Background: Amyloid-β peptide (Aβ) is degraded by different proteases. We recently demonstrated phosphorylation of Aβ.
Results: Phosphorylation of Aβ decreases its clearance by microglial BV-2 cells and selectively inhibits the cleavage by insulin-degrading and angiotensin-converting enzymes.
Conclusion: Phosphorylation at Ser-8 negatively regulates Aβ degradation.
Significance: Phosphorylation could play a dual role in Aβ metabolism. It decreases the clearance by microglial cells and also promotes Aβ aggregation.
Keywords: Alzheimer disease, Amyloid, Phosphorylation, Post-translational Modification, Protein Degradation
Abstract
Accumulation of amyloid-β peptides (Aβ) in the brain is a common pathological feature of Alzheimer disease (AD). Aggregates of Aβ are neurotoxic and appear to be critically involved in the neurodegeneration during AD pathogenesis. Accumulation of Aβ could be caused by increased production, as indicated by several mutations in the amyloid precursor protein or the γ-secretase components presenilin-1 and presenilin-2 that cause familial early-onset AD. However, recent data also indicate a decreased clearance rate of Aβ in AD brains. We recently demonstrated that Aβ undergoes phosphorylation by extracellular or cell surface-localized protein kinase A, leading to increased aggregation. Here, we provide evidence that phosphorylation of monomeric Aβ at Ser-8 also decreases its clearance by microglial cells. By using mass spectrometry, we demonstrate that phosphorylation at Ser-8 inhibited the proteolytic degradation of monomeric Aβ by the insulin-degrading enzyme, a major Aβ-degrading enzyme released from microglial cells. Phosphorylation also decreased the degradation of Aβ by the angiotensin-converting enzyme. In contrast, Aβ degradation by plasmin was largely unaffected by phosphorylation. Thus, phosphorylation of Aβ could play a dual role in Aβ metabolism. It decreases its proteolytic clearance and also promotes its aggregation. The inhibition of extracellular Aβ phosphorylation, stimulation of protease expression and/or their proteolytic activity could be explored to promote Aβ degradation in AD therapy or prevention.
Introduction
Alzheimer disease (AD)3 is characterized by the progressive deposition of amyloid-β peptides (Aβ) in the brain (1, 2). Aβ derives from proteolytic processing of the amyloid precursor protein involving sequential cleavages by enzymes called β- and γ-secretases (3, 4). A critical role of Aβ in the pathogenesis of AD is strongly supported by several mutations within the genes encoding the amyloid precursor protein itself or the two presenilins that represent the proteolytically active components of the γ-secretase complex. All of these mutations affect the production and/or aggregation of Aβ and cause early-onset forms of familial AD (5–7). Although early-onset familial AD appears to be commonly associated with an elevated production of aggregation-prone Aβ variants, it remains unclear whether increased Aβ generation also contributes to the much more common form of late-onset AD. Recent evidence rather indicated a decreased clearance rate of Aβ in AD compared with control brains (8–10).
Several mechanisms for Aβ clearance have been identified, including drainage via the blood-brain barrier (11, 12), internalization of Aβ by phagocytosis and pinocytosis (13–15), and degradation by cell surface-localized or secreted peptidases (16, 17). Major proteases in the degradation of extracellular Aβ are the insulin-degrading enzyme (IDE) and neprilysin (NEP) (18–20), but other proteases, including the angiotensin-converting enzyme (ACE), endothelin-converting enzymes, and plasmin, could also contribute to efficient clearance of Aβ in the brain (21–23).
IDE is localized principally in the cytosol but is also released from cells and found in extracellular fluids and conditioned media of cultured cells (24, 25). However, IDE lacks canonical signal sequences that target the enzyme to the conventional secretory pathway (26). Recent data demonstrated that IDE is secreted via a nonconventional pathway in association with exosomes (27, 28). This nonconventional secretion of IDE is dependent on a hexapeptide amino acid motif in its C-terminal domain (29). The pathophysiological relevance of IDE is demonstrated by the deletion of IDE in mice that showed decreased Aβ degradation and increased cerebral Aβ accumulation (30–32). Conversely, enhancement of IDE activity in neurons effectively reduced Aβ accumulation in AD mouse models (30, 33). Recent data demonstrated that extracellular Aβ could undergo phosphorylation by secreted variants of protein kinase A and that the phosphorylation of Aβ at Ser-8 strongly promoted its aggregation into oligomeric and fibrillar assemblies (34).
Here, we sought to assess the effect of phosphorylation on the clearance of Aβ by microglial BV-2 cells. Our data demonstrate that phosphorylated Aβ (pAβ) has increased stability against microglial degradation compared with non-phosphorylated Aβ (npAβ). Interestingly, phosphorylation significantly decreases its proteolytic degradation by secreted IDE.
EXPERIMENTAL PROCEDURES
Reagents
Synthetic npAβ(1–40) (npAβ) and pAβ(1–40) (pAβ) peptides were purchased from Peptide Specialty Laboratories. Recombinant human IDE, ACE, NEP, and purified human plasmin were procured from R&D Systems. Acetonitrile and α-cyano-4-hydroxycinnamic acid were from Sigma. The SilverQuest silver staining kit and precast 4–12% NuPAGE BisTris minigels were from Invitrogen. Precast 16% Tricine gels were from Anamed. The ZipTip (C18) pipette tips used for mass spectrometric analysis were from Millipore. Primary and secondary antibodies were obtained from the indicated suppliers: anti-Aβ primary antibody 82E1 (IBL Corp.), anti-IDE primary antibody (Abcam), and anti-mouse and anti-rabbit secondary antibodies (Sigma).
Aβ Degradation Assays with BV-2 Cells or Recombinant Enzymes
Synthetic npAβ and pAβ were solubilized in 10 mm NaOH at a concentration of 1 mg/ml (230 μm), sonicated for 5 min, and stored at −80 °C until used. Aliquots were thawed and diluted in the appropriate buffer.
For the cellular degradation assays, BV-2 cells were cultured in Dulbecco's modified Eagle's medium with GlutaMAX (Invitrogen) supplemented with 10% fetal calf serum and 1% penicillin/streptomycin (Invitrogen) in a 24-well plate to 70% confluence. Media were replaced with serum-free media and incubated with 1 μm npAβ or pAβ. Aliquots of the media were taken at the indicated time points and mixed with 4× lithium dodecyl sulfate sample buffer (Invitrogen), incubated at 70 °C for 10 min, and then separated by SDS-PAGE on 16% Tricine gels. Aβ was detected by Western blotting using primary monoclonal antibody 82E1 and HRP-conjugated rabbit anti-mouse secondary antibody.
For Aβ degradation with recombinant enzymes, synthetic npAβ and pAβ were diluted in assay buffer (50 mm Tris and 100 mm NaCl, pH 7.5) to a final concentration of 25 ng/μl (Aβ). Reactions were started by the addition of recombinant enzymes in assay buffer (IDE, 0.3 ng/μl; NEP, 1 ng/μl; ACE, 0.5 ng/μl; and plasmin, 10 ng/μl) and then allowed to proceed for the indicated time points at 37 °C on a block heater at 650 rpm. Sample aliquots (10 μl) were taken, snap-frozen in liquid nitrogen, and stored for further use at −80 °C. Samples were then either analyzed by SDS-PAGE and Western blot analysis or by MALDI-TOF-MS.
