Background: TDP-1 is the worm ortholog of TARDBP, which is a key protein in human neurodegeneration, including ALS and FTLD.
Results: Loss of TDP-1 alleviates proteotoxicity in worm models and extends life span.
Conclusion: TDP-1 has a function in regulating protein homeostasis and aging.
Significance: Learning the novel functions of TDP-1 may advance understanding of the role of RNA processing in protein homeostasis and aging.
Keywords: Aging, Amyotropic Lateral Sclerosis (Lou Gehrig Disease), C. elegans, Neurodegeneration, Protein Misfolding
Abstract
Transactive response DNA-binding protein (TARDBP/TDP-43), a heterogeneous nuclear ribonucleoprotein (hnRNP) with diverse activities, is a common denominator in several neurodegenerative disorders, including amyotrophic lateral sclerosis and frontotemporal lobar degeneration. Orthologs of TDP-43 exist in animals ranging from mammals to invertebrates. Here, we systematically studied mutant Caenorhabditis elegans lacking the nematode TDP-43 ortholog, TDP-1. Heterologous expression of human TDP-43 rescued the defects in C. elegans lacking TDP-1, suggesting their functions are conserved. Although the tdp-1 mutants exhibited deficits in fertility, growth, and locomotion, loss of tdp-1 attenuated defects in several C. elegans models of proteotoxicity. Loss of tdp-1 suppressed defects in transgenic C. elegans expressing TDP-43 or CuZn superoxide dismutase, both of which are associated with proteotoxicity in neurodegenerative diseases. Loss of tdp-1 also reduced defects in mutant animals lacking the heat shock factor HSF-1. Transcriptional profiling demonstrated that the loss of TDP-1 altered expression of genes functioning in RNA processing and protein folding. Furthermore, the absence of tdp-1 extended the life span in C. elegans. The life span extension required a FOXO transcriptional factor DAF-16 but not HSF-1. These results suggest that the C. elegans TDP-1 has a role in the regulation of protein homeostasis and aging.
Introduction
Adult-onset neurodegenerative diseases are characterized by age-dependent and progressive loss of neurons. Proteinaceous inclusions are a common pathological hallmark in many neurodegenerative diseases. From amyloid-β peptides in Alzheimer disease (1) to many aggregation-prone proteins in different neurodegenerative diseases, there is much evidence suggesting that the maintenance of proteostasis is critical for these diseases (2). As a risk factor for neurodegenerative diseases, aging is also associated with regulation of protein homeostasis. In Caenorhabditis elegans, aging is dependent on heat shock factor 1 that controls the expression of molecular chaperones important for protein folding (3, 4). In addition, insulin/IGF-1 signaling, a conserved pathway known to influence life span, has been shown to influence protein aggregation and neurodegeneration in animal models (3, 5, 6). Therefore, alterations in protein quality control appear to be a mechanism that links aging to neurodegeneration.
ALS2 is an age-dependent neurodegenerative disease characterized by progressive degeneration of motor neurons, with ∼10% of all cases considered familial. To date, several genes have been linked to familial ALS, including CuZn superoxide dismutase (SOD1) (7), alsin (8, 9), dynactin (10), senataxin (11), VAMP-associated protein B (12), transactive response DNA-binding protein (TARDBP/TDP-43) (13, 14), fused in sarcoma/translocated in sarcoma (FUS/TLS) (15, 16), optineurin (17), valosin-containing protein (VCP) (18), ubiquilin 2 (19), and a hexanucleotide repeat expansion in the gene C9orf72 (20, 21). Two themes have emerged among these ALS genes. First, perturbation of RNA processing might be implicated in ALS cases linked to TARDBP/TDP-43, FUS, and the hexanucleotide repeat expansion in C9orf72 (20–23). Second, proteinaceous inclusions have been reported as a common pathology in ALS cases linked to SOD1 (24, 25), TARDBP/TDP-43 (13, 14), FUS (15, 16), optineurin (17), VCP (18), and ubiquilin 2 (19).
TDP-43-positive proteinaceous inclusions are also observed in other neurodegenerative disorders, including FTLD, Alzheimer, Parkinson, and Pick disease (22, 23). In ALS and FTLD, the TDP-43-positive inclusions and concurrent depletion of nuclear TDP-43 represent a major pathology of the degenerating tissues (13). Linkage of over 30 TDP-43 mutations to ALS has confirmed the pathogenic role of TDP-43 in neurodegeneration (22). However, it remains unclear whether TDP-43 contributes to neurodegeneration through a gain of toxicity or through loss of its normal function (22, 23).
TDP-43 has protein domains that are characteristic of hnRNP proteins, which were originally defined as a set of proteins that bind to nascent transcripts to form protein-RNA complexes (26). TDP-43 is reported to regulate the transcription of the HIV-1 genome and the SP-10 mouse promoter (27, 28). TDP-43 also plays a role in RNA splicing, including promoting exon skipping during the alternative splicing of the cystic fibrosis transmembrane regulator and apolipoprotein A-II transcripts, as well as promoting exon inclusion during the splicing of the spinal muscular atrophy protein transcript (29, 30). Primarily a nuclear protein, TDP-43 has also been localized to cytoplasmic RNA granules under stress (31–33). TDP-43 has been shown to stabilize the transcripts of low molecular weight neurofilament, histone deacetylase 6, and Atg7 (34–36). TDP-43 also binds to its own transcript and autoregulates its protein levels (37–39). Despite recent studies uncovering roles of TDP-43 in pre-mRNA processing and gene expression regulation (40), the full range of multifaceted TDP-43 functions remains to be elucidated, and their relevance in the neurodegenerative diseases is unclear (22, 23).
Overexpression of TDP-43 causes severe toxicity in many experimental model organisms, including yeast, worm, fly, zebrafish, mouse, and rat (41–53). TDP-43 is highly conserved from mammals to lower metazoans. Knock-out studies suggested that TDP-43 and its orthologs were required in early embryonic development of animals from mouse to Drosophila (35, 54–56). However, the C. elegans ortholog of TDP-43, TDP-1, is dispensable for survival. The physiological function of TDP-1 in C. elegans is currently unknown.
Here, we have systematically characterized loss-of-function tdp-1 mutants to explore the function of TDP-1. We demonstrate that human TDP-43 and C. elegans TDP-1 are functionally conserved. Combining genetic analysis and expression profiling of the loss-of-function tdp-1 mutants, we describe the role of C. elegans TDP-1 in regulating protein homeostasis and aging. In several models of proteotoxicity, loss of TDP-1 alleviated lethality, protein aggregation, and neuronal dysfunction. Loss of TDP-1 also extended life span. These findings may contribute to understanding of the function of this class of hnRNP proteins in protein homeostasis and aging.
EXPERIMENTAL PROCEDURES
C. elegans Strains
N2 Bristol and mutant C. elegans strains were cultured using standard conditions at 20 °C unless indicated otherwise. The mutant strains obtained from the Caenorhabditis Genetics Center were as follows: RB929 (tdp-1(ok803)), VC549 (tdp-1(ok781)), PS3551 (hsf-1(sy441)), CX51 (dyn-1(ky51)), and CF1038 (daf-16(mu86)). The strain FX4439 (fust-1(tm4439)) was received from the National Bioresource Project of Japan. The tdp-1(ok803) and tdp-1(ok781) animals were backcrossed with N2 at least four times. Integrated lines expressing human TDP-C25-YFP (iwIs22), human SOD1-G85R-YFP (iwIs8), or YFP only (iwIs25) driven by the snb-1 promoter have been described previously (6).
The human TDP-43 transgenic iwEx21gf strain was made by injecting 20 μg/ml plasmid Ptdp-1::TDP-43 with 76 μg/ml of a 1-kb ladder DNA and 4 μg/ml Pmyo-2::RFP. The expression of human TDP-43 by Ptdp-1::TDP-43 was driven by the worm tdp-1 promoter, which is defined as a 291-bp genomic DNA fragment between the start codon and the neighboring 5′ gene F44G4.3. This 291-base fragment followed by the human TDP-43 complementary DNA was cloned into the pPD30.38 vector (Fire Lab Vector, Addgene) using HindIII and XhoI sites to generate Ptdp-1::TDP-43. Pmyo-2::RFP was cloned by inserting the red fluorescent protein coding sequence into the pPD132.102 vector (Fire Lab Vector, Addgene) with a myo-2 promoter to drive pharyngeal expression of red fluorescent protein. One line containing the iwEx21gf extrachromosomal array was further treated with 30 μg/ml trimethylpsoralen (Sigma) and two doses of 300 μJ of 365-nm UV light, and the resulting integrant iwIs21gf that stably expressed the transgene was isolated. The iwIs21 integrated line was backcrossed with the N2 strain four times. The iwEx21gf and iwIs21gf strains showed similar phenotypes. Additionally, a Ptdp-1::YFP extrachromosomal array was generated using the same methods and conditions.
