Background: Icsbp represses transcription of PTPN13 (the gene encoding Fap1) and loss of Icsbp induces Fap1-dependent Fas resistance.
Results: Tel and Hdac3 cooperate with Icsbp for PTPN13 repression, but this activity is inhibited by interaction of Tel with Tel-PdgfRβ.
Conclusion: Tel-PdgfRβ antagonizes the effects of Tel and Icsbp on PTPN13 transcription.
Significance: Tyrosine kinase activity of Tel-PdgfRβ is not required for induction of Fap1-dependent Fas resistance.
Keywords: Cell Differentiation, Gene Transcription, Leukemia, Myeloid Cell, Oncogene
Abstract
Icsbp is an interferon regulatory transcription factor with leukemia suppressor activity. In previous studies, we identified the gene encoding Fas-associated phosphatase 1 (Fap1; the PTPN13 gene) as an Icsbp target. In the current study, we determine that repression of PTPN13 by Icsbp requires cooperation with Tel and histone deacetylase 3 (Hdac3). These factors form a multiprotein complex that requires pre-binding of Tel to the PTPN13 cis element with subsequent recruitment of Icsbp and Hdac3. We found that knockdown of Tel or Hdac3 in myeloid cells increases Fap1 expression and results in Fap1-dependent resistance to Fas-induced apoptosis. The TEL gene was initially identified due to involvement in leukemia-associated chromosomal translocations. The first identified TEL translocation partner was the gene encoding platelet-derived growth factor receptor β (PdgfRβ). The resulting Tel-PdgfRβ fusion protein exhibits constitutive tyrosine kinase activity and influences cellular proliferation. In the current studies, we find that Tel-PdgfRβ influences apoptosis in a manner that is independent of tyrosine kinase activity. We found that Tel-PdgfRβ expressing myeloid cells have increased Fap1 expression and Fap1-dependent Fas resistance. We determined that interaction between Tel and Tel-PdgfRβ decreases Tel/Icsbp/Hdac3 binding to the PTPN13 cis element, resulting in increased transcription. Therefore, these studies identify a novel mechanism by which the Tel-PdgfRβ oncoprotein may contribute to leukemogenesis.
Introduction
Fas-associated phosphatase 1 (Fap1)2 is a protein-tyrosine phosphatase (PTP) that is encoded by the PTPN13 gene (1). Fas is a Fap1 substrate, and interaction between Fap1 and the Fas C terminus results in de-phosphorylation and inhibition of Fas (2, 3). In acute myeloid leukemia, increased expression of Fap1 correlates with resistance to Fas-induced apoptosis and decreased response to chemotherapeutic agents (4, 5). Fap1-dependent Fas resistance may also be involved in persistence of the leukemia stem cell clone during treatment of chronic myeloid leukemia (CML) (6).
In previous investigations, we found that the interferon consensus sequence binding protein (Icsbp, also known as interferon regulatory factor 8; Irf8) repressed PTPN13 transcription (7). We determined that Icsbp-induced PTPN13 repression occurred in granulocyte/monocyte progenitor cells, and that this activity increased during myeloid differentiation (7). We found that tyrosine-phosphorylated Icsbp had a greater affinity for the PTPN13 cis element and was a more efficient repressor of transcription (7). Because Icsbp becomes increasingly tyrosine phosphorylated as differentiation proceeds, this provided a mechanism for decreased Fap1 expression during myelopoiesis (8, 9).
We found that decreased Fap1 expression in differentiating phagocytic cells resulted in increased sensitivity to Fas-induced apoptosis (7). Conversely, we found that increased Fap1 expression in myeloid cells with knockdown or knockout of Icsbp resulted in Fap1-dependent Fas resistance (7).
In clinical studies, decreased Icsbp expression was found in the bone marrow of human subjects with uncontrolled CML, during progression of CML to blast crisis, and in the majority of de novo acute myeloid leukemia (10–12). Therefore, decreased Icsbp expression provided a potential mechanism for increased Fap1 in acute myeloid leukemia and CML. In vivo studies in Icsbp knock-out mice and murine CML models demonstrated that bone marrow progenitor cells with decreased Icsbp expression were hypersensitive to cytokine-induced proliferation and survival in comparison to normal progenitor cells (13, 14). We identified a number of target genes that may contribute to Icsbp leukemia suppression activity, including PTPN13. For example, because Icsbp activates genes encoding phagocyte effector proteins (15, 16), decreased Icsbp expression results in differentiation block. Icsbp activates transcription of the gene encoding Neurofibromin 1 (a Ras-Gap), resulting in a sustained proliferative response to cytokines in Icsbp-deficient cells (9, 16–19). Therefore, clarifying mechanisms by which Icsbp regulates gene transcription is important to understanding molecular mechanisms of leukemogenesis.
Icsbp activates some target genes in myeloid progenitor cells and other target genes in mature phagocytes. Icsbp also represses some target genes, and repression activity increases or decreases during differentiation in a gene-specific manner (9, 15, 16, 18, 19). Understanding the leukemia suppressor effect of Icsbp therefore involves better understanding of mechanisms by which Icsbp regulates various genes. Because Icsbp binds to all identified target genes as a member of a multiprotein complex, this involves identifying differences in protein-partnering and binding site selection.
In the current studies, we identified a multiprotein complex that is necessary for Icsbp-induced PTPN13 repression. This repression complex includes Tel and histone deacetylase 3 (Hdac3). Tel is an ets protein that was first identified because the TEL gene is involved in leukemia-associated chromosomal translocations (20). Subsequent studies determined that Tel interacts with Irf proteins, including Icsbp, and represses transcription of various target genes during normal myelopoiesis. Transcriptional repression of previously identified target genes by the Tel-Irf complex involved recruitment of histone deacetylase activity (21, 22).
We considered the possibility that fusion proteins generated by leukemia-associated TEL translocations might alter target gene regulation by Tel. One such translocation involves TEL and the gene encoding platelet-derived growth factor receptor β (PdgfRβ) (20). This translocation was identified in subjects with chronic myelomonocytic leukemia (CMMoL); a myeloproliferative/dysplastic disorder (20). Other investigators demonstrated that the sole expressed transcript resulting from this translocation includes the N terminus of Tel and the C terminus of PdgfRβ; neither mRNA nor protein representing the reciprocal fusion gene were found (20). Wild type Tel is also expressed in these cells from the nonmutated allele.
