Background: Charged residues, abundant in collagen, participate in stabilizing and packing interactions.
Results: Two contact geometries, axial and lateral, are observed in charge pair hydrogen bonding.
Conclusion: Axial salt bridges provide greater stabilization to the triple helical fold than lateral ones.
Significance: Understanding interstrand amino acid interactions in collagen will improve interpretation of natural collagen and will allow more effective design of collagen mimics.
Keywords: Collagen, Nuclear Magnetic Resonance, Peptide Conformation, Peptides, X-ray Crystallography, Self-assembly
Abstract
The collagen triple helix is the most abundant protein fold in humans. Despite its deceptively simple structure, very little is understood about its folding and fibrillization energy landscape. In this work, using a combination of x-ray crystallography and nuclear magnetic resonance spectroscopy, we carry out a detailed study of stabilizing pair-wise interactions between the positively charged lysine and the negatively charged amino acids aspartate and glutamate. We find important differences in the side chain conformation of amino acids in the crystalline and solution state. Structures from x-ray crystallography may have similarities to the densely packed triple helices of collagen fibers whereas solution NMR structures reveal the simpler interactions of isolated triple helices. In solution, two distinct types of contacts are observed: axial and lateral. Such register-specific interactions are crucial for the understanding of the registration process of collagens and the overall stability of proteins in this family. However, in the crystalline state, there is a significant rearrangement of the side chain conformation allowing for packing interactions between adjacent helices, which suggests that charged amino acids may play a dual role in collagen stabilization and folding, first at the level of triple helical assembly and second during fibril formation.
Introduction
The collagen protein superfamily comprises diverse multidomain proteins that constitute a major component of the extracellular matrix in animals (1). All members of this family share a common structural motif known as the collagen triple helix. Triple helical domains have a high content of imino acids, glycine, and charged amino acids (2). The last are important because they participate in molecular recognition events (3), stabilizing pair-wise interactions (4–10) and the packing of triple helices into staggered arrays (11, 12). To understand the mechanism by which ionizable residues stabilize this protein fold and participate in packing interactions, we use model peptides containing an imino acid-rich region at the termini and a guest region that follows the PKGXOG motif, where X is either glutamic or aspartic acid. By a combination of x-ray crystallography and nuclear magnetic resonance (NMR) spectroscopy we are able to study the conformation of the side chains both in solution, where interstrand interactions are important in determining the thermal stability of the helix, and in a crowded macromolecular state, where inter-triple helical contacts play a large role.
The triple helical fold is characterized by long uninterrupted amino acid stretches of XYG triplets, with glycine occupying every third position along the peptide chain. The X and Y positions are commonly proline and 4R-hydroxyproline, a post-translational modification on the proline side chain (single letter code O), respectively. Three such peptide strands, known as α-chains, bundle together into a right-handed super helix held together by a network of hydrogen bonds that runs perpendicular to the helical axis from the glycine amide in one chain to the carbonyl of the amino acid in the X position of an adjacent strand (13). To fulfill this hydrogen-bonding network, the strands assemble staggered by one amino acid differentiating a leading, middle, and lagging chain.
When considering only naturally occurring amino acids, sequences of the form (POG)n self-assemble into the most stable triple helices in an aqueous environment. It is known that any point mutation in the X or Y position from this template will destabilize the resulting helix (14). Particularly, mutations from O to K destabilize the helix by 10 °C, whereas mutations from P to D and P to E by 7 °C and 4 °C, respectively. Despite this fact, previous studies have shown that double mutations will have different effects on the stability of the triple helix. Studying peptides of the form (POG)3XYGX′Y′G(POG)3 Brodsky and co-workers found that some sequences containing double mutations involving oppositely charged amino acids behave as expected from an addition of two-point mutations and some exhibit a higher thermal stability than expected. In particular, the sequences PKGEOG and PKGDOG were found to be highly stabilizing (4), whereas the sequence EKGPOG and DKGPOG were not (15). The difference in thermal stability is hard to explain based on the pairing of oppositely charged residues because molecular models showed that in both cases it is geometrically possible for the charged moieties to come in contact.
Understanding the molecular basis for such stabilizing pair-wise interactions is crucial to further our knowledge on the association of α-chains to form trimers in collagenous proteins. One question that has major biophysical and physiological implications is how the unfolded polypeptide chains choose to form part of a triple helix of a particular composition and, in the case of heterotrimeric helices, a particular register. Globular C-terminal domains that form stable trimers control the composition in some collagen types (1). Nonetheless, their role in choosing a particular register for heterotrimers is not yet understood. When studying the chain recognition problem most of the available studies focus on the role of noncollagenous domains. We think that despite their importance, there is room for cooperativity between the noncollagenous domains and the triple helical domains, especially in cases where the noncollagenous domains are small.
In this study we focus on the structural characterization of host-guest triple helical peptides containing lysine-glutamate (KGE peptide) and lysine-aspartate (KGD peptide) salt bridges as minimalist models for full-length collagenous domains (see Table 2 for full sequences). By means of x-ray crystallography and NMR spectroscopy we are able to study the side chain interactions in two different stages of the life of a collagen molecule and find important differences in their conformation that give insight into the different functions that the same residues can perform during the lifespan of a single triple helix. The NMR experiments serve as a model for individual collagens in solution. Using this technique it is possible to observe preferential pair-wise interactions between the different peptide strands within a single triple helix that serve to stabilize the native state. Two contact geometries, which we will define below as axial and lateral (see Fig. 7), can be observed with different sequence requirements depending on the relative stagger of the interacting peptide chains. We hypothesize that such preferential register-specific interactions may play a role in chain registration process in natural collagens. The x-ray crystallographic studies serve as an example of the triple helix in a crowded molecular environment, akin to the one found in the supramolecular architectures made by tightly packed triple helices in collagen fibrils. In this context we see a residue-dependent change in the side chain conformation that allows for extended hydrogen-bonding networks to form in the interfaces between adjacent triple helices.
