Background: SARS-CoV entry into target cells is a unique and complex process.
Results: Cell entry mediated by SARS-CoV S was strongly inhibited after knockdown of PI4KB.
Conclusion: PI4KB is required for SARS-CoV S-mediated entry, and this function is dependent on its kinase activity.
Significance: Our results indicate a new function for PI4KB and suggest a new drug target for preventing SARS-CoV infection.
Keywords: Endocytosis, Phosphatidylinositol Kinase, RNA viruses, siRNA, Virus Entry, ACE2, PI4KB, PI4P, SARS-CoV, VSVG
Abstract
Phosphatidylinositol kinases (PI kinases) play an important role in the life cycle of several viruses after infection. Using gene knockdown technology, we demonstrate that phosphatidylinositol 4-kinase IIIβ (PI4KB) is required for cellular entry by pseudoviruses bearing the severe acute respiratory syndrome-coronavirus (SARS-CoV) spike protein and that the cell entry mediated by SARS-CoV spike protein is strongly inhibited by knockdown of PI4KB. Consistent with this observation, pharmacological inhibitors of PI4KB blocked entry of SARS pseudovirions. Further research suggested that PI4P plays an essential role in SARS-CoV spike-mediated entry, which is regulated by the PI4P lipid microenvironment. We further demonstrate that PI4KB does not affect virus entry at the SARS-CoV S-ACE2 binding interface or at the stage of virus internalization but rather at or before virus fusion. Taken together, these results indicate a new function for PI4KB and suggest a new drug target for preventing SARS-CoV infection.
Introduction
Phosphoinositide (PI)3 lipids play crucial roles in membrane trafficking. PIs can be tightly regulated both spatially and temporally by the many PI kinases and phosphatases that are distributed throughout the different intracellular compartments (1). The potential roles for PIs and PI kinases in the life cycle of viruses have been studied extensively. For example, PI3Ks appear to play a predominant role in regulating Ebola virus entry (2). Several other viruses also activate PI3K during infection, such as VSV and influenza virus (3, 4). In addition, several groups identified PI4KA as a cofactor for hepatitis C virus (HCV) replication (5–7). The HCV nonstructural protein NS5A was demonstrated to interact with PI4KA and stimulate its kinase activity (8, 9). Recently, PI4KB was shown to be a key cellular protein exploited by several plus-strand RNA viruses for replication, and the PI4P-rich lipid microenvironment is essential for enteroviral RNA replication (10).
In April 2003, a novel coronavirus, SARS coronavirus (SARS-CoV), was identified as the etiological agent of severe acute respiratory syndrome (SARS) (11). SARS-CoV is an enveloped, positive-strand RNA virus. Angiotensin-1-converting enzyme-2 (ACE2) is the functional receptor of SARS-CoV, which is responsible for mediating SARS-CoV entry into host cells (12, 13). Understanding the molecular basis by which SARS-CoV targets cells has been critical to unraveling the mechanisms of its pathogenesis. SARS-CoV enters cells by direct fusion at the plasma membrane (14–16). Further studies primarily using pseudotyped virus systems have shown that SARS-CoV entry is pH-dependent and have suggested that the endosome protease cathepsin L may be involved in entry (17–19). Apart from these observations, no other key cellular factors have been shown to be specifically involved in the SARS-CoV life cycle.
In this study, we reveal the role of PI4KB in SARS-CoV S-mediated entry. This is the first study to demonstrate that PI4KB is required for cell entry mediated by SARS-CoV S and that its function is dependent upon its kinase activity. Interestingly, we found that PI4P, the product of PI4KB catalysis, creates a lipid microenvironment that is required for SARS-CoV S-mediated entry. Our results provide important insights into the life cycle of SARS-CoV and suggest that PI4KB may be a potential therapeutic target in the near future.
MATERIALS AND METHODS
Cell Culture
VeroE6 cells (African green monkey kidney cell line) and HEK293T cells (human embryonic kidney cell line) from the American Type Culture Collection were propagated in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 units/ml penicillin, and 100 units/ml streptomycin at 37 °C with 5% CO2.
Reagents and Antibodies
Anti-PI4KB polyclonal antibodies were obtained from Millipore (Billerica, MA). Anti-β-actin monoclonal antibodies, anti-FLAG antibodies, and anti-human Fc antibodies were purchased from Sigma. Anti-ACE2 polyclonal antibodies were purchased from R&D Systems (Minneapolis, MN). Anti-GFP antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-PI4P mouse monoclonal antibodies were obtained from Echelon Biosciences (Salt Lake City, UT). LY294002, wortmannin, DMSO, Hoechst 33342, TPCK-trypsin, and proteinase K were purchased from Sigma.
Pseudovirus Production
Pseudoviruses were produced as described previously (20). Briefly, HEK293T cells were co-transfected with pQCXIX vector, a spike-encoding plasmid (Sh-2, provided by Dr. Michael Farzan) or a control plasmid (VSV-G) and a gag/pol expression plasmid. The enhanced GFP gene was inserted into the pQCXIX vector. At 48 h post-transfection, virus supernatants were harvested and filtered through a 0.45-μm filter. If required, virions were concentrated by ultracentrifugation at 40,000 × g in an SW41 rotor (Beckman Instruments) for 2 h at 4 °C. The pellets were resuspended in DMEM and stored at −80 °C. Viral titers were determined as described previously (21).
Drug Inhibition of Pseudovirus Entry into VeroE6 Cells
VeroE6 cells were treated with the indicated concentration of LY294002 or DMSO for 30 min at 37 °C. Next, the cells were incubated with pseudovirus for 2 h at 37 °C in the presence of drug before fresh medium was added, and the cells were incubated at 37 °C for 48 h. Then the cells were fixed with 4% paraformaldehyde in PBS at room temperature (RT) for 20 min. Cell nuclei were stained with Hoechst 33342 diluted in PBS for 10 min at RT. Images were captured using a Nikon Eclipse TE2000-U inverted fluorescence microscope and analyzed with Image-Pro Plus software (Media Cybernetics).
siRNA Transfections
All siRNAs used in this study were obtained from Ribobio, Guanzhou, China. For transfection, VeroE6 cells were seeded at 5 × 104 cells/well in 24-well plates. The following day, the cells were transfected with 1 μl of Lipofectamine RNAiMax reagent (Invitrogen) at 50 nm siRNA in Opti-MEM (Invitrogen). After 24 h, the cells were trypsinized and seeded into a 96-well plate. At 48 h post-siRNA transfection, the experimental virus infections were performed.
Western Blot Analysis
Protein samples were separated on 4–20% SDS-polyacrylamide gels and transferred to nitrocellulose. The membranes were probed with primary antibodies. The proteins were visualized by HRP-conjugated secondary antibodies and a chemiluminescent substrate (Santa Cruz Biotechnology) and exposed to film.