Tel-PdgfRβ fusion protein includes the basic helix loop helix domain from Tel, but does not include the Tel DNA-binding domain. The fusion protein also includes the transmembrane and kinase domains of PdgfRβ, but not the extracellular domain (20). Additional studies determined that homodimerization of Tel-PdgfRβ through Tel-basic helix loop helix domains resulted in constitutive tyrosine kinase activity (23). Dimerization between Tel-PdgfRβ and the product of the normal Tel allele was also observed (20, 23). It is additionally possible that Tel-PdgfRβ could interact with other normal Tel protein partners, such as Irf proteins, although this has not been investigated.
In the current study, we found that expression of Tel-PdgfRβ in myeloid cells increased Fap1 expression and induced Fas resistance. We found that this was due to decreased binding of the Tel-Icsbp-Hdac3 complex to the PTPN13 cis element in Tel-PdgfRβ expressing cells. Therefore, these studies identified a mechanism, independent of Tel-PdgfRβ tyrosine kinase activity, by which this fusion protein may influence the pathogenesis of leukemia.
MATERIALS AND METHODS
Plasmids
Protein Expression Vectors
The Icsbp cDNA was obtained from Dr. Ben Zion-Levi (Technion, Haifa, Israel), the full-length cDNA was generated by PCR and subcloned into the mammalian expression vector pcDNA and the pMSCVpuro retroviral vector (Stratagene, La Jolla, CA) (24). The cDNAs for Tel and Hdac3 were generated using mRNA isolated from U937 cells by reverse transcription followed by PCR. Sequence analysis was performed to verify identity with published sequences and the cDNAs were subcloned into expression vectors. The Tel-PdgfRβ cDNA was a generous gift from Dr. Jonathan Licht (Northwestern University, Chicago, IL). A kinase-inactive form of Tel-PdgfRβ was very kindly provided by Dr. Jean-Baptiste Demoulin (De Duve Institute, University of Louvain, Brussels) (25).
shRNA Expression Vectors
shRNAs and scrambled control sequences for Icsbp, Tel, and Hdac3 were designed with the assistance of the Promega website. Double-stranded oligonucleotides representing the complementary sequences separated by a hairpin loop were subcloned into the pLKO.1puro vector (a gift from Dr. Kathy Rundell, Northwestern University). Several sequences were tested and the most efficient for suppression were used individually or in combinations, as previously reported (22).
PTPN13 Reporter Constructs
Oligonucleotides were synthesized with 3 copies of the −587 to −627 bp sequence from the PTPN13 promoter (previously identified Icsbp-binding site) (7). This double-stranded oligonucleotide was subcloned into a minimal promoter/luciferase reporter vector (pGL3-promoter, Stratagene). Reporter constructs were also generated with 2.0 kb or 500 bp of PTPN13 5′ flank. These constructs were described previously (7).
Oligonucleotides
Oligonucleotides were custom synthesized by MWG Biotech (Piedmont, NC). Double-stranded oligonucleotides used in DNA affinity purification experiments represented the −587 to −627-bp sequence of the PTPN13 promoter (5′-CTCCCGGAGTCTGTTTCTAATTTCTGCAAATGATTGTGG-3′: the ISRE-like sequence is underlined), or a mutant form of the PRDI-consensus (which also binds ets/Irf heterodimers), which does not bind Icsbp (5′-TGTCTTTGTCTTTGTCTT-3′).
Myeloid Cell Line Culture
The human myelomonocytic leukemia cell line U937 (26) was obtained from Andrew Kraft (Hollings Cancer Center, Medical University of South Carolina, Charleston, SC). The human myeloblastic cell line KG1 was obtained from Dr. Leonidas Platanias (Robert H. Lurie Comprehensive Cancer Center, Northwestern University). Cells were maintained as described (7).
Murine Bone Marrow Culture
Animal studies were approved by the Animal Care and Use Committees of Northwestern University and Jesse Brown Veterans Affairs Medical Center. Bone marrow mononuclear cells were obtained from the femurs of WT C57/BL6 mice. Sca1+ cells were separated using the Miltenyi magnetic bead system (Miltenyi Biotechnology, Auburn, CA). Bi-potential myeloid progenitor cells (granulocyte monocyte progenitors) were cultured (at 2 × 105 cells/ml) in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 1% penicillin-streptomycin, 20 ng/ml of murine GM-CSF (R & D Systems Inc., Minneapolis, MN), 100 ng/ml of stem cell factor (SCF, R & D Systems Inc.), and 10 ng/ml of murine recombinant IL3 (R & D Systems Inc.). Cells were maintained in GM-CSF/SCF/IL3 for 48 h, or differentiated over 48 h in 10 ng/ml of G-CSF or M-CSF, as described (7, 22). Some myeloid progenitors were transduced with a retroviral vector to express Tel-specific shRNAs, Hdac3-specific shRNAs, or scrambled control shRNAs, as described (22).
Chromatin Co-immunoprecipitation
U937 cells were incubated briefly in medium supplemented with formaldehyde and cell lysates were sonicated to generate chromatin fragments with an average size of <200 bp (27). Lysates were immunoprecipitated with antibody to Tel or irrelevant control antibody, as described (22).
Quantitative Real Time PCR
Determining mRNA Expression
RNA was isolated using the TRIzol reagent (Invitrogen) and tested for integrity by denaturing gel electrophoresis. Primers were designed with Applied Biosystems software and real time PCR was performed using SYBR Green according to the “standard curve” method. Results were normalized to 18 S and are reported as mRNA abundance relative to abundance in the cDNA sample used for standard curve.
Quantifying Chromatin Immunoprecipitation
Chromatin that co-precipitated with Tel antibody or preimmune serum was amplified with primers flanking the Icsbp-binding cis element in the PTPN13 promoter (−669 to −646: 5′-GTCGTGCTTGCACAGCTCCGCTCT-3′ and −512 to −489: 5′-ACCTTGCATCAGACAGTGTCTCTC-3′). Results were normalized to PCR with total, nonprecipitated chromatin to control for differences in DNA abundance between samples.
