Background: ERdj4 is an endoplasmic reticulum chaperone co-factor that assists with destruction of misfolded secretory proteins.
Results: ERdj4 is a mobile soluble luminal protein that is recruited to the misfolded secretory protein quality control machinery.
Conclusion: The role of ERdj4 in secretory protein destruction is specified by association with the degradation machinery.
Significance: These studies provide a new insight into how different ERdj family members regulate the fates of their clients.
Keywords: Cell Biology, Chaperone Chaperonin, Endoplasmic Reticulum (ER), ER-associated Degradation, Protein Misfolding, Protein Processing, Protein Sorting, DnaJ, FRAP, Signal Sequence
Abstract
Protein localization within cells regulates accessibility for interactions with co-factors and substrates. The endoplasmic reticulum (ER) BiP co-factor ERdj4 is up-regulated by ER stress and has been implicated in ER-associated degradation (ERAD) of multiple unfolded secretory proteins. Several other ERdj family members tend to interact selectively with nascent proteins, presumably because those ERdj proteins associate with the Sec61 translocon that facilitates entry of nascent proteins into the ER. How ERdj4 selects and targets terminally misfolded proteins for destruction remains poorly understood. In this study, we determined properties of ERdj4 that might aid in this function. ERdj4 was reported to retain its signal sequence and to be resistant to mild detergent extraction, suggesting that it was an integral membrane protein. However, live cell photobleaching analyses of GFP-tagged ERdj4 revealed that the protein exhibits diffusion coefficients uncommonly high for an ER integral membrane protein and more similar to the mobility of a soluble luminal protein. Biochemical characterization established that the ERdj4 signal sequence is cleaved to yield a soluble protein. Importantly, we found that both endogenous and overexpressed ERdj4 associate with the integral membrane protein, Derlin-1. Our findings now directly link ERdj4 to the ERAD machinery and suggest a model in which ERjd4 could help recruit clients from throughout the ER to ERAD sites.
Introduction
Within the environment of a cell, the ability of a protein to perform its function depends on the “availability” of the protein. The probability of encountering and interacting with substrates reflects a combination of subcellular localization, abundance, and the mobilities of both the protein and its substrates. For example, a freely and rapidly diffusing protein can readily encounter a substrate, even if the substrate is relatively immobile. In contrast, if both protein and substrate diffuse slowly and are expressed at low levels, then encounters will be rare events. The latter situation could represent an inefficient reaction or a cellular regulatory mechanism. In this study, we describe how ERdj4 availability is regulated.
In the endoplasmic reticulum (ER),5 nascent proteins fold and assemble with the assistance of an array of chaperones (1, 2). The ER Hsp70 chaperone, BiP, participates in both protein folding and quality control of unfoldable proteins (3, 4). Similar to all of the Hsp70 family members, the ability of BiP to perform both of these roles depends on its interactions with co-factors, including the DnaJ-like proteins, ERdjs, that stimulate the ATPase activity of BiP and stabilize its binding to substrates. In many cases, ERdjs can bind directly to unfolded proteins and hand the proteins off to BiP (5, 6). By stimulating the ATPase activity of BiP, ERdjs transition BiP from a low to high affinity substrate binding form (7). In other cases, DnaJ-like proteins can stimulate the ATPase activity of their Hsp70 partner without interacting directly with substrates (8, 9). The nucleotide states of Hsp70 family members regulate their conformation and substrate affinity. Nucleotide-free and ATP-bound Hsp70 family members have low affinity for substrates, whereas ADP-bound Hsp70s bind substrates with high affinity (10). Similar to all Hsp70s, the intrinsic ATP hydrolysis rate of BiP is inefficient, and ERdjs accelerate the reaction 2–5-fold (5, 11–15).
The ER DnaJ-like, termed ERdj, proteins include seven members (ERdj1/Mtj1, ERdj2/Sec63, ERdj3/HEDJ, ERdj4/Mdg1, ERdj5/JPDI, ERdj6/p58IPK, and ERdj7), four (ERdj1, 2, 4, and 7) of which have been reported to be integral membrane proteins (3). ERdj1–3 and ERdj6 have been linked to folding of nascent proteins, often at the translocation channel (3, 9, 16). In contrast, ERdj4 and 5 play important roles in the ER-associated degradation (ERAD) of unfolded secretory proteins (17–19). Interestingly, ERdj4 is reported to be membrane-associated (11), whereas ERdj5 is a soluble protein (18).
Subcompartment localization may help define substrate or functional specificity for ERdj proteins, similar to the way that lectin chaperones, membrane-associated calnexin, and soluble calreticulin interact with different sets of substrates (20). ERdj4 levels are exceptionally low in homeostatic cells (15) but rise significantly when the ER unfolded protein response is activated (11). Together, these findings suggest that ERdj4 would normally encounter substrates at relatively slow rates because of both the low mobility caused by membrane association and its low abundance. Unfolded protein response-mediated up-regulation could compensate for the slow membrane diffusion and increase encounters by simply increasing ERdj4 levels. Dong et al. (17) reported that overexpression of ERdj4, indeed, increases turnover of mutants of the surfactant protein SP-C.
In this study, we investigated regulation of ERdj4 availability in live cells through photobleaching analysis. We found that ERdj4 is mobile throughout the ER and, surprisingly, that it is not an integral membrane protein. However, ERdj4 does associate with at least one ER integral membrane protein, the ERAD component Derlin-1. This finding suggests a mechanism for how differences in ERdj substrate specificity are achieved. Just as ERdj1–3 preferably interact with nascent proteins, at least in part because of association with the ER translocon, ERdj4 interactions with clients slated for ERAD could be specified through direct association with ERAD machinery.
EXPERIMENTAL PROCEDURES
Plasmid Construction and Transfection
To construct ERdj4-GFP and ERdj4sfGFP, mouse ERdj4 cDNA (11) was PCR-amplified with primers: 5′-GATCGCTAGCGCCACCATGGCTACTCCACAGTCAG (forward) and 5′-GATCACCGGTCCTTGTCCTGAACAATCAG (reverse) that include a Kozak sequence (bold type) and NheI and AgeI sites (underlined type) for ease of subcloning into a monomeric GFP (21) or sfGFP (22, 23) vector based on the Clontech N1-GFP backbone. VSV G-GFP (24) was mutated to increase ER retention without incubating cells at the restrictive temperature of 40 °C. The enhance ER exit motif of DXE was mutated to EXE, which dramatically slows ER exit (25). All of the constructs were confirmed by sequencing. ER-RFP (26), ERdj4 (11) and BiP (27) constructs have been previously described. All of the constructs were transiently transfected for 16–48 h into cells using either Lipofectamine 2000 (Invitrogen) or Gene Cellin (BioCellChallenge SAS, Toulon, France) according to the manufacturer's instructions.
