Google Scholar]
- 11. Barrett T., Suresh C. G., Tolley S. P., Dodson E. J., Hughes M. A. (1995) The crystal structure of a cyanogenic β-glucosidase from white clover, a family 1 glycosyl hydrolase. Structure 3, 951–960 [DOI] [PubMed] [Google Scholar]
- 12. Jacobson R. H., Zhang X. J., DuBose R. F., Matthews B. W. (1994) Three-dimensional structure of β-galactosidase from E. coli. Nature 369, 761–766 [DOI] [PubMed] [Google Scholar]
- 13. Dominguez R., Souchon H., Spinelli S., Dauter Z., Wilson K. S., Chauvaux S., Béguin P., Alzari P. M. (1995) A common protein fold and similar active site in two distinct families of β-glycanases. Nat. Struct. Biol. 2, 569–576 [DOI] [PubMed] [Google Scholar]
- 14. Ducros V., Czjzek M., Belaich A., Gaudin C., Fierobe H. P., Belaich J. P., Davies G. J., Haser R. (1995) Crystal structure of the catalytic domain of a bacterial cellulase belonging to family 5. Structure 3, 939–949 [DOI] [PubMed] [Google Scholar]
- 15. Derewenda U., Swenson L., Green R., Wei Y., Morosoli R., Shareck F., Kluepfel D., Derewenda Z. S. (1994) Crystal structure, at 2.6-Å resolution, of the Streptomyces lividans xylanase A, a member of the F family of β-1,4-d-glycanases. J. Biol. Chem. 269, 20811–20814 [PubMed] [Google Scholar]
- 16. White A., Withers S. G., Gilkes N. R., Rose D. R. (1994) Crystal structure of the catalytic domain of the β-1,4-glycanase cex from Cellulomonas fimi. Biochemistry 33, 12546–12552 [DOI] [PubMed] [Google Scholar]
- 17. Harris G. W., Jenkins J. A., Connerton I., Cummings N., Lo Leggio L., Scott M., Hazlewood G. P., Laurie J. I., Gilbert H. J., Pickersgill R. W. (1994) Structure of the catalytic core of the family F xylanase from Pseudomonas fluorescens and identification of the xylopentaose-binding sites. Structure 2, 1107–1116 [DOI] [PubMed] [Google Scholar]
- 18. Dominguez R., Souchon H., Alzari P. M. (1994) Characterization of two crystal forms of Clostridium thermocellum endoglucanase CelC. Proteins 19, 158–160 [DOI] [PubMed] [Google Scholar]
- 19. Varghese J. N., Garrett T. P., Colman P. M., Chen L., Høj P. B., Fincher G. B. (1994) Three-dimensional structures of two plant β-glucan endohydrolases with distinct substrate specificities. Proc. Natl. Acad. Sci. U.S.A. 91, 2785–2789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Søgaard M., Kadziola A., Haser R., Svensson B. (1993) Site-directed mutagenesis of histidine 93, aspartic acid 180, glutamic acid 205, histidine 290, and aspartic acid 291 at the active site and tryptophan 279 at the raw starch binding site in barley α-amylase 1. J. Biol. Chem. 268, 22480–22484 [PubMed] [Google Scholar]
- 21. McCarter J. D., Withers S. G. (1994) Mechanisms of enzymatic glycoside hydrolysis. Curr. Opin. Struct. Biol. 4, 885–892 [DOI] [PubMed] [Google Scholar]
- 22. Domínguez R., Souchon H., Lascombe M., Alzari P. M. (1996) The crystal structure of a family 5 endoglucanase mutant in complexed and uncomplexed forms reveals an induced fit activation mechanism. J. Mol. Biol. 257, 1042–1051 [DOI] [PubMed] [Google Scholar]
- 23. Vieille C., Zeikus G. J. (2001) Hyperthermophilic enzymes: sources, uses, and molecular mechanisms for thermostability. Microbiol. Mol. Biol. Rev. 65, 1–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhaxybayeva O., Swithers K. S., Lapierre P., Fournier G. P., Bickhart D. M., DeBoy R. T., Nelson K. E., Nesbø C. L., Doolittle W. F., Gogarten J. P., Noll K. M. (2009) On the chimeric nature, thermophilic origin, and phylogenetic placement of the Thermotogales. Proc. Natl. Acad. Sci. U.S.A. 106, 5865–5870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang Y., Wang X., Tang R., Yu S., Zheng B., Feng Y. (2010) A novel thermostable cellulase from Fervidobacterium nodosum. J. Mol. Catal. B Enzym. 66, 294–301 [Google Scholar]
- 26. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Macromol. Crystallogr. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 27. Sheldrick G. M. (2008) A short history of SHELX. Acta Crystallogr. A 64, 112–122 [DOI] [PubMed] [Google Scholar]