Myeloid Cell Line Transfections and Reporter Gene Assays
Stable Transfectant Cell Lines
U937 cells were transfected by electroporation with vectors to express Icsbp, Tel, Hdac3, Icsbp + Tel + Hdac3, or empty pcDNAamp vector plus a vector with a neomycin phosphotransferase cassette (pSRα) (30 μg each). U937 or KG1 cells were also transfected by electroporation with a construct to express Tel-PdgfRβ or empty pcDNAamp control plus pSRα (30 μg each). Stable pools of cells were selected in G418 (0.5 mg/ml) and aliquots were tested for Icsbp, Tel, and Hdac3 expression by real time PCR and Western blot.
Transient Transfections for Reporter Gene Assays
U937 cells (32 × 106/ml) were transfected with a vector to express various combinations of Icsbp, Tel, HDAC3, Tel-PdgfRβ, a kinase inactive form of Tel-PdgfRβ, Tel-specific shRNAs, Hdac3-specific shRNAs, or relevant control vectors and minimal promoter/firefly luciferase reporter constructs with 3 copies of the −587 to −627 bp PTPN13 sequence (ptpn13GL3-p) or minimal promoter/reporter control (p-GL3-p) (70 μg). Cells were co-transfected with a CMV/Renilla luciferase vector to control for transfection efficiency. Some cells were analyzed after treatment with imatinib (IM) (at 1 μm).
Transfectants were assayed for firefly and Renilla luciferase expression after 48 h, as described (7, 22). Results of Renilla assays were used to normalize firefly luciferase values between samples and between experiments. Luciferase assay results are reported in light intensity units.
Apoptosis Assays
Cells were incubated with Fas-agonist antibody (CH11) or control antibody, as described (28). In some experiments, cells were incubated for the last 12 h with SLV peptide (or control VLS peptide) to disrupt the Fas/Fap-1 interaction (29). Apoptosis assays were performed using annexin V/propidium iodide double staining. The cells were washed with culture medium, adjusted to a concentration of 1 × 106 cells/ml, incubated with Annexin-V/FITC solution (2.5 μg/ml) and propidium iodide (12.5 μg/ml) on ice for 15 min, and analyzed on a BD Biosciences FACScan flow cytometry (Cambridge, MA). Results are reported as % apoptotic cells out of total cells.
Immunoprecipitation and Western Blots
Western Blots of Lysates Proteins
Cells were lysed by boiling in 2× SDS sample buffer. Lysate proteins (50 μg) were separated by SDS-PAGE (8% acrylamide) and transferred to nitrocellulose. Western blots were serially probed with various antibodies plus GAPDH or tubulin (to control for loading). Each experiment was repeated at least three times with different batches of lysate proteins. A representative blot is shown.
Immunoprecipitation and Western Blots
Lysate proteins were isolated from control and Tel-PdgfRβ KG1 transfectants and immunoprecipitated under denaturing conditions with an anti-phosphotyrosine (phospho-Tyr) antibody (clone 4G10, Upstate Biotechnology, Charlottesville, VA) or irrelevant control antibody. Precipitated proteins were collected with Staph protein A-agarose, separated by SDS-PAGE, and transferred to nitrocellulose. Western blots were probed with anti-Icsbp antibody.
In other experiments, lysate proteins were immunoprecipitated under nondenaturing conditions with an antibody to PdgfRβ or irrelevant control antibody. Western blots of precipitated proteins were probed with antibodies to Icsbp, Tel, Hdac3, and PdgfRβ (from Santa Cruz Biotechnology, Santa Cruz, CA). Each experiment was repeated at least three times with different batches of lysate proteins. A representative blot is shown.
Isolation of Nuclear Proteins
Nuclear proteins were extracted from U937 cells as described in previous studies (7, 22).
In Vitro DNA-binding Assays
DNA Affinity Purification Assays
Nuclear proteins (300 μg) were incubated with biotin-labeled double-stranded oligonucleotide probe representing the −587 to −627 bp PTPN13 promoter sequence or a non-Icsbp-binding PRDI mutant sequence overnight in DNA affinity purification assay buffer (25 mm HEPES (pH 7.6), 60 mm KCl, 5 mm MgCl2, 7.5% glycerol, 0.1 mm EDTA, 1 mm DTT, and 0.25% Triton X-100). The DNA-protein complexes were precipitated with 50 μl of a 50% slurry of neutravidin-coated agarose beads (Pierce). Proteins bound to the beads were eluted, separated by SDS-PAGE (10% acrylamide), and transferred to nitrocellulose. Western blots were probed with an antibody to Icsbp, Tel, Hdac3, or PdgfRβ.
Statistical Analysis
Statistical significance was determined by Student's t test and analysis of variance methods using SigmaPlot and SigmaStat software. Statistical significance is reported as standard error. For transfection, apoptosis, and real time PCR experiments, n values represent the number of independent experiments, each of which was performed in triplicate. Differences are considered statistically significant at p < 0.05 (95% confidence interval). Bars that are not statistically significantly different are indicated by a lowercase letter in all figures.
RESULTS
Icsbp, Tel, and Hdac3 Cooperate to Repress a PTPN13 Promoter cis Element
In previous investigations, we identified a cis element in the PTPN13 promoter located between −587 and −627 bp, repressed by Icsbp, and homologous to the Irf/ets DNA-binding consensus sequence (7, 30). We demonstrated binding of Icsbp to this cis element in vitro and in vivo (7). In the current investigations, we generated an artificial promoter/reporter plasmid with three copies of this PTPN13 cis element (referred to as ptpn13GL3-p). In initial experiments, this construct (or empty pGL3-p control vector) was transfected into U937 myeloid leukemia cells along with a dose titration of a vector to express Icsbp (or empty control expression vector).
As anticipated, these studies demonstrated that overexpressed Icsbp induced a dose-dependent decrease in activity of the isolated PTPN13 cis element (Fig. 1A). We found that reporter expression in experiments with this PTPN13 cis element containing construct was decreased significantly in transfectants with 30 or 50 μg of Icsbp expression plasmid in comparison to transfectants with empty vector control (p = 0.0001, n = 7 and p = 0.000001, n = 7, respectively). In contrast, there was no significant decrease in PTPN13 cis element activity in transfectants with 10 μg of Icsbp expression plasmid in comparison to control transfectants (p = 0.4, n = 8). For all reporter gene assays, statistical significance is reported as standard error and n indicates the number of independent transfection experiments (performed in triplicate).