Cells and Reagents
MDCK and U2OS cells (obtained from ATCC, Manassas, VA) were all grown in RPMI lacking phenol red plus GlutaMAX (Invitrogen), 10% heat-inactivated fetal bovine serum, and penicillin/streptomycin at 37 °C in 5% CO2. We limited imaging to low to moderate expressing cells. For all imaging experiments, the cells were grown in 8-well LabTek coverglass chambers (Nalge Nunc International, Rochester, NY). 0.2 μm pactamycin (a gift from E. Steinbrecher, Pharmacia Corp., Peapack, NJ) was dissolved in Me2SO and applied for 1 h (28). The anti-rodent BiP polyclonal antibody has been previously described (27). The Hrd1 antibody was a gift from Dr. Richard Wojcikiewicz (SUNY Upstate Medical University). The polyclonal Derlin-1 antiserum was kindly provided by Dr. Yihong Ye (NIDDK, National Institutes of Health, Bethesda, MD), although the polyclonal p97 antibody was a generous gift from Dr. George DeMartino (University of Texas Southwestern, Dallas, TX). Production of a monoclonal antibody against ERdj4 is described in a study about to be submitted by our group.6 The Hsc70 antibody was from Santa Cruz Biotechnology. Anti-PDI (SPA 890; Stressgen, now Enzo Life Sciences International, Plymouth Meeting, PA) and used at 1:500 for immunofluorescence. Monoclonal anti-p115 was a gift from the late Dr. Dennis Shields (Albert Einstein College of Medicine) and used at 1:300 for immunofluorescence. Rabbit anti-GFP was a gift from Ramanujan Hegde (Medical Research Council Laboratory of Molecular Biology, Cambridge, UK) and used at 1:10,000 for immunoblotting.
Immunofluorescence and Imaging of Live and Fixed Cells
For immunofluorescence, the cells were fixed with freshly diluted 3.7% formaldehyde in PBS for 15 min at room temperature, permeabilized with PBS with 0.1% Triton X-100. Blocking was performed in 10% fetal bovine serum in 1× PBS. Subsequently, the cells were labeled with primary antibodies, followed by Alexa 555-conjugated anti-rabbit or anti-mouse IgG secondary antibodies (Invitrogen). Live cells were imaged in phenol red-free RPMI supplemented with 10 mm Hepes, GlutaMAX, and 10% fetal bovine serum. Live cells were imaged on a 37 °C environmentally controlled chamber of a confocal microscope system (Duoscan; Carl Zeiss MicroImaging, Inc.) with a 63×/1.4 NA oil objective and a 489-nm 100-milliwatt diode laser with a 500–550-nm band pass filter for GFP and a 40-milliwatt 561-nm diode laser with a 565-nm long pass filter for mRFP and Cy3. Composite figures were prepared using Photoshop CS4 and CS5 and Illustrator CS4 software (Adobe Systems, San Jose, CA).
Photobleaching Analysis
Fluorescence recovery after photobleaching (FRAP) and fluorescence loss in photobleaching (FLIP) were performed by photobleaching a small region of interest (ROI) and monitoring either fluorescence recovery or loss with repeated bleaching in the same ROI over time, as described previously (29, 30). Fluorescence intensity plots and effective diffusion coefficient (Deff) measurements were calculated as described previously (29, 30). To create the fluorescence recovery curves, the fluorescence intensities were transformed into a 0–100% scale and were plotted using Kaleidagraph 3.5 (Synergy Software). The p values were calculated using a Student's two-tailed t test in Excel (Microsoft) or Prism 5.0c (Graphpad Software Inc., La Jolla, CA).
Immunoblots and Immunoprecipitations
Total cell lysates for immunoblotting were prepared by lysing COS-1 or MDCK cells in (1% SDS, 0.1 m Tris, pH 8.0) 6-well plates at 80–90% confluence. The lysates were separated on 12% Tris-Tricine gels, transferred to nitrocellulose, probed with the indicated antibodies, developed using Pierce enhanced chemiluminescent reagents (Thermo Scientific, Rockford, IL), and exposed to x-ray film. Anti-GFP, horseradish peroxidase-labeled anti-rabbit, and anti-mouse reagents were purchased (Jackson Immunoresearch Laboratories). To isolate protein from cells overexpressing ERdj4, 2 × 107 COS-1 cells were transfected with pSGmERdj4 and harvested 24 h later in Nonidet P-40 lysis buffer (50 mm Tris, 150 mm NaCl, 0.5% Nonidet P-40, 0.5% deoxycholate, 1 mm PMSF, and Roche complete protease inhibitor mixture EDTA free (Roche Applied Science)). Cell lysates were divided evenly into three aliquots, each of which was immunoprecipitated with the indicated antibodies. IgG was used as negative control. Immunoprecipitated proteins were separated to SDS-PAGE and immunoblotted with the indicated antibodies. To isolate endogenous ERdj4 complexes, 1 × 108 P3U.1 cells were incubated with the membrane-permeable cross-linking agent dithiobis(succinimidyl propionate) as previously described (5). The cells were incubated in Hepes homogenization buffer (25 mm Hepes, 125 mm KCl, 1 mm PMSF, and Roche complete protease inhibitor mixture EDTA free) and broken in a Dounce homogenizer, and membranes were pelleted by centrifugation at 500 × g. The isolated membranes, which contained the ER, were lysed in Nonidet P-40 buffer, divided into equal fractions, and immunoprecipitated with the indicated antisera. Rabbit IgG was used as negative control. Immunoprecipitated proteins were separated by SDS-PAGE and immunoblotted.
Membrane Fractionation of Cells Overexpressing Murine ERdj4
Nontransfected P3U.1 cells or COS-1 cells transfected with pSGmERdj4 and harvested at 24 h were fractionated. Membranes fractions were prepared by homogenizing 2.6 × 107 cells in 5 ml of Hepes buffer (25 mm Hepes, 125 mm KCl), and a postnuclear supernatant containing ER membranes was obtained by centrifugation at 500 × g for 10 min. The resulting supernatant was divided evenly into six 250-μl aliquots. After higher speed centrifugation (10,000 × g), the pelleted membranes were resuspended in 250 μl of PBS alone or PBS containing the indicated amount of digitonin or 0.1 m Na2CO3. Samples were rocked at 4 °C for 1 h. In all cases, the samples were then centrifuged at 16,000 × g to repellet the membranes and attached proteins. Cell equivalents of the membrane pellets and supernatants were subjected to electrophoresis and immunoblotting.
Mass Spectrometry and Edman Degradation
Exogenously expressed ERdj4 was immunoprecipitated from COS-1 cells 40 h post-transfection, separated on SDS-polyacrylamide gels and stained with Sypro Ruby. Excised protein was treated with trypsin and subjected to electron spray ionization mass spectrometry (Hartwell Center for Bioinformatics). For N-terminal sequencing of ERdj4, ∼6 × 107 COS-1 cells transiently transfected with pSGmERdj4 were lysed in Nonidet P-40 lysis buffer and immunoprecipitated with a monoclonal ERdj4 antibody. The immunoprecipitated material was separated by SDS-PAGE, transferred to a PVDF membrane, stained with Coomassie Blue, excised, and analyzed by Edman degradation in a ABI 494 protein sequencer (Tufts University Core Facility).
Statistics
To minimize the cell-to-cell variables such as cell cycle stage or contact inhibition, we always selected flat, mononucleate, nonmitotic cells in cultures at between 40 and 70% confluence and expressing low to modest levels of fluorescent proteins for analysis. The data were analyzed using a two-tailed Student's t test (using Graphpad Prism 5.0c; Graphpad Software La Jolla, CA) to compare the different conditions. The variances of data sets were compared using an F-test (Prism) to establish whether to use equal or nonequal variance t tests. Significance was tested using α ≤ 0.01.