Cell Viability
VeroE6 cells were seeded in 96-well plates at 1 × 105/ml. DMSO or LY294002 diluted in DMEM was added to the cells the following day. After a 3-h incubation at 37 °C and 5% CO2, the cell culture medium was removed, and new DMEM was added to the cells. Each experimental group included triplicate wells. Then 20 μl of CellTiter 96 AQueous One Solution cell proliferation assay buffer (Promega) was added to each well, and the cultures were incubated for an additional 2 h. Absorbance was recorded at 490 nm. For siRNA treatment, cell viability was determined 48 h after siRNA transfection.
RNA Isolation and Quantitative PCR
Total RNA was harvested from cells using TRIzol reagent (Invitrogen) for analysis of host gene expression. Cellular RNAs were reverse-transcribed and amplified by PCR using the SuperScriptTM III Platinum One-Step quantitative RT-PCR system with Platinum Taq (Invitrogen) and TaqMan gene expression assays (Applied Biosystems). Cellular RNAs were normalized to GAPDH levels. Data were analyzed relative to siControl-treated cells. All assays were performed on an ABI 7500 system and analyzed with SDS 1.3 (Applied Biosystems).
Fluorescence-activated Cell Sorting Analysis
At 48 h post-transfection of siRNAs, VeroE6 cells were trypsinized and collected in 1.5-ml Eppendorf tubes. Then the cultures were incubated with ACE2 antibodies for 2 h at 37 °C. After three washes in PBS, the cells were incubated with Alexa Fluor 488-labeled secondary antibodies. After fixation in 0.5% paraformaldehyde, the samples were analyzed on a Beckman Coulter EPICS Elite ESP instrument.
Immunofluorescence Microscopy
Cells grown on glass coverslips were rinsed with PBS and fixed in 4% paraformaldehyde in PBS for 15 min at RT followed by quenching in 50 mm NH4Cl in PBS for 10 min at RT. Then the cells were blocked and permeabilized in 0.2% saponin, 10% FBS in PBS for 1 h at RT. The cells were incubated with primary antibody against PI4P in blocking buffer at 4 °C overnight followed by three 5-min washes in PBS. Alexa 488-conjugated secondary antibodies (Invitrogen) were used at a dilution of 1:500 in blocking buffer. After three washes with PBS, the cell nuclei were stained with Hoechst 33342 (Sigma) in PBS for 10 min at RT. For FLAG-Sac1 immunostaining, after PI4P immunostaining, the cells were incubated with anti-FLAG primary antibodies in blocking buffer for 1 h at RT followed by three 5-min washes with PBS. Alexa 568-conjugated secondary antibodies (Invitrogen) were used at a dilution of 1:500 in blocking buffer. Images were captured using confocal laser-scanning microscopy (Leica TCS SP2) and analyzed with Leica confocal software.
Binding and Internalization
The SARS-CoV RBR (S318–510) was expressed as a chimeric protein with human IgG-Fc, and proteins were expressed in 293T cells. The recombinant proteins were purified on protein G-agarose beads (GE Healthcare). Virus, S318–510Fc, or Fc proteins were bound to cells at the indicated concentrations at 4 °C. After 2 h, unbound virus or protein was removed by washing. The cells were either lysed to measure bound virus or protein or were incubated at 37 °C for 2 h to allow for internalization and treated with 1 mg/ml proteinase K prior to lysis to remove uninternalized virus. For neutralization experiments, SARS pseudovirus was preincubated with anti-SARS spike-neutralizing antibodies (50 μg/ml) or with an irrelevant isotype-matched control antibody (anti-CD81) for 1 h at RT. Then SARS pseudovirus was added to VeroE6 cells for virus binding or internalization assays.
Transient Transfection with SAC1
VeroE6 cells were transfected at 50–70% confluence with a FLAG-Sac1 expression vector or empty vector (pCDNA3.1). Transfections were performed with X-tremeGENE HP reagents following the manufacturer's instructions (Roche Applied Science). Virus infection experiments were performed at 48 h post-transfection.
Trypsin Bypass
VeroE6 cells were preincubated at 37 °C for 30 min with DMEM in the presence or absence of LY294002 (30 μm). Then the cells were spin-infected with pseudovirus at 2000 × g for 30 min at 4 °C. The medium was changed, and the cells were incubated with 5 μg/ml TPCK-trypsin or with PBS alone at 37 °C for 13 min. The trypsin was removed, and DMEM with or without LY294002 (30 μm) was added. After 2 h, the medium was replaced with fresh DMEM containing 10% FBS. The cells were analyzed for GFP expression after 48 h.
PI4P Mass Strips
The cells were seeded in duplicate six-well plates, one for lipid extractions and one for cell counting on a hemocytometer. Equivalent numbers of cells (4 million) were collected, and PI4P was extracted according to a detailed protocol from a PI4P mass strips kit (Echelon Biosciences). Lipids were spotted onto nitrocellulose membranes pre-spotted with PI standards and visualized with the PI4P-specific detectors and chemiluminescent reagent (Santa Cruz Biotechnology).
Statistical Analysis
All data shown are the means ± S.E., and statistical analyses were performed using Student's t test.
RESULTS
LY294002 and Wortmannin Treatment Inhibit SARS-CoV S-mediated Entry into VeroE6 Cells
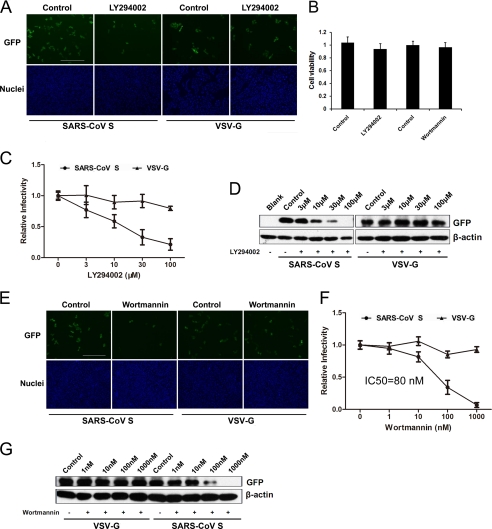
To assess the efficiency of virus entry, we produced GFP-containing pseudovirus with SARS-CoV envelope glycoproteins (spike) and VSV-G protein (VSVP), with infected cells fluorescing green during microscopy. We first screened several chemicals known to be kinase inhibitors to determine their effects on SARS-CoV S-mediated entry. We found that LY294002 (PI kinase inhibitor) significantly inhibited the entry of SARS pseudovirions into VeroE6 cells, whereas entry of VSV-G pseudovirus was not affected (Fig. 1A). A cell viability assay demonstrated that LY294002 had no significant effect on VeroE6 growth during the experiments (Fig. 1B). LY294002 inhibited entry in a dose-dependent manner (Fig. 1C). LY294002 treatment at 30 μm strongly inhibited SARS-CoV S-mediated entry by more than 50%. In addition, we observed similar results using Western blot analysis to quantify GFP expression to detect virus entry (Fig. 1D). Another PI kinase inhibitor, wortmannin, also inhibited SARS-CoV S-mediated entry into VeroE6 cells with no effect on VSV-G pseudovirus entry (Fig. 1E). We found that entry of SARS pseudovirions into VeroE6 cells was inhibited by wortmannin with an IC50 of 80 nm (Fig. 1F), which was consistent with the results reported for type III PI4Ks (PI4KA and PI4KB) (22, 23). Wortmannin treatment at 10 nm, a concentration that efficiently inhibits PI 3-kinases, had no significant effect on SARS-CoV S-mediated entry. There was no significant cytotoxicity with wortmannin treatment at concentrations up to 1 μm (Fig. 1B). Finally, total GFP expression after SARS-CoV S-mediated entry also decreased when the cells were pretreated with wortmannin (Fig. 1G).