TABLE 2.
Petide sequences and melting temperatures
| Peptide | Sequencea | Tm |
|---|---|---|
| °C | ||
| KGE | (POG)3PKGEOG(POG)3 | 51 |
| KGD | (POG)3PKGDOG(POG)3 | 48 |
a Highlighted amino acids are 15N-labeled.
b The melting temperature is defined here as the minimum in the derivative of the circular dichroism melting curve.
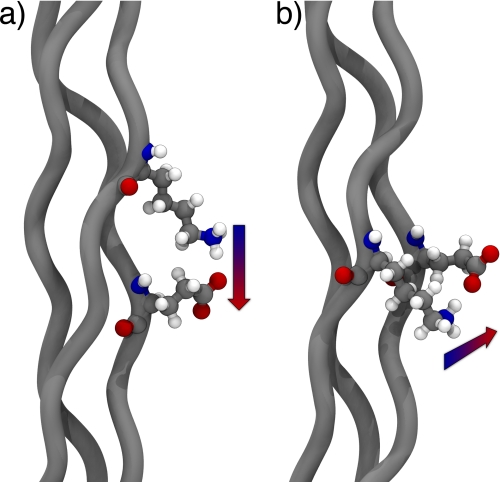
FIGURE 7.
Molecular models highlighting the different contact geometries observed in triple helical proteins. a, axial interaction between lysine and glutamate. b, lateral interaction between lysine and glutamate.
EXPERIMENTAL PROCEDURES
Peptide Synthesis and Purification
The peptides were synthesized using standard chemistry for solid phase peptide synthesis and purified by high pressure liquid chromatography (HPLC). Experimental details of the synthesis and mass spectra are available in the supplemental Experimental Procedures and supplemental Fig. S1.
Circular Dichroism
Spectra were acquired between 215 and 250 nm, and the maximum around 222 nm, was monitored during unfolding curves. Melting experiments were performed from 5 to 85 °C with a heating rate of 10 °C/h. Additional details are provided in the supplemental Experimental Procedures.
Crystallization and Data Collection
KGE and KGD were crystallized using the hanging drop method, and data were acquired at 100 K. The crystals diffracted to 1.68 Å and 2.00 Å, respectively, and were indexed to a triclinic unit cell, space group P1 using the hkl2000 software (16).
Structure Determination and Refinement
The structures were solved by molecular replacement with the epmr software (17) using a modified version of the structure 1QSU (12) for KGE and a modified version of the KGE structure for KGD. Initial phases were improved by rigid body refinement followed by rounds of simulated annealing and anisotropic B-factor refinement using the CNS suite (18). Model rebuilding was done in COOT (19). The refinement was performed starting at a 3.0 Å resolution and gradually increasing to the limiting value. Water picking was started at 1.8 Å for KGE and 2.2 Å for KGD at which point simulated annealing was replaced by atomic position refinement. The final CNS models were subjected to TLS refinement in refmac (20).
NMR Spectroscopy
All NMR experiments were recorded in an 800-MHz Varian spectrometer equipped with a triple resonance probe at 5 °C. Each sample was characterized using two-dimensional total correlated spectroscopy, NOESY, 1H,5N heteronuclear single quantum coherence (HSQC),2 and three-dimensional NOESY-15N HSQC experiments (supplemental Tables S4 and S5).
NMR Ensemble Calculation
The KGE ensemble was generated starting from the crystal structure, and for the KGD ensemble appropriate modifications were done using PyMOL (21). Conformational sampling was achieved by running langevin dynamic simulations in implicit solvent using the AMBER99 (22) force field followed by a constrained minimization subject including constraints derived from NOE data. A list of NOE constraints is presented in supplemental Tables S2 and S3. The 100 lowest energy structures after the minimization step were selected for the final ensemble. More details are available in the supplemental Experimental Procedures.
RESULTS
Crystal Structure of KGE Peptide
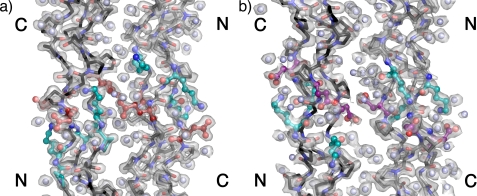
KGE folds into a stable homotrimeric triple helix with a melting temperature of 51 °C in 10 mm phosphate buffer at pH 7 (supplemental Fig. S2) and readily crystallizes around neutral pH in a tacsimate buffer. The structure of the peptide was solved by molecular replacement and contains two antiparallel triple helices in the asymmetric unit packed in a quasi-hexagonal lattice. As is commonly observed in triple helical peptides the structure shows some disorder at the termini (23), evidenced by the B-factors obtained for the terminal triplets (supplemental Fig. S4). This is particularly pronounced at the C terminus, where poor density prohibited the modeling of the Gly24(C). The final model contains 935 peptide atoms and 219 water molecules and was refined to an Rwork/Rfree value of 18.9/20.7. Fig. 1a shows the contents of the asymmetric unit, highlighting the charged residues in the guest region by coloring lysine residues cyan, glutamic acid residues red, and water molecules light blue. The 2Fo − Fc map contoured at 1.2σ is also shown as a transparent surface to illustrate the accuracy of the phases. A more traditional wireframe model is available in the supporting information, and the data acquisition and refinement statistics are available in Table 1. Supplemental Table S1 shows the average dihedral values calculated from the structure. The values observed in (POG)10 and expected for an idealized 7/2- and 10/3-helix are also shown for comparison. Overall, the host region agrees with the values reported for (POG)10, which approximates the 7/2-helix. The guest region shows only small deviations from the host region dihedrals; however, because it comprises so few amino acids, the calculated averages may not be significant.