Prediction of Protein Domains
Predictions of nuclear localization signal and nuclear export signal for C. elegans TDP-1 were made based upon the criteria of bipartite nuclear localization signal with two clusters of basic amino acids separated by a linker of greater than 10 amino acids and the class 2 nuclear export signal consensus pattern φXφX2φXφ, in which the hydrophobic residue (φ) may be Leu, Ile, Val, Met, or Phe and preferably at least two of the four hydrophobic residues are Leu or Ile, respectively (see detailed information in the supplemental material). Prediction of RRM domains was based on their homology to human TDP-43 RRM domains (22).
Egg Laying at 25 °C and Hatching
C. elegans strains were cultured at 20 °C until they grew to the L4 larval stage. L4 larvae were individually transferred to new plates and then cultured further at 25 °C. These adults were allowed to lay eggs at 25 °C and were then transferred to new plates every 24 h thereafter until they stopped producing eggs (3 days). The number of hatched larvae and dead eggs was counted after 24 h of incubation at 20 °C following the transfer of adults.
Growth Speed at 25 °C
Twenty five synchronized C. elegans eggs laid within a 3-h period at 20 °C were transferred to 25 °C. The number of animals that had reached adulthood was counted after 48, 72, and 96 h.
Locomotor Assay
C. elegans at the L4 larval stage were transferred to M9 buffer (3 mg/ml KH2PO4, 6 mg/ml Na2HPO4, 5 mg/ml NaCl, and 1 mm MgSO4) to observe their thrashing movements. To quantitate their relative motility, the number of the thrashes was counted for 1 min after 1 min of adaptation. A thrash was counted when both the head and the tail bent more than 45° away from the anterior-posterior axis.
Microscopy
For C. elegans low magnification imaging, Leica M165 FC stereo microscope, Leica DFC310 FX camera, and Leica Application Suite were used. For high magnification imaging, animals were immobilized with 100 mm muscimol and examined with a Zeiss AxioObserver Z1 with Apotome imaging system. For nuclear staining, C. elegans were fixed in 500 μl of methanol on dry ice for 5 min. Following three washes of PBS with 0.1% Tween 20 (PBST), the fixed worms were mounted onto microscope slides in a solution of 2.5% 1,4-diazobicyclo[2,2,2]-octane in 100 mm Tris, pH 8.8, with 50% glycerol and 0.2 μg/ml 4′,6′-diamidino-2-phenylindole dihydrochloride (DAPI). For the quantitation of the fluorescent protein aggregates, the overall fluorescence of the inclusion bodies was measured by ImageJ software (National Institutes of Health).
Protein Aggregate Extraction Assay
C. elegans were collected by washing them off the NGM plates using M9 buffer. After five further washes with M9 buffer, worm pellets were resuspended in 200 μl of extraction buffer (10 mm Tris-HCl, pH 8.0, with 1 mm EDTA, 100 mm NaCl, and 0.5% Nonidet P-40) supplemented with mini-EDTA protease inhibitor mixture (Roche Applied Science) and 50 mm iodoacetamide (Sigma) and then homogenized by sonication on ice. The lysates were then transferred to an Airfuge (Beckman Coulter) and centrifuged at 25 p.s.i. (∼130,000 × g) for 5 min. The supernatant was saved as the “S1” fraction. The remaining pellets were sonicated again in the extraction buffer and ultracentrifuged (∼130,000 × g) for 5 min. The final pellet “P2” samples were resuspended in 100 μl of buffer containing 10 mm Tris-HCl, pH 8.0, with 1 mm EDTA, 100 mm NaCl, 0.5% Nonidet P-40, 0.5% deoxycholic acid, and 2% SDS. The S1 and P2 fractions were subjected to SDS-PAGE. The immunoblotting analyses were performed using 1:2,000 anti-YFP (BD Biosciences) and 1:1,000 anti-TDP-43 (Proteintech Group, Chicago) antibodies. Proteins were visualized using enhanced chemiluminescence.
Microarray
Total RNAs were extracted with TRIzol (Invitrogen) from triplicates of N2 and outcrossed tdp-1(ok803lf) C. elegans and purified with the RNeasy kit (Qiagen). The RNAs were labeled using the 3′ IVT Express labeling protocol described by Affymetrix. Briefly, 100 ng of total RNA was used to synthesize first strand cDNA using T7 oligo(dT) oligonucleotides and first strand enzyme mix (Affymetrix). The resulting single strand cDNA was subsequently converted into double strand cDNA using DNA polymerase and RNase H. The double strand cDNA was used to generate and label linearly amplified RNA through in vitro transcription followed by purification using magnetic beads. 15 μg of the labeled amplified RNA was fragmented and hybridized to the Affymetrix C. elegans genome array. Affymetrix Fluidics Station 450 was used to wash and stain the chips, removing the nonhybridized target and incubating with a streptavidin-phycoerythrin conjugate to stain the biotinylated amplified RNA. The staining was further amplified using a biotinylated anti-streptavidin antibody, followed by a second staining step with a streptavidin-phycoerythrin conjugate. Fluorescence was detected using the Affymetrix G3000 GeneArray Scanner, and image analysis of each GeneChip was performed through the Affymetrix GeneChip Command Console version 3.4 software.
The management and statistical analysis of the microarray data were performed using the Partek Genomic Suite (Partek Inc., St. Louis) and Spotfire DecisionSite software (TIBCO Software Inc., Palo Alto, CA). For the Gene Ontology analysis, the annotation file for the C. elegans genome was downloaded from the website of the Gene Ontology Consortium August, 2011.
The microarray data were also analyzed through the use of Ingenuity Pathways Analysis (Ingenuity Systems). For the network analysis, the microarray data set containing C. elegans gene identifiers and expression values was uploaded into the application. Each identifier was mapped to its corresponding gene product in the Ingenuity Knowledge Base. The molecules that met the selection criteria (e.g. expression fold changes above a threshold) were overlaid onto a global molecular network developed from information contained in the Ingenuity Knowledge Base. The relevant networks of selected molecules were then algorithmically generated based on their connectivity. Similarly, for the functional analysis, selected molecules that were associated with biological functions in the Ingenuity Knowledge Base were analyzed to identify the biological functions that were most significant to the data set. The raw data of the present microarray analysis can be found at the NCBI gene expression and hybridization array data repository (GEO, www.ncbi.nlm.nih.gov, accession number GSE34113).
Quantitative Reverse Transcription and Q-PCR
C. elegans was harvested, and RNA was isolated using a phenol/chloroform extraction with TRIzol reagent (Invitrogen), followed by purification using RNeasy mini kit (Qiagen). A two-step RT-PCR was employed to assess relative changes in transgenic transcripts using iScript cDNA synthesis kit and SYBR Green Supermix (Bio-Rad). Standard curves were generated for all primers used, and the worm gdh-1 gene was used as the control.
Life Span Assay
Synchronized C. elegans eggs were isolated within a 3-h period of egg laying for life span assay. Animals were considered dead if they showed no response when probed with a platinum pick. At least 90 animals were used for each experiment. The animals were censored if they crawled out of the plate, had a ruptured vulva, or died as “bags of worms” with larvae hatching inside the adults. For hsf-1(sy441lf) mutants, which die at a high frequency as bags of worms, 100 μg/ml 5-fluorodeoxyuridine (Sigma) was included in the medium to prevent reproduction. Synchronized late L4 larvae were transferred from normal NGM plates to NGM plates containing 5-fluorodeoxyuridine. The life span data were analyzed with Prism 3 software.
Statistical Analysis
p values for all phenotypic analyses were obtained using Student's t test, with the exception of the life span data, for which the log rank test was used. For the microarray data, Student's t test was used to analyze the gene expressions. For the analysis of the transcriptome profiles using Gene Ontology and Ingenuity Pathways Analysis, Fisher's exact test was used.
RESULTS
C. elegans tdp-1 Is Required for Optimal Fertility, Growth, and Locomotion
Our bioinformatic analyses and searches in gene homology databases suggested that tdp-1 (sequence name F44G4.4) is the sole ortholog of human TDP-43 in the C. elegans genome (Fig. 1A and supplemental Fig. S1). C. elegans tdp-1 encodes a protein that has the same length, 414 amino acids, as its human ortholog. Human TDP-43 and C. elegans TDP-1 share the same alignment of protein domains characteristic of an hnRNP protein, including two RRM domains, a nuclear localization signal proximate to the N terminus, and a nuclear export signal embedded in the second RRM domain.
FIGURE 1.
C. elegans TDP-1 and human TDP-43 are functionally conserved. A, schematic diagram of C. elegans TDP-1 and human TDP-43 proteins. Predictions of domains are described under “Experimental Procedures.” Loss of TDP-1 protein regions caused by tdp-1 mutant alleles, ok803 and ok781, are indicated. NLS, nuclear localization signal; NES, nuclear export signal. B–F, loss of C. elegans TDP-1 (ok803 or ok781) and expression of human TDP-43 (iwIs21) rescue each other's phenotypic defects in egg-laying, growth, and locomotion. B, mean number of eggs (solid bars) and corresponding larvae (hatched bars) produced when parental hermaphrodites at the L4 stage were transferred from 20 to 25 °C (± S.E.; n = 10). *, p < 0.0001. C, mean fraction of eggs laid at 20 °C reaching adulthood after 48 (solid bars) or 72 h (hatched bars) of growth at 25 °C (± S.E.; n = 5). *, p < 0.02. D, mean locomotor activity of L4 larvae, as indicated by the number of thrashes per min in liquid (± S.E.; n = 25) following culture at 20 °C (solid bars) and 25 °C (hatched bars). *, p < 0.0001. E and F, representative images of animals grown at 20 and 25 °C for 3 days. G, total TDP-43 protein level in TDP-43(iwIs21gf) animals is not affected by tdp-1(ok803lf).