RESULTS
The ERdj4-GFP Fusion Is Functional
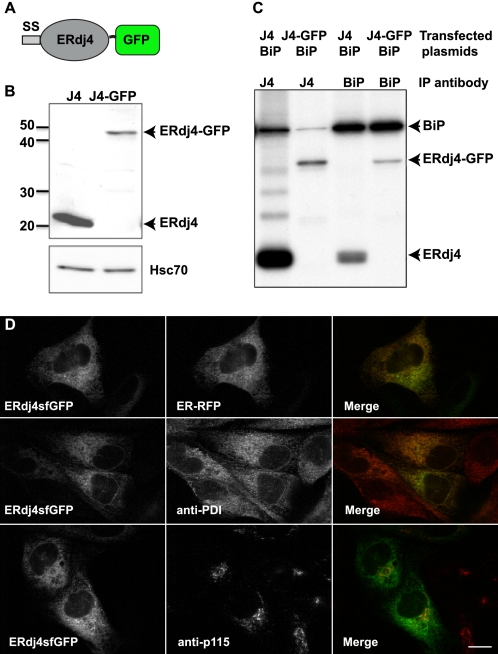
To better understand how ERdj4 encounters ERAD substrates and participates in the ERAD process, we created a live cell reporter for ERdj4. To visualize ERdj4 behavior in live cells, we generated a fusion of mouse ERdj4 with superfolder GFP (sfGFP) (Fig. 1A). The sfGFP-tagged ERdj4 had a mobility on SDS gels of ∼50 kDa, which is consistent with a 27-kDa increase in size caused by the addition of GFP (Fig. 1B). The presence of a 5-nm sfGFP did not obviously impair ERdj4 functionality, as determined by comparisons with untagged or HA-tagged variants of ERdj4. The sfGFP fusion associated as well as untagged ERdj4 with its Hsp70 partner, BiP, in co-immunoprecipitation experiments (Fig. 1C). In addition, we observed co-localization of ERdj4-sfGFP with the ER marker (ER-RFP) by live cell imaging and with the endogenous ER resident folding enzyme protein disulfide isomerase by immunofluorescence staining (Fig. 1D). Similar ER localization was observed when ERdj4, fused to a much smaller HA tag, was expressed in MDCK and COS-1 cells (supplemental Fig. S1). In addition, ERdj4sfGFP did not co-localize with the Golgi complex marker p115. These results confirmed that the sfGFP tag does not interrupt known characteristics of ERdj4 and argue that mobility studies performed with this construct are likely to be informative. Notably, we consistently observe lower expression levels of the sfGFP fusion compared with the overexpressed untagged protein. This is possibly due to differences in the vectors used to express these two proteins. The untagged construct vector contains a β-globin intron that significantly stabilizes and thus increases mRNA levels (31), whereas the sfGFP fusion does not.
FIGURE 1.
ERdj4-GFP behaves like the untagged ERdj4. A, schematic illustration of the ERdj4-GFP fusion construct. B, immunoblot of COS-1 cells transiently transfected with ERdj4 or ERdj4-GFP and lysates were subjected to immunoblotting with anti-ERdj4. Anti-Hsc70 staining indicates that similar amounts of cell lysates were loaded. C, immunoprecipitated ERdj4-GFP and ERdj4 interact with BiP in vivo. COS-1 cells were transiently transfected with plasmids expressing ERdj4 or ERdj4-GFP in combination with BiP. The cells were labeled 24 h post-transfection with [35S]methionine for 1 h, and the proteins were cross-linked by treating with 150 μg/ml dithiobis(succinimidyl propionate). The cell lysates were immunoprecipitated with either an anti-ERdj4 or anti-BiP antibody, as indicated. D, co-localization of ERdj4sfGFP with ER-RFP and anti-protein disulfide isomerase (PDI) by immunofluorescence staining, but not with the Golgi complex marker, p115. Scale bar, 10 μm.
ERdj4sfGFP Is More Mobile than Predicted in Cells
Using our new live cell reporter of ERdj4 behavior, we asked how mobile ERdj4 was in cells, which would affect its ability to encounter substrates. ERdj1 and 2 are integral membrane proteins that associate with the essentially immobile Sec61 translocation channel (32–34). In these cases, substrate interactions should be relatively restricted to nascent substrates. As a membrane protein, ERdj4 association with ERAD machinery could similarly restrict the mobility of the co-chaperone. In this case, either substrates would have to be brought to ERdj4, or the association of ERdj4 with ERAD components could be triggered by substrate interaction. To investigate ERdj4 availability, we performed photobleaching experiments on live cells expressing the reporter.
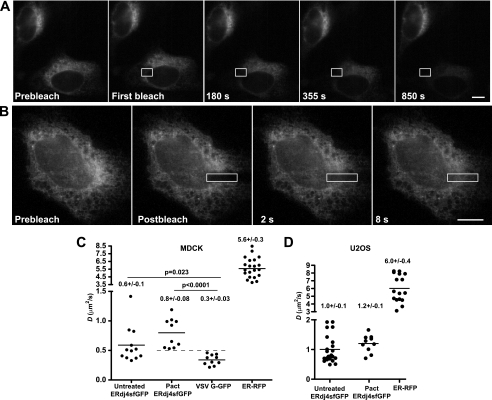
First, we asked whether ERdj4sfGFP is at all mobile. Using a technique termed FLIP, a discrete region of the cell was subjected to high intensity laser light to destroy GFP fluorescence. Repeated photobleaching of the same ROI homogenously depleted fluorescence from the entire ER, demonstrating that GFP-tagged ERdj4 is mobile throughout the ER lumen. Although informative data on mobility are obtained with FLIP, this assay does not quantitate the degree of protein mobility. To quantitate mobility, we performed FRAP (Fig. 2B) and then determined the effective diffusion coefficient (Deff) of the protein. In MDCK cells, the ERdj4sfGFP Deff value (0.6 ± 0.1 μm2/s) was much lower than a freely diffusing fluorescent protein, ER-RFP, but similar to that of the binding partner of ERdj4, BiP (∼0.6 μm2/s (35), as well as other ER chaperones calreticulin and GRP94 (26, 36) (Fig. 2C). We then blocked the synthesis of nascent proteins in the ER to determine whether ERdj4 mobility was sensitive to decreased availability of substrates. Translational inhibition with pactamycin slightly increased the Deff value for ERdj4, although this was not statistically significant (p = 0.1124) compared with that obtained in untreated cells (Fig. 2C). Similar results were also observed in a second cell type, U2OS (Fig. 2D). Although the mobilities of other chaperones, calreticulin and BiP, have been shown to be dependent on substrate levels (26, 35), ERdj4 mobility appears to be relatively insensitive to translational inhibition. This difference in behavior could be due to association of ERdj4 with the ER membrane or an ER membrane protein complex that is relatively stable. Alternatively, it is conceivable that ERdj4 client levels are inherently low at steady state, and/or client interactions are fleetingly transient.
FIGURE 2.
ERdj4sfGFP is a dynamic ER protein. A, FLIP experiment of an MDCK cell transiently expressing ERdj4sfGFP. The cell was repeatedly photobleached in the ROI (white box), and images are collected between bleaches. If the fluorescent proteins are mobile and can pass through the ROI, all fluorescence will be depleted. B, FRAP of U2OS cell transiently expressing ERdj4sfGFP. The cells were photobleached a single time in the ROI (white box), and recovery of the signal was monitored over the indicated times. Scale bars, 10 μm. C and D, FRAP Deff values with ERdj4sfGFP, VSV G-GFP, or ER-RFP transiently transfected in MDCK (left) or U2OS cells (right). Each dot represents the Deff value for a single cell and solid lines indicate mean Deff values. Translational inhibition was performed with 0.2 μm pactamycin (Pact) for 1 h to deplete cells of nascent substrates for ERdj4. The mean Deff value ± S.E. is reported over each column. The dashed line in C was added to emphasize the lack of overlap between the Deff of a well established integral membrane protein (VSV G) and the reported transmembrane protein ERdj4 (plus pactamycin (Pact)). The overall higher D values in U2OS cells likely reflect either decreased ER tortuosity (58) and/or decreased viscosity, because molecular diffusion is dependent on environmental viscosity (59).