FIGURE 1.
Atomic structure of KGE (a) and KGD (b). Contents of the asymmetric unit of the crystals highlighting the two antiparallel triple helices are shown. The position of the N and C terminus of each helix is noted. The 1.68 Å 2Fo − Fc map in a and 2.00 Å 2Fo − Fc map in b are contoured at 1.2σ and depicted as transparent surfaces. Image were generated using PyMOL (21).
TABLE 1.
Data collection and refinement statistics
Data in parentheses correspond to the highest resolution shell.
| Crystal parameters | 3T4F (KGE) | 3U29 (KGD) |
| Space group | P1 | P1 |
| Unit cell dimensions | ||
| α, β, γ (°) | 95.0, 94.7, 94.9 | 94.3, 94.7, 93.3 |
| Data collection | ||
| Resolution (Å) | 1.68 (1.74–1.68) | 2.00 (2.05–2.00) |
| Completeness | 80.1% | 76.9% |
| Redundancy | 1.1 | 1.1 |
| Rmerge | 5.3 (12.0) | 9.8(17.1) |
| I/σI | 19.4 | 21.0 |
| Refinement | ||
| Resolution (Å) | 1.68 (1.74–1.68) | 2.00 (2.05–2.00) |
| Total reflections | 7,957 | 4,220 |
| Rwork/Rfreea | 19.5/20.8 | 23.9/25.0 |
| No. atoms | ||
| Protein | 935 | 915 |
| Water | 218 | 180 |
| r.m.s.d.b | ||
| Bond lengths (Å) | 0.07 | 0.08 |
| Bond angles (°) | 1.4 | 1.5 |
a 5% of reflections were included in the test set.
b Root mean square deviation.
In general, three types of interactions are possible for each of the charged moieties: (i) a salt bridge or ionic hydrogen bond, where there is a direct contact between two oppositely charged residues; (ii) a hydrogen bond, where the charged side chain group shares a hydrogen with a neutral peptidic polar atom; and (iii) a water-mediated contact, where a water molecule forms a bridge between the charged side chain moiety and another polar atom. The extensive network of hydrogen bonds observed in the quasi-hexagonal crystal packing of our structure shows all three of these cases, and selected examples are available in the supplemental Experimental Procedures (supplemental Fig. S5).
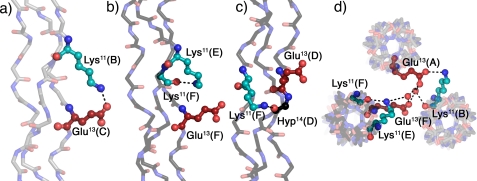
The contacts observed in the structure can be intrastrand, interstrand, or interhelical depending on the relative positions of the interacting amino acids. Fig. 2 shows different interstrand and interhelical contacts involving the amino acids in the guest region of the KGE crystal structure with symmetry equivalent positions denoted by color: triple helices depicted gray are looking down from the N terminus in Fig. 1 (chains A, leading; B, middle; and C, trailing), and triple helices depicted black are looking down from the C terminus in Fig. 1 (chains D, leading; E, middle; and F, trailing). (For abbreviations of peptide chains please refer to Table 2.) Fig. 2a shows an ionic hydrogen bond between Lys11(B) and Glu13(C). Fig. 2b shows the equivalent interaction in the second helix of the asymmetric unit. In this case Lys11(E) forms a hydrogen bond with the backbone carbonyl of Lys11(F) instead of a direct salt bridge with Glu13(F). In Fig. 2c the Lys11(F) side chain also prefers a backbone hydrogen bond, in this case to the Hyp14(D) carbonyl, instead of the direct hydrogen bond to the negatively charged Glu13(A) carboxylate. Fig. 2d shows a top view of the packing interactions involving the residues in a and b to highlight the fact that although the Glu13(F) side chain does not interact directly with Lys11(E) there is a water-mediated hydrogen bond between both residues. Further water-mediated contacts are observed between Glu13(F) and Lys11(A) and Glu13(A), with the latter two amino acids coming from distinct symmetry-related helices.
FIGURE 2.
Interstrand and interhelical interactions in KGE. a–c, interstrand hydrogen bonds involving the charged side chains. d, top view of the packing interactions involving the residues depicted in b. Triple helices oriented N to C terminus in Fig. 1 are shown in gray (A, leading chain; B, middle chain; C, lagging chain), and triple helices oriented C to N terminus in Fig. 1 are depicted in black (D, leading chain; E, middle chain; F, lagging chain). Lysines are shown in cyan and glutamates in red. Amino acids are labeled using their three-letter code, sequence position, and chain. Images were generated using PyMOL (21).
Crystal Structure of KGD Peptide
A similar peptide in which the guest sequence is PKGDOG (Table 2) was also studied. The peptide, KGD, folds into a stable homotrimeric triple helix with a melting temperature of 48 °C in 10 mm phosphate buffer at pH 7 and crystallizes around neutral pH in a tacsimate buffer and (supplemental Fig. S2). The structure of the peptide was solved by molecular replacement, and it is similar to the KGE structure, with two antiparallel triple helices in the asymmetric unit packed in a quasi-hexagonal lattice. The final model contains 915 peptide atoms and 180 water molecules and was refined to an Rwork/Rfree value of 23.9/25.0. Fig. 1b shows the contents of the asymmetric unit, highlighting the charged residues in the guest region by coloring lysines cyan and aspartates purple with waters depicted in light blue. The 2.00 Å 2Fo − Fc map contoured at 1.2σ is depicted as a transparent surface. Supplemental Fig. S3 includes a more traditional wireframe model. Table 1 summarizes the refinement statistics.