We obtained two mutant alleles of C. elegans tdp-1, ok803 and ok781, each of which remove ∼1.2 kb from tdp-1, including the two RRM domains and the nuclear export signal (Fig. 1A and supplemental Fig. S1). Both deletions are probably null mutations. These C. elegans tdp-1 loss-of-function (lf) mutants exhibited discernible defects in fertility, growth, and locomotion. Most of the tdp-1(lf) data shown here were obtained using ok803, and the results were verified using the other allele ok781. First, loss of tdp-1 led to lower fertility in C. elegans. The tdp-1(ok803lf) hermaphrodites generated only half as many eggs as N2 wild-type (WT) animals, likely as a result of an early depletion of sperm (Fig. 1B and supplemental Fig. S2). Second, loss of function of tdp-1 led to slower growth. At 48 h after egg laying, ∼75% of tdp-1(ok803lf) animals had reached adulthood, as compared with ∼91% of N2 WT animals (Fig. 1C). Finally, loss of tdp-1 led to a locomotor deficit, which was exacerbated when the animals were cultured at 25 °C. Specifically the rate of thrashing by tdp-1(ok803lf) animals was ∼74% that of N2 WT animals (Fig. 1D). Thus, intact tdp-1 is required for optimal developmental and physiological processes in C. elegans, including fertility, growth, and locomotion.
Human TDP-43 and C. elegans TDP-1 Are Functionally Conserved
To examine whether human TDP-43 could functionally replace TDP-1 in C. elegans, we expressed human TDP-43 under the control of the endogenous tdp-1 promoter. Following generation of extrachromosomal transgenic arrays expressing human TDP-43, we further developed a stable integrated transgene iwIs21. Next, we constructed strains containing both the gain-of-function TDP-43(iwIs21gf) transgene and tdp-1(lf) mutations. The TDP-43(iwIs21gf) transgene moderately improved the fertility defects in the tdp-1(ok803lf) mutant (Fig. 1B, p = 0.10). Moreover, the human TDP-43(iwIs21gf) transgene rescued the slow growth phenotype in loss-of-function mutant tdp-1(ok803) (Fig. 1C). In addition, the TDP-43(iwIs21gf) transgene rescued the locomotor deficit caused by the tdp-1(ok803lf) mutation at 25 °C (Fig. 1D). These data demonstrated that human TDP-43 could substitute for C. elegans TDP-1, indicating that they are functional orthologs.
Transgenic expression of human TDP-43 in C. elegans can induce locomotor and developmental defects. The TDP-43(iwIs21gf) transgenic C. elegans exhibited locomotor defects as compared with WT animals. The rate of thrashing by the TDP-43(iwIs21) animals was ∼70% that of WT animals at 25 °C (Fig. 1D). Notably, the TDP-43(iwIs21gf) transgene caused profound temperature-dependent developmental defects. When parental hermaphrodites harboring the TDP-43(iwIs21gf) transgene were transferred from 20 to 25 °C, most offspring died during embryogenesis (Fig. 1B), and many surviving larvae failed to grow to adulthood (Fig. 1E). By comparison, if the temperature elevation occurred after the parental hermaphrodites laid eggs at 20 °C, most of the offspring could grow to adulthood, albeit at a slower rate than the N2 WT animals (Fig. 1C). For example, when eggs laid at 20 °C were hatched subsequently at 25 °C, ∼60% of TDP-43(iwIs21) animals reached adulthood at 48 h after egg laying, as compared with ∼91% of WT animals (Fig. 1C). By a series of 25 °C incubations of varying duration administered at several time points during development, elevation of temperature for 2–3 h near the time of fertilization was found to be sufficient and necessary to induce embryonic arrest (supplemental Fig. S3). This temperature-dependent phenotype was solely dependent on the TDP-43 transgene but not its chromosomal integration site, because examination of extrachromosomal array transgenic animals demonstrated similar defects. The profound temperature-sensitive developmental defect in TDP-43(iwIs21gf) animals is consistent with the notion that the toxicity of TDP-43 protein is dependent on its misfolding (6), which can be exacerbated by rapid temperature elevation.
Next, we examined the expression pattern of the tdp-1 gene. The promoter region of the tdp-1 gene was cloned to drive expression of a YFP reporter. With the engineered tdp-1 promoter, the YFP reporter was expressed in embryos before hatching. In both larvae and adults, the reporter expression was found in multiple C. elegans tissues, including body wall muscles, pharynx, and neurons (supplemental Fig. S4). It should be noted that the endogenous expression of tdp-1 that is not recapitulated by the engineered promoter is still possible.
A diverse set of model organisms was shown to be sensitive to elevated levels of TDP-43 expression (41–53). If human TDP-43 is functionally redundant with TDP-1, lowering the level of endogenous TDP-1 would attenuate the toxicity of the TDP-43 transgene in C. elegans. Indeed, the tdp-1(ok803lf) mutation suppressed the observed defects in the TDP-43(iwIs21gf) animals. First, loss of tdp-1 attenuated the embryonic lethality and slow growth phenotype observed in transgenic TDP-43(iwIs21gf) animals at 25 °C (Fig. 1, B, C, and E). Second, loss of tdp-1 also suppressed the locomotor deficit observed in TDP-43(iwIs21gf) animals (Fig. 1D). Finally, another independent loss-of-function mutation tdp-1(ok781lf) suppressed the developmental defects caused by TDP-43(iwIs21gf) at 25 °C (Fig. 1F). The protein expression level of TDP-43 was not changed by the presence of the tdp-1(ok803lf) mutation (Fig. 1G). The insoluble protein aggregates in TDP-43(iwIs21) were not detected. These results showed that human TDP-43 can substitute for TDP-1 and that the removal of TDP-1 alleviates the toxicity of elevated levels of TDP-43, suggesting that human TDP-43 and C. elegans TDP-1 are functionally conserved.
Loss of tdp-1 Attenuates Neurotoxicity of Aggregation-prone Proteins
Next, we investigated whether loss of tdp-1 confers any protection against the toxicity of TDP-C25, which is a 25-kDa carboxyl fragment of TDP-43 and a signature component of the TDP-43 positive inclusions in brain tissues from ALS and FTLD patients (13). TDP-C25 exhibits an unusually high propensity to form protein aggregates (6). In transgenic TDP-C25(iwIs22gf) C. elegans stably expressing the TDP-C25-YFP fusion protein under the pan-neuronal promoter of the snb-1 synaptobrevin gene, TDP-C25-YFP forms discrete fluorescent protein aggregates in neuronal somas and axons (Fig. 2B) (6). TDP-C25(iwIs22gf) transgenic C. elegans exhibited severe locomotor deficits, which resulted from neurotoxicity of the misfolded proteins (6). Interestingly, loss of tdp-1 significantly ameliorated the locomotor defects caused by TDP-C25(iwIs22gf) (Fig. 2A), suggesting a strong suppression of the neurotoxicity. In addition, the amount of fluorescent TDP-C25-YFP aggregates in neurons was reduced in TDP-C25(iwIs22gf);tdp-1(ok803lf) animals, as compared with animals carrying the TDP-C25(iwIs22gf) transgene alone (Fig. 2, C and D). Similar amelioration of TDP-C25 protein aggregation was also observed in animals carrying the independent loss-of-function mutation tdp-1(ok781) and the TDP-C25(iwIs22gf) transgene (supplemental Fig. S5). To examine the effect of loss of tdp-1 on TDP-C25 aggregation, an aggregate extraction assay was used as described previously (6). When compared with the protein aggregates in animals carrying only the TDP-C25(iwIs22gf) transgene, insoluble TDP-C25 aggregates were significantly reduced in animals carrying both the TDP-C25(iwIs22gf) transgene and the loss-of-function mutation tdp-1(ok803) (Fig. 2E), but no change in soluble TDP-C25 protein levels was detected. These data suggest that loss of tdp-1 may alleviate TDP-C25-associated neurotoxicity by suppressing protein aggregation.
FIGURE 2.