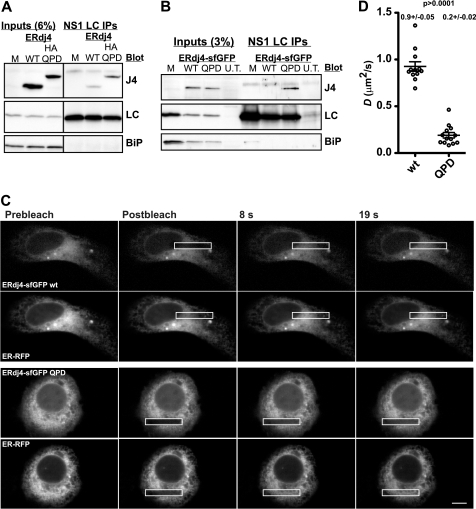
To investigate these possibilities, we exploited a previously described ERdj4 mutant (H54Q) that inhibits the ability of ERdj4 to stimulate BiP ATPase activity (11) and also appears to decrease release of clients from ERdj4 (17). Note that the mutant did not significantly bind correctly folding proteins (17). In keeping with this, both HA- and GFP-tagged wild-type ERdj4 bound much less efficiently than the corresponding QPD mutants to the client NS1 LC (Fig. 3, A and B). Taken together with the data from Dong et al. (17), ERdj4 interactions with clients appear to be normally highly transient. We then asked whether there are significant levels of endogenous clients in unstressed cells. FRAP analysis of chaperone mobility can report on changes in global client levels (35). FRAP analysis of the QPD mutant revealed significantly slower fluorescence recovery compared with WT cells (Fig. 3, C and D). The decreased recovery was not due to disruption of the ER, because the inert reporter ER-RFP still readily recovered in co-transfected cells (Fig. 3). Thus, ERdj4 is constantly engaging and releasing clients in cells, but this flux is so rapid that only trapping mutants can reveal the extent of the client burden.
FIGURE 3.
Mutant ERdj4sfGFP is significantly less mobile. Mutant ERdj4 binds with high affinity to substrates as revealed in co-immunoprecipitation of ERdj4 and substrate experiments (A and B). A, COS-1 cells were transfected with ERdj4 or an HA-tagged ERdj4 QPD (H54Q) mutant in addition to NS1 κ LC and BiP and harvested at 24 h. The cells not transfected with ERdj4 (M), and untransfected cells (U.T.) were used as controls. Cell lysates were immunoprecipitated with anti-κ light chain, subjected to electrophoresis, and immunoblotted with the indicated antibodies. B, conditions were identical as in A, except cells were transfected with the indicated ERdj4-sfGFP variants. In both A and B, minimal amounts of WTERdj4 are detected, whereas substantial levels of mutant associate with LC. C, FRAP images series of HeLa cells co-expressing WT or QPD ERdj4-sfGFP and ER-RFP. Note the poor recovery of QPD ERdj4-sfGFP. D, plot of D values WT and QPD ERdj4-sfGFP expressed in U2OS cells and analyzed by FRAP.
The FRAP results for ERdj4 were surprising, because it was previously reported to be an integral membrane protein (11), and the ER integral membrane proteins that have been characterized by this method have Deff values of between ∼0.3 and 0.5 μm2/s (37–40). The higher mobility of ERdj4sfGFP raised an important question: was the mobility of ERdj4sfGFP distinguishable from a typical integral membrane protein in the ER? To answer this, FRAP analysis was performed on an ER-retained mutant VSV G-GFP chimera (25, 39), which contains a single transmembrane domain. We observed a striking difference between the mobilities of ERdj4 in both untreated and translationally inhibited cells compared with VSV G-GFP (p = 0.0243 for untreated cells and p > 0.0001 for pactamycin-treated cells) (Fig. 2C). In both cases, ERdj4 diffused at substantially higher rates (0.6 and 0.8 μm2/s versus 0.3 μm2/s) than VSV G. Similarly, in U2OS cells, the single membrane spanning Lull protein diffuses at the much slower Deff of 0.3 μm2/s (40). Furthermore, the experimental variation in observed Deff values was substantially tighter for VSV G-GFP, consistent with a protein constrained in mobility by a high viscosity environment. Together, these data suggested that the mobility of ERdj4 was either atypical for an integral membrane protein or that ERdj4 might not be an integral membrane protein.
Previous data for endogenous ERdj4 suggested the protein tends to co-fractionate in a detergent extraction assay similar to at least one ER membrane protein, calnexin (11). We therefore revisited this point. ERdj4 has been reported to be a nonsecreted, type II resident ER protein that is anchored in the ER membrane via an uncleaved N-terminal signal sequence (11). However, ERdj4 does not possess an NH2-domain double arginine (RR) motif (41) that is typically required for retention of Type II integral membrane proteins in the ER. A closer inspection of the signal sequence of ERdj4 revealed that it has a predicted hydrophobic α-helical transmembrane domain of only 11 amino acids (underlined) (NH2-MATPQSVFVFAICILMITELILAS), which is shorter than reported for most resident ER integral membrane proteins, which are typically 14–17 hydrophobic amino acids in length (42, 43). In fact, in a survey of 40% of the human integral membrane proteome, no transmembrane domains shorter than 14 hydrophobic amino acids were reported on known ER membrane proteins (43). Presumably, the minimum transmembrane length is constrained by the thickness of the ER lipid bilayer (44). These features raise the possibility that ERdj4 is not stably integrated into the ER membrane as originally proposed.
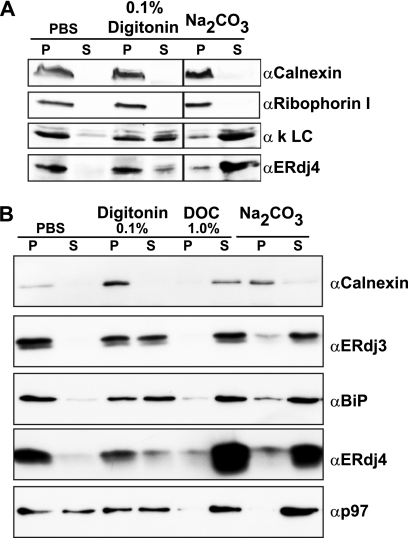
We investigated the membrane association and signal sequence (SS) processing of both endogenous and transiently transfected ERdj4 by several biochemical methods. First, the cells were extracted with the detergent digitonin, which selectively binds cholesterol. Because the ER membrane has low concentrations of cholesterol, integral membrane and membrane-associated proteins tend to remain associated with pellet fractions. Our control integral membrane protein, calnexin, fractionated primarily in the pellet fraction (Fig. 4). The soluble proteins ERdj3 and BiP (luminal) and p97 (cytoplasmic) were more readily extracted into the supernatant when 0.1% digitonin was used. In contrast, ERdj4 fractionates primarily in the pellet fraction, as had been previously observed (11). Extraction with a membrane-disrupting detergent, deoxycholate, released all of the proteins into the soluble fraction (Fig. 4). To determine whether ERdj4 is integrated into the ER membrane, we performed sodium carbonate extraction (45). Sodium carbonate opens microsome membranes into sheets and disrupts many protein-protein interactions, primarily leaving only integral membrane proteins associated in the pellet fraction. After treating microsomes with sodium carbonate, the control membrane protein calnexin was retained in the pellet fraction, and the control soluble proteins, as well as ERdj4, were quantitatively released from the membranes (Fig. 4). Together, these results argue ERdj4 is not an integral membrane protein.