FIGURE 1.
LY294002 inhibits SARS-CoV S-mediated entry. A, VeroE6 cells were pretreated with LY294002 at 30 μm or DMSO (control) and then infected with SARS-CoV S or VSV-G pseudovirus. After 48 h, the cells were fixed, and the nuclei were stained with Hoechst 33342. Images were captured on a fluorescence microscope. Scale bar, 400 μm. B, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay of VeroE6 cells treated with 100 or 1 μm wortmannin for 3 h. C, VeroE6 cells were treated with the indicated concentrations of LY294002 (3–100 μm) or DMSO (control). Statistical analysis was performed on the proportion of GFP-positive cells 48 h after virus infection. For quantification, >1000 cells were scored in three independent experiments. D, effect of LY294002 (3–100 μm) or DMSO (control) treatment or untreated (blank) on SARS pseudovirus entry was determined by assessing the GFP expression level by immunoblotting with the indicated antibodies. Cell lysates were prepared after virus infection for 48 h. E, VeroE6 cells were pretreated with wortmannin at 100 nm or DMSO (control) and then infected with SARS-CoV S or VSV-G pseudovirus. After 48 h, the cells were fixed, and the nuclei were stained with Hoechst 33342. Images were captured on a fluorescence microscope. Scale bar, 400 μm. F, VeroE6 cells were treated with the indicated concentrations of wortmannin or DMSO. Statistical analysis was performed to assess the proportion of GFP-positive cells 48 h after virus infection. For quantification, >1000 cells were scored in three independent experiments. G, effect of wortmannin (1–1000 nm), DMSO, or no treatment (blank) on SARS pseudovirion entry was determined by GFP expression levels by immunoblotting with the indicated antibodies. Cell lysates were prepared after virus infection for 48 h.
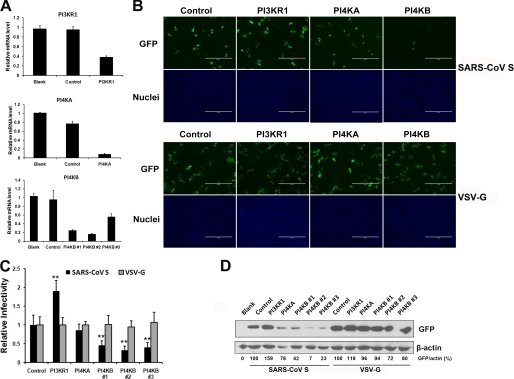
Knockdown of PI4KB Inhibits SARS-CoV S-mediated Entry into VeroE6 Cells
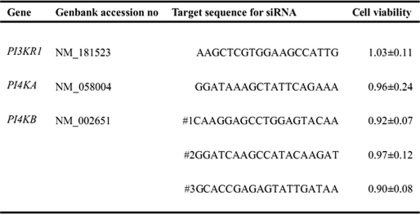
LY294002 and wortmannin are PI kinase inhibitors that target several PI kinases, such as PI3Ks and type III PI4Ks (PI4KA and PI4KB) (23). To our knowledge, the functions of PI4Ks in mammalian cells have not been well defined. In this study, we sought to identify the type of PI kinase that was affected by LY294002 and wortmannin. Given that p85α is the major and most important subunit among the PI3K subunits (24), this subunit was assessed in the following experiments. We designed a series of corresponding siRNAs to test the effect of knocking down each individual gene on virus entry without significant effects on cell viability (Table 1). Quantitative RT-PCR results demonstrated that the siRNAs we used efficiently inhibited target gene expression (PI3KR1, PI4KA, and PI4KB), whereas the control siRNA had little effect (Fig. 2A). Fluorescence microscopy showed a significant inhibition of SARS-CoV S-mediated entry only after PI4KB siRNA transfection compared with control siRNA-treated cells, whereas the PI4KA knockdown had no significant effect on SARS-CoV S-mediated entry. Conversely, SARS-CoV S-mediated entry increased after transfection of PI3KR1 (p85α) siRNA into VeroE6 cells (Fig. 2B). In contrast, PI kinase siRNAs had no effect on VSV-G entry into VeroE6 cells (Fig. 2B). Statistical analysis demonstrated the significance of the above observation (Fig. 2C). PI4KB silencing by three individual siRNA duplexes strongly inhibited SARS-CoV S-mediated entry, as confirmed by Western blot analysis (Fig. 2D). Moreover, siRNA duplex 2 was the most potent for both PI4KB transcript silencing and for the inhibition of SARS-CoV S-mediated entry. From these results, we conclude that LY294002 and wortmannin inhibit SARS pseudovirus entry by targeting PI4KB and that PI4KB is involved in SARS-CoV S-mediated entry into VeroE6 cells.
TABLE 1.
Genes and siRNAs assessed in RNA interference experiments
Cell viability values represent fold-change in cell viability in specific siRNA-treated cells compared with the corresponding control (cells transfected with a nontargeting siRNA). Each data point represents the mean ± S.D. of three independent experiments.
FIGURE 2.