Loss of tdp-1 alleviates locomotor deficits and protein aggregation in C. elegans expressing neuronal TDP-C25(iwIs22gf) or SOD1-G85R(iwIs8gf). A and F, relative locomotor activity of 1-day-old adults as indicated by thrashing rates in liquid normalized against control strains expressing YFP only (± S.E.; n = 32). *, p < 0.0001. B and G, cytoplasmic protein aggregates indicated by YFP fluorescence and nuclei by DAPI staining (blue) in neurons expressing TDP-C25-YFP (iwIs22gf) or SOD1-G85R-YFP (iwIs8gf). Scale bar, 5 μm. C and H, protein aggregates indicated by YFP fluorescence in head neurons expressing TDP-C25-YFP or SOD1-G85R-YFP from live C. elegans animals are compared between WT and loss-of-function tdp-1(ok803lf) backgrounds. Scale bar, 5 μm. D and I, protein aggregation was quantified by measuring the YFP fluorescence intensity of the inclusions (± S.E.; n = 3). *, p < 0.05. E and J, protein levels of TDP-C25-YFP and SOD1-G85R-YFP in soluble (supernatant) and insoluble (pellet) fractions of differentially extracted tissues from animals in the WT or tdp-1(ok803lf) mutant background.
We then asked whether the protective effect of loss of tdp-1 on neurotoxicity associated with protein misfolding and aggregation was specific to the TDP-43 polypeptides. Mutations in CuZn superoxide dismutase (SOD1) have been implicated in ∼20% of familial ALS cases, and many of these mutations lead to increased protein aggregation (57). A glycine-to-arginine substitution at amino acid 85 (G85R) renders SOD1 particularly aggregation-prone, and the C. elegans-stable transgene iwIs8 expressing an SOD1-G85R-YFP fusion protein under the snb-1 pan-neuronal promoter causes pronounced locomotor defects and formation of protein aggregates in neurons (Fig. 2G) (58). The loss-of-function tdp-1(ok803) mutation was found to significantly improve locomotor activity and reduce fluorescent aggregates in animals carrying the SOD1-G85R(iwIs8gf) transgene (Fig. 2, F, H, and I). Detergent extraction assay further indicated reduction in SOD1-G85R-YFP protein aggregate levels in C. elegans carrying both the transgene SOD1-G85R(iwIs8gf) and the loss-of-function mutation tdp-1(ok803), as compared with animals carrying the SOD1-G85R(iwIs8gf) transgene alone (Fig. 2J). Thus, loss of tdp-1 may alleviate the neurotoxicity caused by TDP-43 and SOD1 through a shared mechanism.
We also tested whether the effects of TDP-1 on protein aggregation could be seen in another RNA-binding protein. Similar to TDP-43, FUS is also an hnRNP protein that has been implicated in the neurodegenerative diseases ALS and FTLD. Mutations in FUS were linked to familial ALS, and its protein products were found in ubiquitin-positive inclusions in the central nervous system of a subset of ALS and FTLD patients (15, 16). The C. elegans ortholog of FUS is fust-1(C27H5.3), and a large deletion allele, tm4439, is probably a null allele of fust-1. We generated C. elegans carrying both the loss-of-function mutation fust-1(tm4439) and the transgenic TDP-C25(iwIs22gf). Our examination indicated no alleviating effects of fust-1(tm4439) on the protein aggregation pathology. The amount of fluorescent TDP-C25-YFP aggregates in neurons was similar between animals carrying TDP-C25(iwIs22gf);tdp-1(ok803lf) and those carrying the TDP-C25(iwIs22gf) transgene alone (supplemental Fig. S6). Although fust-1 may potentially differ from tdp-1 in many aspects, including their temporal and spatial expressions, this result suggested that the role in suppressing protein aggregation may be specific for tdp-1.
Loss of tdp-1 Alleviates Defects in the Absence of Heat Shock Factor 1
The observation that loss of tdp-1 suppressed proteotoxicity and aggregation of distinct proteins such as TDP-43 and SOD1 suggested that tdp-1(lf) might suppress defects caused by mutations that interfere with overall protein quality control. In response to heat-induced stress, heat shock factor 1 induces transcription of many genes, including molecular chaperones responsible for protection from protein misfolding and aggregation (59). A loss-of-function mutation of the C. elegans heat shock factor 1, hsf-1(sy441lf), causes temperature-dependent growth and egg-laying defects (60). Therefore, we used the mutant hsf-1(sy441lf) animals as a model for compromised protein quality control independent of ectopic transgene expressions. To test whether loss of tdp-1 improves the compromised protein quality control in the absence of hsf-1, animals carrying both hsf-1(sy441lf) and tdp-1(ok803lf) were constructed and examined. The hsf-1(sy441lf);tdp-1(ok803lf) double mutant animals showed significant improvement in growth and egg laying as compared with the hsf-1(sy441lf) single mutant animals (Fig. 3, A and B). Next, to address whether the protective effect of tdp-1(ok803lf) was general to all temperature-sensitive mutants, we examined another temperature-sensitive mutant, the dynamin GTPase dyn-1(ky51lf), previously shown to be influenced by proteotoxicity (61). Unlike hsf-1(sy441lf), the locomotor and egg-laying defects of dyn-1(ky51lf) were not alleviated but worsened by tdp-1(ok803lf) (supplemental Fig. S7). These data indicated that loss of tdp-1 specifically protected against the defects caused by the absence of normal HSF-1 in C. elegans.
FIGURE 3.
tdp-1(ok803lf) mutation alleviates defects in loss-of-function mutant hsf-1(sy441lf). A, relative number of eggs (± S.E.; n = 20) laid per animal as normalized against the WT N2 strain following culture at 25 °C. *, p < 0.0001. B, mean fraction of eggs laid at 20 °C reaching adulthood after 72 (solid bars) or 96 h (hatched bars) of growth at 25 °C (± S.E.; n = 5). *, p < 0.05. C, protein aggregates indicated by YFP fluorescence in head neurons expressing hemizygous SOD1-G85R-YFP (iwIs8gf) from live C. elegans L4 larvae are compared between the hsf-1(sy441lf) and double mutant tdp-1(ok803If);hsf-1(sy441lf) backgrounds. Scale bar, 5 μm. D, protein aggregation was quantified by measuring the YFP fluorescence intensity of the inclusions (± S.E.; n = 3). *, p < 0.05. E, relative locomotor activity of L4 larvae as indicated by thrashing rates in liquid normalized against control strains expressing hemizygous SOD1G85R-YFP (iwIs8gf) only (± S.E.; n = 25). *, p < 0.0001.
To further test the effects of loss of tdp-1 on hsf-1-dependent proteotoxicity, we used the transgene SOD1-G85R(iwIs8gf) as a reporter. Loss-of-function hsf-1(sy441lf) mutant alone significantly worsened the protein aggregation and locomotor defects caused by the transgene SOD1-G85R(iwIs8gf). In the presence of homozygous hsf-1(sy441lf), most animals homozygous for the transgene SOD1-G85R(iwIs8gf) were too sick to grow to adulthood, so we used animals hemizygous for the transgene SOD1-G85R(iwIs8gf) for our analysis. Specifically, we constructed and compared a triple mutant that is hemizygous for SOD1-G85R(iwIs8gf) and homozygous for hsf-1(sy441lf);tdp-1(ok803lf) with a double mutant that is hemizygous for SOD1-G85R(iwIs8gf) and homozygous for hsf-1(sy441lf). The presence of tdp-1(ok803lf) attenuated the protein aggregation and locomotor defects in the triple mutant animals, as compared with the double mutant animals (Fig. 3, C–E).Thus, tdp-1 may act downstream of hsf-1 or in a parallel pathway to influence proteotoxicity. Together, these results support a role for C. elegans tdp-1 in regulating protein quality control.
Transcriptional Profiling of tdp-1 Loss-of-function Mutant
To examine the global gene expression changes induced by loss of tdp-1, C. elegans transcriptomes were compared between N2 WT animals and tdp-1(ok803lf) mutant animals. Messenger RNAs were isolated from triplicate samples of WT or tdp-1(ok803lf) C. elegans, and linearly amplified RNA probes were used to hybridize to the Affymetrix C. elegans genome array that probes over 22,500 gene transcripts. Q-PCR was used to verify the gene expression data from the microarrays. Correlation analysis indicated that the microarray data were highly reproducible with the Q-PCR assay. Overall, the microarray and Q-PCR values exhibited a good linear correlation for 29 representative genes tested (R > 0.90). In the absence of TDP-1, there were more genes down-regulated than those up-regulated. If the threshold was set at a fold change (FC) of 1.5 by the microarray measurement, 712 genes (3.15%) were differentially regulated (485 down and 227 up) in the tdp-1(lf) mutant. If the threshold was set at FC of 1.2, 4381 genes (19.3%) were differentially regulated (2600 down and 1781 up) in the tdp-1(lf) mutant (supplemental Table S1).
To examine biological processes that are significantly affected in the tdp-1(ok803lf) mutant, we conducted gene ontology analysis on the transcriptome profiling results using annotation from the Gene Ontology Consortium (62). We asked which biological processes as defined by Gene Ontology terms were most significantly represented by the top 712 differentially regulated genes in the C. elegans genome. This analysis indicated that the most significantly affected biological processes in tdp-1(ok803lf) were those involving molting cycle, growth, locomotion, determination of adult life span, and endoplasmic reticulum unfolded protein response (Fig. 4B and supplemental Table S2). These top-ranked biological processes matched well with the phenotypes observed in the tdp-1(lf) mutant C. elegans. For example, 98 out of 1466 genes related to locomotion were represented in the differentially expressed set (p < 10−9) (supplemental Table S2). Consistently, locomotor defects were observed in the tdp-1(lf) mutant (Fig. 1). Also, 132 out of 2329 genes related to growth were represented in the differentially expressed set (p < 10−7) (supplemental Table S2), and loss of tdp-1 caused growth defects in the mutant C. elegans (Fig. 1). Finally, there was a trend of down-regulation for the genes that encode molecular chaperones (supplemental Table S2). Q-PCR assays confirmed the trend of down-regulation for a few molecular chaperones in the tdp-1(lf) mutant (supplemental Fig. S8A), suggesting that there is a reduced burden on protein quality control in these mutants.