FIGURE 4.
ERdj4 is not integrated in the ER membrane. A, fractionation of endogenous ERdj4 from P3U.1 cells. The membranes were prepared from 2.5 × 107 cells and divided evenly into five aliquots. After pelleting by centrifugation, the membranes were resuspended in either PBS or PBS containing 0.1% digitonin, 1% deoxycholate (DOC), or 0.1 m Na2CO3. The samples were rocked at 4 °C for 1 h and then centrifuged to pellet the membranes. Pellets (P) and supernatants (S) were subjected to electrophoresis and immunoblotting with the indicated antibodies. B, COS-1 cells were transfected for 24 h with pSGmERdj4. The membranes were processed, separated by gel electrophoresis, and immunoblotted with the indicated antibodies as in A.
The Signal Sequence of ERdj4 Is Cleaved
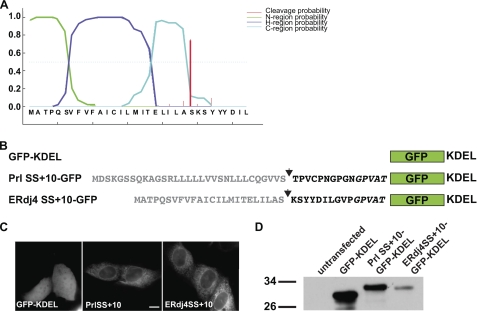
Since the original study describing ERdj4 was reported (11), new and improved computational tools have become available for predicting ER targeting SSs and their potential to be cleaved (46). We submitted the mouse and human ERdj4 protein sequences to the SignalP 3.0 server, which predicted that the SS is cleaved with very high probability (0.964 for mouse and 0.953 for human) and that this most likely occurs between amino acids Ala-23 and Ser-24 (Fig. 5A). The small difference in the probability scores is likely due to the fact that the human signal sequence differs from mouse by two conservative amino acids, VI7I and EI9V, respectively. Although the SignalP 3.0 algorithm is robust, we sought biochemical evidence that the SS is cleaved. We expressed a series of ER-targeted GFP reporters (Fig. 5, B and C) that were predicted to have defined sizes following SS cleavage. Using 12% Tricine-SDS-PAGE, we found that the ERdj4 reporter migrated at a size indistinguishable from the similarly sized Prl SS+10 reporter (Fig. 5D). The prolactin SS is an efficiently cleaved SS, and we could demonstrate this with additional control proteins. This confirmed that our SDS-PAGE achieves sufficient resolution to distinguish protein species that differ by 2–13 amino acids (supplemental Fig. S2). Together, these data strongly argue the ERdj4 SS also can be efficiently cleaved.
FIGURE 5.
Cleavage of the ERdj4 SS upon translocation of a GFP reporter. A, SignalP 3.0 analysis of the mouse ERdj4 N terminus predicts the presence of a cleavable signal peptide. The predicted N, H, and C domains are colored green, purple, and aqua, respectively. The bold red line indicates the predicted cleavage site. B, SS constructs (gray letters) plus first 10 amino acids of mature domain (black letters) fused to GFP-KDEL. Italicized black letters indicate multicloning site linker amino acids. Arrows mark confirmed Prl (bovine prolactin) and predicted (ERdj4) cleavage sites. C, constructs were transiently transfected in MDCK cells and imaged by fluorescence microscopy. The core GFP-KDEL construct lacks a SS and displays a cytoplasmic and nuclear distribution, whereas Prl SS+10 and ERdj4 SS+10 both robustly exhibit an ER localization pattern. Scale bar, 10 μm. D, transiently transfected cells were lysed and immunoblotted with anti-GFP. The Prl SS is efficiently cleaved from the GFP-KDEL, leaving only 15 amino acids fused to GFP-KDEL, which is larger than GFP-KDEL alone on a 12% Tricine gel. ERdj4 SS migrates comparably to Prl SS+10 (see supplemental Fig. S2 for additional immunoblot evidence of SS cleavage).
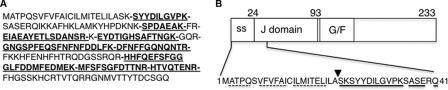
To test for the cleavage of the predicted SS, we first performed a tryptic digest coupled with mass spectrometry on a transfected ERdj4, because the expression of the endogenous protein is too low to perform this experiment. This analysis detected peptides comprising nearly all of the protein, but importantly, the predicted SS was absent from the sequences identified (Fig. 6A). Although trypsin typically cleaves peptides after lysines or arginines, we note that neither amino acid is present in the predicted SS. Our inability to detect the N-terminal peptide in this analysis is consistent with cleavage of the predicted SS but is inconclusive, because hydrophobic peptides are not always identifiable by this method. To unambiguously determine whether the predicted SS is cleaved, we isolated ERdj4 and subjected it to Edman degradation (Fig. 6B). Consistent with the SignalP 3.0 prediction, the predominant detectable species began at serine 24. There are no other sequences in ERdj4 that resemble a transmembrane domain. Thus, ERdj4, like ERdj3, 5, and 6, is actually a soluble luminal protein.
FIGURE 6.
The ERdj4 SS is cleaved in the ER. A, mass spectrometry analysis of purified transfected murine ERdj4. ERdj4 was immunoprecipitated from COS-1 cells 40 h post-transfection, separated on SDS-polyacrylamide gels, and excised. Protein was treated with trypsin and subjected to electron spray ionization mass spectrometry. The identified peptides are indicated with underlined, bold letters, and spaces are inserted between sequential peptides that were identified. B, N-terminal sequencing of ERdj4. ERdj4 was immunoprecipitated from lysates of COS-1 cells transiently transfected with ERdj4, purified, and analyzed by Edman degradation. The amino acids detected by sequencing are underlined. The sequence underlined with the solid line was the predominant species, whereas that underlined with the dashed line represents a very minor detected species. The downward facing triangle represents the signal sequence cleavage site.