Mass Spectrometry
The molecular masses of intact peptides and proteolytic products of npAβ and pAβ were determined using MALDI-TOF-MS. Samples were purified through reversed-phase ZipTip C18 tips following the manufacturer's instructions and analyzed by a MALDI-TOF-MS system (BIFLEX III, Bruker Daltonics GmbH). Samples were mixed with α-cyano-4-hydroxycinnamic acid and spotted onto a ground-steel MALDI target plate (Bruker Daltonics GmbH). Spectra were recorded in the linear mode at a laser frequency of 20 Hz within a mass range of 1000–6000 Da. Each spectrum is the result of an average outcome of at least 300 laser shots collected in 30-shot steps. FlexAnalysis 1.0 software (Bruker Daltonics GmbH) was used for visual estimation, smoothening and base-line substraction of the mass spectra.
siRNA-mediated Knockdown of IDE
BV-2 cells were transfected with 22.5 nm siRNA (target sequence GCCTGTTGTCAGAACTCAA) using HiPerFect transfection reagent (Qiagen) according to the supplier's instructions. Knockdown of IDE was analyzed after 24 h in cell supernatants and cell lysates by Western immunoblotting. For Aβ degradation experiments, cells were then incubated for 12 h in serum-free medium. Synthetic npAβ and pAβ variants were added to conditioned media, and aliquots were taken at the indicated time points. Aβ was then detected by Western immunoblotting.
RESULTS
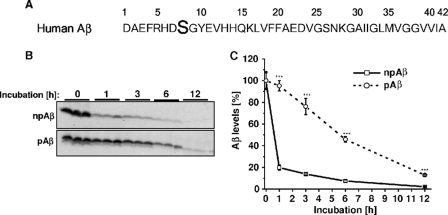
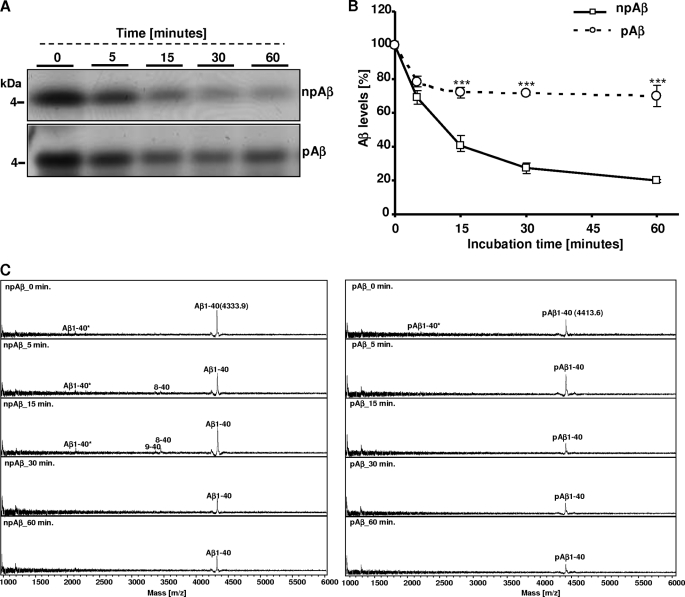
Recent data demonstrated phosphorylation of extracellular Aβ at Ser-8 by secreted or cell surface-localized variants of PKA (Fig. 1A) (34, 35). To test whether this phosphorylation affects the clearance of Aβ by microglia, mouse microglial BV-2 cells were incubated with synthetic pAβ or npAβ, and stability was assessed by detection of the peptides after different time periods of incubation by Western blotting (Fig. 1B). Consistent with previous data (28, 36), extracellular monomeric Aβ was efficiently cleared in the conditioned media of BV-2 cells. About 80% of npAβ was cleared within the first hour of incubation. In contrast, only ∼5% of pAβ was cleared after the first hour. Even after 6 h, ∼50% of pAβ was detected in the cell supernatant, a time point at which only residual amounts (<10%) of npAβ were left (Fig. 1C). The calculated half-life times of npAβ and pAβ in these experiments were 45 min and 6 h, respectively. These data demonstrate that phosphorylation of Aβ strongly decreases the clearance of extracellular Aβ by microglial cells.
FIGURE 1.
Decreased clearance of pAβ by microglial BV-2 cells. A, primary amino acid sequence of human Aβ40/42 with the phosphorylation site at Ser-8, indicated in boldface. B, BV-2 cells were incubated with synthetic npAβ or pAβ (1 μm), and aliquots of the conditioned media were taken at the indicated time points. Samples were separated by SDS-PAGE, and Aβ was detected by Western immunoblotting. C, quantification of Western blots in B was performed by densitometric analysis. Aβ levels at time 0 were set as 100%. Values represent means ± S.D. of three independent experiments (n = 3). ***, p < 0.001 (t test).
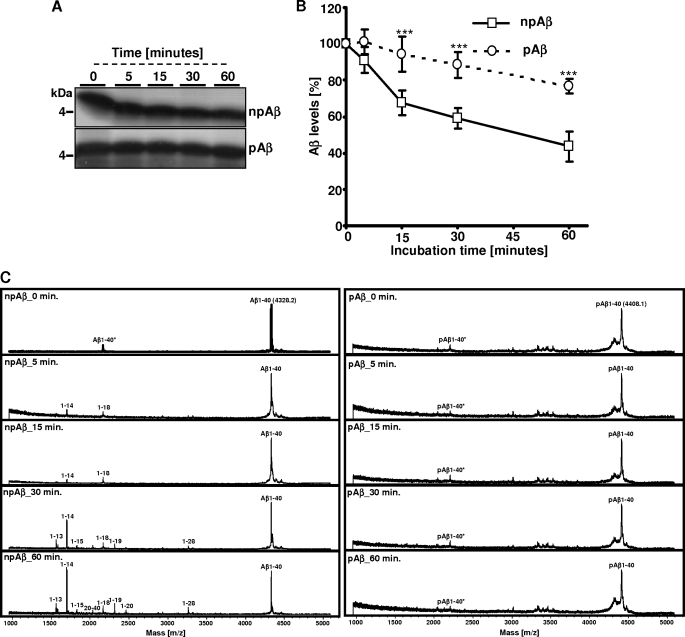
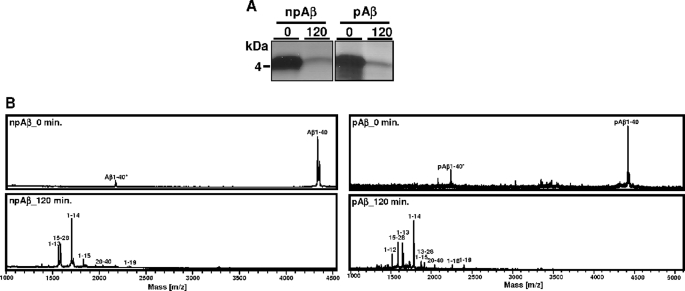
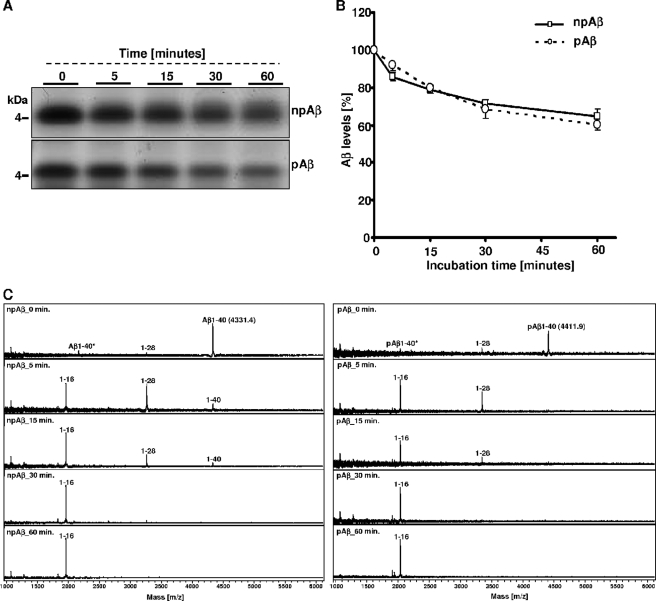
As shown previously (24, 28, 36), microglial BV-2 cells efficiently degrade extracellular monomeric Aβ by secreted IDE. To specifically analyze the effect of Aβ phosphorylation on IDE-mediated cleavage, we next performed in vitro experiments using recombinant IDE. Synthetic pAβ or npAβ was incubated with IDE, and time-dependent cleavage was examined by SDS-PAGE and subsequent silver staining or by MALDI-TOF-MS (Fig. 2, A–C). Although the amount of npAβ gradually decreased during the incubation, pAβ appeared to be very stable under these conditions (Fig. 2, A and B). MALDI-TOF-MS analysis demonstrated that, at the start of incubation, only full-length Aβ variants were detected (Fig. 2C). Already after 5 and 15 min, an additional peak was detected in the samples with npAβ, demonstrating cleavage of Aβ by IDE. The m/z ratio of this peak corresponds to Aβ(1–14), indicating cleavage between amino acids 14 and 15. This peak increases over time and was the predominant peak detected after 30 and 60 min of incubation. However, some additional peptides were detected that represent minor degradation products of less efficient cleavages between amino acids 13 and 14, 15 and 16, 18 and 19, 19 and 20, 20 and 21, and 28 and 29, corresponding to Aβ(1–13), Aβ(1–14), Aβ(1–15), Aβ(1–18), Aβ(1–19), Aβ(1–20), and Aβ(1–28) (Fig. 2C and Table 1). Importantly, very few (if any) cleavage products of pAβ were detected even after 60 min of incubation with recombinant IDE. These results demonstrate that phosphorylation of Aβ strongly decreases its degradation by IDE. Because we observed clearance by microglial cells also of the pAβ variant, although strongly decreased, we tested whether phosphorylation completely blocks IDE-mediated degradation. pAβ and npAβ were incubated with higher amounts of recombinant IDE, and degradation was analyzed by SDS-PAGE and silver staining or by MALDI-TOF-MS. Notably, under these conditions, we observed almost complete degradation of both Aβ variants (Fig. 3, A and B). The resulting pattern of degradation products of pAβ and npAβ was very similar, except for the increased masses due to the phosphoryl group (Fig. 3B and Table 1). These data indicate that phosphorylation of Aβ does not completely block but rather decreases the efficiency of cleavage by IDE. In addition, the cleavage specificity of IDE is also not affected by phosphorylation of Aβ. The detection of phosphorylated degradation products also indicates that IDE-mediated cleavage of Aβ does not require dephosphorylation.