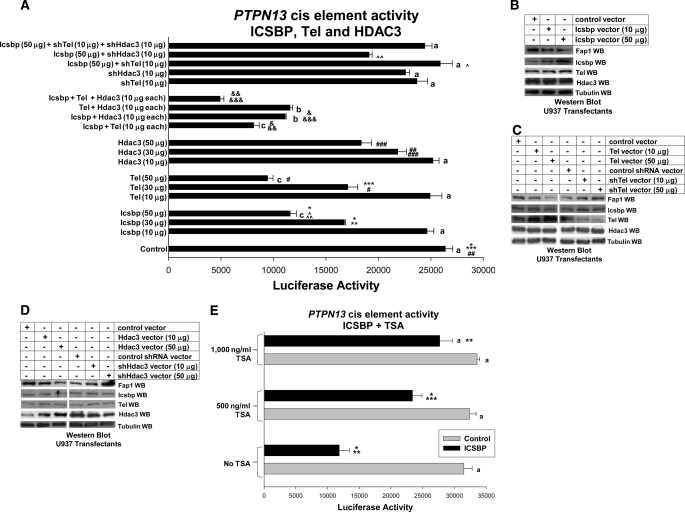
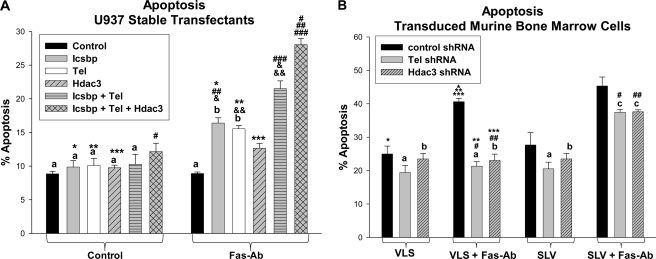
FIGURE 1.
Tel and Hdac3 cooperate with Icsbp to repress the Icsbp-binding cis element from the PTPN13 promoter. A, Icsbp, Tel, and Hdac3 cooperate to repress a PTPN13 cis element. U937 myeloid cells were co-transfected with a reporter vector with three copies of the Icsbp-binding PTPN13 cis element linked to a minimal promoter (ptpn13GL3-p) or with empty vector control (p-GL3p) along with various combinations of vectors to overexpress Icsbp, Tel, and Hdac3 (or empty expression vector, indicated as control). Other cells were co-transfected with these reporter constructs plus various combinations of vectors to express Icsbp (or empty vector control), and Tel- or Hdac3-specific shRNAs (or scrambled control shRNA). Activity of the empty p-GL3p plasmid was less than 10% of the activity of the ptpn13GL3-p construct and was subtracted as background. Activity of the PTPN13 cis element was not influenced by co-transfection with control expression vector or vectors expressing scrambled control shRNA, so the latter is not shown. Sets of experiments with reporter gene activity that is not significantly different (p > 0.2 for all comparisons) are indicated by a lowercase letter (i.e. all bars indicated by a are not significantly different, etc.). Statistically significant difference in reporter gene activity between pairs of experiments (p < 0.01 for all comparisons) are indicated by *, **, ***, #, ##, ###, &, &&, &&&, ∧ or ∧∧ (i.e. the bars labeled by * are significantly different from each other, etc.). B, overexpression of Icsbp decreases Fap1 expression in a dose-dependent manner. U937 myeloid cells were transfected with an Icsbp expression vector or with empty control vector, as indicated. Western blots of cell lysates were probed with antibodies to Icsbp, Tel, Hdac3, Fap1, or Gapdh (as a loading control). This blot represents transfectants performed at the same time as one of the reporter gene assays from A. C, overexpression or knockdown of Tel influences Fap1 expression. U937 myeloid cells were transfected with a Tel expression vector (or empty control vector), or a vector to express a Tel-specific shRNAs (or scrambled shRNA control vector). Western blots of cell lysates were probed with antibodies to Icsbp, Tel, Hdac3, Fap1, or Gapdh (as a loading control). This blot represents transfectants performed at the same time as one of the reporter gene assays from A. D, overexpression or knockdown of Hdac3 influences Fap1 expression. U937 myeloid cells were transfected with an Hdac3 expression vector (or empty control vector), or a vector to express an Hdac3-specific shRNAs (or scrambled shRNA control vector). Western blots of cell lysates were probed with antibodies to Icsbp, Tel, Hdac3, Fap1, or Gapdh (as a loading control). This blot represents transfectants performed at the same time as one of the reporter gene assays from A. E, the histone deacetylase inhibitor trichostatin A blocks repression of the Icsbp-binding PTPN13 promoter cis element. U937 cells were co-transfected with the PTPN13 cis element containing the reporter construct (as above) and a vector to overexpress Icsbp. Samples were treated with TSA. Reporter assays were performed and sets of experiments without a statistically significant difference (p > 0.2) are indicated by a lowercase a. Statistically significant differences in reporter activity (p < 0.01 for all comparisons) are indicated by *, **, or ***.
In control experiments, we found that overexpression of the Icsbp protein was increased by increasing amounts of Icsbp expression plasmid in the transfectants (Fig. 1B). This correlated with decreased expression of endogenous Fap1 protein. These experiments were performed using lysate proteins from reporter gene assays presented in Fig. 1A.
Because Icsbp cooperates with Tel and Hdac3 to repress other target genes, we tested the ability of Tel and Hdac3 to repress the PTPN13 cis element-containing reporter construct (22). We found that overexpressed Tel repressed this cis element in a dose-dependent manner (Fig. 1A). Activity of the PTPN13 cis element was significantly less in transfection experiments with 30 or 50 μg of Tel expression plasmid in comparison to transfectants with empty control expression vector (p = 0.0001, n = 8, and p < 0.0001, n = 8, respectively). However, PTPN13 cis element activity was not significantly repressed in transfectants with 10 μg of Tel expression plasmid (p = 0.8, n = 8 for comparison with control transfectants).
In control experiments, overexpression of Tel was increased in the transfectants with increasing doses of Tel expression plasmid (Fig. 1C). This correlated with decreased endogenous Fap1 protein expression in these cells. These experiments were performed with cell lysate proteins from reporter gene assays from Fig. 1A.