PI4KB knockdown inhibits SARS-CoV S-mediated entry, whereas PI3KR1 knockdown increases SARS-CoV S-mediated entry. A, VeroE6 cells were transfected with a nontarget siRNA (control) or siRNAs specific for PI3KR1, PI4KA, or PI4KB. Untreated cells were used as a blank control. The transcripts of target genes were quantified by quantitative RT-PCR at 48 h post-transfection. mRNA levels were normalized to GAPDH levels. Values are presented as the means ± S.D. B, VeroE6 cells were transfected with corresponding siRNAs and then infected with SARS-CoV S or VSV-G pseudovirus. After 48 h, the cells were fixed, and the nuclei were stained with Hoechst 33342. Images were captured on a fluorescence microscope. Scale bar, 400 μm. C, VeroE6 cells were transfected with the corresponding siRNAs and then infected with SARS-CoV S or VSV-G pseudovirus. Statistical analysis was performed on the proportion of GFP-positive cells 48 h after virus infection. For quantification, >1000 cells were scored in three independent experiments. **, p < 0.001 compared with the control. D, siRNA treatment affected SARS-CoV S-mediated entry as determined by GFP expression levels measured by immunoblotting with the indicated antibodies. Cell lysates were prepared after virus infection for 48 h. GFP band intensities were quantitated by densitometric analysis using Quantity One (Bio-Rad) and normalized to β-actin levels.
Ectopic Expression of Sac1 Phosphatase Inhibits SARS-CoV S-mediated Entry
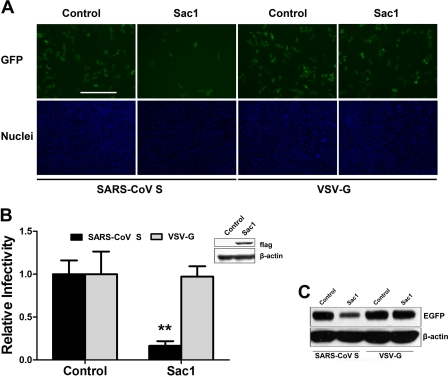
Interestingly, the opposing roles of PI3KR1 and PI4KB during SARS-CoV S-mediated entry into VeroE6 cells suggest that their different roles in this process may be linked to their specific kinase activities. Previous studies have established that PI4KB is responsible for the production of most of the PI4P present in cells (25), whereas PI3K can convert PI4P into phosphatidylinositol 3,4-bisphosphate, thereby decreasing PI4P concentrations (26). These previous results suggest that the level of PI4P may be the main reason that PI3KR1 and PI4KB demonstrate different functions in SARS-CoV S-mediated entry. To investigate whether kinase activity could account for the different effects of the PI kinases on SARS-CoV S-mediated entry and to test whether PI4P lipids directly regulate viral entry independent of PI4KB, VeroE6 cells were transiently transfected with the SAC1 gene, a PI phosphatase that specifically converts PI4P lipids back to PI (27). This phosphatase has been shown to oppose PI4KB in vivo. As demonstrated by the virus entry assay (Fig. 3, A and B), SARS-CoV S-mediated entry was reduced by ∼80% in VeroE6 cells that transiently overexpressed recombinant FLAG-Sac1 compared with control VeroE6 cells, whereas VSV-G pseudovirus entry was not affected by recombinant Sac1 expression. In transiently transfected cells, recombinant Sac1 was indeed present, as shown by the detection of FLAG expression at the N terminus of Sac1 (Fig. 3B). Furthermore, Western blot analysis showed that GFP expression after SARS-CoV S-mediated entry decreased in the recombinant Sac1-expressing cells compared with vector-transfected VeroE6 cells (Fig. 3C). These results indicate that PI4P is indispensable for SARS-CoV S-mediated entry and suggest that PI4KB mediates SARS-CoV S entry by regulating the level of cellular PI4P.
FIGURE 3.
Overexpression of Sac1 inhibits SARS-CoV S-mediated entry. A, normal VeroE6 cells or cells transiently transfected with FLAG-Sac1 were infected with SARS-CoV S or VSV-G pseudovirus. After 48 h, the cells were fixed, and the nuclei were stained with Hoechst 33342. Images were captured on a fluorescence microscope. Scale bar, 400 μm. B, statistical analysis of the proportion of GFP-positive cells at 48 h post-virus infection in A. Inset, Western blot showing recombinant FLAG-Sac1 expression at 48 h post-transfection. **, p < 0.001. C, GFP expression levels were determined by immunoblotting with the antibodies indicated in A.
PI4P Levels in VeroE6 Cells Are Regulated by PI4KB and Sac1
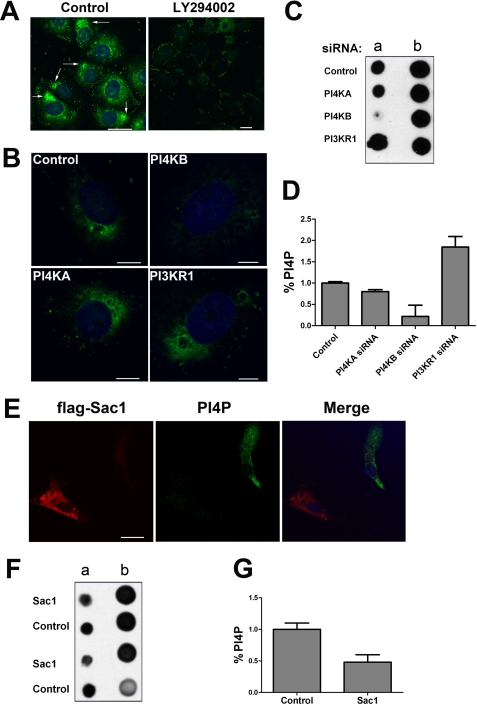
To further confirm that PI4KB regulates SARS-CoV S-mediated entry by controlling the production of PI4P, we monitored the levels of cellular PI4P during PI4KB inhibition or ectopic expression of Sac1. Immunostaining with an anti-PI4P monoclonal antibody revealed the enrichment of PI4P within the cytoplasm of normal cells (Fig. 4A, left panel). However, most PI4P immunofluorescence was lost after a 30-min treatment with LY294002 (Fig. 4A, right panel). Additionally, the reduction of PI4KB by siRNA knockdown greatly affected PI4P levels in VeroE6 cells (Fig. 4B). In parallel, lipid extracts were prepared from siRNA-transfected cells and probed for PI4P content using PI4P mass strips that contained known loading standards. As shown, PI4KB inhibition had a greater effect on lowering PI4P levels in VeroE6 cells compared with PI4KA inhibition, whereas PI3KR1 inhibition increased the level of PI4P (Fig. 4, C and D). The effect of PI3KR1, PI4KA, and PI4KB on PI4P levels in VeroE6 cells was in accordance with their effect on SARS-CoV S-mediated entry. These results suggest that PI4KB is responsible for most of the production of PI4P. Next, we monitored the effect of recombinant Sac1 expression on PI4P levels. VeroE6 cells were transiently transfected with a FLAG-Sac1 expression plasmid. Transfected cells (Fig. 4, red) were visualized by indirect immunofluorescence using an anti-FLAG antibody, and PI4P (green) was detected with an anti-PI4P antibody. Confocal images show that PI4P levels in FLAG-Sac1-positive cells were lower than in nontransfected cells (Fig. 4E). Detection of PI4P from lipid extracts of VeroE6 cells that were transfected with FLAG-Sac1 or a control plasmid revealed that ectopic expression of Sac1 reduced the levels of PI4P in VeroE6 (Fig. 4, F and G). These results demonstrate that Sac1 phosphatase is a key factor in regulating PI4P levels in VeroE6 cells.