FIGURE 2.
Phosphorylation of Aβ inhibits degradation by recombinant IDE. A–C, synthetic npAβ and pAβ were incubated with recombinant IDE at 37 °C for various time intervals (0, 5, 15, 30, and 60 min). Aliquots of the reaction mixture were analyzed by SDS-PAGE and silver staining (A and B) or by MALDI-TOF-MS (C). Cleavage products were detected with npAβ but not with pAβ. Peaks at m/z 4408.1 and 2204.6 correspond to single- and double-ionized full-length pAβ. The masses of the peptide fragments are provided in Table 1. Values represent means ± S.D. of three independent experiments (n = 3). ***, p < 0.001 (t test).
TABLE 1.
MALDI-TOF-MS analysis of cleavage products of npAβ and pAβ
Shown are the various peptide fragments generated upon cleavage of npAβ and pAβ variants by different proteases and their calculated mass (Da) and observed mass (Da) by MALDI-TOF. 1–40* represents the double-ionized full-length peptide (m/2z instead of normal m/z). The indicated masses of the full-length peptide (1–40) and the double-ionized full-length peptide (1–40*) of both peptide variants are the mean values (n = 10). The difference between the observed mass and the calculated mass is ∼1–5 Da. The proteolytic products with a mass below 1000 kDa and C-terminal fragments of Aβ generated after the proteolytic cleavage were barely detectable even at spectrum recordings at low mass range (between 100 and 1000 kDa). ND, peaks that were not detected by MALDI-TOF-MS.
| Peptide fragment | npAβ |
pAβ |
||
|---|---|---|---|---|
| Calculated mass | Observed mass | Calculated mass | Observed mass | |
| Da | Da | |||
| 1–7 | 888.4 | ND | 889.4 | ND |
| 1–8 | 975.4 | ND | 1055.4 | ND |
| 1–12 | 1423.6 | ND | 1503.6 | 1508.3 |
| 1–13 | 1560.7 | 1560.5 | 1640.7 | 1644.3 |
| 1–14 | 1697.7 | 1697.1 | 1777.7 | 1782.5 |
| 1–15 | 1825.8 | 1826.3 | 1905.8 | 1910.7 |
| 1–16 | 1953.9 | 1957.7 | 2033.9 | 2038.2 |
| 1–17 | 2067.0 | 2069.4 | 2147.8 | ND |
| 1–18 | 2166.0 | 2164.6 | 2246.0 | 2251.8 |
| 1–19 | 2313.1 | 2316.3 | 2393.1 | 2399.4 |
| 1–20 | 2460.2 | 2460.2 | 2540.2 | 2545.8 |
| 1–28 | 3260.5 | 3261.2 | 3340.5 | 3340.3 |
| 1–29 | 3317.6 | 3321.8 | 3397.6 | ND |
| 8–40 | 3456.8 | 3451.2 | 3536.8 | ND |
| 9–40 | 3369.8 | 3367.7 | 3369.8 | ND |
| 15–28 | 1580.8 | 1584.7 | 1580.8 | 1584.7 |
| 15–40 | 2647.4 | 2646.8 | 2647.4 | ND |
| 16–40 | 2519.4 | ND | 2519.4 | ND |
| 18–40 | 2278.2 | ND | 2278.2 | ND |
| 19–40 | 2179.1 | ND | 2179.1 | ND |
| 20–40 | 2032.1 | ND | 2032.1 | ND |
| 30–40 | 1028.3 | ND | 1028.3 | ND |
| 1–40 | 4327.2 | 4328.4 | 4407.2 | 4414.6 |
| 1–40* | 2163.6 | 2164.5 | 2203.6 | 2204.3 |
FIGURE 3.
Similar cleavage products of npAβ and pAβ by recombinant IDE. A and B, synthetic npAβ and pAβ were incubated with a higher concentration of recombinant IDE (0.9 ng/μl) for 120 min at 37 °C. Samples obtained before (0) and after (120) the incubation period were analyzed by SDS-PAGE and silver staining (A) and by MALDI-TOF-MS (B). Degradation products from both peptide variants are clearly evident after 120 min of incubation. Notably, the resulting pattern of degradation products of pΑβ is very similar to that of npAβ except for the increased masses due to the phosphoryl group.
In addition, we also carried out similar experiments with human cerebrospinal fluid. At similar concentrations of IDE, the degradation of Aβ was lower in cerebrospinal fluid samples compared with the IDE assay buffer. These data indicate that cerebrospinal fluid contains proteins or other factors that negatively affect the degradation of Aβ. However, the inhibitory effect of phosphorylation on Aβ degradation was still evident (supplemental Fig. 1).
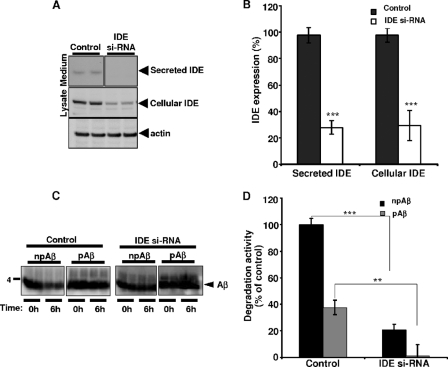
Recently, we demonstrated that IDE is the main protease contributing to Aβ degradation in BV-2 cells (28). Consistent with these results, siRNA-mediated knockdown of IDE strongly reduced the expression of cellular as well as secreted IDE in control cells. After knockdown, IDE levels were strongly decreased in the conditioned media and cell lysates by ∼75% (Fig. 4, A and B). Importantly, siRNA-mediated knockdown of IDE in these cells decreased the degradation of Aβ also by ∼75% (Fig. 4, C and D). Consistent with the previous results, the degradation of pAβ was also lower compared with npAβ upon siRNA-mediated knockdown of IDE. Together, these data strongly indicate that secreted IDE is the major protease in the degradation of extracellular Aβ in this cell system.