Repression of the PTPN13 cis element-containing construct by overexpressed Hdac3 was significant at the highest levels of Hdac3 expression plasmid tested (p = 0.00001, n = 8 for transfectants with 50 μg of Hdac3 expression plasmid versus control plasmid, and p = 0.04, n = 8 for transfectants with 30 μg of Hdac3 expression plasmid) (Fig. 1A). PTPN13 cis element activity was not significantly different in control transfectants versus transfectants with 10 μg of Hdac3 expression plasmid (p = 0.9, n = 8).
In control experiments, we found that increasing doses of Hdac3 expression plasmid in the transfectants resulted in increased Hdac3 expression (Fig. 1D). This correlated with decreased expression of endogenous Fap1 protein. These experiments were also performed with cell lysates from reporter gene assays presented in Fig. 1A.
To address the specificity of these results, we also tested activity of the PTPN13 cis element in transfection experiments with overexpressed Irf1 or Irf2, Pu.1 (an ets protein that also partners with Icsbp), and Hdac2. None of these proteins repressed the cis element (not shown).
We next investigated whether Icsbp, Tel, and Hdac3 cooperate to repress the Icsbp-binding PTPN13 cis element. For these experiments, U937 cells were co-transfected with PTPN13 cis element-containing reporter vector (or control) and vectors to overexpress both Icsbp and Tel. Because we were interested in cooperative rather than additive effects, amounts of Icsbp and Tel expression vectors were used that did not individually influence PTPN13 cis element activity (10 μg for each). We found that activity of the PTPN13 cis element was significantly less in cells co-expressing Icsbp + Tel at these levels in comparison to transfectants with empty expression vectors (p < 0.00001, n = 12) (Fig. 1A), suggesting a greater than additive effect (because 10 μg of either Icsbp or Tel plasmid did not alone alter PTPN13 cis element activity).
We performed similar experiments to determine whether Hdac3 cooperated with either Icsbp or Tel for repression of the PTPN13 cis element. Combinations of Hdac3, Icsbp, and Tel expression vectors were transfected into U937 cells using amounts of expression plasmid that did not individually repress the PTPN13 cis element. We found that activity of the PTPN13 cis element-containing construct was significantly less in transfectants co-expressing Hdac3 with either Icsbp or Tel in comparison to transfectants with control expression vectors (p < 0.000001, n = 12, and p < 0.00001, n = 11, respectively) (Fig. 1A). We also found that PTPN13 cis element activity was significantly less in transfectants co-overexpressing Icsbp + Tel + Hdac3 (10 μg each) in comparison to Icsbp + Tel (p = 0.0006, n = 12) or Icsbp + Hdac3 (p = 0.00001, n = 12) (Fig. 1A).
Because Tel and Hdac3 are constitutively expressed in U937 cells, we determined the effect of decreased expression of these proteins on repression of the PTPN13 cis element by Icsbp. This provided an additional method to examine cooperation. For these experiments, U937 cells were co-transfected with the PTPN13 cis element-containing reporter vector (or control vector) and various combinations of vectors to overexpress Icsbp (or vector control) and a vector to express Tel or Hdac3 specific shRNAs (or scrambled shRNAs).
A dose titration of Tel- and Hdac3-specific shRNA vectors was performed to identify the amount of vector that produced significant knockdown of the protein (Fig. 1, C and D), but did not influence activity of the PTPN13 cis element-containing construct (Fig. 1A). For these experiments, we used an amount of Icsbp expression vector that induced maximal PTPN13 cis element repression (50 μg). We found that PTPN13 cis element activity was significantly greater in transfectants with Icsbp overexpression and Tel knockdown, or Icsbp overexpression and Hdac3 knockdown in comparison to transfectants with Icsbp overexpression alone (p < 0.000001, n = 6 for both sets). Therefore, Icsbp-induced repression of the PTPN13 cis element was impaired by knockdown of either Tel or Hdac3.
In control experiments, expression of Tel protein was decreased in the transfectants with increasing doses of a Tel-specific shRNA expression plasmid and this correlated with increased endogenous Fap1 protein (Fig. 1C). Similarly, we found that increasing doses of an Hdac3-specific shRNA expression plasmid decreased Hdac3 expression and increased expression of endogenous Fap1 protein (Fig. 1D). These blots were performed with cell lysates from reporter gene assays presented in Fig. 1A.
These results suggested that Icsbp depends upon partner proteins for binding to and/or repression of the PTPN13 cis element. To determine whether histone deacetylase activity was required for PTPN13 repression, additional studies were performed using trichostatin A (TSA; a histone deacetylase inhibitor). U937 cells were co-transfected with the PTPN13 cis element-containing artificial promoter construct (ptpn13GL3-p; or control vector), and an Icsbp expression vector (or control expression vector), and analyzed with or without TSA treatment. We found that TSA reversed the repression activity of overexpressed Icsbp in a dose-dependant manner (Fig. 1E). The dose of TSA that was required to inhibit Hdac3 in these studies was consistent with our published investigations of repression of the GAS2 promoter by Icsbp, Tel, and Hdac3 in U937 cells (22).
For the experiments above (Fig. 1, A and D), activity of the empty pGL3-p reporter vector was less than 10% of the activity of the PTPN13 cis element-containing reporter vector. This control vector activity was not influenced by overexpression or knockdown of Tel, Icsbp, or Hdac3, or by treatment with TSA, and was subtracted as background. Activity of the transfected ptpn13GL3-p reporter plasmid was not altered by transfection with empty control expression vector or control vectors with various scrambled shRNAs. “Control” lanes in Fig. 1A represent transfection with empty pcDNA expression vector.
These studies suggested that Icsbp, Tel, and Hdac3 all influenced the same PTPN13 cis element. However, it was possible that this might be an artifact of the artificial promoter/reporter system used in these studies, and that Tel and Hdac3 might not influence activity of this cis element in the context of the intact promoter. To investigate this, additional transfection experiments were performed using a construct with 670 bp of the PTPN13 5′ flank. In previous studies, we demonstrated that 670 bp was the minimal PTPN13 promoter sequence that included the Icsbp-binding cis element (7).