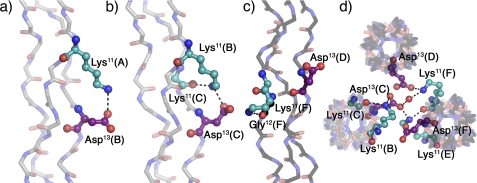
Fig. 3 shows different interstrand, intrastrand, and interhelical contacts involving the amino acids in the guest region of the KGD crystal structure with symmetry equivalent positions denoted by color as described previously. Fig. 3a shows a salt bridge between Lys11(A) and Asp13(B). Fig. 3b shows a similar interaction between Lys11(B) and Asp13(C); however, because of differences in the lysine side chain conformation a second hydrogen bond between the amino group and the Lys11(C) backbone carbonyl is also possible. Fig. 3c depicts the Lys11(F) side chain engaging in an intrastrand backbone hydrogen bond to the Gly12(F) carbonyl instead of a direct interaction with its nearest neighbor carboxylate. Fig. 3d shows a top view of the packing interactions involving the residues from Figs. 3, b and c, to highlight that even though Glu13(D) does not participate in interstrand interactions it engages in several interhelical contacts, including an ionic hydrogen bond and a water-mediated contact to Lys11(F) in an adjacent, symmetry-related helix. Furthermore, it shows Glu13(C) participating both in interstrand hydrogen bonds, as described above, and in interhelical contacts with an ionic hydrogen bond to Lys11(E) and a water-mediated hydrogen bond to Lys11(F).
FIGURE 3.
Interstrand, intrastrand, and interhelical interactions in KGD. a and b, interstrand hydrogen bonds involving the charged side chains. c, intrastrand hydrogen bond. d, top view of packing interactions involving the residues depicted in b and c. Triple helices oriented N to C terminus in Fig. 1 are shown in gray (A, leading chain; B, middle chain; C, lagging chain), and triple helices oriented C to N terminus in Fig. 1 are depicted in black (D, leading chain; E, middle chain; F, lagging chain). Lysines are shown in cyan and aspartates in purple. Amino acids are labeled using their three-letter code, sequence position, and chain. Images were generated using PyMOL (21).
NMR Ensemble of KGE Peptide
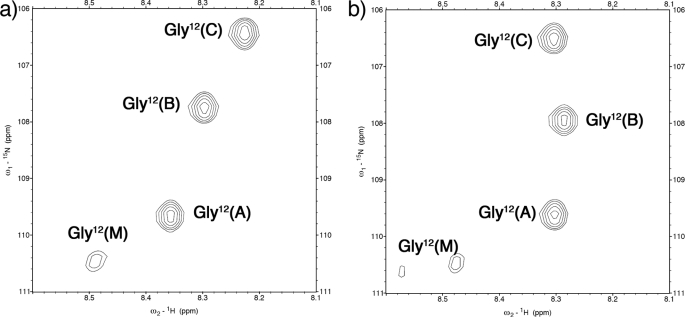
The solution conformation of the KGE peptide was also investigated by means of multidimensional NMR experiments. To facilitate the analysis, a 15N-labeled glycine residue was included in position 12 (Table 2). The 1H,15N HSQC spectrum of the system (Fig. 4a) shows three distinct peaks for the triple helical assembly and also a monomer peak. Despite having a homotrimeric composition, the one-residue stagger characteristic of this protein fold lifts the degeneracy associated with symmetry-equivalent positions in the guest region of our triple helix, which is observed for the host region (supplemental Fig. S6). Because of the degeneracy observed in this region, the NMR analysis will focus on amino acids Lys11-Glu13. In the following text when referring to a particular proton the superscript next to the three-letter code will denote its chain and the atom name will be given in parentheses.
FIGURE 4.
1H,15N HSQC spectra. a, KGE spectrum. b, KGD spectrum. Amino acids are labeled using their three-letter code, sequence position, and chain (A, leading chain; B, middle chain; C, lagging chain; and M, monomer).
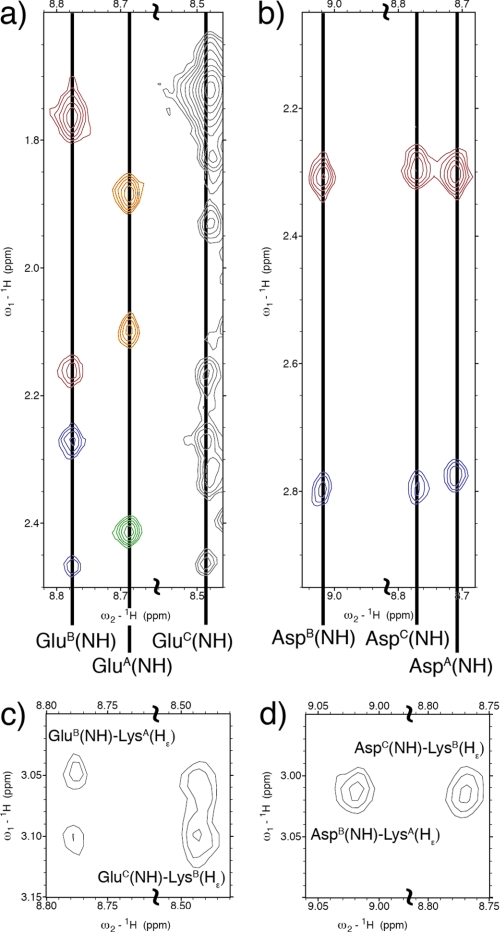
The side chain conformation of the charged residues can be studied using the NOESY spectrum of KGE (Fig. 5). The glutamic acid residues in both chain B and C show a rather rigid conformation with distinct chemical shifts for each of the four side chain methylene protons (Fig. 5a). Besides the intraresidue NOEs each of these Glu(NH) protons shows cross-peaks to the Lys(Hϵ) methylene in the preceding chain, which shows distinct resonances for each of its diasteriotopic protons (Fig. 5, a and c). However, the glutamate in the leading chain shows distinct chemical shifts only for its β-protons whereas the γ-methylene presents a single chemical shift for both hydrogens, indicating a more dynamic conformation. This residue lacks a resonance to the lysine ϵ-methylene of the lagging strand, which shows degenerate shifts for both ϵ-protons.