FIGURE 4.
siRNA-mediated knockdown of endogenous IDE decreases degradation of Aβ in BV-2 cells. A and B, BV-2 cells were transfected with IDE-targeting siRNA and incubated for 24 h. After 24 h, expression of secreted and cellular IDEs was analyzed by Western immunoblotting. Cellular actin was detected as a loading control. IDE levels were strongly decreased in the conditioned media and cell lysates in IDE siRNA-transfected cells compared with non-transfected controls. Quantification by densitometric analysis of Western blots indicated the percentage of reduction of secreted and cellular IDE expression in siRNA-transfected cells (IDE si-RNA) compared with the control. Values represent means ± S.D. (n = 4). ***, p < 0.001 (t test). The values are normalized to actin. C and D, conditioned media collected from control and IDE siRNA-transfected cells were incubated with npAβ or pAβ (1 μm) for 6 h. Aβ levels at the beginning (0h) and end (6h) of the incubation period were analyzed by Western immunoblotting (C). Densitometric analysis of the Western blots indicated that an ∼75% reduction in degradation activity was observed after siRNA-mediated knockdown of IDE in BV-2 cells for both npAβ and pAβ (D). Values represent means ± S.D. (n = 4). ***, p < 0.001; **, p < 0.01 (t test).
Because other proteases also can contribute to the degradation of Aβ in other cell systems or in vivo, we further tested the effect of phosphorylation on Aβ degradation by known Aβ-cleaving enzymes, including ACE, NEP, and plasmin. Although NEP showed comparably low Aβ-degrading activity under the experimental conditions and even at higher enzyme concentrations (supplemental Fig. 2), plasmin and ACE efficiently degraded the synthetic Aβ variants. The main cleavage products generated by plasmin had m/z ratios of ∼3265 and ∼1958, representing Aβ(1–28), and Aβ(1–16), respectively (Fig. 5, A–C, and Table 1). However, phosphorylation had little (if any) inhibitory effect on plasmin-mediated degradation of Aβ, which was also indicated by the similar appearance of phosphorylated degradation products (Fig. 5C). In contrast, the degradation of pAβ by ACE was strongly decreased compared with that of npAβ (Fig. 6, A–C). Interestingly, cleavage of Aβ by ACE occurred directly N- or C-terminal of Ser-8, either between Asp-7 and Ser-8 or between Ser-8 and Gly-9, as indicted by the m/z ratios of C-terminal cleavage products of ∼3451 (Aβ(8–40)) and ∼3370 (Aβ(9–40)).
FIGURE 5.
Plasmin degrades Aβ and pAβ. A–C, synthetic npAβ and pAβ were incubated with purified plasmin at 37 °C for various time intervals (0, 5, 15, 30, and 60 min). Aliquots of the reaction mixture were analyzed by SDS-PAGE and silver staining (A and B) or by MALDI-TOF-MS (C). The main cleavage products representing Aβ(1–28) and Aβ(1–16) fragments were generated in both peptide variants. The corresponding masses of the cleavage products are given in Table 1. Values represent means ± S.D. (n = 3).
FIGURE 6.
Phosphorylation of Aβ decreases degradation by recombinant ACE. A–C, synthetic npAβ and pAβ were incubated with recombinant ACE at 37 °C for various time intervals (0, 5, 15, 30, and 60 min). Sample aliquots of the reaction mixture collected at different time intervals were analyzed by SDS-PAGE and silver staining (A and B) or by MALDI-TOF-MS (C). The rate of degradation of pAβ was significantly reduced (A and B). The peaks representing cleavage products Aβ(8–40) and Aβ(9–40) were detected only with npAβ, whereas no peaks were detected with pAβ (C). The corresponding masses of the cleavage products are provided in Table 1. Values represent means ± S.D. (n = 3). ***, p < 0.001 (t test).
DISCUSSION
This study revealed a novel mechanism in the regulation of Aβ metabolism. The phosphorylation at Ser-8 inhibits the degradation by IDE, a major Aβ-degrading enzyme secreted by microglial BV-2 cells.
Increased concentrations of Aβ favor its aggregation and deposition in the form of β-amyloid plaques in the human brain. This is well supported by rare mutations in amyloid precursor protein and the presenilin proteins that cause autosomal dominant early-onset AD and commonly increase the production of Aβ and/or its aggregation (37). However, mechanisms that alter the metabolism of Aβ in the pathogenesis of the much more common late-onset form of AD are largely unclear. Interestingly, recent data indicated a decreased rate of Aβ clearance in the AD brain rather than increased production (8, 9). The half-life time of Aβ is ∼8–12 h in human cerebrospinal fluid and 3–4 h in the interstitial fluid of mouse brain, indicating efficient clearance mechanisms that counteract the production of Aβ (9, 10, 38). The clearance of Aβ from the brain involves internalization via pinocytosis or receptor-mediated endocytosis/phagocytosis (13–15, 39) and subsequent degradation in the endosomal/lysosomal compartments (40), transcytosis and drainage via the blood-brain barrier to the vasculature (12, 41), and proteolytic degradation of extracellular Aβ by cell surface-localized and secreted proteases (21, 28, 36).
Microglial cells could contribute to the clearance of Aβ, as they are the main phagocytes in the brain and also express several proteases at the cell surface that could degrade extracellular and internalized Aβ (27). Recent evidence demonstrated that microglia secrete substantial amounts of IDE, thereby allowing efficient degradation of monomeric Aβ variants also at some distance from the microglial cell (19, 42). IDE is a zinc metalloprotease with broad substrate specificity that is involved in the degradation of several peptides, including insulin, glucagon, transforming growth factor, and other peptide hormones (43). The different peptide substrates have little (if any) sequence homology, but many of the substrates share a propensity to form a β-sheet-rich conformation under certain conditions (e.g. Aβ, insulin, glucagon, amylin, atrial natriuretic factor, and calcitonin) (44–47). Accordingly, substrate selection of IDE might be determined mainly by the size and secondary and tertiary conformation of the peptides (46). Co-crystallization indicated that, after being engulfed into the catalytic center of IDE, Aβ could undergo conformational changes that might be important for the subsequent hydrolysis reaction (48). We recently showed that the phosphorylation of Aβ affects its conformation (34). Thus, the conformational change induced by phosphorylation might decrease the efficiency of hydrolysis by IDE. The introduction of a negatively charged phosphoryl group could also directly impair enzyme-substrate interaction. Similar effects of phosphorylation have been shown previously for caspase-mediated processing of several other AD-related proteins, including presenilin-1, presenilin-2, and Tau (49–51). Thus, it would be interesting to co-crystallize IDE with pAβ or npAβ to further elucidate the structural basis of the inhibitory effect of phosphorylation on Aβ degradation.
In addition to IDE, several other proteases involved in Aβ degradation that contribute to the regulation of Aβ levels in the human brain have been identified (52). In particular, NEP, ACE, and plasmin are considered to be physiologically and pathologically relevant in sporadic AD. NEP is a membrane-bound zinc metallopeptidase localized at the cell surface and in cytoplasmic vesicles preferentially hydrolyzing extracellular oligopeptides on the amino side of hydrophobic residues (53) and has been shown to be capable of degrading Aβ both in vivo (54, 55) and in vitro (55–57). NEP is also reported to be expressed on neuronal pre- and postsynaptic membranes (52). However, only very little concentrations of NEP are found in extracellular fluids (58). In the current experimental paradigm, NEP showed low Aβ-degrading activity even at higher enzyme concentrations. Furthermore, the expression of NEP is very low, and its activity is hardly detectable in microglial BV-2 cells (28). In addition, we observed a very strong correlation between IDE expression after RNAi-mediated knockdown and residual Aβ-degrading activity by BV-2 cells (Fig. 4). Thus, a major contribution of NEP to the degradation of extracellular Aβ by this cell type is unlikely. However, it will be interesting to further investigate the effect of Aβ phosphorylation on NEP-dependent degradation.