FIGURE 4.
Transcriptional profile analysis of mutant C. elegans lacking tdp-1. A, results from C. elegans genome microarrays are validated with quantitative PCR. The relative fold changes in gene expression of 29 representative genes between tdp-1(ok803lf) and the WT N2 animals are shown. The values from the microarray and the quantitative PCR experiments show a linear relationship. R is the Pearson correlation coefficient. B, biological processes that are most affected in tdp-1(ok803lf) according to the Gene Ontology analysis. ER, endoplasmic reticulum. C, molecular networks that are most affected in tdp-1(ok803lf) according to the Ingenuity Pathways Analysis. D and E, molecular and cellular functions that are most affected in tdp-1(ok803lf) according to the Ingenuity Pathways Analysis, with the threshold at FC >1.5 (D) or FC >1.2 (E). P is the probability value from Fisher's exact test.
To further understand the molecular consequences of loss of tdp-1 at the molecular level, we analyzed the differentially regulated genes using the Ingenuity Pathways Analysis from Ingenuity Systems, which is supported by a repository of evidence-based biological interactions and functional annotations. The differentially regulated genes were highly enriched in several functional categories that involve protein post-translational and RNA post-transcriptional modifications (Fig. 4, C–E). For the top 712 differentially regulated genes (FC >1.5), the most relevant molecular network included proteins that function in protein conformational modification and mRNA processing (Fig. 4C and supplemental Table S3). Additionally, the Ingenuity Pathways Analysis of over-represented molecular functions pointed to a few cellular functions, including protein folding (Fig. 4D and supplemental Table S4). To gain a full picture of expression of genes involved in protein quality control, the genes involved in protein folding and ubiquitination were selected with a lower threshold of expression fold changes (FC >1.2) (Table S5). At this threshold (FC >1.2), RNA modification was the top-ranked molecular function represented by the differentially regulated genes (Fig. 4E and supplemental Table S6). The loss of tdp-1 had pleiotrophic consequences on the expression of diverse RNA-processing proteins, including those functioning in the processing and splicing of messenger RNA, ribosomal RNA, transfer RNA, and small noncoding RNA (supplemental Table S6). For example, the loss of tdp-1 altered the expression of hnRNP genes, including the C. elegans homolog of hnRNPA1, hrp-2 (supplemental Fig. S8B). The expression of genes involved in RNA splicing, such as snr-5, which encodes a small nuclear ribonucleoprotein, was also affected (supplemental Fig. S8B). Together, the transcriptome profiling of the tdp-1 loss-of-function mutant showed a prominent change in the expression of genes involved in protein post-translational and RNA post-transcriptional modifications.
TDP-1 Regulates Life Span via a Pathway Requiring DAF-16 but Independent of HSF-1
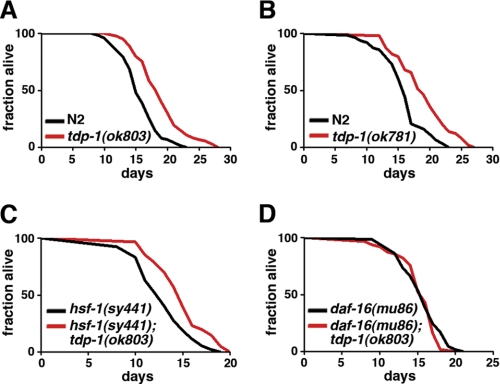
The transcriptional profiling analysis also showed that genes functioning in the determination of adult life span were differentially regulated in the tdp-1(lf) mutant (Fig. 4B and supplemental Table S2). Like neurodegeneration, aging has been linked to deleterious effects of protein misfolding (3, 4). Reasoning that the improved protein homeostasis resulting from tdp-1(lf) might be associated with altered life span, we examined the life span of tdp-1(lf) mutants. Interestingly, the loss-of-function tdp-1(ok803) mutants lived ∼20% longer than the N2 wild-type animals when cultured at 20 °C (Fig. 5A). An independent loss-of-function allele, tdp-1(ok781), also extended the life span of C. elegans to a similar extent (Fig. 5B). Our combined results indicate that C. elegans TDP-1 is a negative regulator of aging.
FIGURE 5.
tdp-1(lf) mutation leads to a life span extension that is dependent on daf-16 but not on hsf-1. A, survival of the WT N2 (black) and tdp-1(ok803lf) (red) animals. p < 0.0001. B, survival of N2 WT (black) and tdp-1(ok781lf) (red) animals. p < 0.0001. C, survival of hsf-1(sy441lf) animals (black) and hsf-1(sy441lf);tdp-1(ok803lf) double mutants (red) at 25 °C using 100 μg/ml of 5-fluorodeoxyuridine. D, survival of daf-16(mu86lf) animals (black) and daf-16(mu86); tdp-1(ok803lf) double mutants (red). p < 0.0001.
There are two characterized transcriptional factors, encoded by hsf-1 and daf-16, that regulate both aging and protein homeostasis (3, 4). Because hsf-1 and daf-16 are two key genes that positively regulate life span, we asked whether the life span extension conferred by tdp-1(lf) requires the normal function of hsf-1 or daf-16. We observed that hsf-1(sy441);tdp-1(ok803lf) double mutants lived longer than the single mutant hsf-1(sy441), suggesting that tdp-1(lf) influenced life span at least through some mechanisms independent of hsf-1 (Fig. 5C). In contrast, a loss-of-function mutant daf-16(mu86lf) completely blocked the life span-extending effects of tdp-1(ok803lf) (Fig. 5D), indicating that the long lived phenotype conferred by loss of tdp-1 required intact daf-16.
DISCUSSION
In this study, we systematically investigated the function of TDP-1 in C. elegans. TDP-1 shares significant homology with its mammalian ortholog TDP-43. The genetic interactions between TDP-1 and TDP-43 suggest that they are functionally conserved. Loss of TDP-1 appeared to increase the tolerance of C. elegans to proteotoxicity. Furthermore, loss of TDP-1 extended life span in C. elegans. Together, these results suggest a novel role for a conserved RNA-processing protein in the regulation of protein homeostasis and life span.
C. elegans TDP-1 is highly homologous to mammalian TDP-43. TDP-1 has the same protein domains as mammalian TDP-43 (Fig. 1). Although the C-terminal region of TDP-1 is not as glycine-rich as mammalian TDP-43, there is significant homology in this region, and the C-terminal part of TDP-1 can replace its mammalian counterpart and maintain the RNA splicing activity of the mammalian protein (46). The genetic interactions between C. elegans TDP-1 and human TDP-43 suggest that they are functionally conserved. For example, heterologous expression of human TDP-43 in C. elegans is able to suppress the defects caused by loss of tdp-1 (Fig. 1). C. elegans TDP-1 is not required for survival, unlike other TDP-43 orthologs in more complex metazoans from Drosophila to mice (35, 54–56). Although it is likely that TDP-43 in higher animals may have acquired distinct properties compared with TDP-1, it is also possible that their conserved function is required for survival of complex organisms but not for survival of relatively simple C. elegans. Such low stringency of requirement for survival in C. elegans as compared with mammals has been observed in loss-of-function studies of many other genes. For example, loss of the type III RNase Dicer1 leads to lethality early in mouse development (63), but C. elegans lacking the ortholog DCR-1 are viable (64). Thus, the nonlethal phenotype of C. elegans lacking TDP-1 provides a simple system to study the conserved functions of this family of proteins.
RNA-processing proteins such as hnRNPs have multiple roles in RNA metabolism, including transcription, splicing, and nucleocytoplasmic transport of RNAs (26). The function of TDP-1, like its mammalian ortholog TDP-43 (40), is probably multifaceted. Loss of TDP-1 caused changes in expression levels of many RNA-processing genes, including orthologs of hnRNP proteins and splicing factors (supplemental Table S6 and Fig. S8). The effect of loss of TDP-1 on RNA processing could lead to the observed pleiotrophic phenotypes in C. elegans, including defects in fertility, growth, and locomotion (Fig. 1). Consistent with these observations, gene ontology analysis of the transcriptome profiles indicated altered expression of genes that have related functions such as growth and locomotion (Fig. 4B).