- 28. Terwilliger T. C., Berendzen J. (1999) Automated MAD and MIR structure solution. Acta Crystallogr. D. Biol. Crystallogr. 55, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Terwilliger T. C. (2000) Maximum-likelihood density modification. Acta Crystallogr. D. Biol. Crystallogr. 56, 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Perrakis A., Harkiolaki M., Wilson K. S., Lamzin V. S. (2001) ARP/wARP and molecular replacement. Acta Crystallogr. D. Biol. Crystallogr. 57, 1445–1450 [DOI] [PubMed] [Google Scholar]
- 31. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 32. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 33. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zheng B., Yang W., Wang Y., Feng Y., Lou Z. (2009) Crystallization and preliminary crystallographic analysis of thermophilic cellulase from Fervidobacterium nodosum Rt17-B1. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 65, 219–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Violot S., Aghajari N., Czjzek M., Feller G., Sonan G. K., Gouet P., Gerday C., Haser R., Receveur-Bréchot V. (2005) Structure of a full length psychrophilic cellulase from Pseudoalteromonas haloplanktis revealed by x-ray diffraction and small angle x-ray scattering. J. Mol. Biol. 348, 1211–1224 [DOI] [PubMed] [Google Scholar]
- 36. Chan M. K., Mukund S., Kletzin A., Adams M. W., Rees D. C. (1995) Structure of a hyperthermophilic tungstopterin enzyme, aldehyde ferredoxin oxidoreductase. Science 267, 1463–1469 [DOI] [PubMed] [Google Scholar]
- 37. Russell R. J., Hough D. W., Danson M. J., Taylor G. L. (1994) The crystal structure of citrate synthase from the thermophilic archaeon, Thermoplasma acidophilum. Structure 2, 1157–1167 [DOI] [PubMed] [Google Scholar]
- 38. Leszczynski J. F., Rose G. D. (1986) Loops in globular proteins: a novel category of secondary structure. Science 234, 849–855 [DOI] [PubMed] [Google Scholar]
- 39. Daggett V., Levitt M. (1993) Protein unfolding pathways explored through molecular dynamics simulations. J. Mol. Biol. 232, 600–619 [DOI] [PubMed] [Google Scholar]
- 40. Sadeghi M., Naderi-Manesh H., Zarrabi M., Ranjbar B. (2006) Effective factors in thermostability of thermophilic proteins. Biophys. Chem. 119, 256–270 [DOI] [PubMed] [Google Scholar]
- 41. Sinnot M. L. (1990) Catalytic mechanism of enzymic glycosyl transfer. Chem. Rev. 90, 1171–1202 [Google Scholar]
- 42. Barras F., Bortoli-German I., Bauzan M., Rouvier J., Gey C., Heyraud A., Henrissat B. (1992) Stereochemistry of the hydrolysis reaction catalyzed by endoglucanase Z from Erwinia chrysanthemi. FEBS Lett. 300, 145–148 [DOI] [PubMed] [Google Scholar]
- 43. Baird S. D., Hefford M. A., Johnson D. A., Sung W. L., Yaguchi M., Seligy V. L. (1990) The Glu residue in the conserved Asn-Glu-Pro sequence of two highly divergent endo-β-1,4-glucanases is essential for enzymatic activity. Biochem. Biophys. Res. Commun. 169, 1035–1039 [DOI] [PubMed] [Google Scholar]
- 44. Py B., Bortoli-German I., Haiech J., Chippaux M., Barras F. (1991) Cellulase EGZ of Erwinia chrysanthemi: structural organization and importance of His98 and Glu133 residues for catalysis. Protein Eng. 4, 325–333 [DOI] [PubMed] [Google Scholar]
- 45. Clarke A. J., Yaguchi M. (1985) The role of carboxyl groups in the function of endo-β-1,4-glucanase from Schizophyllum commune. Eur. J. Biochem. 149, 233–238 [DOI] [PubMed] [Google Scholar]
- 46. Tomme P., Claeyssens M. (1989) Identification of a functionally important carboxyl group in cellobiohydrolase I from Trichoderma reesei: A chemical modification study. FEBS Lett. 243, 239–243 [Google Scholar]
- 47. Withers S. G., Antony R., Warren J., Street I. P., Rupitz K., Kempton. J. P., Aebersold R. (1990) Unequivocal demonstration of the involvement of a glutamate residue as a nucleophile in the mechanism of a retaining glycosidase. J. Am. Chem. Soc. 112, 5887–5889 [Google Scholar]
- 48. Bortoli-German I., Haiech J., Chippaux M., Barras F. (1995) Informational suppression to investigate structural functional and evolutionary aspects of the Erwinia chrysanthemi cellulase EGZ. J. Mol. Biol. 246, 82–94 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.