Plasmin, a serine protease released after cleavage of its zymogen plasminogen, can also modulate the clearance of Aβ (59). In the nervous system, plasmin/plasminogen is expressed in neurons, whereas plasminogen activator is synthesized by neurons and microglial cells (60). The plasmin system is involved in many neural functions, such as neuronal plasticity, learning, and memory (61). The plasmin-dependent degradation of Aβ decreases its neurotoxicity in rat cortical cultures (62, 63). Our mass spectrometric analysis revealed main cleavage sites between Lys-16 and Leu-17 and between Arg-28 and Gly-29. However, the phosphorylation of Aβ had a negligible (if any) inhibitory effect on plasmin-mediated degradation.
ACE, also known as dipeptidyl carboxypeptidase (EC 3.4.15.1), is a membrane-bound zinc metalloprotease (64) and is widely expressed in peripheral tissues and the brain. The involvement of ACE in the pathogenesis of AD was suggested by the genetic association of polymorphisms in the ACE-1 gene (DCP1) with an increased risk of AD (65–67). However, recent large genome-wide association studies did not identify ACE as a significant genetic risk factor (68–70). However, recombinant ACE is capable of cleaving both Aβ40 and Aβ42 in vitro and thereby decreases Aβ aggregation and toxicity (71–73). We also observed efficient cleavage of Aβ by ACE. This cleavage occurs directly before or after Ser-8. Interestingly, the phosphorylation at Ser-8 strongly inhibited ACE-mediated degradation of Aβ.
In this study, we have demonstrated that phosphorylation at Ser-8 could selectively affect the degradation of Aβ by different proteases. Although the degradation of Aβ by plasmin was independent of phosphorylation, the cleavage of Aβ by IDE and ACE was strongly decreased upon phosphorylation at Ser-8. These data could have important implications for the accumulation of Aβ observed during the pathogenesis of AD. The decreased degradation of phosphorylated Aβ by IDE and ACE would eventually result in increased concentrations of this peptide in the brain. As shown recently, ∼20–30% of the total extractable pool of Aβ was phosphorylated at Ser-8 and found in plaques and oligomeric aggregates (34). Thus, phosphorylation not only decreases the clearance of Aβ but also increases its propensity to aggregate (35). This modification would promote the formation of neurotoxic variants in a dual way. As the phosphorylation of Aβ at Ser-26 has also been described (74, 75), it will be interesting to also assess the effect of phosphorylation at this site on Aβ metabolism. The targeting of extracellular Aβ phosphorylation by inhibition of extracellular kinases or stimulation of Aβ dephosphorylation could be explored to facilitate Aβ clearance in the brain for prevention or therapy of AD.
Supplementary Material
Acknowledgment
We thank Dr. Sebastian Franken (Institute of Biochemistry and Molecular Biology, University of Bonn) for helpful suggestions and technical assistance in MALDI-TOF measurements.
This work was supported by Deutsche Forschungsgemeinschaft Grants WA1477/6, SFB645, and KFO177/WA1477/4 and the BONFOR Program.

This article contains supplemental Figs. 1 and 2.
- AD
- Alzheimer disease
- Aβ
- amyloid-β peptide(s)
- IDE
- insulin-degrading enzyme
- NEP
- neprilysin
- ACE
- angiotensin-converting enzyme
- pAβ
- phosphorylated Aβ
- npAβ
- non-phosphorylated Aβ
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1. Masters C. L., Beyreuther K. (2006) Alzheimer's centennial legacy: prospects for rational therapeutic intervention targeting the Aβ amyloid pathway. Brain 129, 2823–2839 [DOI] [PubMed] [Google Scholar]
- 2. Selkoe D. J. (2001) Alzheimer disease results from the cerebral accumulation and cytotoxicity of amyloid-β protein. J. Alzheimers Dis. 3, 75–80 [DOI] [PubMed] [Google Scholar]
- 3. De Strooper B., Vassar R., Golde T. (2010) The secretases: enzymes with therapeutic potential in Alzheimer disease. Nat. Rev. Neurol. 6, 99–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Haass C., De Strooper B. (1999) The presenilins in Alzheimer disease: proteolysis holds the key. Science 286, 916–919 [DOI] [PubMed] [Google Scholar]
- 5. Tanzi R. E., St George-Hyslop P. H., Gusella J. F. (1989) Molecular genetic approaches to Alzheimer disease. Trends Neurosci. 12, 152–158 [DOI] [PubMed] [Google Scholar]
- 6. Kamino K., Sato S., Sakaki Y., Yoshiiwa A., Nishiwaki Y., Takeda M., Tanabe H., Nishimura T., Ii K., St George-Hyslop P. H., Miki T., Ogihara T. (1996) Three different mutations of presenilin-1 gene in early-onset Alzheimer disease families. Neurosci. Lett. 208, 195–198 [DOI] [PubMed] [Google Scholar]
- 7. Rossor M. N., Newman S., Frackowiak R. S., Lantos P., Kennedy A. M. (1993) Alzheimer disease families with amyloid precursor protein mutations. Ann. N.Y. Acad. Sci. 695, 198–202 [DOI] [PubMed] [Google Scholar]
- 8. Mawuenyega K. G., Sigurdson W., Ovod V., Munsell L., Kasten T., Morris J. C., Yarasheski K. E., Bateman R. J. (2010) Decreased clearance of CNS β-amyloid in Alzheimer disease. Science 330, 1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bateman R. J., Munsell L. Y., Morris J. C., Swarm R., Yarasheski K. E., Holtzman D. M. (2006) Human amyloid-β synthesis and clearance rates as measured in cerebrospinal fluid in vivo. Nat. Med. 12, 856–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cirrito J. R., May P. C., O'Dell M. A., Taylor J. W., Parsadanian M., Cramer J. W., Audia J. E., Nissen J. S., Bales K. R., Paul S. M., DeMattos R. B., Holtzman D. M. (2003) In vivo assessment of brain interstitial fluid with microdialysis reveals plaque-associated changes in amyloid-β metabolism and half-life. J. Neurosci. 23, 8844–8853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Alvira-Botero X., Carro E. M. (2010) Clearance of amyloid-β peptide across the choroid plexus in Alzheimer disease. Curr. Aging Sci. 3, 219–229 [DOI] [PubMed] [Google Scholar]
- 12. Deane R., Bell R. D., Sagare A., Zlokovic B. V. (2009) Clearance of amyloid-β peptide across the blood-brain barrier: implication for therapies in Alzheimer disease. CNS Neurol. Disord. Drug Targets 8, 16–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mandrekar S., Jiang Q., Lee C. Y., Koenigsknecht-Talboo J., Holtzman D. M., Landreth G. E. (2009) Microglia mediate the clearance of soluble Aβ through fluid-phase macropinocytosis. J. Neurosci. 29, 4252–4262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fiala M., Lin J., Ringman J., Kermani-Arab V., Tsao G., Patel A., Lossinsky A. S., Graves M. C., Gustavson A., Sayre J., Sofroni E., Suarez T., Chiappelli F., Bernard G. (2005) Ineffective phagocytosis of amyloid-β by macrophages of Alzheimer disease patients. J. Alzheimers Dis. 7, 221–232 [DOI] [PubMed] [Google Scholar]