FIGURE 5.
1H,1H NOESY spectra. a and c, strips of the KGE NOESY spectrum. b and d, strips of the KGD NOESY spectrum. a shows intraresidue Glu(NH)-Glu(Hβ,γ) resonances in KGE. Three columns can be observed corresponding to Glu(NH) in chains B, A, and C. GluB(NH) shows cross-peaks with GluB(Hβ) protons (red) and GluB(Hγ) protons (blue). GluA(NH) shows cross-peaks with GluA(Hβ) protons (orange) and GluA(Hγ) protons (green). A similar pattern is observed for the GluC(NH) column, although the peak corresponding to GluC(Hβ3) overlaps with the intense LysA,B(NH)-LysA,B(Hγ3) resonance. b shows intraresidue Asp(NH)-Asp(Hβ) resonances in KGD. Three columns can be observed corresponding to Asp(NH) in chains B, C, and A. The peaks near 2.78 (blue) and 2.30 ppm (red) correspond to the correlation with Asp(Hβ3) and Asp(Hβ2) protons, respectively. c shows interchain Glu(NH)-Lys(Hϵ) resonances in KGE. d shows interchain Asp(NH)-Lys(Hϵ) resonances in KGD.
To illustrate the side chain conformation observed in solution for the guest region of the KGE peptide a family of structures was generated to approximate the native ensemble of the triple helical assembly. Molecular dynamics simulations were carried out starting from the crystal structure to sample alternate conformations, which were then subjected to a constrained minimization using distance restraints extracted from the NOE cross-peak intensities. This methodology allows for efficient sampling of the relevant conformational space by biasing the geometry of the structures visited during the molecular dynamics simulations toward the native free energy basin using experimental constraints derived from the NOESY spectra (a list of NOE constraints is provided in supplemental Tables S2 and S3). It has been shown that molecular dynamic simulations can generate ensembles that show significant overlap with those obtained by a traditional NMR structure determination process (24). Thus, by using a combination of both techniques we are striving to merge their strengths (25) and obtain an accurate representation of the side chain behavior in solution. It should be noted that we are interested in a qualitative structure-based understanding of the interactions. We do not attempt to treat these structures as a quantitative thermodynamic ensemble due to the difficulties associated with accurate computation of electrostatic contributions to protein stability (26).
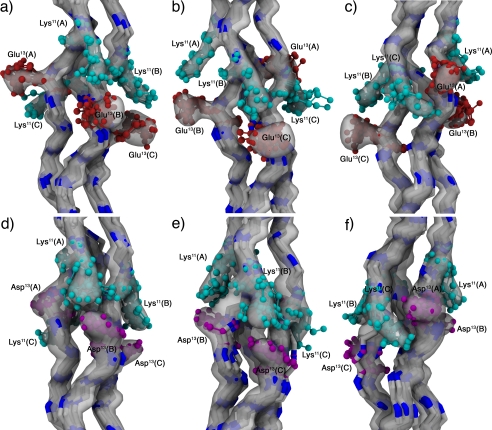
The 100 lowest energy structures were selected for the final ensemble with an overall backbone root mean square deviation of 0.83 Å. Fig. 6, a–c, shows the ensemble highlighting interactions between chains A-B (a), B-C (b), and C-A (c). It is possible to divide the observed contacts into two subsets based on their geometry. The first one includes contacts between chains A-B and chains B-C, specifically Lys11(A)-Glu13(B) and Lys11(B)-Glu13(C) and will be referred to as an axial interaction because the interacting side chains are arranged along the helical axis (Fig. 7a), leading to a geometry that facilitates the formation of ionic hydrogen bonds between the charged side chains. Furthermore, there is a slight difference in the interaction between chains A-B and B-C, which can be observed in the NMR ensemble of the assembly as depicted in Fig. 6, a and b, with the B-C interaction presenting a higher degree of conformational flexibility. In terms of sequence, this interaction occurs between a lysine residue in position n and a glutamic acid in position n+2 in a subsequent chain of a triple helix, provided that the lysine is either in chain A or B. The axial interaction between chains C-A, although possible, requires glutamic acid to be in position n+5 in chain A if lysine occupies position n in chain C. Instead, we observe a lateral interaction between the two remaining chains (Figs. 6c and 7b), which is characterized by a larger degree of conformational flexibility. In this case, there is a competing interaction between the Lys11(C)-Glu13(A) salt bridges and a Lys11(C)-Hyp14(A) hydrogen bond involving the lysine Hζ protons and the hydroxyproline backbone carbonyl. Although it is possible to satisfy both contacts simultaneously, some conformers sampled interacted with neither (supplemental Fig. S7). The corresponding lateral interactions between chains A-B or B-C are also possible, but would require for glutamic acid to be in position n−1 (in chain B or C) if lysine is in position n (in chain A or B, respectively). NMR ensemble statistics are shown in supplemental Table S6.
FIGURE 6.