Furthermore, our characterization of tdp-1 loss-of-function mutant suggested a role for TDP-1 in the regulation of protein homeostasis and life span in C. elegans. Loss of TDP-1 increased the solubility of an aggregation-prone C-terminal fragment of TDP-43 as well as that of a mutant SOD1. Loss of TDP-1 also conferred resistance to HSF-1 depletion in C. elegans. These data suggested that long term reduction in the function of TDP-1 led to a resistance to stress on proteostasis. In accordance with our whole-animal observations from C. elegans, a recent study showed that knockdown of the Drosophila ortholog of TDP-43 protected against the neurotoxicity of overexpressed mutant VCP, an AAA+ ATPase functioning in protein quality control, in a transgenic fly model (65). The molecular mechanisms through which TDP-1 regulates proteostasis in C. elegans remain to be elucidated. We propose three potential models that are not mutually exclusive. First, the removal of the TDP-1 protein itself may lessen the burden on the protein folding machinery in the cell. This model is supported by recent observations that the TDP-43 protein is highly prone to misfold and aggregate in vitro and in vivo (6, 66) and that a TDP-43 fragment can form amyloid fibrils (67). Second, diverse activities of TDP-1 in RNA processing may allow it to act as an orchestrator to adapt to proteotoxic stress through the regulation of RNA. RNA processing could affect all aspects of protein homeostasis, from protein synthesis to degradation. Loss of TDP-1 may alter global RNA levels and in turn protein homeostasis, resulting in adaptation of the cell to stress on protein quality control systems. Consistent with this model, the microarray analysis showed a trend of decreasing mRNA levels (supplemental Table S1). Finally, TDP-1 may regulate proteostasis through specific pathways. Our analysis of the life span extension in the tdp-1 loss-of-function mutant suggested that the phenotype required intact DAF-16 but not HSF-1 (Fig. 5). Therefore, the transcriptional factor DAF-16, which is known to promote protein homeostasis (3, 6), may be an important player mediating the effect of TDP-1 on proteostasis.
Our finding that loss of TDP-1 reduces proteotoxicity is consistent with the observation that the loss-of-function mutant animals had extended life span. Although the molecular connections between aging and protein homeostasis are not fully understood, two pathways have been found to regulate both aging and protein homeostasis, i.e. the heat shock response and the insulin/IGF-1 signaling (3, 4). HSF-1 and DAF-16 are the master transcriptional factors that mediate the heat shock response and the insulin/IGF-1 signaling, respectively. Whereas reduced activity in TDP-1 reversed the defects in a loss-of-function mutant of HSF-1 (Fig. 3), the depletion of HSF-1 did not block the life span-extending effects of the TDP-1 reduction (Fig. 5). By contrast, the loss of DAF-16 completely blocked the life span-extending effects of the TDP-1 reduction. In simplest genetic interpretations, TDP-1 acts upstream of DAF-16 but independently of HSF-1 to influence life span (supplemental Fig. S9). However, complex networks rather than the linear pathways are possible. Nevertheless, the life span extension in the absence of TDP-1 requires intact DAF-16, an evolutionarily conserved FOXO family transcriptional factor at the intersection of regulations of aging, immunity, and stress responses (68).
In summary, we identified a new function of TDP-1 in regulating protein homeostasis and life span. Given the established functions of the TDP-1 homologs in RNA processing, this study suggests a layer of regulation of proteostasis and aging imparted by RNA-processing proteins. Further studies on these new functions of TDP-1 may help understand the conserved function of this class of RNA-processing proteins in metazoan evolution.
Supplementary Material
Acknowledgments
We thank D. Drummond-Barbossa, M. Matunis, P. Coulombe, and members of Wang laboratory for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants NS062089 and NS07432 (to J. W.). This work was also supported by the Robert Packard Center for ALS Research at Johns Hopkins, the Muscular Dystrophy Association, and The Johns Hopkins Claude D. Pepper Older Americans Independence Center.
This article was selected as a Paper of the Week.

This article contains supplemental Tables 1–6, Figs. S1–S9, and additional references.
- ALS
- amyotrophic lateral sclerosis
- FTLD
- frontotemporal lobar degeneration
- RRM
- RNA recognition motif
- lf
- loss of function
- FC
- fold change
- Q-PCR
- quantitative PCR
- hnRNP
- heterogeneous nuclear ribonucleoprotein.
REFERENCES
- 1. Glenner G. G., Wong C. W. (1984) Alzheimer disease. Initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem. Biophys. Res. Commun. 120, 885–890 [DOI] [PubMed] [Google Scholar]
- 2. Balch W. E., Morimoto R. I., Dillin A., Kelly J. W. (2008) Adapting proteostasis for disease intervention. Science 319, 916–919 [DOI] [PubMed] [Google Scholar]
- 3. Hsu A. L., Murphy C. T., Kenyon C. (2003) Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science 300, 1142–1145 [DOI] [PubMed] [Google Scholar]
- 4. Morley J. F., Morimoto R. I. (2004) Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol. Biol. Cell 15, 657–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cohen E., Bieschke J., Perciavalle R. M., Kelly J. W., Dillin A. (2006) Opposing activities protect against age-onset proteotoxicity. Science 313, 1604–1610 [DOI] [PubMed] [Google Scholar]
- 6. Zhang T., Mullane P. C., Periz G., Wang J. (2011) TDP-43 neurotoxicity and protein aggregation modulated by heat shock factor and insulin/IGF-1 signaling. Hum. Mol. Genet. 20, 1952–1965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rosen D. R., Siddique T., Patterson D., Figlewicz D. A., Sapp P., Hentati A., Donaldson D., Goto J., O'Regan J. P., Deng H. X.. (1993) Mutations in CuZn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 362, 59–62 [DOI] [PubMed] [Google Scholar]
- 8. Yang Y., Hentati A., Deng H. X., Dabbagh O., Sasaki T., Hirano M., Hung W. Y., Ouahchi K., Yan J., Azim A. C., Cole N., Gascon G., Yagmour A., Ben-Hamida M., Pericak-Vance M., Hentati F., Siddique T. (2001) The gene encoding alsin, a protein with three guanine-nucleotide exchange factor domains, is mutated in a form of recessive amyotrophic lateral sclerosis. Nat. Genet. 29, 160–165 [DOI] [PubMed] [Google Scholar]
- 9. Hadano S., Hand C. K., Osuga H., Yanagisawa Y., Otomo A., Devon R. S., Miyamoto N., Showguchi-Miyata J., Okada Y., Singaraja R., Figlewicz D. A., Kwiatkowski T., Hosler B. A., Sagie T., Skaug J., Nasir J., Brown R. H., Jr., Scherer S. W., Rouleau G. A., Hayden M. R., Ikeda J. E. (2001) A gene encoding a putative GTPase regulator is mutated in familial amyotrophic lateral sclerosis 2. Nat. Genet. 29, 166–173 [DOI] [PubMed] [Google Scholar]
- 10. Puls I., Jonnakuty C., LaMonte B. H., Holzbaur E. L., Tokito M., Mann E., Floeter M. K., Bidus K., Drayna D., Oh S. J., Brown R. H., Jr., Ludlow C. L., Fischbeck K. H. (2003) Mutant dynactin in motor neuron disease. Nat. Genet. 33, 455–456 [DOI] [PubMed] [Google Scholar]
- 11. Chen Y. Z., Bennett C. L., Huynh H. M., Blair I. P., Puls I., Irobi J., Dierick I., Abel A., Kennerson M. L., Rabin B. A., Nicholson G. A., Auer-Grumbach M., Wagner K., De Jonghe P., Griffin J. W., Fischbeck K. H., Timmerman V., Cornblath D. R., Chance P. F. (2004) DNA/RNA helicase gene mutations in a form of juvenile amyotrophic lateral sclerosis (ALS4). Am. J. Hum. Genet. 74, 1128–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nishimura A. L., Mitne-Neto M., Silva H. C., Richieri-Costa A., Middleton S., Cascio D., Kok F., Oliveira J. R., Gillingwater T., Webb J., Skehel P., Zatz M. (2004) A mutation in the vesicle-trafficking protein VAPB causes late-onset spinal muscular atrophy and amyotrophic lateral sclerosis. Am. J. Hum. Genet. 75, 822–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Neumann M., Sampathu D. M., Kwong L. K., Truax A. C., Micsenyi M. C., Chou T. T., Bruce J., Schuck T., Grossman M., Clark C. M., McCluskey L. F., Miller B. L., Masliah E., Mackenzie I. R., Feldman H., Feiden W., Kretzschmar H. A., Trojanowski J. Q., Lee V. M. (2006) Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314, 130–133 [DOI] [PubMed] [Google Scholar]