ERdj4 Associates with ERAD Component
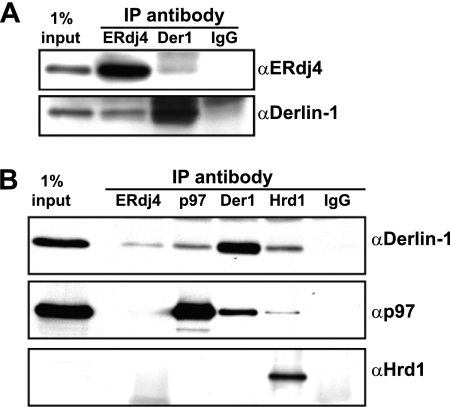
ERdj4 participates in ERAD-L, a form of ERAD in which misfolded luminal domains of soluble and single membrane-spanning proteins are recognized and retrotranslocated by ERAD machinery (47). The machinery includes integral membrane ER proteins Derlin-1 and Hrd1 (48, 49). We hypothesized that these proteins could be potential binding partners for ERdj4. Immunoprecipitation of overexpressed ERdj4 failed to reveal interactions with Hrd1 (not shown). However, significant amounts of Derlin-1 were observed to co-immunoprecipitate with ERdj4 (Fig. 7A). Because the levels of expression in this type of experiment are so much higher than the endogenous ERdj4 levels, which are quite low in unstressed cells (11), we were concerned that the interaction with Derlin-1 could be an overexpression artifact. To test the physiologic relevance of this interaction, we performed co-IPs in nontransfected PU3.1 cells and then immunoblotted against ERAD components. Once again, anti-ERdj4 co-immunoprecipitated Derlin-1, but not p97 or Hrd1 (Fig. 7B). As a positive control, the ERAD-L components p97, Derlin-1, and Hrd1 all co-immunoprecipitated with each other to varying degrees, consistent with previous reports (50).
FIGURE 7.
ERdj4 interacts with ERAD component Derlin-1. A, COS-1 cells were transfected with pSGmERdj4 and harvested at 24 h. Cell lysates were divided evenly into four aliquots, each of which was immunoprecipitated with the indicated antibodies. IgG was used as negative control. Immunoprecipitated proteins were subjected to electrophoresis and immunoblotting with the indicated antibodies. B, immunoprecipitation from cells expressing endogenous ERdj4. 1 × 108 P3U.1 cells were homogenized. The membranes were pelleted by centrifugation and lysed. The lysates were divided into equal fractions and immunoprecipitated with the indicated antibodies. IgG was used as negative control. Immunoprecipitates were subjected to electrophoresis and immunoblotting with the indicated antibodies. IP, immunoprecipitation.
DISCUSSION
Together, our findings reveal several new insights into the availability of ERdj4 in the ER lumen. First, ERdj4 is now reclassified as a soluble ERdj protein, and therefore it is not necessarily confined to the ER membrane. In practical terms, this means ERdj4 can diffuse to its substrates, and the diffusion is comparable with other chaperones. Given the much lower level of ERdj4 relative to other ER chaperones, the probability of a substrate encountering ERdj4 is likely to be quite low, regardless of the ERdj4 D value. However, during ER stress, when ERdj4 levels increase significantly (11), encounter probabilities will increase and can enhance ERAD of substrates (17). A co-chaperone that can diffuse more rapidly than integral membrane proteins can more efficiently sample the ER and enhance clearance of ERAD substrates.
The demonstration that the ERdj4 SS is cleaved helps resolve the biochemical problem of reconciling the short hydrophobic helix with the previous conclusion that it was integrated in the ER lipid bilayer. However, cleavage of the SS creates a new problem. How are ERdj4 levels maintained in the ER lumen? ERdj4, similar to ERdj3, lacks a known ER retention motif, such as a C-terminal -KDEL sequence, which can serve as a retrieval sequence that returns secretory pathway proteins from the intermediate compartment of the cis-Golgi complex to the ER by retrograde trafficking (51, 52). Retention of ERdj4 in the ER could occur through at least three general types of mechanisms: 1) binding to a protein that provides an ER retrieval motif similar to the way glucosidase IIβ provides a -KDEL sequence for its binding partner glucosidase IIα (53), 2) exclusion from ER exit sites, or 3) the presence of a previously uncharacterized ER retention/retrieval motif (54). Our data and previous reports provide evidence for mechanism 1 but do not exclude the possibilities of 2 and 3. First, ERdj4 reversibly binds BiP, a KDEL containing chaperone. The transient interactions with BiP could significantly retard secretion from cells. Second, ERdj4 can interact with at least one component of the ERAD machinery, the ER-localized integral membrane protein Derlin-1. In future studies, it will be important to identify any other binding partners of ERdj4 that might participate in ER retention or function. We cannot exclude the possibility of a novel ER retention motif or an exclusionary mechanism to prevent entry into ER exit sites. These details will be an important focus for future studies.
Why ERdj3, ERdj4, and p58IPK lack the -KDEL sequence found on most resident ER luminal chaperones is an intriguing mystery. This is an evolutionarily conserved phenomenon, because even the yeast luminal ERdj protein Scj1p lacks a -KDEL-related sequence. We speculate that the levels of these co-chaperones, as regulators of BiP activity, may need to be tightly controlled by the cell. ER chaperones typically have long half-lives, and the ER lumen lacks many of the reversible post-translational modifications of the cytoplasm, such as phosphorylation and ubiquitylation. Thus, there may be few options for regulating co-chaperone activities in the ER. One possibility is constitutive secretion of a protein out of the ER, possibly exiting the cell. If more ERdj molecules are required, the synthesis of these proteins can be up-regulated, as happens for all three of these ERdj proteins during the unfolded protein response (3). If less protein is needed, synthesis could decrease, and either regulated secretion or ERAD could deplete soluble ERdj levels over relatively short times. For example, the half-time for secretion of a small inert soluble protein out of the ER is calculated to be ∼40 min (55). Our labs are actively investigating the mechanisms for regulating the levels or activity of ERdj proteins.
The most significant finding of our study comes from the co-immunoprecipitation of ERdj4 with Derlin-1. This result suggests a simple mechanism for specifying whether an individual ERdj regulates events during protein synthesis or as part of quality control of unfoldable proteins. ERdj1 and 2 directly associate with the nascent protein translocation channel (33) (the Sec61 translocon). ERdj3 has recently been reported to interact with the translocation channel protein Sec61α (16). Thus, all three of these ERdj proteins are positioned to specifically and immediately engage nascent proteins as they enter the ER. In contrast, ERdj4 association with the ERAD machinery helps explain why it appears to be an ERAD-specific ERdj (17, 56). Given the mobility of ERdj4-sfGFP, we anticipate that interactions with Derlin-1 are likely to be transient, and we hypothesize that ERdj4 can bind clients throughout the ER and potentially release them to BiP at ERAD retrotranslocon sites. Future biochemistry studies should help resolve the dynamics of ERdj4 association with Derlin-1 and the importance of this interaction in recruiting BiP to ERAD retrotranslocons. In our tissue culture cells overexpressing HA- or GFP-tagged ERdj4, we observed the protein throughout the ER. In cells expressing the extremely low endogenous levels (15) of this protein, we cannot rule out the possibly discrete localization of ERdj proteins to ER functional subdomains (57). Given the proclivities of the different ERdj proteins to bind distinctly rough ER proteins (the translocon) or not, it is tempting to speculate that ERdj functions may be segregated spatially, as well as biochemically.
Supplementary Material
Acknowledgments
We thank Ramanujan Hegde for the anti-GFP antibody, the Albert Einstein College of Medicine Analytical Imaging Facility, and Mike Berne at the Tufts University Core Facility. We thank Yuichiro Shimizu and Beata Lizak for assistance with sample preparation for mass spectrometry and helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants R01GM086530 and 1R21AG032544 (to E. S. and C. W. L.) and R01GM054068 (to L. M. H. and J. H. O.).

This article contains supplemental Figs. S1 and S2.
Beate Lizak, John F. Kearney, and Linda M. Hendershot, manuscript in preparation.