NMR ensembles. a–c, KGE ensemble. d–f, KGD ensemble. Interactions between the charged residues in chains A-B (a and d), B-C (b and e), and C-A (c and f) are highlighted. The reweighted atomic density for each ensemble (33) is depicted as a semitransparent surface at a value of 40%, and 10 representative structures are shown as a backbone trace with the lysine (cyan), glutamic acid (red), and aspartic acid (purple) residues depicted in a cpk model. Amino acids are labeled using their three-letter code, sequence position, and chain (A, leading chain; B, middle chain; and C, lagging chain). Images were generated using vmd-xplor (34).
NMR Ensemble of KGD Peptide
The solution structure of the KGD peptide was also studied by multidimensional NMR experiments. The 1H,15N HSQC spectrum of the system (Fig. 4b) also shows three distinct peaks for the triple helical assembly and two monomer peaks, which we ascribe to cis-trans isomerization of the prolyl amide bonds surrounding the guest region (27). As for KGE, the host region (supplemental Fig. S6b) shows mostly degenerate peaks and thus further analysis will focus in the guest region (Lys11-Asp13).
Again, the side chain conformation of the charged residues is studied using the NOESY spectrum (Fig. 5, b and d). In this case, all three aspartic acid residues show similar conformations with distinct chemical shifts for the Asp(Hβ2) and Asp(Hβ3) protons (Fig. 5b). Interchain NOEs are also observed between AspB(NH) and AspC(NH) and the LysA,B(Hϵ) protons, which show a single chemical shift for both methylene protons. Based on structural constraints, it is possible to assign the resonances to be LysA(Hϵ)-AspB(NH) and LysB(Hϵ)-AspC(NH). There is no comparable resonance between chains C and A (Fig. 5d).
An NMR ensemble was also generated for this peptide sequence using the same methodology described earlier with modified coordinates to include aspartic acid. Fig. 6, d–f, shows the 100 lowest energy structures, which have an overall backbone root mean square deviation of 0.91 Å. The conformations observed mirror those of the KGE ensemble, presenting both axial interactions between chains A-B and B-C and lateral interactions between C-A. Unlike KGE, the KGD ensemble shows no significant difference between the two axial interactions. Furthermore, there are few geometries with salt bridges between chains C-A, instead the Hζ atoms of lysine interact preferentially with the backbone carbonyl of Hyp14(A) and Asp13(A) (supplemental Fig. S8).
DISCUSSION
Most atomic resolution data concerning charge pair interactions in collagens come from the crystal structures of homotrimeric triple helical peptides (11,12). However, because their large surface-exposed area (two thirds of all amino acids) can lead to extensive interprotein contacts, crystallography may not the best analytical tool to study events that concern individual triple helices in solution. This fact is illustrated in our studies by the different rotamers observed for the same charged amino acid in different triple helices of the KGE and KGD asymmetric units. The NMR spectra, however, suggest a more homogeneous ensemble for both peptides. After analyzing the rotamers observed in the crystal structure it seems clear that they adopt conformations strongly influenced by the crystal packing. Furthermore, even the residues that are involved in intrahelical contacts also present interactions with other helices either directly or through water-mediated hydrogen bonds. It is interesting to note that the packing interactions observed in our quasi-hexagonal lattice are able to accommodate different interaction networks for the same charged amino acids. This finding supports the idea that lysine and glutamic acid may rearrange between different sets of possible interactions in collagen type I fibers (28). Because we have two antiparallel triple helices in the unit cell, we observe interfaces between antiparallel triple helices, which are relevant as chemical models for some collagen assemblies. For instance, type II collagen fibers, which pack in a hexagonal array (29), are known to be decorated in the surface by type IX molecules in an antiparallel fashion (30), a collagen type that has a particularly high content of glutamic acid residues (31).
When trying to understand the molecular basis of stabilizing interactions in triple helical proteins it is necessary to study the amino acid interactions in the appropriate environment. As mentioned previously, the charged amino acids in the guest region engage in extensive networks of inter-triple helical interactions in the crystal structure. However, in solution such interactions are minimal and therefore do not contribute to the side chain conformation. This leads to a different arrangement of the charged amino acids. Despite these fundamental differences the crystallographic study is vital because it allows us to obtain information on the overall backbone structure of the assemblies and shows some examples of conformation that allow for intra- and interstrand hydrogen bonds. Comparing the two methods, we feel that solution NMR experiments are the most accurate way to study the amino acid interactions that lead toward an increase in the thermal stability of triple helical proteins.
Our structural study shows that it possible to divide the observed interactions in solution into two categories: the first one comprises interactions from the leading chain to the middle chain as well as interactions from the middle chain to the lagging chain. These interactions we have labeled as axial contacts. The second type of interaction comprises interactions from the lagging chain to the leading chain which we label a lateral contact (Fig. 7). Despite having identical sequences these differences arise from the relative stagger of the peptide strands within the triple helix and lead to different conformations of the interacting amino acids.
In the axial contact the lysine side chain extends down the helical axis reaching toward the acidic residue in the neighboring strand. In terms of amino acid composition, the lysine-glutamate pair shows a more rigid conformation for the lysine side chain than the lysine-aspartate one. This can be rationalized by noting that lysine and glutamate are only able to make a direct ionic hydrogen bond, which we deem the most stable interaction, if all the lysine χ dihedrals adopt a trans conformation. However, the shorter aspartate side chain allows for more flexibility forming salt bridges with different rotamers, including the all-trans conformation and a related rotamer where the χ4 dihedral adopts a gauche conformation.
The lateral interaction is characterized by an overall increase in side chain flexibility. An analysis of the solution conformation of this interaction shows that there are several hydrogen bonds and salt bridges that are possible for this contact geometry leading to less rigid conformation for both interacting amino acids.