Therefore, U937 cells were co-transfected with the 670-bp PTPN13 promoter construct (or empty reporter vector) and vectors to overexpress Tel (50 μg), Hdac3 (50 μg), or Icsbp + Tel + Hdac3 (10 μg each) (or empty expression vectors). These studies demonstrated repression by overexpressed Tel or Hdac3, consistent with repression of the isolated cis element by these proteins (Fig. 2A). Also similar to the studies with the isolated cis element, the three proteins together repressed the 670-bp promoter more efficiently than each of them individually (Fig. 2A). We found similar results in transfection experiments with a 2.0-kb PTPN13 promoter construct (not shown).
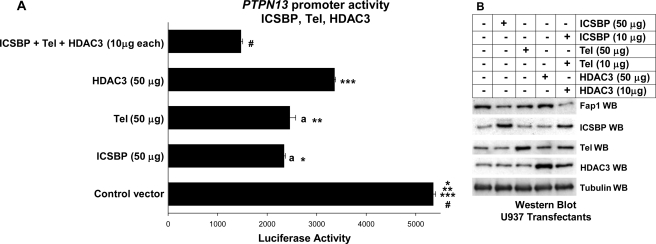
FIGURE 2.
Icsbp, Tel, and Hdac3 cooperate to repress the PTPN13 promoter. A, Icsbp, Tel, and Hdac3 cooperate to repress the PTPN13 promoter. U937 myeloid cells were co-transfected with a reporter vector with 670 bp of the PTPN13 promoter (ptpn13GL3) or with empty vector control (p-GL3) and a vector to express Icsbp (50 μg), Tel (50 μg), Hdac3 (50 μg), all three (10 μg each), or empty expression vector (indicated as control). Activity of the empty p-GL3 plasmid was ∼10% of the activity of the ptpn13GL3 construct and was subtracted as background. Results of reporter assays that are not statistically significantly different are indicated by lowercase a. Statistically significant differences in reporter gene activity are indicated by *, **, ***, or #. B, overexpression of the Icsbp, Tel, and Hdac3 decrease in Fap1 expression in a dose-dependent manner. U937 myeloid cells were transfected with vectors to express Icsbp, Tel, Hdac3, or all three together (or with empty expression vector). Western blots (WB) of cell lysates were probed with antibodies to Icsbp, Tel, Hdac3, Fap1, or Gapdh (as a loading control). This blot represents transfectants performed at the same time as one of the reporter gene assays from A.
In control experiments, we found that overexpression of Icsbp, Tel, or Hdac3 each decreased expression of endogenous Fap1 protein, and the three together had an even greater effect (Fig. 2B). These experiments were performed with cell lysate proteins from reporter gene assays from Fig. 2A. We found that the activity of empty reporter vector (pGL3) was less than 10% the activity of the PTPN13 promoter/reporter vector. Activity of the control vector was not influenced by overexpression of Tel, Icsbp, or Hdac3 and was subtracted as background.
Icsbp Binding to PTPN13 Cis Element Requires Tel
These studies demonstrated that Icsbp, Tel, and Hdac3 cooperate to repress the PTPN13 cis element. However, they did not specifically demonstrate Tel or Hdac3 interaction with this cis element. We investigated these potential interactions using an in vitro DNA-affinity purification assay. For these experiments, nuclear proteins from U937 cells were incubated with a biotin-labeled, double-stranded oligonucleotide probe representing the PTPN13 cis element, or a probe representing a mutant form of the ets/Irf consensus sequence. Proteins that bound to the oligonucleotide probes were purified by affinity of the probe for avidin beads, separated by SDS-PAGE, and identified by Western blot. Because we previously demonstrated increased Icsbp binding to this PTPN13 cis element during differentiation, some nuclear proteins were isolated from IFNγ-treated U937 cells (for monocyte differentiation; 500 units/ml for 48 h). We found that Icsbp, Tel, and Hdac3 all bound to the PTPN13 probe in this assay, but not to the mutant probe (Fig. 3A). In the case of Tel, binding was increased by differentiation (Fig. 3A).
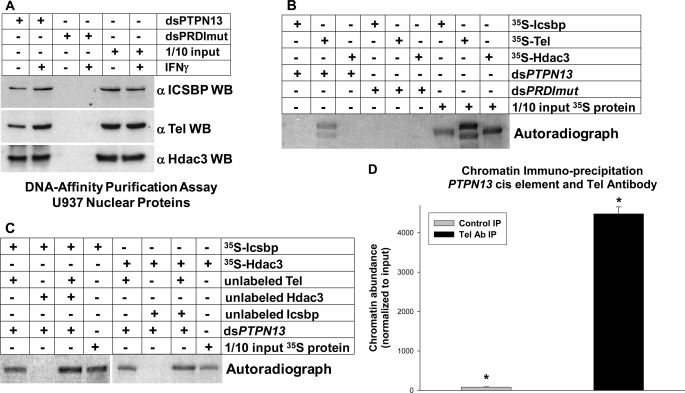
FIGURE 3.
Tel and Hdac3 bind to the PTPN13 cis element. A, Tel and Hdac3 interact with the Icsbp-binding PTPN13 cis element in vitro. DNA affinity purification assays were performed with U937 nuclear proteins and biotin-labeled, double-stranded oligonucleotide probes representing the PTPN13 cis element (dsPTPN13) or a mutant form of the ets/Irf DNA-binding consensus sequence (dsPRDImut). Proteins that bound to the probes were purified by affinity probe for avidin-conjugated beads, separated by SDS-PAGE, and identified by serially probing Western blots with antibodies to Icsbp, Tel, and Hdac3. Nonaffinity purified protein was included on the SDS-PAGE to control for loading (indicated as 1/10 input protein). B, Tel, but not Icsbp or Hdac3, bound to the PTPN13 cis element in vitro. DNA affinity purification assays were also performed using the dsPTPN13 and dsPRDImut probes and in vitro translated Icsbp, Tel, or Hdac3. For these experiments, recombinant proteins were labeled with [35S]methionine. Untreated, in vitro translated proteins were included on the SDS-PAGE as a loading control and to demonstrate size of the proteins (indicated as 1/10 input protein). C, Tel binding facilitated binding of Icsbp and Hdac3 to the PTPN13 cis element in vitro. DNA-affinity purification assays were also performed with combinations of in vitro translated Tel, Icsbp, and Hdac3. For these studies Tel synthesized in vitro in the absence of [35S]methionine (unlabeled) was combined with 35S-labeled Icsbp or Hdac3. For other experiments, unlabeled Tel and Hdac3 were combined with 35S-labeled Icsbp, or unlabeled Tel and Icsbp were combined with labeled 35S-labeled Hdac3. Probes used in these experiments were dsPTPN13 and dsPRDImut, as above, and unbound in vitro translated proteins were run in control lanes (indicated as 1/10 input protein). D, Tel binds to the PTPN13 cis element in vivo. Chromatin immunoprecipitation experiments were performed using U937 myeloid leukemia cells and an antibody to Tel (or irrelevant control antibody). Cell lysates were sonicated to generate chromatin fragments of <200 bp prior to immunoprecipitation. Co-precipitating chromatin was amplified by quantitative real time PCR using primers that flank the Icsbp-binding PTPN13 cis element (generating an 80-bp product). Results are normalized to total input chromatin. A statistically significant increase in co-precipitation with Tel antibody is indicated by *.