FIGURE 4.
PI4P lipid levels are regulated by PI4KB and Sac1 in VeroE6 cells. A, PI4P levels decreased after LY294002 treatment. VeroE6 cells were treated with 100 μm LY294002 or DMSO (control) for 30 min. PI4P lipid content was determined by immunostaining with anti-PI4P antibodies. The white arrows indicate the PI4P distribution (green). Nuclei were stained with Hoechst 33342. Scale bar, 25 μm. B, PI4KB is responsible for a significant fraction of PI4P lipids in VeroE6. The cells were transfected with control, PI4KB, PI4KA, or PI3KR1 siRNAs for 48 h. Then the cells were immunostained with anti-PI4P antibodies. The nuclei were counterstained with Hoechst 33342. Scale bar, 7 μm. C, detection of PI4P from lipid extracts of VeroE6 cells. At 48 h post-transfection of siRNAs, the PI4P lipid levels in VeroE6 cells were determined using PI4P mass strips. Column a consists of PI4P lipid extractions from experimental samples. Column b consists of pre-spotted PI4P standards as follows (top to bottom): 20, 15, 10, and 5 pmol. D, statistical analysis of the relative levels of PI4P in C. Error bars represent the S.E. from two independent experiments. E, recombinant Sac1 expression decreased PI4P lipid levels in VeroE6. The cells were transfected with a FLAG-Sac1 expression plasmid. After 48 h, the transfected cells were detected by indirect immunofluorescence with an anti-FLAG antibody (red). PI4P lipid (green) was detected with anti-PI4P antibody. The nuclei were counterstained with Hoechst 33342. Scale bar, 23 μm. F, detection of PI4P lipids from extracts of VeroE6 cells expressing recombinant Sac1. Column a consists of PI4P lipid extracts from two independent experimental samples. Column b consists of pre-spotted PI4P standards as follows (top to bottom): 4, 2, 1, and 0.5 pmol. G, statistical analysis of the relative levels of PI4P in F. Error bars are S.E. from two independent experiments.
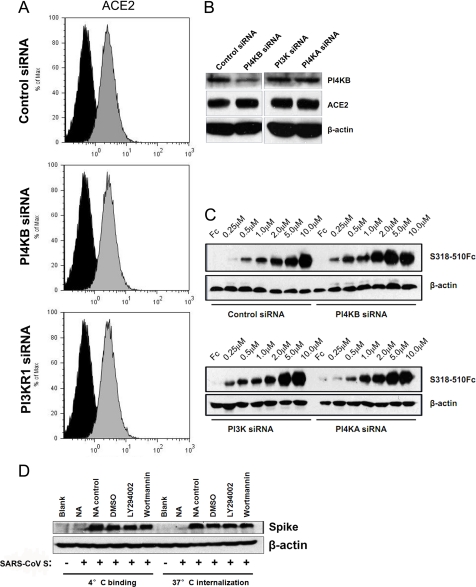
PI4KB Facilitates SARS-CoV S-mediated Entry after Virus Binding and Internalization
Previous studies have shown that virus entry is a multistep process. The primary step is often initiated by low affinity binding to attachment sites, which promotes the concentration of virions on the cell surface. The subsequent binding to a high affinity receptor triggers cell entry (28). After virus binding, the entry of enveloped viruses into cells is known to occur via two primary pathways, membrane fusion or endocytosis (29). To determine whether PI4KB facilitates SARS-CoV entry via several stages or via one specific stage, the ACE2 protein on the cell surface was characterized by FACS. After siRNA transfection with control siRNA or PI kinase siRNA for 48 h, the fluorescence from ACE2 on the cell surface did not decrease compared with the control group, irrespective of the siRNA that was transfected (Fig. 5A), indicating that the level of cell surface ACE2 remained constant. In addition, total ACE2 protein levels in cells were not affected by the inhibition of PI kinases, although the synthesis of the PI4KB protein was severely inhibited after transfection of PI4KB siRNA (Fig. 5B). In addition, we compared the binding of increasing quantities of a recombinant SARS-CoV RBR (S318–510) to siRNA-treated cells. Consistent with the above results, there was no significant difference in RBR binding between control siRNA cells or those treated with PI kinase siRNAs (Fig. 5C). To confirm these results, the binding of SARS-CoV S to VeroE6 cells was quantified by Western blot analysis. Equivalent pseudovirus binding was observed in DMSO-, LY294002- or wortmannin-treated VeroE6 cells at 4 °C (Fig. 5D). Furthermore, we detected equivalent quantities of internalized SARS-CoV pseudovirions in VeroE6 cells at 37 °C before and after LY294002 or wortmannin treatment (Fig. 5D). The association of SARS-CoV S with cells was specific because neutralizing antibody treatment against SARS coronavirus spike protein almost completely inhibited SARS-CoV S binding and internalization (Fig. 5D). Therefore, our results suggest that PI4KB plays a role after virus binding and internalization.
FIGURE 5.
PI4KB facilitates SARS-CoV S-mediated entry after virus internalization into cells. A, flow cytometric analysis of surface levels of ACE2 (gray histograms) in siRNA-transfected VeroE6 cells. Background fluorescence levels (black histograms) were determined by labeling cells with an irrelevant isotype-matched control antibody. B, VeroE6 cells were transfected with the indicated siRNAs, and total cell lysates were analyzed by Western blotting with anti-PI4KB antibodies, anti-ACE2 antibodies, and anti-β-actin antibodies. C, increasing concentrations of recombinant SARS-CoV RBR (S318–510Fc) or control Fc (10 μm) were bound to VeroE6 siRNA cell lines and analyzed by Western blotting. D, VeroE6 cells were pretreated with DMSO (control), LY294002 (30 μm), or wortmannin (100 nm) for 30 min, and SARS-CoV S was added to the cells. After 2 h at 4 °C, the quantity of bound virus was determined by Western blot using anti-SARS spike antibodies or warming to 37 °C to allow the virus to internalize for another 2 h. Uninternalized virus was removed by treatment with 1 mg/ml proteinase K for 10 min. The internalized virus was assayed by Western blot using anti-SARS spike antibodies. For neutralization experiments, SARS-CoV S pseudovirus was preincubated with anti-SARS spike neutralizing antibody (NA) or an irrelevant control antibody (NA control) and then used for binding or internalization assays.