The stabilizing effect of the sequences present in this study were first noted by Brodsky and co-workers (4). Using molecular modeling the authors hypothesized the presence of contacts similar to the ones described in the previous sections. The authors also carried out a statistical analysis of human collagen type I, II, III, V, and type X that showed a higher occurrence of KGE/D sequences than that expected based on the number of occurrences for each of the individual amino acids, indicating that this motif may have been selected through evolution as an alternate stabilization mechanism for triple helical proteins. However, no difference was made between axial and lateral interactions in that analysis.
A computational study of the KGE and KGD peptides carried out by Stultz and co-workers (32) shows asymmetry in the salt bridges between the different peptide chains, which agrees with our interpretation of the structural data. Their methodology allowed for an estimation of the free energy contribution of each pair using an explicitly solvated model and assigns low free energy contributions to the lateral pairs of both peptides. This study allows us to make a direct link between the observed structural differences in the contact geometry and their effect on the stability of the triple helix. In this context, it is known that the EKG sequence does not provide any thermal stabilization to triple helical peptides (15). In the EKG arrangement where K is in position n and E is position n−1, the sequence alignment resulting from the one-residue stagger characteristic of collagenous domains allows for only lateral interactions from leading to middle and middle to lagging chains and does not allow for any efficient interactions between the lagging and leading chains because the residues are too far apart (4). It should be noted that the structure of the peptide with the sequence EKG in its guest region (12), which packs into a staggered parallel array, does not exhibit any salt bridges. Additionally, it shows conformers for the lysine residues in its leading and middle chains similar to the one observed in lagging chain F of our crystal structure. More generally, it agrees with the conformations observed for the lagging chain of our NMR ensemble, which participates in a lateral interaction. Given that no increased thermal stability is observed for the sequence containing only lateral interactions we propose that they contribute marginally, if at all, to the stability of triple helical proteins. In contrast, the KGE and KGD peptides have thermal stabilities comparable with unsubstituted (POG)8 triple helices which can only be explained by invoking a pair-wise inter-amino acid contribution that does not exist for peptides with only lateral interactions. By this logic, the increased thermal stability observed in the KGE/D sequences comes from the two axial interactions that we observe in the both NMR ensembles as well as the x-ray structure of both peptides.
Assuming that there is an energetic difference between the lateral and axial interactions one can speculate about the assembly and energy landscape of type I collagen, an AAB heterotrimer, particularly the segments that have the sequences presented in this study. Further analysis of the collagen type I sequence shows that the KGD sequence occurs only in the α1 chain whereas the KGE sequence appears in both the α1 and α2 chains. However, all but one of the KGD occurrences in the α1 chain correspond to KGE triplets in the α2 chain. Given the asymmetry observed in the different charge pair interactions this sequence arrangement suggests that type I collagen may utilize this as a form of register-specific stabilization.
The structural study presented in this paper follows the conformation of charged amino acids in different environments that are relevant for the function of collagenous proteins. We are able to identify how the charge pairs stabilize this fold and identify what amino acid interactions are favorable. Using these ideas we are able to speculate on the registration process of natural collagens, an important question that remains unanswered. Furthermore, we are able to characterize the differences between the solution conformation of ionizable residues and their conformation in a crowded macromolecular state in which interhelical contacts are important. In this state we find hydrogen-bonding networks that can direct the packing of antiparallel triple helices into quasi-hexagonal arrays. Similar packing of triple helices is found in collagen type II heterotypical fibers decorated with FACIT type IX collagen.
Supplementary Material
This work was supported in part by National Science Foundation Career Award DMR-0645474, Robert A. Welch Foundation Grants C1557 and C1565, the Norman Hackerman Advanced Research Program of Texas, the Shared University Grid at Rice funded by National Science Foundation Grant EIA-0216467, and a partnership among Rice University, Sun Microsystems, and Sigma Solutions, Inc.

This article contains supplemental Figs. S1–S8, Tables S1–S6, Experimental Procedures, and additional references.
The atomic coordinates and structure factors (codes 3T4F and 3U29) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- HSQC
- heteronuclear single quantum coherence.
REFERENCES
- 1. Khoshnoodi J., Cartailler J. P., Alvares K., Veis A., Hudson B. G. (2006) Molecular recognition in the assembly of collagens: terminal noncollagenous domains are key recognition modules in the formation of triple helical protomers. J. Biol. Chem. 281, 38117–38121 [DOI] [PubMed] [Google Scholar]
- 2. Emsley J., Knight C. G., Farndale R. W., Barnes M. J. (2004) Structure of the integrin α2β1-binding collagen peptide. J. Mol. Biol. 335, 1019–1028 [DOI] [PubMed] [Google Scholar]
- 3. Emsley J., Knight C. G., Farndale R. W., Barnes M. J., Liddington R. C. (2000) Structural basis of collagen recognition by integrin α2β1. Cell 101, 47–56 [DOI] [PubMed] [Google Scholar]