We next investigated whether binding order was important for assembly of the Icsbp-Tel-Hdac3 protein complex on the PTPN13 cis element. For these experiments, 35S-labeled Icsbp, Tel, and Hdac3 were individually in vitro translated in rabbit reticulocyte lysate and incubated with the PTPN13 cis element probe or mutant control probe. Proteins that bound to the probes were co-purified by affinity to avidin beads, separated by SDS-PAGE, and identified by autoradiography. We found that only in vitro translated Tel bound independently to the PTPN13 probe (Fig. 3B).
Therefore, we investigated whether Tel binding to the PTPN13 cis element facilitated Icsbp or Hdac3 binding. For these experiments, 35S-labeled in vitro translated Icsbp was incubated with the PTPN13 probe in the presence of in vitro translated (but unlabeled) Tel, Hdac3, or Tel + Hdac3. Affinity purified proteins were analyzed by SDS-PAGE. We found that Icsbp bound in the presence of Tel, but not Hdac3 (Fig. 3C). We also found reproducibly more Icsbp binding in the presence of Tel + Hdac3 in comparison to Tel alone. We performed similar studies of Hdac3 binding. We found that Hdac3 bound in the presence of Tel, but not Icsbp alone (Fig. 3C). These studies suggested that assembly of the repression complex required pre-binding of the Tel for Icsbp and Hdac3 recruitment.
We initially identified PTPN13 as an Icsbp target gene by coupling chromatin immunoprecipitation (with an Icsbp antibody) with screening a CpG island microarray (7). Therefore, we used chromatin immunoprecipitation to investigate in vivo Tel binding to this PTPN13 cis element. For these experiments, U937 cells were treated with formaldehyde to form DNA-protein cross-links, cell lysates were sonicated under conditions that sheared the DNA to fragments of less than 200 bp, and chromatin was co-immunoprecipitated with an antibody to Tel (or irrelevant control antibody). Co-precipitating chromatin was amplified by quantitative real time PCR using primers flanking the PTPN13 cis element, as described in Ref. 7. This assay identified specific in vivo Tel binding to the PTPN13 cis element (Fig. 3D).
Icsbp, Tel, and Hdac3 Cooperate to Influence Fap1 Expression
Our previous studies determined that expression of Fap1 protein and mRNA was decreased by Icsbp overexpression and increased by Icsbp knockdown in myeloid cell lines and primary murine myeloid progenitor cells (7). In the current investigations, we determined whether the expression level of Tel and/or Hdac3 also influenced Fap1 expression. For initial studies, U937 cells were stably transfected with vectors to overexpress Icsbp, Tel, Hdac3, or Icsbp + Tel + Hdac3 (or empty control vectors). Cell lysates were analyzed by Western blot for Fap1 expression. We found that overexpression of any of these proteins individually decreased Fap1 protein (Fig. 4A). Simultaneous overexpression of the three proteins resulted in a greater decrease in Fap1 protein in comparison to overexpression of any individual protein. In control experiments, we found that each of the proteins were appropriately overexpressed (Fig. 4A). We also found that overexpression of Icsbp, Tel, or Hdac3 did not influence the expression level of the other two.
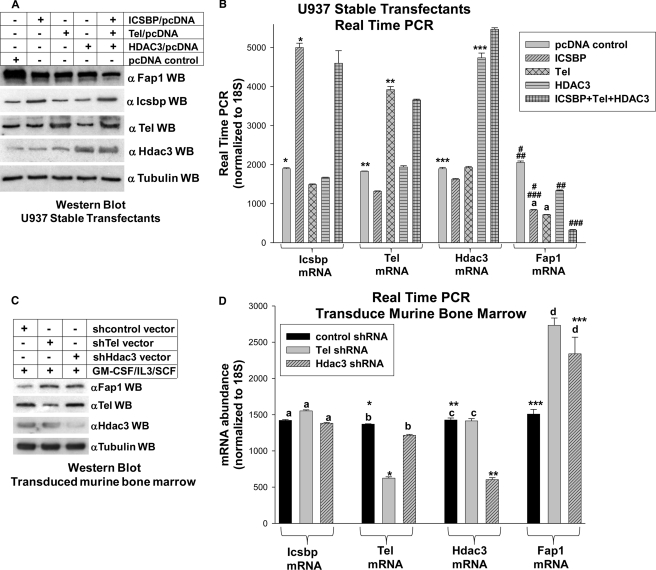
FIGURE 4.