- 15. DeWitt D. A., Perry G., Cohen M., Doller C., Silver J. (1998) Astrocytes regulate microglial phagocytosis of senile plaque cores of Alzheimer disease. Exp. Neurol. 149, 329–340 [DOI] [PubMed] [Google Scholar]
- 16. Panchal M., Lazar N., Munoz N., Fahy C., Clamagirand C., Brouard J. P., Dubost L., Cohen P., Brakch N., Rholam M. (2007) Clearance of amyloid-β peptide by neuronal and non-neuronal cells: proteolytic degradation by secreted and membrane-associated proteases. Curr. Neurovasc. Res. 4, 240–251 [DOI] [PubMed] [Google Scholar]
- 17. Yin K. J., Cirrito J. R., Yan P., Hu X., Xiao Q., Pan X., Bateman R., Song H., Hsu F. F., Turk J., Xu J., Hsu C. Y., Mills J. C., Holtzman D. M., Lee J. M. (2006) Matrix metalloproteinases expressed by astrocytes mediate extracellular amyloid-β peptide catabolism. J. Neurosci. 26, 10939–10948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fukami S., Watanabe K., Iwata N., Haraoka J., Lu B., Gerard N. P., Gerard C., Fraser P., Westaway D., St George-Hyslop P., Saido T. C. (2002) Aβ-degrading endopeptidase, neprilysin, in mouse brain: synaptic and axonal localization inversely correlating with Aβ pathology. Neurosci. Res. 43, 39–56 [DOI] [PubMed] [Google Scholar]
- 19. Vekrellis K., Ye Z., Qiu W. Q., Walsh D., Hartley D., Chesneau V., Rosner M. R., Selkoe D. J. (2000) Neurons regulate extracellular levels of amyloid-β protein via proteolysis by insulin-degrading enzyme. J. Neurosci. 20, 1657–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Qiu W. Q., Folstein M. F. (2006) Insulin, insulin-degrading enzyme, and amyloid-β peptide in Alzheimer disease: review and hypothesis. Neurobiol. Aging 27, 190–198 [DOI] [PubMed] [Google Scholar]
- 21. Iwata N., Saido T. C. (2003) Amyloid-β peptide metabolism and Alzheimer disease. Nippon Yakurigaku Zasshi 122, 5–14 [DOI] [PubMed] [Google Scholar]
- 22. Turner A. J., Fisk L., Nalivaeva N. N. (2004) Targeting amyloid-degrading enzymes as therapeutic strategies in neurodegeneration. Ann. N.Y. Acad. Sci. 1035, 1–20 [DOI] [PubMed] [Google Scholar]
- 23. Eckman E. A., Eckman C. B. (2005) Aβ-degrading enzymes: modulators of Alzheimer disease pathogenesis and targets for therapeutic intervention. Biochem. Soc. Trans. 33, 1101–1105 [DOI] [PubMed] [Google Scholar]
- 24. Qiu W. Q., Ye Z., Kholodenko D., Seubert P., Selkoe D. J. (1997) Degradation of amyloid-β protein by a metalloprotease secreted by microglia and other neural and non-neural cells. J. Biol. Chem. 272, 6641–6646 [DOI] [PubMed] [Google Scholar]
- 25. Roth R. A., Mesirow M. L., Cassell D. J., Yokono K., Baba S. (1985) Characterization of an insulin-degrading enzyme from cultured human lymphocytes. Diabetes Res. Clin. Pract. 1, 31–39 [DOI] [PubMed] [Google Scholar]
- 26. Zhao J., Li L., Leissring M. A. (2009) Insulin-degrading enzyme is exported via an unconventional protein secretion pathway. Mol. Neurodegener. 4, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bulloj A., Leal M. C., Xu H., Castano E. M., Morelli L. (2009) Insulin-degrading enzyme sorting in exosomes: A secretory pathway for a key brain amyloid-β degrading protease. J. Alzheimers Dis. 19, 79–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tamboli I. Y., Barth E., Christian L., Siepmann M., Kumar S., Singh S., Tolksdorf K., Heneka M. T., Lütjohann D., Wunderlich P., Walter J. (2010) Statins promote the degradation of extracellular amyloid-β peptide by microglia via stimulation of exosome-associated insulin-degrading enzyme (IDE) secretion. J. Biol. Chem. 285, 37405–37414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Glebov K., Schütze S., Walter J. (2011) Functional relevance of a novel SlyX motif in nonconventional secretion of insulin-degrading enzyme. J. Biol. Chem. 286, 22711–22715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Farris W., Mansourian S., Chang Y., Lindsley L., Eckman E. A., Frosch M. P., Eckman C. B., Tanzi R. E., Selkoe D. J., Guenette S. (2003) Insulin-degrading enzyme regulates the levels of insulin, amyloid-β protein, and the β-amyloid precursor protein intracellular domain in vivo. Proc. Natl. Acad. Sci. U.S.A. 100, 4162–4167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Farris W., Mansourian S., Leissring M. A., Eckman E. A., Bertram L., Eckman C. B., Tanzi R. E., Selkoe D. J. (2004) Partial loss-of-function mutations in insulin-degrading enzyme that induce diabetes also impair degradation of amyloid-β protein. Am. J. Pathol. 164, 1425–1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Miller B. C., Eckman E. A., Sambamurti K., Dobbs N., Chow K. M., Eckman C. B., Hersh L. B., Thiele D. L. (2003) Amyloid-β peptide levels in brain are inversely correlated with insulysin activity levels in vivo. Proc. Natl. Acad. Sci. U.S.A. 100, 6221–6226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Leissring M. A., Farris W., Chang A. Y., Walsh D. M., Wu X., Sun X., Frosch M. P., Selkoe D. J. (2003) Enhanced proteolysis of β-amyloid in APP transgenic mice prevents plaque formation, secondary pathology, and premature death. Neuron 40, 1087–1093 [DOI] [PubMed] [Google Scholar]
- 34. Kumar S., Rezaei-Ghaleh N., Terwel D., Thal D. R., Richard M., Hoch M., Mc Donald J. M., Wüllner U., Glebov K., Heneka M. T., Walsh D. M., Zweckstetter M., Walter J. (2011) Extracellular phosphorylation of the amyloid-β peptide promotes formation of toxic aggregates during the pathogenesis of Alzheimer disease. EMBO J. 30, 2255–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kumar S., Walter J. (2011) Phosphorylation of amyloid-β (Aβ) peptides: a trigger for formation of toxic aggregates in Alzheimer disease. Aging 3, 803–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Qiu W. Q., Walsh D. M., Ye Z., Vekrellis K., Zhang J., Podlisny M. B., Rosner M. R., Safavi A., Hersh L. B., Selkoe D. J. (1998) Insulin-degrading enzyme regulates extracellular levels of amyloid-β protein by degradation. J. Biol. Chem. 273, 32730–32738 [DOI] [PubMed] [Google Scholar]
- 37. Haass C., Selkoe D. J. (2007) Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer amyloid-β peptide. Nat. Rev. Mol. Cell Biol. 8, 101–112 [DOI] [PubMed] [Google Scholar]
- 38. Abramowski D., Wiederhold K. H., Furrer U., Jaton A. L., Neuenschwander A., Runser M. J., Danner S., Reichwald J., Ammaturo D., Staab D., Stoeckli M., Rueeger H., Neumann U., Staufenbiel M. (2008) Dynamics of Aβ turnover and deposition in different β-amyloid precursor protein transgenic mouse models following γ-secretase inhibition. J. Pharmacol. Exp. Ther. 327, 411–424 [DOI] [PubMed] [Google Scholar]