- 4. Persikov A. V., Ramshaw J. A., Kirkpatrick A., Brodsky B. (2005) Electrostatic interactions involving lysine make major contributions to collagen triple-helix stability. Biochemistry 44, 1414–1422 [DOI] [PubMed] [Google Scholar]
- 5. Gauba V., Hartgerink J. D. (2007) Surprisingly high stability of collagen ABC heterotrimer: evaluation of side chain charge pairs. J. Am. Chem. Soc. 129, 15034–15041 [DOI] [PubMed] [Google Scholar]
- 6. Gauba V., Hartgerink J. D. (2007) Self-assembled heterotrimeric collagen triple helices directed through electrostatic interactions. J. Am. Chem. Soc. 129, 2683–2690 [DOI] [PubMed] [Google Scholar]
- 7. Gauba V., Hartgerink J. D. (2008) Synthetic collagen heterotrimers: structural mimics of wild-type and mutant collagen type I. J. Am. Chem. Soc. 130, 7509–7515 [DOI] [PubMed] [Google Scholar]
- 8. Fallas J. A., Gauba V., Hartgerink J. D. (2009) Solution structure of an ABC collagen heterotrimer reveals a single-register helix stabilized by electrostatic interactions. J. Biol. Chem. 284, 26851–26859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Russell L. E., Fallas J. A., Hartgerink J. D. (2010) Selective assembly of a high stability AAB collagen heterotrimer. J. Am. Chem. Soc. 132, 3242–3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. O'Leary L. E., Fallas J. A., Hartgerink J. D. (2011) Positive and negative design leads to compositional control in AAB collagen heterotrimers. J. Am. Chem. Soc. 133, 5432–5443 [DOI] [PubMed] [Google Scholar]
- 11. Boudko S. P., Engel J., Okuyama K., Mizuno K., Bächinger H. P., Schumacher M. A. (2008) Crystal structure of human type III collagen Gly991-Gly1032 cystine knot-containing peptide shows both 7/2 and 10/3 triple helical symmetries. J. Biol. Chem. 283, 32580–32589 [DOI] [PubMed] [Google Scholar]
- 12. Kramer R. Z., Venugopal M. G., Bella J., Mayville P., Brodsky B., Berman H. M. (2000) Staggered molecular packing in crystals of a collagen-like peptide with a single charged pair. J. Mol. Biol. 301, 1191–1205 [DOI] [PubMed] [Google Scholar]
- 13. Ramachandran G. N., Kartha G. (1955) Structure of collagen. Nature 176, 593–595 [DOI] [PubMed] [Google Scholar]
- 14. Persikov A. V., Ramshaw J. A., Brodsky B. (2005) Prediction of collagen stability from amino acid sequence. J. Biol. Chem. 280, 19343–19349 [DOI] [PubMed] [Google Scholar]
- 15. Persikov A. V., Ramshaw J. A., Kirkpatrick A., Brodsky B. (2002) Peptide investigations of pairwise interactions in the collagen triple helix. J. Mol. Biol. 316, 385–394 [DOI] [PubMed] [Google Scholar]
- 16. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Method Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 17. Kissinger C. R., Gehlhaar D. K., Fogel D. B. (1999) Rapid automated molecular replacement by evolutionary search. Acta Crystallogr. D 55, 484–491 [DOI] [PubMed] [Google Scholar]
- 18. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 19. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of COOT. Acta Crystallogr. D 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 21. DeLano W. L. (2010) The PyMOL Molecular Graphics System, version 1.3r1, DeLano LLC, New York [Google Scholar]
- 22. Case D. A., Cheatham T. E., 3rd, Darden T., Gohlke H., Luo R., Merz K. M., Jr., Onufriev A., Simmerling C., Wang B., Woods R. J. (2005) The Amber biomolecular simulation programs. J. Comput. Chem. 26, 1668–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schumacher M. A., Mizuno K., Bächinger H. P. (2006) The crystal structure of a collagen-like polypeptide with 3(S)-hydroxyproline residues in the Xaa position forms a standard 7/2 collagen triple helix. J. Biol. Chem. 281, 27566–27574 [DOI] [PubMed] [Google Scholar]
- 24. Abseher R., Horstink L., Hilbers C. W., Nilges M. (1998) Essential spaces defined by NMR structure ensembles and molecular dynamics simulation show significant overlap. Proteins 31, 370–382 [PubMed] [Google Scholar]
- 25. Kumar S., Nussinov R. (2002) Relationship between ion pair geometries and electrostatic strengths in proteins. Biophys. J. 83, 1595–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Morozov A. V., Kortemme T., Baker D. (2003) Evaluation of models of electrostatic interactions in proteins. J. Phys. Chem. B 107, 2075–2090 [Google Scholar]
- 27. Buevich A. V., Dai Q. H., Liu X., Brodsky B., Baum J. (2000) Site-specific NMR monitoring of cis-trans isomerization in the folding of the proline-rich collagen triple helix. Biochemistry 39, 4299–4308 [DOI] [PubMed] [Google Scholar]
- 28. Jelinski L. W., Torchia D. A. (1980) Investigation of labeled amino acid side chain motion in collagen using 13C nuclear magnetic resonance. J. Mol. Biol. 138, 255–272 [DOI] [PubMed] [Google Scholar]
- 29. Antipova O., Orgel J. P. (2010) In situ D-periodic molecular structure of type II collagen. J. Biol. Chem. 285, 7087–7096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wu J. J., Woods P. E., Eyre D. R. (1992) Identification of cross-linking sites in bovine cartilage type IX collagen reveals an antiparallel type II-type IX molecular relationship and type IX to type IX bonding. J. Biol. Chem. 267, 23007–23014 [PubMed] [Google Scholar]
- 31. Pihlajamaa T., Perälä M., Vuoristo M. M., Nokelainen M., Bodo M., Schulthess T., Vuorio E., Timpl R., Engel J., Ala-Kokko L. (1999) Characterization of recombinant human type IX collagen: association of α-chains into homotrimeric and heterotrimeric molecules. J. Biol. Chem. 274, 22464–22468 [DOI] [PubMed] [Google Scholar]
- 32. Gurry T., Nerenberg P. S., Stultz C. M. (2010) The contribution of interchain salt bridges to triple-helical stability in collagen. Biophys. J. 98, 2634–2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schwieters C. D., Clore G. M. (2002) Reweighted atomic densities to represent ensembles of NMR structures. J. Biomol. NMR 23, 221–225 [DOI] [PubMed] [Google Scholar]
- 34. Schwieters C. D., Clore G. M. (2001) The VMD-XPLOR visualization package for NMR structure refinement. J. Magn. Reson. 149, 239–244 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.