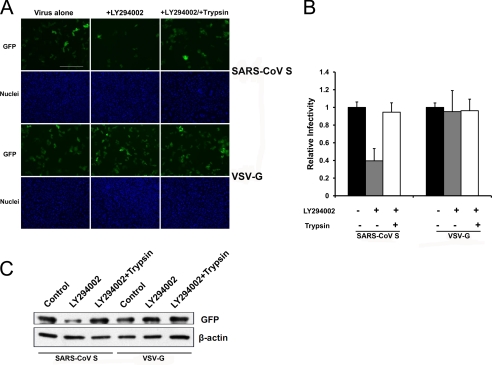
Trypsin Treatment Bypasses LY294002 Restriction of Entry Mediated by SARS-CoV S
After virus binding and internalization, SARS-CoV must fuse with the membrane of the late endosome. Previous studies have shown that cathepsin L is sufficient to mediate the fusion process and that the addition of exogenous trypsin to cell surface-bound virions can bypass the cathepsin L-dependent entry of SARS-CoV (18). Trypsin promotes fusion at or near the plasma membrane. To localize PI4KB-mediated virus entry, we investigated the effect of trypsin treatment on SARS pseudovirion entry into PI4KB-inhibited cells. VeroE6 cells pre-incubated with or without LY294002 were infected with SARS-CoV S or VSV-G pseudovirions. As expected, LY294002 treatment restricted SARS-CoV S-mediated entry but not entry of VSV-G. In contrast, when SARS-CoV S was bound to cells at 4 °C and then incubated briefly with TPCK-trypsin, entry into LY294002-treated cells was restored (Fig. 6A). The percentage of GFP-positive cells and total GFP expression confirmed our observations (Fig. 6, B and C). These data suggest that PI4KB inhibition cannot restrict trypsin-induced fusion at or near the plasma membrane and that PI4KB functions before the viral spike proteins are cleaved by cathepsin L in acidic cellular compartments. Taken together, these data indicate that PI4KB is not required for virus binding or internalization but is required at or before virus fusion.
FIGURE 6.
TPCK-trypsin treatment bypasses LY294002 restriction of SARS-CoV S-mediated entry. A, VeroE6 cells were treated with LY294002 (30 μm) or DMSO (control). After 30 min, the cells were spin-infected at 4 °C with the indicated pseudoviruses and then treated with trypsin or PBS. After 30 °C for 13 min, infected cells were maintained in growth medium for 48 h. The cells were fixed, and nuclei were stained with Hoechst 33342. Images were captured with a fluorescence microscope. Scale bar, 400 μm. B, statistical analysis of the proportion of GFP-positive cells in A.
DISCUSSION
SARS-CoV entry into target cells is a unique and complex process (30). Currently, several cellular factors have been demonstrated to be involved with SARS-CoV entry, including TNF-α-converting enzyme, cathepsin L, and IFITM (19, 32, 33). To our knowledge, TNF-α-converting enzyme is activated during SARS-CoV entry and modulates viral entry by inducing ACE2 receptor shedding, whereas the cathepsin L and IFITM-mediated restrictions are localized to late stages of the endocytic pathway. Cathepsin L and IFITM are intracellular factors that play opposing roles in SARS-CoV entry. Cathepsin L activates the membrane fusion function of the SARS-CoV spike protein, whereas IFITM inhibits that process. These two factors are both independent of viral receptor expression. Apart from this knowledge, little is known about the cellular factors involved in SARS-CoV entry.
This study is the first to implicate PI4KB and the PI4P microenvironment in SARS-CoV entry. Recently, the independent functions of the PI4P lipids, the most abundant monophosphorylated inositol phospholipids in mammalian cells, have been described, showing that PI4P lipids can alter local membrane curvature and regulate selective autophagy and endoplasmic reticulum exit site biogenesis (1). In addition, PI4P lipids can specifically bind to some host proteins (1). Moreover, the relationship between phosphatidylinositol 4-kinase and PI4P in the lipid microenvironment of a virus is gradually being revealed. For example, both enteroviruses and flaviviruses exploit host phosphatidylinositol 4-kinase enzymes and replicate their respective viral RNAs on PI4P lipid-enriched membranes (5, 10). More recently, Reiss et al. (8) found that elevated levels of the PI4KA product PI4P, which is detected in HCV-infected cultured hepatocytes and liver tissues from chronic hepatitis C patients, exhibited enzymatic activity via PI4KA that was critical for HCV replication. HCV NS5A was found to interact with PI4KA and stimulate its kinase activity. In our study, we examined the contradictory roles of PI3KR1 and PI4KB, and we investigated the mechanisms of SARS-CoV S-mediated entry to determine that PI4P is an indispensable element for entry. Interestingly, this hypothesis was demonstrated by mimicking a depleted PI4P microenvironment by transiently introducing Sac1 into VeroE6 cells. However, it is still not clear whether other RNA viruses, in addition to SARS-CoV, share similar intracellular molecular events that are important for viral entry.
An important event during endocytosis of SARS-CoV is cathepsin L proteolysis, which activates the fusion potential of the spike protein in late endosomes (18). Previous studies have shown that trypsin treatment can mediate virus and cell fusion directly at or near the plasma membrane, thereby bypassing the dependence on cathepsin L for entry (35). In our experiments, trypsin could also bypass the inhibition by LY294002, suggesting that PI4KB assists SARS-CoV entry before virions encounter cathepsin L.
Previous studies have shown that SARS-CoV entry into cells utilizes a novel endocytic pathway and that its entry is not dependent on clathrin or caveolin-mediated endocytosis (36, 37). Therefore, some specific cellular factors may provide important clues to elucidate this novel entry pathway. PI4KB may be a key factor involved in SARS-CoV entry, although currently, limited evidence is available to identify the virus entry stage or the cellular organelle where PI4KB is present and participates in SARS-CoV entry. In consideration of the crucial roles of cathepsin L, IFITM, and PI4KB in SARS-CoV entry, further work is required to determine how these intracellular factors mediate SARS-CoV entry at each step and whether they interact.
Interestingly, in this study, PI3KR1, the p85α subunit of PI3K, may function as an inhibitor of SARS-CoV S-mediated entry at an early stage. Whether the other subunits of PI3K function in the same way as p85α remains to be determined. Because there are only two forms of PI3K regulatory subunits and the p85α subunit accounts for the majority of its activity, we believe that a knockdown of p85α would inhibit most PI3K functions. However, we were unable to determine whether the p85β subunit was also involved in SARS-CoV entry; therefore, further research is needed.