- 39. Deane R., Sagare A., Zlokovic B. V. (2008) The role of the cell-surface LRP and soluble LRP in blood-brain barrier Aβ clearance in Alzheimer disease. Curr. Pharm. Des. 14, 1601–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cole G. M., Bell L., Truong Q. B., Saitoh T. (1992) An endosomal-lysosomal pathway for degradation of amyloid precursor protein. Ann. N.Y. Acad. Sci. 674, 103–117 [DOI] [PubMed] [Google Scholar]
- 41. Mooradian A. D. (1988) Effect of aging on the blood-brain barrier. Neurobiol. Aging 9, 31–39 [DOI] [PubMed] [Google Scholar]
- 42. Eikelenboom P., Veerhuis R., van Exel E., Hoozemans J. J., Rozemuller A. J., van Gool W. A. (2011) The early involvement of the innate immunity in the pathogenesis of late-onset Alzheimer disease: neuropathological, epidemiological, and genetic evidence. Curr. Alzheimer Res. 8, 142–150 [DOI] [PubMed] [Google Scholar]
- 43. Authier F., Posner B. I., Bergeron J. J. (1996) Insulin-degrading enzyme. Clin. Invest. Med. 19, 149–160 [PubMed] [Google Scholar]
- 44. Bennett R. G., Duckworth W. C., Hamel F. G. (2000) Degradation of amylin by insulin-degrading enzyme. J. Biol. Chem. 275, 36621–36625 [DOI] [PubMed] [Google Scholar]
- 45. Fawcett J., Hamel F. G., Bennett R. G., Vajo Z., Duckworth W. C. (2001) Insulin and analogue effects on protein degradation in different cell types. Dissociation between binding and activity. J. Biol. Chem. 276, 11552–11558 [DOI] [PubMed] [Google Scholar]
- 46. Duckworth W. C., Bennett R. G., Hamel F. G. (1998) Insulin degradation: progress and potential. Endocr. Rev. 19, 608–624 [DOI] [PubMed] [Google Scholar]
- 47. Kurochkin I. V. (2001) Insulin-degrading enzyme: embarking on amyloid destruction. Trends Biochem. Sci. 26, 421–425 [DOI] [PubMed] [Google Scholar]
- 48. Shen Y., Joachimiak A., Rosner M. R., Tang W. J. (2006) Structures of human insulin-degrading enzyme reveal a new substrate recognition mechanism. Nature 443, 870–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Walter J., Schindzielorz A., Grünberg J., Haass C. (1999) Phosphorylation of presenilin-2 regulates its cleavage by caspases and retards progression of apoptosis. Proc. Natl. Acad. Sci. U.S.A. 96, 1391–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fluhrer R., Friedlein A., Haass C., Walter J. (2004) Phosphorylation of presenilin-1 at the caspase recognition site regulates its proteolytic processing and the progression of apoptosis. J. Biol. Chem. 279, 1585–1593 [DOI] [PubMed] [Google Scholar]
- 51. Johnson G. V. (2006) Tau phosphorylation and proteolysis: insights and perspectives. J. Alzheimers Dis. 9, 243–250 [DOI] [PubMed] [Google Scholar]
- 52. Wang D. S., Dickson D. W., Malter J. S. (2006) β-Amyloid degradation and Alzheimer disease. J. Biomed. Biotechnol. 2006, 58406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Carson J. A., Turner A. J. (2002) Beta-amyloid catabolism: roles for neprilysin (NEP) and other metallopeptidases? J. Neurochem. 81, 1–8 [DOI] [PubMed] [Google Scholar]
- 54. Iwata N., Mizukami H., Shirotani K., Takaki Y., Muramatsu S., Lu B., Gerard N. P., Gerard C., Ozawa K., Saido T. C. (2004) Presynaptic localization of neprilysin contributes to efficient clearance of amyloid-β peptide in mouse brain. J. Neurosci. 24, 991–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Liu Y., Guan H., Beckett T. L., Juliano M. A., Juliano L., Song E. S., Chow K. M., Murphy M. P., Hersh L. B. (2007) In vitro and in vivo degradation of Aβ peptide by peptidases coupled to erythrocytes. Peptides 28, 2348–2355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Howell S., Nalbantoglu J., Crine P. (1995) Neutral endopeptidase can hydrolyze β-amyloid(1–40) but shows no effect on β-amyloid precursor protein metabolism. Peptides 16, 647–652 [DOI] [PubMed] [Google Scholar]
- 57. Betts V., Leissring M. A., Dolios G., Wang R., Selkoe D. J., Walsh D. M. (2008) Aggregation and catabolism of disease-associated intra-Aβ mutations: reduced proteolysis of Aβ A21G by neprilysin. Neurobiol. Dis. 31, 442–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Iwata N., Higuchi M., Saido T. C. (2005) Metabolism of amyloid-β peptide and Alzheimer disease. Pharmacol. Ther. 108, 129–148 [DOI] [PubMed] [Google Scholar]
- 59. Barker R., Love S., Kehoe P. G. (2010) Plasminogen and plasmin in Alzheimer disease. Brain Res. 1355, 7–15 [DOI] [PubMed] [Google Scholar]
- 60. Melchor J. P., Strickland S. (2005) Tissue plasminogen activator in central nervous system physiology and pathology. Thromb Haemost. 93, 655–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Madani R., Nef S., Vassalli J. D. (2003) Emotions are building up in the field of extracellular proteolysis. Trends Mol. Med. 9, 183–185 [DOI] [PubMed] [Google Scholar]
- 62. Tucker H. M., Kihiko M., Caldwell J. N., Wright S., Kawarabayashi T., Price D., Walker D., Scheff S., McGillis J. P., Rydel R. E., Estus S. (2000) The plasmin system is induced by and degrades amyloid-β aggregates. J. Neurosci. 20, 3937–3946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tucker H. M., Kihiko-Ehmann M., Wright S., Rydel R. E., Estus S. (2000) Tissue plasminogen activator requires plasminogen to modulate amyloid-β neurotoxicity and deposition. J. Neurochem. 75, 2172–2177 [DOI] [PubMed] [Google Scholar]
- 64. Miners J. S., van Helmond Z., Raiker M., Love S., Kehoe P. G. (2010) ACE variants and association with brain Aβ levels in Alzheimer disease. Am. J. Transl. Res. 3, 73–80 [PMC free article] [PubMed] [Google Scholar]
- 65. Kehoe P. G., Russ C., McIlory S., Williams H., Holmans P., Holmes C., Liolitsa D., Vahidassr D., Powell J., McGleenon B., Liddell M., Plomin R., Dynan K., Williams N., Neal J., Cairns N. J., Wilcock G., Passmore P., Lovestone S., Williams J., Owen M. J. (1999) Variation in DCP1, encoding ACE, is associated with susceptibility to Alzheimer disease. Nat. Genet. 21, 71–72 [DOI] [PubMed] [Google Scholar]
- 66. Miners S., Ashby E., Baig S., Harrison R., Tayler H., Speedy E., Prince J. A., Love S., Kehoe P. G. (2009) Angiotensin-converting enzyme levels and activity in Alzheimer disease: differences in brain and cerebrospinal fluid ACE and association with ACE1 genotypes. Am. J. Transl. Res. 1, 163–177 [PMC free article] [PubMed] [Google Scholar]