- 14. Sreedharan J., Blair I. P., Tripathi V. B., Hu X., Vance C., Rogelj B., Ackerley S., Durnall J. C., Williams K. L., Buratti E., Baralle F., de Belleroche J., Mitchell J. D., Leigh P. N., Al-Chalabi A., Miller C. C., Nicholson G., Shaw C. E. (2008) TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science 319, 1668–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kwiatkowski T. J., Jr., Bosco D. A., Leclerc A. L., Tamrazian E., Vanderburg C. R., Russ C., Davis A., Gilchrist J., Kasarskis E. J., Munsat T., Valdmanis P., Rouleau G. A., Hosler B. A., Cortelli P., de Jong P. J., Yoshinaga Y., Haines J. L., Pericak-Vance M. A., Yan J., Ticozzi N., Siddique T., McKenna-Yasek D., Sapp P. C., Horvitz H. R., Landers J. E., Brown R. H., Jr. (2009) Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science 323, 1205–1208 [DOI] [PubMed] [Google Scholar]
- 16. Vance C., Rogelj B., Hortobágyi T., De Vos K. J., Nishimura A. L., Sreedharan J., Hu X., Smith B., Ruddy D., Wright P., Ganesalingam J., Williams K. L., Tripathi V., Al-Saraj S., Al-Chalabi A., Leigh P. N., Blair I. P., Nicholson G., de Belleroche J., Gallo J. M., Miller C. C., Shaw C. E. (2009) Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science 323, 1208–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Maruyama H., Morino H., Ito H., Izumi Y., Kato H., Watanabe Y., Kinoshita Y., Kamada M., Nodera H., Suzuki H., Komure O., Matsuura S., Kobatake K., Morimoto N., Abe K., Suzuki N., Aoki M., Kawata A., Hirai T., Kato T., Ogasawara K., Hirano A., Takumi T., Kusaka H., Hagiwara K., Kaji R., Kawakami H. (2010) Mutations of optineurin in amyotrophic lateral sclerosis. Nature 465, 223–226 [DOI] [PubMed] [Google Scholar]
- 18. Johnson J. O., Mandrioli J., Benatar M., Abramzon Y., Van Deerlin V. M., Trojanowski J. Q., Gibbs J. R., Brunetti M., Gronka S., Wuu J., Ding J., McCluskey L., Martinez-Lage M., Falcone D., Hernandez D. G., Arepalli S., Chong S., Schymick J. C., Rothstein J., Landi F., Wang Y. D., Calvo A., Mora G., Sabatelli M., Monsurrò M. R., Battistini S., Salvi F., Spataro R., Sola P., Borghero G., ITALSGEN Consortium, Galassi G., Scholz S. W., Taylor J. P., Restagno G., Chiò A., Traynor B. J. (2010) Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron 68, 857–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Deng H. X., Chen W., Hong S. T., Boycott K. M., Gorrie G. H., Siddique N., Yang Y., Fecto F., Shi Y., Zhai H., Jiang H., Hirano M., Rampersaud E., Jansen G. H., Donkervoort S., Bigio E. H., Brooks B. R., Ajroud K., Sufit R. L., Haines J. L., Mugnaini E., Pericak-Vance M. A., Siddique T. (2011) Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature 477, 211–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. DeJesus-Hernandez M., Mackenzie I. R., Boeve B. F., Boxer A. L., Baker M., Rutherford N. J., Nicholson A. M., Finch N. A., Flynn H., Adamson J., Kouri N., Wojtas A., Sengdy P., Hsiung G. Y., Karydas A., Seeley W. W., Josephs K. A., Coppola G., Geschwind D. H., Wszolek Z. K., Feldman H., Knopman D. S., Petersen R. C., Miller B. L., Dickson D. W., Boylan K. B., Graff-Radford N. R., Rademakers R. (2011) Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72, 245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Renton A. E., Majounie E., Waite A., Simón-Sánchez J., Rollinson S., Gibbs J. R., Schymick J. C., Laaksovirta H., van Swieten J. C., Myllykangas L., Kalimo H., Paetau A., Abramzon Y., Remes A. M., Kaganovich A., Scholz S. W., Duckworth J., Ding J., Harmer D. W., Hernandez D. G., Johnson J. O., Mok K., Ryten M., Trabzuni D., Guerreiro R. J., Orrell R. W., Neal J., Murray A., Pearson J., Jansen I. E., Sondervan D., Seelaar H., Blake D., Young K., Halliwell N., Callister J. B., Toulson G., Richardson A., Gerhard A., Snowden J., Mann D., Neary D., Nalls M. A., Peuralinna T., Jansson L., Isoviita V. M., Kaivorinne A. L., Hölttä-Vuori M., Ikonen E., Sulkava R., Benatar M., Wuu J., Chiò A., Restagno G., Borghero G., Sabatelli M., Heckerman D., Rogaeva E., Zinman L., Rothstein J. D., Sendtner M., Drepper C., Eichler E. E., Alkan C., Abdullaev Z., Pack S. D., Dutra A., Pak E., Hardy J., Singleton A., Williams N. M., Heutink P., Pickering-Brown S., Morris H. R., Tienari P. J., Traynor B. J., et al. (2011) A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72, 257–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lagier-Tourenne C., Polymenidou M., Cleveland D. W. (2010) TDP-43 and FUS/TLS. Emerging roles in RNA processing and neurodegeneration. Hum. Mol. Genet. 19, R46–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen-Plotkin A. S., Lee V. M., Trojanowski J. Q. (2010) TAR DNA-binding protein 43 in neurodegenerative disease. Nat. Rev. Neurol. 6, 211–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang J., Xu G., Borchelt D. R. (2002) High molecular weight complexes of mutant superoxide dismutase 1. Age-dependent and tissue-specific accumulation. Neurobiol. Dis. 9, 139–148 [DOI] [PubMed] [Google Scholar]
- 25. Bosco D. A., Morfini G., Karabacak N. M., Song Y., Gros-Louis F., Pasinelli P., Goolsby H., Fontaine B. A., Lemay N., McKenna-Yasek D., Frosch M. P., Agar J. N., Julien J. P., Brady S. T., Brown R. H., Jr. (2010) Wild-type and mutant SOD1 share an aberrant conformation and a common pathogenic pathway in ALS. Nat. Neurosci. 13, 1396–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dreyfuss G., Matunis M. J., Piñol-Roma S., Burd C. G. (1993) hnRNP proteins and the biogenesis of mRNA. Annu. Rev. Biochem. 62, 289–321 [DOI] [PubMed] [Google Scholar]
- 27. Abhyankar M. M., Urekar C., Reddi P. P. (2007) A novel CpG-free vertebrate insulator silences the testis-specific SP-10 gene in somatic tissues. Role for TDP-43 in insulator function. J. Biol. Chem. 282, 36143–36154 [DOI] [PubMed] [Google Scholar]
- 28. Ou S. H., Wu F., Harrich D., García-Martínez L. F., Gaynor R. B. (1995) Cloning and characterization of a novel cellular protein, TDP-43, that binds to human immunodeficiency virus type 1 TAR DNA sequence motifs. J. Virol. 69, 3584–3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bose J. K., Wang I. F., Hung L., Tarn W. Y., Shen C. K. (2008) TDP-43 overexpression enhances exon 7 inclusion during the survival of motor neuron pre-mRNA splicing. J. Biol. Chem. 283, 28852–28859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mercado P. A., Ayala Y. M., Romano M., Buratti E., Baralle F. E. (2005) Depletion of TDP 43 overrides the need for exonic and intronic splicing enhancers in the human apoA-II gene. Nucleic Acids Res. 33, 6000–6010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dewey C. M., Cenik B., Sephton C. F., Dries D. R., Mayer P., 3rd, Good S. K., Johnson B. A., Herz J., Yu G. (2011) TDP-43 is directed to stress granules by sorbitol, a novel physiological osmotic and oxidative stressor. Mol. Cell. Biol. 31, 1098–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McDonald K. K., Aulas A., Destroismaisons L., Pickles S., Beleac E., Camu W., Rouleau G. A., Vande Velde C. (2011) TAR DNA-binding protein 43 (TDP-43) regulates stress granule dynamics via differential regulation of G3BP and TIA-1. Hum. Mol. Genet. 20, 1400–1410 [DOI] [PubMed] [Google Scholar]
- 33. Liu-Yesucevitz L., Bilgutay A., Zhang Y. J., Vanderweyde T., Vanderwyde T., Citro A., Mehta T., Zaarur N., McKee A., Bowser R., Sherman M., Petrucelli L., Wolozin B. (2010) Tar DNA binding protein-43 (TDP-43) associates with stress granules. Analysis of cultured cells and pathological brain tissue. PLoS One 5, e13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Strong M. J., Volkening K., Hammond R., Yang W., Strong W., Leystra-Lantz C., Shoesmith C. (2007) TDP43 is a human low molecular weight neurofilament (hNFL) mRNA-binding protein. Mol. Cell. Neurosci. 35, 320–327 [DOI] [PubMed] [Google Scholar]