- ER
- endoplasmic reticulum
- ERAD
- endoplasmic reticulum-associated degradation
- FLIP
- fluorescence loss in photobleaching
- FRAP
- fluorescence recovery after photobleaching
- Prl
- bovine prolactin
- RFP
- monomeric red fluorescent protein
- ROI
- region of interest
- sfGFP
- superfolder GFP
- SS
- signal sequence
- VSV G
- vesicular stomatitis virus G protein
- MDCK
- Madin-Darby canine kidney
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1. van Anken E., Braakman I. (2005) Versatility of the endoplasmic reticulum protein folding factory. Crit. Rev. Biochem. Mol. Biol. 40, 191–228 [DOI] [PubMed] [Google Scholar]
- 2. Hebert D. N., Molinari M. (2007) In and out of the ER. Protein folding, quality control, degradation, and related human diseases. Physiol. Rev. 87, 1377–1408 [DOI] [PubMed] [Google Scholar]
- 3. Otero J. H., Lizák B., Hendershot L. M. (2010) Life and death of a BiP substrate. Semin. Cell Dev. Biol. 21, 472–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dudek J., Benedix J., Cappel S., Greiner M., Jalal C., Müller L., Zimmermann R. (2009) Functions and pathologies of BiP and its interaction partners. Cell. Mol. Life Sci. 66, 1556–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shen Y., Hendershot L. M. (2005) ERdj3, a stress-inducible endoplasmic reticulum DnaJ homologue, serves as a cofactor for BiP's interactions with unfolded substrates. Mol. Biol. Cell 16, 40–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jin Y., Awad W., Petrova K., Hendershot L. M. (2008) Regulated release of ERdj3 from unfolded proteins by BiP. EMBO J. 27, 2873–2882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Marcinowski M., Höller M., Feige M. J., Baerend D., Lamb D. C., Buchner J. (2011) Substrate discrimination of the chaperone BiP by autonomous and cochaperone-regulated conformational transitions. Nat. Struct. Mol. Biol. 18, 150–158 [DOI] [PubMed] [Google Scholar]
- 8. Ziegelhoffer T., Lopez-Buesa P., Craig E. A. (1995) The dissociation of ATP from hsp70 of Saccharomyces cerevisiae is stimulated by both Ydj1p and peptide substrates. J. Biol. Chem. 270, 10412–10419 [DOI] [PubMed] [Google Scholar]
- 9. Hennessy F., Nicoll W. S., Zimmermann R., Cheetham M. E., Blatch G. L. (2005) Not all J domains are created equal. Implications for the specificity of Hsp40-Hsp70 interactions. Protein Sci. 14, 1697–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liberek K., Marszalek J., Ang D., Georgopoulos C., Zylicz M. (1991) Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proc. Natl. Acad. Sci. U.S.A. 88, 2874–2878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shen Y., Meunier L., Hendershot L. M. (2002) Identification and characterization of a novel endoplasmic reticulum (ER) DnaJ homologue, which stimulates ATPase activity of BiP in vitro and is induced by ER stress. J. Biol. Chem. 277, 15947–15956 [DOI] [PubMed] [Google Scholar]
- 12. Chevalier M., Rhee H., Elguindi E. C., Blond S. Y. (2000) Interaction of murine BiP/GRP78 with the DnaJ homologue MTJ1. J. Biol. Chem. 275, 19620–19627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Corsi A. K., Schekman R. (1997) The lumenal domain of Sec63p stimulates the ATPase activity of BiP and mediates BiP recruitment to the translocon in Saccharomyces cerevisiae. J. Cell Biol. 137, 1483–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Petrova K., Oyadomari S., Hendershot L. M., Ron D. (2008) Regulated association of misfolded endoplasmic reticulum lumenal proteins with P58/DNAJc3. EMBO J. 27, 2862–2872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Weitzmann A., Baldes C., Dudek J., Zimmermann R. (2007) The heat shock protein 70 molecular chaperone network in the pancreatic endoplasmic reticulum: A quantitative approach. FEBS J 274, 5175–5187 [DOI] [PubMed] [Google Scholar]
- 16. Dejgaard K., Theberge J. F., Heath-Engel H., Chevet E., Tremblay M. L., Thomas D. Y. (2010) Organization of the Sec61 translocon, studied by high resolution native electrophoresis. J. Proteome Res. 9, 1763–1771 [DOI] [PubMed] [Google Scholar]
- 17. Dong M., Bridges J. P., Apsley K., Xu Y., Weaver T. E. (2008) ERdj4 and ERdj5 are required for endoplasmic reticulum-associated protein degradation of misfolded surfactant protein C. Mol. Biol. Cell 19, 2620–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ushioda R., Hoseki J., Araki K., Jansen G., Thomas D. Y., Nagata K. (2008) ERdj5 is required as a disulfide reductase for degradation of misfolded proteins in the ER. Science 321, 569–572 [DOI] [PubMed] [Google Scholar]
- 19. Hagiwara M., Maegawa K., Suzuki M., Ushioda R., Araki K., Matsumoto Y., Hoseki J., Nagata K., Inaba K. (2011) Structural basis of an ERAD pathway mediated by the ER-resident protein disulfide reductase ERdj5. Mol. Cell 41, 432–444 [DOI] [PubMed] [Google Scholar]
- 20. Molinari M., Eriksson K. K., Calanca V., Galli C., Cresswell P., Michalak M., Helenius A. (2004) Contrasting functions of calreticulin and calnexin in glycoprotein folding and ER quality control. Mol. Cell 13, 125–135 [DOI] [PubMed] [Google Scholar]
- 21. Snapp E. L., Hegde R. S., Francolini M., Lombardo F., Colombo S., Pedrazzini E., Borgese N., Lippincott-Schwartz J. (2003) Formation of stacked ER cisternae by low affinity protein interactions. J. Cell Biol. 163, 257–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Aronson D. E., Costantini L. M., Snapp E. L. (2011) Superfolder GFP is fluorescent in oxidizing environments when targeted via the Sec translocon. Traffic 12, 543–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pédelacq J. D., Cabantous S., Tran T., Terwilliger T. C., Waldo G. S. (2006) Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 24, 79–88 [DOI] [PubMed] [Google Scholar]
- 24. Presley J. F., Cole N. B., Schroer T. A., Hirschberg K., Zaal K. J., Lippincott-Schwartz J. (1997) ER-to-Golgi transport visualized in living cells. Nature 389, 81–85 [DOI] [PubMed] [Google Scholar]
- 25. Nishimura N., Balch W. E. (1997) A di-acidic signal required for selective export from the endoplasmic reticulum. Science 277, 556–558 [DOI] [PubMed] [Google Scholar]
- 26. Snapp E. L., Sharma A., Lippincott-Schwartz J., Hegde R. S. (2006) Monitoring chaperone engagement of substrates in the endoplasmic reticulum of live cells. Proc. Natl. Acad. Sci. U.S.A. 103, 6536–6541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hendershot L. M., Wei J. Y., Gaut J. R., Lawson B., Freiden P. J., Murti K. G. (1995) In vivo expression of mammalian BiP ATPase mutants causes disruption of the endoplasmic reticulum. Mol. Biol. Cell 6, 283–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Snapp E. L., Reinhart G. A., Bogert B. A., Lippincott-Schwartz J., Hegde R. S. (2004) The organization of engaged and quiescent translocons in the endoplasmic reticulum of mammalian cells. J. Cell Biol. 164, 997–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Siggia E. D., Lippincott-Schwartz J., Bekiranov S. (2000) Diffusion in inhomogeneous media. Theory and simulations applied to whole cell photobleach recovery. Biophys. J. 79, 1761–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Snapp E., Altan-Bonnet N., Lippincott-Schwartz J. (2003) Measuring protein mobility by photobleaching GFP-chimeras in living cells, in Current Protocols in Cell Biology (Bonafacino J. S., Dasso M., Harford J., Lippincott-Schwartz J., Yamada K., eds) Unit 21.21, John Wiley & Sons, Inc., New York: [DOI] [PubMed] [Google Scholar]