In this study, we propose the following model for SARS coronavirus spike-mediated entry (Fig. 7). PI4KB facilitates SARS-CoV S-mediated entry without affecting the ACE2 receptor, whereas PI3KR1 plays an opposing role, with both regulating the PI4P microenvironment to promote entry at or before cathepsin L-mediated virus fusion and potentially further replication. Future work will be necessary to determine the other factors, including host cell proteins, that are involved in this process and to determine how the PI4P microenvironment initiates the next steps for replication.
FIGURE 7.
Model of the PI kinases and PI4P involved in SARS-CoV S-mediated entry. After SARS-CoV binds to ACE2, PI4P, the catalytic product of PI4KB, creates a lipid microenvironment or PI4P-enriched organelle required for the steps leading to fusion. Pharmacological inhibitors of PI4KB, such as LY294002 or wortmannin, suppress PI4KB activity and thereby inhibit SARS-CoV S-mediated entry. Cellular factors, such as PI3Ks or Sac1, that negatively regulate PI4P generation can also inhibit SARS-CoV S-mediated entry.
Based on these discoveries of cell-SARS-CoV interactions, especially the SARS-CoV entry process, several types of inhibitors have been implicated for SARS treatment. TNF-α-converting enzyme antagonists that block ACE2 shedding are candidate antiviral compounds (31). Small molecule oxocarbazate inhibitors of human cathepsin L have also been shown to block SARS pseudotyped virus infection in HEK293T cells (34). In our study, PI4KB is identified as a key factor involved in SARS-CoV S-mediated entry, which may provide a potential target for SARS-CoV treatment in the future. Furthermore, small molecules targeting PI4KB may provide a basis for the design of new classes of therapeutics against SARS-CoV infection.
Acknowledgments
We thank Michael Farzan (Harvard Medical School) for providing the Sh-2 plasmid and Peter Mayinger (Oregon Health and Science University) for the generous gift of the Sac1 plasmid.
This work was supported by Ministry of Science and Technology of China Grant 2009CB522105 and Ministry of Education 111 Project Grant B08007.
- PI
- phosphoinositide
- RBR
- receptor-binding region
- VSV
- vesicular stomatitis virus
- HCV
- hepatitis C virus
- PI4KB
- phosphatidylinositol 4-kinase IIIβ
- SARS-CoV
- SARS coronavirus
- SARS-CoV S
- SARS-CoV spike
- TPCK
- l-1-tosylamido-2-phenylethyl chloromethyl ketone
- PI4P
- phosphatidylinositol 4-phosphate
- IFITM
- interferon-inducible transmembrane protein.
REFERENCES
- 1. Vicinanza M., D'Angelo G., Di Campli A., De Matteis M. A. (2008) Function and dysfunction of the PI system in membrane trafficking. EMBO J. 27, 2457–2470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Saeed M. F., Kolokoltsov A. A., Freiberg A. N., Holbrook M. R., Davey R. A. (2008) Phosphoinositide 3-kinase-Akt pathway controls cellular entry of Ebola virus. PLoS Pathog. 29, e1000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dunn E. F., Connor J. H. (2011) Dominant inhibition of Akt/protein kinase B signaling by the matrix protein of a negative-strand RNA virus. J. Virol. 85, 422–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ehrhardt C., Wolff T., Pleschka S., Planz O., Beermann W., Bode J. G., Schmolke M., Ludwig S. (2007) Influenza A virus NS1 protein activates the PI3K/Akt pathway to mediate antiapoptotic signaling responses. J. Virol. 81, 3058–3067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Berger K. L., Cooper J. D., Heaton N. S., Yoon R., Oakland T. E., Jordan T. X., Mateu G., Grakoui A., Randall G. (2009) Roles for endocytic trafficking and phosphatidylinositol 4-kinase IIIα in hepatitis C virus replication. Proc. Natl. Acad. Sci. U.S.A. 106, 7577–7582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Trotard M., Lepère-Douard C., Régeard M., Piquet-Pellorce C., Lavillette D., Cosset F. L., Gripon P., Le Seyec J. (2009) Kinases required in hepatitis C virus entry and replication highlighted by small interference RNA screening. FASEB J. 23, 3780–3789 [DOI] [PubMed] [Google Scholar]
- 7. Tai A. W., Benita Y., Peng L. F., Kim S. S., Sakamoto N., Xavier R. J., Chung R. T. (2009) A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host Microbe 5, 298–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Reiss S., Rebhan I., Backes P., Romero-Brey I., Erfle H., Matula P., Kaderali L., Poenisch M., Blankenburg H., Hiet M. S., Longerich T., Diehl S., Ramirez F., Balla T., Rohr K., Kaul A., Bühler S., Pepperkok R., Lengauer T., Albrecht M., Eils R., Schirmacher P., Lohmann V., Bartenschlager R. (2011) Recruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartment. Cell Host Microbe 9, 32–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lim Y. S., Hwang S. B. (2011) Hepatitis C virus NS5A protein interacts with phosphatidylinositol 4-kinase type IIIα and regulates viral propagation. J. Biol. Chem. 286, 11290–11298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hsu N. Y., Ilnytska O., Belov G., Santiana M., Chen Y. H., Takvorian P. M., Pau C., van der Schaar H., Kaushik-Basu N., Balla T., Cameron C. E., Ehrenfeld E., van Kuppeveld F. J., Altan-Bonnet N. (2010) Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell 141, 799–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Drosten C., Günther S., Preiser W., van der Werf S., Brodt H. R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R. A., Berger A., Burguière A. M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J. C., Müller S., Rickerts V., Stürmer M., Vieth S., Klenk H. D., Osterhaus A. D., Schmitz H., Doerr H. W. (2003) Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348, 1967–1976 [DOI] [PubMed] [Google Scholar]