- 35. Fiesel F. C., Voigt A., Weber S. S., Van den Haute C., Waldenmaier A., Görner K., Walter M., Anderson M. L., Kern J. V., Rasse T. M., Schmidt T., Springer W., Kirchner R., Bonin M., Neumann M., Baekelandt V., Alunni-Fabbroni M., Schulz J. B., Kahle P. J. (2010) Knockdown of transactive response DNA-binding protein (TDP-43) down-regulates histone deacetylase 6. EMBO J. 29, 209–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bose J. K., Huang C. C., Shen C. K. (2011) Regulation of autophagy by neuropathological protein TDP-43. J. Biol. Chem. 286, 44441–44448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Polymenidou M., Lagier-Tourenne C., Hutt K. R., Huelga S. C., Moran J., Liang T. Y., Ling S. C., Sun E., Wancewicz E., Mazur C., Kordasiewicz H., Sedaghat Y., Donohue J. P., Shiue L., Bennett C. F., Yeo G. W., Cleveland D. W. (2011) Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat. Neurosci. 14, 459–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ayala Y. M., De Conti L., Avendaño-Vázquez S. E., Dhir A., Romano M., D'Ambrogio A., Tollervey J., Ule J., Baralle M., Buratti E., Baralle F. E. (2011) TDP-43 regulates its mRNA levels through a negative feedback loop. EMBO J. 30, 277–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Igaz L. M., Kwong L. K., Lee E. B., Chen-Plotkin A., Swanson E., Unger T., Malunda J., Xu Y., Winton M. J., Trojanowski J. Q., Lee V. M. (2011) Dysregulation of the ALS-associated gene TDP-43 leads to neuronal death and degeneration in mice. J. Clin. Invest. 121, 726–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Buratti E., Baralle F. E. (2010) The multiple roles of TDP-43 in pre-mRNA processing and gene expression regulation. RNA Biol. 7, 420–429 [DOI] [PubMed] [Google Scholar]
- 41. Johnson B. S., McCaffery J. M., Lindquist S., Gitler A. D. (2008) A yeast TDP-43 proteinopathy model. Exploring the molecular determinants of TDP-43 aggregation and cellular toxicity. Proc. Natl. Acad. Sci. U.S.A. 105, 6439–6444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wegorzewska I., Bell S., Cairns N. J., Miller T. M., Baloh R. H. (2009) TDP-43 mutant transgenic mice develop features of ALS and frontotemporal lobar degeneration. Proc. Natl. Acad. Sci. U.S.A. 106, 18809–18814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kabashi E., Lin L., Tradewell M. L., Dion P. A., Bercier V., Bourgouin P., Rochefort D., Bel Hadj S., Durham H. D., Vande Velde C., Rouleau G. A., Drapeau P. (2010) Gain and loss of function of ALS-related mutations of TARDBP (TDP-43) cause motor deficits in vivo. Hum. Mol. Genet. 19, 671–683 [DOI] [PubMed] [Google Scholar]
- 44. Li Y., Ray P., Rao E. J., Shi C., Guo W., Chen X., Woodruff E. A., 3rd, Fushimi K., Wu J. Y. (2010) A Drosophila model for TDP-43 proteinopathy. Proc. Natl. Acad. Sci. U.S.A. 107, 3169–3174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hanson K. A., Kim S. H., Wassarman D. A., Tibbetts R. S. (2010) Ubiquilin modifies TDP-43 toxicity in a Drosophila model of amyotrophic lateral sclerosis (ALS). J. Biol. Chem. 285, 11068–11072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ash P. E., Zhang Y. J., Roberts C. M., Saldi T., Hutter H., Buratti E., Petrucelli L., Link C. D. (2010) Neurotoxic effects of TDP-43 overexpression in C. elegans. Hum. Mol. Genet. 19, 3206–3218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liachko N. F., Guthrie C. R., Kraemer B. C. (2010) Phosphorylation promotes neurotoxicity in a Caenorhabditis elegans model of TDP-43 proteinopathy. J. Neurosci. 30, 16208–16219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wils H., Kleinberger G., Janssens J., Pereson S., Joris G., Cuijt I., Smits V., Ceuterick-de Groote C., Van Broeckhoven C., Kumar-Singh S. (2010) TDP-43 transgenic mice develop spastic paralysis and neuronal inclusions characteristic of ALS and frontotemporal lobar degeneration. Proc. Natl. Acad. Sci. U.S.A. 107, 3858–3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Stallings N. R., Puttaparthi K., Luther C. M., Burns D. K., Elliott J. L. (2010) Progressive motor weakness in transgenic mice expressing human TDP-43. Neurobiol. Dis. 40, 404–414 [DOI] [PubMed] [Google Scholar]
- 50. Xu Y. F., Gendron T. F., Zhang Y. J., Lin W. L., D'Alton S., Sheng H., Casey M. C., Tong J., Knight J., Yu X., Rademakers R., Boylan K., Hutton M., McGowan E., Dickson D. W., Lewis J., Petrucelli L. (2010) Wild-type human TDP-43 expression causes TDP-43 phosphorylation, mitochondrial aggregation, motor deficits, and early mortality in transgenic mice. J. Neurosci. 30, 10851–10859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shan X., Chiang P. M., Price D. L., Wong P. C. (2010) Altered distributions of Gemini of coiled bodies and mitochondria in motor neurons of TDP-43 transgenic mice. Proc. Natl. Acad. Sci. U.S.A. 107, 16325–16330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Swarup V., Phaneuf D., Bareil C., Robertson J., Rouleau G. A., Kriz J., Julien J. P. (2011) Pathological hallmarks of amyotrophic lateral sclerosis/frontotemporal lobar degeneration in transgenic mice produced with TDP-43 genomic fragments. Brain 134, 2610–2626 [DOI] [PubMed] [Google Scholar]
- 53. Zhou H., Huang C., Chen H., Wang D., Landel C. P., Xia P. Y., Bowser R., Liu Y. J., Xia X. G. (2010) Transgenic rat model of neurodegeneration caused by mutation in the TDP gene. PLoS Genet. 6, e1000887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wu L. S., Cheng W. C., Hou S. C., Yan Y. T., Jiang S. T., Shen C. K. (2010) TDP-43, a neuro-pathosignature factor, is essential for early mouse embryogenesis. Genesis 48, 56–62 [DOI] [PubMed] [Google Scholar]
- 55. Sephton C. F., Good S. K., Atkin S., Dewey C. M., Mayer P., 3rd, Herz J., Yu G. (2010) TDP-43 is a developmentally regulated protein essential for early embryonic development. J. Biol. Chem. 285, 6826–6834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chiang P. M., Ling J., Jeong Y. H., Price D. L., Aja S. M., Wong P. C. (2010) Deletion of TDP-43 down-regulates Tbc1d1, a gene linked to obesity, and alters body fat metabolism. Proc. Natl. Acad. Sci. U.S.A. 107, 16320–16324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wang J., Slunt H., Gonzales V., Fromholt D., Coonfield M., Copeland N. G., Jenkins N. A., Borchelt D. R. (2003) Copper-binding site-null SOD1 causes ALS in transgenic mice. Aggregates of non-native SOD1 delineate a common feature. Hum. Mol. Genet. 12, 2753–2764 [DOI] [PubMed] [Google Scholar]
- 58. Wang J., Farr G. W., Hall D. H., Li F., Furtak K., Dreier L., Horwich A. L. (2009) An ALS-linked mutant SOD1 produces a locomotor defect associated with aggregation and synaptic dysfunction when expressed in neurons of Caenorhabditis elegans. PLoS Genet. 5, e1000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sorger P. K. (1991) Heat shock factor and the heat shock response. Cell 65, 363–366 [DOI] [PubMed] [Google Scholar]
- 60. Hajdu-Cronin Y. M., Chen W. J., Sternberg P. W. (2004) The L-type cyclin CYL-1 and the heat-shock factor HSF-1 are required for heat-shock-induced protein expression in Caenorhabditis elegans. Genetics 168, 1937–1949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gidalevitz T., Ben-Zvi A., Ho K. H., Brignull H. R., Morimoto R. I. (2006) Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science 311, 1471–1474 [DOI] [PubMed] [Google Scholar]
- 62. Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., Harris M. A., Hill D. P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J. C., Richardson J. E., Ringwald M., Rubin G. M., Sherlock G. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bernstein E., Kim S. Y., Carmell M. A., Murchison E. P., Alcorn H., Li M. Z., Mills A. A., Elledge S. J., Anderson K. V., Hannon G. J. (2003) Dicer is essential for mouse development. Nat. Genet. 35, 215–217 [DOI] [PubMed] [Google Scholar]
- 64. Knight S. W., Bass B. L. (2001) A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science 293, 2269–2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ritson G. P., Custer S. K., Freibaum B. D., Guinto J. B., Geffel D., Moore J., Tang W., Winton M. J., Neumann M., Trojanowski J. Q., Lee V. M., Forman M. S., Taylor J. P. (2010) TDP-43 mediates degeneration in a novel Drosophila model of disease caused by mutations in VCP/p97. J. Neurosci. 30, 7729–7739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Johnson B. S., Snead D., Lee J. J., McCaffery J. M., Shorter J., Gitler A. D. (2009) TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J. Biol. Chem. 284, 20329–20339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Guo W., Chen Y., Zhou X., Kar A., Ray P., Chen X., Rao E. J., Yang M., Ye H., Zhu L., Liu J., Xu M., Yang Y., Wang C., Zhang D., Bigio E. H., Mesulam M., Shen Y., Xu Q., Fushimi K., Wu J. Y. (2011) An ALS-associated mutation affecting TDP-43 enhances protein aggregation, fibril formation and neurotoxicity. Nat. Struct. Mol. Biol. 18, 822–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Yen K., Narasimhan S. D., Tissenbaum H. A. (2011) DAF-16/Forkhead box O transcription factor. Many paths to a single Fork(head) in the road. Antioxid. Redox. Signal. 14, 623–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.