- 31. Buchman A. R., Berg P. (1988) Comparison of intron-dependent and intron-independent gene expression. Mol. Cell. Biol. 8, 4395–4405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Meyer H. A., Grau H., Kraft R., Kostka S., Prehn S., Kalies K. U., Hartmann E. (2000) Mammalian Sec61 is associated with Sec62 and Sec63. J. Biol. Chem. 275, 14550–14557 [DOI] [PubMed] [Google Scholar]
- 33. Dudek J., Greiner M., Müller A., Hendershot L. M., Kopsch K., Nastainczyk W., Zimmermann R. (2005) ERj1p has a basic role in protein biogenesis at the endoplasmic reticulum. Nat. Struct. Mol. Biol. 12, 1008–1014 [DOI] [PubMed] [Google Scholar]
- 34. Nikonov A. V., Snapp E., Lippincott-Schwartz J., Kreibich G. (2002) Active translocon complexes labeled with GFP-Dad1 diffuse slowly as large polysome arrays in the endoplasmic reticulum. J. Cell Biol. 158, 497–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lai C. W., Aronson D. E., Snapp E. L. (2010) BiP availability distinguishes states of homeostasis and stress in the endoplasmic reticulum of living cells. Mol. Biol. Cell 21, 1909–1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ostrovsky O., Makarewich C. A., Snapp E. L., Argon Y. (2009) An essential role for ATP binding and hydrolysis in the chaperone activity of GRP94 in cells. Proc. Natl. Acad. Sci. U.S.A. 106, 11600–11605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Marguet D., Spiliotis E. T., Pentcheva T., Lebowitz M., Schneck J., Edidin M. (1999) Lateral diffusion of GFP-tagged H2Ld molecules and of GFP-TAP1 reports on the assembly and retention of these molecules in the endoplasmic reticulum. Immunity 11, 231–240 [DOI] [PubMed] [Google Scholar]
- 38. Ellenberg J., Siggia E. D., Moreira J. E., Smith C. L., Presley J. F., Worman H. J., Lippincott-Schwartz J. (1997) Nuclear membrane dynamics and reassembly in living cells. Targeting of an inner nuclear membrane protein in interphase and mitosis. J. Cell Biol. 138, 1193–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nehls S., Snapp E. L., Cole N. B., Zaal K. J., Kenworthy A. K., Roberts T. H., Ellenberg J., Presley J. F., Siggia E., Lippincott-Schwartz J. (2000) Dynamics and retention of misfolded proteins in native ER membranes. Nat. Cell Biol. 2, 288–295 [DOI] [PubMed] [Google Scholar]
- 40. Vander Heyden A. B., Naismith T. V., Snapp E. L., Hodzic D., Hanson P. I. (2009) LULL1 retargets TorsinA to the nuclear envelope revealing an activity that is impaired by the DYT1 dystonia mutation. Mol. Biol. Cell 20, 2661–2672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schutze M. P., Peterson P. A., Jackson M. R. (1994) An N-terminal double-arginine motif maintains type II membrane proteins in the endoplasmic reticulum. EMBO J. 13, 1696–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bulbarelli A., Sprocati T., Barberi M., Pedrazzini E., Borgese N. (2002) Trafficking of tail-anchored proteins. Transport from the endoplasmic reticulum to the plasma membrane and sorting between surface domains in polarised epithelial cells. J. Cell Sci. 115, 1689–1702 [DOI] [PubMed] [Google Scholar]
- 43. Sharpe H. J., Stevens T. J., Munro S. (2010) A comprehensive comparison of transmembrane domains reveals organelle-specific properties. Cell 142, 158–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Munro S. (1998) Localization of proteins to the Golgi apparatus. Trends Cell Biol. 8, 11–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fujiki Y., Hubbard A. L., Fowler S., Lazarow P. B. (1982) Isolation of intracellular membranes by means of sodium carbonate treatment. Application to endoplasmic reticulum. J. Cell Biol. 93, 97–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bendtsen J. D., Nielsen H., von Heijne G., Brunak S. (2004) Improved prediction of signal peptides. SignalP 3.0. J. Mol. Biol. 340, 783–795 [DOI] [PubMed] [Google Scholar]
- 47. Brodsky J. L., Skach W. R. (2011) Protein folding and quality control in the endoplasmic reticulum: Recent lessons from yeast and mammalian cell systems. Curr. Opin. Cell Biol. 23, 464–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lilley B. N., Ploegh H. L. (2004) A membrane protein required for dislocation of misfolded proteins from the ER. Nature 429, 834–840 [DOI] [PubMed] [Google Scholar]
- 49. Shimizu Y., Okuda-Shimizu Y., Hendershot L. M. (2010) Ubiquitylation of an ERAD substrate occurs on multiple types of amino acids. Mol. Cell 40, 917–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ye Y., Meyer H. H., Rapoport T. A. (2001) The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature 414, 652–656 [DOI] [PubMed] [Google Scholar]
- 51. Munro S., Pelham H. R. (1987) A C-terminal signal prevents secretion of luminal ER proteins. Cell 48, 899–907 [DOI] [PubMed] [Google Scholar]
- 52. Semenza J. C., Hardwick K. G., Dean N., Pelham H. R. (1990) ERD2, a yeast gene required for the receptor-mediated retrieval of luminal ER proteins from the secretory pathway. Cell 61, 1349–1357 [DOI] [PubMed] [Google Scholar]
- 53. Drenth J. P., Martina J. A., Te Morsche R. H., Jansen J. B., Bonifacino J. S. (2004) Molecular characterization of hepatocystin, the protein that is defective in autosomal dominant polycystic liver disease. Gastroenterology 126, 1819–1827 [DOI] [PubMed] [Google Scholar]
- 54. Vander Heyden A. B., Naismith T. V., Snapp E. L., Hanson P. I. (2011) Static retention of the lumenal monotopic membrane protein torsinA in the endoplasmic reticulum. EMBO J. 30, 3217–3231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Thor F., Gautschi M., Geiger R., Helenius A. (2009) Bulk flow revisited. Transport of a soluble protein in the secretory pathway. Traffic 10, 1819–1830 [DOI] [PubMed] [Google Scholar]
- 56. Buck T. M., Kolb A. R., Boyd C. R., Kleyman T. R., Brodsky J. L. (2010) The endoplasmic reticulum-associated degradation of the epithelial sodium channel requires a unique complement of molecular chaperones. Mol. Biol. Cell 21, 1047–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hegde R. S., Ploegh H. L. (2010) Quality and quantity control at the endoplasmic reticulum. Curr. Opin. Cell Biol. 22, 437–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sbalzarini I. F., Mezzacasa A., Helenius A., Koumoutsakos P. (2005) Effects of organelle shape on fluorescence recovery after photobleaching. Biophys. J. 89, 1482–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Einstein A. (1905) Über die von der molekularkinetischen Theorie der Wärme geforderte Bewegung von in ruhenden Flüssigkeiten suspendierten Teilchen. Ann. Phys. 17, 549–560 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.