- 67. Thornton-Wells T. A., Moore J. H., Martin E. R., Pericak-Vance M. A., Haines J. L. (2008) Confronting complexity in late-onset Alzheimer disease: application of two-stage analysis approach addressing heterogeneity and epistasis. Genet. Epidemiol. 32, 187–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hollingworth P., Harold D., Sims R., Gerrish A., Lambert J. C., Carrasquillo M. M., Abraham R., Hamshere M. L., Pahwa J. S., Moskvina V., Dowzell K., Jones N., Stretton A., Thomas C., Richards A., Ivanov D., Widdowson C., Chapman J., Lovestone S., Powell J., Proitsi P., Lupton M. K., Brayne C., Rubinsztein D. C., Gill M., Lawlor B., Lynch A., Brown K. S., Passmore P. A., Craig D., McGuinness B., Todd S., Holmes C., Mann D., Smith A. D., Beaumont H., Warden D., Wilcock G., Love S., Kehoe P. G., Hooper N. M., Vardy E. R., Hardy J., Mead S., Fox N. C., Rossor M., Collinge J., Maier W., Jessen F., Rüther E., Schürmann B., Heun R., Kolsch H., van den B. H., Heuser I., Kornhuber J., Wiltfang J., Dichgans M., Frolich L., Hampel H., Gallacher J., Hull M., Rujescu D., Giegling I., Goate A. M., Kauwe J. S., Cruchaga C., Nowotny P., Morris J. C., Mayo K., Sleegers K., Bettens K., Engelborghs S., De Deyn P. P., Van B. C., Livingston G., Bass N. J., Gurling H., McQuillin A., Gwilliam R., Deloukas P., Al-Chalabi A., Shaw C. E., Tsolaki M., Singleton A. B., Guerreiro R., Muhleisen T. W., Nothen M. M., Moebus S., Jockel K. H., Klopp N., Wichmann H. E., Pankratz V. S., Sando S. B., Aasly J. O., Barcikowska M., Wszolek Z. K., Dickson D. W., Graff-Radford N. R., Petersen R. C., van Duijn C. M., Breteler M. M., Ikram M. A., DeStefano A. L., Fitzpatrick A. L., Lopez O., Launer L. J., Seshadri S., Berr C., Campion D., Epelbaum J., Dartigues J. F., Tzourio C., Alperovitch A., Lathrop M., Feulner T. M., Friedrich P., Riehle C., Krawczak M., Schreiber S., Mayhaus M., Nicolhaus S., Wagenpfeil S., Steinberg S., Stefansson H., Stefansson K., Snaedal J., Bjornsson S., Jonsson P. V., Chouraki V., Genier-Boley B., Hiltunen M., Soininen H., Combarros O., Zelenika D., Delepine M., Bullido M. J., Pasquier F., Mateo I., Frank-Garcia A., Porcellini E., Hanon O., Coto E., Alvarez V., Bosco P., Siciliano G., Mancuso M., Panza F., Solfrizzi V., Nacmias B., Sorbi S., Bossu P., Piccardi P., Arosio B., Annoni G., Seripa D., Pilotto A., Scarpini E., Galimberti D., Brice A., Hannequin D., Licastro F., Jones L., Holmans P. A., Jonsson T., Riemenschneider M., Morgan K., Younkin S. G., Owen M. J., O'Donovan M., Amouyel P., Williams J. (2011) Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33, and CD2AP are associated with Alzheimer disease. Nat. Genet. 43, 429–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Seshadri S., Fitzpatrick A. L., Ikram M. A., DeStefano A. L., Gudnason V., Boada M., Bis J. C., Smith A. V., Carassquillo M. M., Lambert J. C., Harold D., Schrijvers E. M., Ramirez-Lorca R., Debette S., Longstreth W. T., Jr., Janssens A. C., Pankratz V. S., Dartigues J. F., Hollingworth P., Aspelund T., Hernandez I., Beiser A., Kuller L. H., Koudstaal P. J., Dickson D. W., Tzourio C., Abraham R., Antunez C., Du Y., Rotter J. I., Aulchenko Y. S., Harris T. B., Petersen R. C., Berr C., Owen M. J., Lopez-Arrieta J., Varadarajan B. N., Becker J. T., Rivadeneira F., Nalls M. A., Graff-Radford N. R., Campion D., Auerbach S., Rice K., Hofman A., Jonsson P. V., Schmidt H., Lathrop M., Mosley T. H., Au R., Psaty B. M., Uitterlinden A. G., Farrer L. A., Lumley T., Ruiz A., Williams J., Amouyel P., Younkin S. G., Wolf P. A., Launer L. J., Lopez O. L., van Duijn C. M., Breteler M. M. (2010) Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA 303, 1832–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Naj A. C., Jun G., Beecham G. W., Wang L. S., Vardarajan B. N., Buros J., Gallins P. J., Buxbaum J. D., Jarvik G. P., Crane P. K., Larson E. B., Bird T. D., Boeve B. F., Graff-Radford N. R., De Jager P. L., Evans D., Schneider J. A., Carrasquillo M. M., Ertekin-Taner N., Younkin S. G., Cruchaga C., Kauwe J. S., Nowotny P., Kramer P., Hardy J., Huentelman M. J., Myers A. J., Barmada M. M., Demirci F. Y., Baldwin C. T., Green R. C., Rogaeva E., St George-Hyslop P., Arnold S. E., Barber R., Beach T., Bigio E. H., Bowen J. D., Boxer A., Burke J. R., Cairns N. J., Carlson C. S., Carney R. M., Carroll S. L., Chui H. C., Clark D. G., Corneveaux J., Cotman C. W., Cummings J. L., DeCarli C., DeKosky S. T., az-Arrastia R., Dick M., Dickson D. W., Ellis W. G., Faber K. M., Fallon K. B., Farlow M. R., Ferris S., Frosch M. P., Galasko D. R., Ganguli M., Gearing M., Geschwind D. H., Ghetti B., Gilbert J. R., Gilman S., Giordani B., Glass J. D., Growdon J. H., Hamilton R. L., Harrell L. E., Head E., Honig L. S., Hulette C. M., Hyman B. T., Jicha G. A., Jin L. W., Johnson N., Karlawish J., Karydas A., Kaye J. A., Kim R., Koo E. H., Kowall N. W., Lah J. J., Levey A. I., Lieberman A. P., Lopez O. L., Mack W. J., Marson D. C., Martiniuk F., Mash D. C., Masliah E., McCormick W. C., McCurry S. M., McDavid A. N., McKee A. C., Mesulam M., Miller B. L., Miller C. A., Miller J. W., Parisi J. E., Perl D. P., Peskind E., Petersen R. C., Poon W. W., Quinn J. F., Rajbhandary R. A., Raskind M., Reisberg B., Ringman J. M., Roberson E. D., Rosenberg R. N., Sano M., Schneider L. S., Seeley W., Shelanski M. L., Slifer M. A., Smith C. D., Sonnen J. A., Spina S., Stern R. A., Tanzi R. E., Trojanowski J. Q., Troncoso J. C., Van D., V, Vinters H. V., Vonsattel J. P., Weintraub S., Welsh-Bohmer K. A., Williamson J., Woltjer R. L., Cantwell L. B., Dombroski B. A., Beekly D., Lunetta K. L., Martin E. R., Kamboh M. I., Saykin A. J., Reiman E. M., Bennett D. A., Morris J. C., Montine T. J., Goate A. M., Blacker D., Tsuang D. W., Hakonarson H., Kukull W. A., Foroud T. M., Haines J. L., Mayeux R., Pericak-Vance M. A., Farrer L. A., Schellenberg G. D. (2011) Common variants at MS4A4/MS4A6E, CD2AP, CD33, and EPHA1 are associated with late-onset Alzheimer disease. Nat. Genet. 43, 436–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Oba R., Igarashi A., Kamata M., Nagata K., Takano S., Nakagawa H. (2005) The N-terminal active center of human angiotensin-converting enzyme degrades Alzheimer amyloid-β peptide. Eur. J. Neurosci. 21, 733–740 [DOI] [PubMed] [Google Scholar]
- 72. Hemming M. L., Selkoe D. J. (2005) Amyloid-β protein is degraded by cellular angiotensin-converting enzyme (ACE) and elevated by an ACE inhibitor. J. Biol. Chem. 280, 37644–37650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hu J., Igarashi A., Kamata M., Nakagawa H. (2001) Angiotensin-converting enzyme degrades Alzheimer amyloid-β peptide (Aβ); retards Aβ aggregation, deposition, fibril formation; and inhibits cytotoxicity. J. Biol. Chem. 276, 47863–47868 [DOI] [PubMed] [Google Scholar]
- 74. Milton N. G. (2005) Phosphorylated amyloid-β: the toxic intermediate in Alzheimer disease neurodegeneration. Subcell. Biochem. 38, 381–402 [DOI] [PubMed] [Google Scholar]
- 75. Milton N. G. (2001) Phosphorylation of amyloid-β at the serine 26 residue by human Cdc2 kinase. Neuroreport 12, 3839–3844 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.