- 12. Kuba K., Imai Y., Rao S., Gao H., Guo F., Guan B., Huan Y., Yang P., Zhang Y., Deng W., Bao L., Zhang B., Liu G., Wang Z., Chappell M., Liu Y., Zheng D., Leibbrandt A., Wada T., Slutsky A. S., Liu D., Qin C., Jiang C., Penninger J. M. (2005) A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 11, 875–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li W., Moore M. J., Vasilieva N., Sui J., Wong S. K., Berne M. A., Somasundaran M., Sullivan J. L., Luzuriaga K., Greenough T. C., Choe H., Farzan M. (2003) Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426, 450–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ng M. L., Tan S. H., See E. E., Ooi E. E., Ling A. E. (2003) Early events of SARS coronavirus infection in Vero cells. J. Med. Virol. 71, 323–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Qinfen Z., Jinming C., Xiaojun H., Huanying Z., Jicheng H., Ling F., Kunpeng L., Jingqiang Z. (2004) The life cycle of SARS coronavirus in VeroE6 cells. J. Med. Virol. 73, 332–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Simmons G., Reeves J. D., Rennekamp A. J., Amberg S. M., Piefer A. J., Bates P. (2004) Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc. Natl. Acad. Sci. U.S.A. 101, 4240–4245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yang Z. Y., Huang Y., Ganesh L., Leung K., Kong W. P., Schwartz O., Subbarao K., Nabel G. J. (2004) pH-dependent entry of severe acute respiratory syndrome coronavirus is mediated by the spike glycoprotein and enhanced by dendritic cell transfer through DC-SIGN. J. Virol. 78, 5642–5650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Simmons G., Gosalia D. N., Rennekamp A. J., Reeves J. D., Diamond S. L., Bates P. (2005) Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. U.S.A. 102, 11876–11881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huang I. C., Bosch B. J., Li F., Li W., Lee K. H., Ghiran S., Vasilieva N., Dermody T. S., Harrison S. C., Dormitzer P. R., Farzan M., Rottier P. J., Choe H. (2006) SARS coronavirus, but not human coronavirus NL63, utilizes cathepsin L to infect ACE2-expressing cells. J. Biol. Chem. 281, 3198–3203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Moore M. J., Dorfman T., Li W., Wong S. K., Li Y., Kuhn J. H., Coderre J., Vasilieva N., Han Z., Greenough T. C., Farzan M., Choe H. (2004) Retroviruses pseudotyped with the severe acute respiratory syndrome coronavirus spike protein efficiently infect cells expressing angiotensin-converting enzyme 2. J. Virol. 78, 10628–10635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nègre D., Mangeot P. E., Duisit G., Blanchard S., Vidalain P. O., Leissner P., Winter A. J., Rabourdin-Combe C., Mehtali M., Moullier P., Darlix J. L., Cosset F. L. (2000) Characterization of novel safe lentiviral vectors derived from simian immunodeficiency virus (SIVmac251) that efficiently transduce mature human dendritic cells. Gene Ther. 7, 1613–1623 [DOI] [PubMed] [Google Scholar]
- 22. Fruman D. A., Meyers R. E., Cantley L. C. (1998) Phosphoinositide kinases. Annu. Rev. Biochem. 67, 481–507 [DOI] [PubMed] [Google Scholar]
- 23. Balla A., Balla T. (2006) Phosphatidylinositol 4-kinases. Old enzymes with emerging functions. Trends Cell Biol. 16, 351–361 [DOI] [PubMed] [Google Scholar]
- 24. Carpenter C. L., Duckworth B. C., Auger K. R., Cohen B., Schaffhausen B. S., Cantley L. C. (1990) Purification and characterization of phosphoinositide 3-kinase from rat liver. J. Biol. Chem. 265, 19704–19711 [PubMed] [Google Scholar]
- 25. D'Angelo G., Vicinanza M., Di Campli A., De Matteis M. A. (2008) The multiple roles of PtdIns(4)P, Not just the precursor of PtdIns(4,5)P2. J. Cell Sci. 121, 1955–1963 [DOI] [PubMed] [Google Scholar]
- 26. Foster F. M., Traer C. J., Abraham S. M., Fry M. J. (2003) The phosphoinositide (PI) 3-kinase family. J. Cell Sci. 116, 3037–3040 [DOI] [PubMed] [Google Scholar]
- 27. Blagoveshchenskaya A., Cheong F. Y., Rohde H. M., Glover G., Knödler A., Nicolson T., Boehmelt G., Mayinger P. (2008) Integration of Golgi trafficking and growth factor signaling by the lipid phosphatase SAC1. J. Cell Biol. 180, 803–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Leistner C. M., Gruen-Bernhard S., Glebe D. (2008) Role of glycosaminoglycans for binding and infection of hepatitis B virus. Cell. Microbiol. 10, 122–133 [DOI] [PubMed] [Google Scholar]
- 29. Marsh M., Helenius A. (2006) Virus entry. Open sesame. Cell 124, 729–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lang J., Yang N., Deng J., Liu K., Yang P., Zhang G., Jiang C. (2011) Inhibition of SARS pseudovirus cell entry by lactoferrin binding to heparan sulfate proteoglycans. PLoS One 6, e23710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Haga S., Nagata N., Okamura T., Yamamoto N., Sata T., Yamamoto N., Sasazuki T., Ishizaka Y. (2010) TACE antagonists blocking ACE2 shedding caused by the spike protein of SARS-CoV are candidate antiviral compounds. Antiviral Res. 85, 551–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Huang I. C., Bailey C. C., Weyer J. L., Radoshitzky S. R., Becker M. M., Chiang J. J., Brass A. L., Ahmed A. A., Chi X., Dong L., Longobardi L. E., Boltz D., Kuhn J. H., Elledge S. J., Bavari S., Denison M. R., Choe H., Farzan M. (2011) Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog. 7, e1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Haga S., Yamamoto N., Nakai-Murakami C., Osawa Y., Tokunaga K., Sata T., Yamamoto N., Sasazuki T., Ishizaka Y. (2008) Modulation of TNF-α-converting enzyme by the spike protein of SARS-CoV and ACE2 induces TNF-α production and facilitates viral entry. Proc. Natl. Acad. Sci. U.S.A. 105, 7809–7814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shah P. P., Wang T., Kaletsky R. L., Myers M. C., Purvis J. E., Jing H., Huryn D. M., Greenbaum D. C., Smith A. B., 3rd, Bates P., Diamond S. L. (2010) A small molecule oxocarbazate inhibitor of human cathepsin L blocks severe acute respiratory syndrome and Ebola pseudotype virus infection into human embryonic kidney 293T cells. Mol. Pharmacol. 78, 319–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bosch B. J., Bartelink W., Rottier P. J. (2008) Cathepsin L functionally cleaves the severe acute respiratory syndrome coronavirus class I fusion protein upstream of rather than adjacent to the fusion peptide. J. Virol. 82, 8887–8890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang H., Yang P., Liu K., Guo F., Zhang Y., Zhang G., Jiang C. (2008) SARS coronavirus entry into host cells through a novel clathrin- and caveolae-independent endocytic pathway. Cell Res. 18, 290–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang S., Guo F., Liu K., Wang H., Rao S., Yang P., Jiang C. (2008) Endocytosis of the receptor-binding domain of SARS-CoV spike protein together with virus receptor ACE2. Virus Res. 136, 8–15 [DOI] [PMC free article] [PubMed] [Google Scholar]