Icsbp, Tel, and Hdac3 cooperate to decrease Fap1 expression. A, Icsbp, Tel, and Hdac3 cooperated to decrease Fap1 protein expression in U937 transfectants. U937 cells were stably transfected with vectors to express Icsbp, Tel, Hdac3, Icsbp + Tel + Hdac3, or empty control vectors. Protein expression was analyzed by Western blots (WB) of total cell lysates serially probed with antibodies to Fap1, Icsbp, Tel, HDac3, and tubulin (as a loading control). B, Icsbp, Tel, and Hdac3 cooperated to decrease Fap1 mRNA expression in U937 transfectants. The stable U937 transfectants described above were also analyzed for expression of Fap1, Icsbp, Tel, and Hdac3 mRNA by quantitative real time PCR. Results were normalized to 18 S. Lack of statistically significant difference (p > 0.4) in Fap1 mRNA expression are indicated with a lowercase a. Statistically significant differences in mRNA expression (p < 0.001 for all comparisons) are indicated by *, **, ***, #, ##, and ###. C, knockdown of Tel or Hdac3 expression increased Fap1 protein in primary murine myeloid progenitor cells. Western blots were performed using cell lysates from transduced murine bone marrow cells, as described above. Blots were serially probed with antibodies to Icsbp, Tel, Hdac3, Fap1, and tubulin (as a loading control). D, knockdown of Tel or Hdac3 expression increased Fap1 mRNA expression in primary murine myeloid progenitor cells. Bone marrow was harvested from WT mice and transduced with retroviral vectors to express Tel-specific shRNAs, Hdac3-specific shRNAs, or scrambled control shRNAs. Cells were cultured in GM-CSF, IL3, and SCF followed by CD34 separation (granulocyte/monocyte progenitor conditions). Expression of Fap1, Icsbp, Tel, and Hdac3 mRNAs were determined by real time PCR. Differences in mRNA expression that are not statistically significantly different (p > 0.4 for all comparisons) are indicated by lowercase letters (a, b, c, and d). Statistically significant differences (p < 0.01 for all comparisons) are indicated by *, **, or ***.
These stable transfectants were analyzed for Fap1 mRNA expression by real time PCR. Consistent with protein expression results, we found significantly less Fap1 mRNA in transfectants overexpressing Icsbp, Tel, or Hdac3 in comparison to control vector transfectants (p < 0.0001, n = 3) (Fig. 4B). The effect of Hdac3 overexpression on Fap1 mRNA abundance was somewhat less than the effects of Icsbp or Tel. However, co-overexpression of all three proteins decreased Fap1 mRNA expression to a significantly greater extent than each protein individually (p < 0.00001, n = 3) (Fig. 4B). In control studies, Icsbp, Tel, and Hdac3 were each significantly overexpressed in cells stably transfected with the relevant expression vector (p < 0.000001 for all proteins, n = 3). For all real time PCR experiments, n represents the number of independent experiments performed in triplicate.
We previously found that Fap1 expression in primary murine myeloid progenitors and differentiating cells from Icsbp−/− mice was greater than expression in WT mice (7). Therefore, we investigated the impact of Tel or Hdac3 knockdown on Fap1 expression in primary murine myeloid cells. For these experiments, bone marrow cells were isolated from WT mice and transduced with a retroviral vector to express Tel-specific shRNAs, Hdac3-specific shRNAs, or scrambled control shRNAs. Cells were cultured in GM-CSF, IL3, and SCF and CD34+ cells were separated. Cells cultured under these conditions are enriched for the granulocyte/monocyte progenitor population (31).
These transduced cells were analyzed for Fap1 protein expression by Western blot of total cell lysates. Consistent with results of studies in U937 cells, we found increased Fap1 protein in cells with Tel or Hdac3 knockdown (Fig. 4C). Control experiments demonstrated knockdown of the target proteins by the respective shRNA expression vectors. In these studies, Hdac3 knockdown did not influence Tel protein expression, and Tel knockdown did not influence Hdac3 protein expression.
Transduced cells were also analyzed for Fap1 mRNA expression by real time PCR. We found significantly more Fap1 mRNA in cells with Tel or Hdac3 knockdown in comparison to cells expressing a scrambled control shRNA (p < 0.002, n = 3) (Fig. 4D). In control experiments, the abundance of Tel or Hdac3 mRNA were appropriately decreased by these vectors. However, knockdown of Tel did not significantly alter mRNA expression for Icsbp or Hdac3 in comparison to expression of scrambled, control shRNA (p ≥ 0.2, n = 3) (Fig. 4D). Similarly, knockdown of Hdac3 did not significantly alter mRNA expression for Icsbp or Tel in comparison to expression of scrambled, control shRNA (p ≥ 0.1, n = 3) (Fig. 4D).
Expression of Fap1, Tel, Hdac3, and Icsbp mRNA was equivalent in sham transduced cells and in cells transduced with scrambled control shRNA expression vectors specific to Tel or Hdac3. Therefore, data are shown only from the scrambled shRNA control for shTel.
Tel and Hdac3 Influence Fas-induced Apoptosis in a Fap1-dependent Manner
Overexpression of Icsbp in U937 cells increases the sensitivity of this relatively resistant cell line to Fas-induced apoptosis (7). In the current studies, we used stable transfectants, described above, to investigate the impact of Tel and Hdac3 overexpression on Fas sensitivity, and the possibility of functional cooperation between proteins. For these studies, cells were treated with a Fas agonist antibody (CH11) or irrelevant control antibody, and apoptosis was determined by annexin V staining and flow cytometry. For all apoptosis studies, n represents the number of independent experiments, performed in triplicate.
Control cells were resistant to induction of apoptosis by Fas-agonist antibody in this assay (no significant difference in apoptosis with versus without Fas-Ab, p = 0.9, n = 3) (Fig. 5A). Icsbp overexpression significantly increased the % of cells undergoing apoptosis in response to Fas antibody in comparison to control U937 cells (p < 0.0001, n = 3), as in our previous studies (7). We found that overexpression of Tel or Hdac3 also significantly increased the % of cells undergoing apoptosis in response to Fas-Ab in comparison to control vector transfected cells (for Tel, p = 0.0002, n = 3, and for Hdac3, p = 0.006, n = 3) (Fig. 5A). Cells that were co-overexpressing Icsbp and Tel were significantly more Fas sensitive in comparison to cells that were overexpressing Icsbp or Tel alone (p = 0.02 and 0.01, respectively, n = 3). Cells co-overexpressing Icsbp and Tel and Hdac3 were more Fas sensitive than cells co-overexpressing Icsbp and Tel (p = 0.01, n = 3) (Fig. 5A). These studies suggested that decreased Fap1 expression, caused by overexpression of the three proteins, increased Fas sensitivity.
FIGURE 5.
Icsbp, Tel, and Hdac3 cooperated to increase sensitivity to Fas-induced apoptosis. A, Icsbp, Tel, and Hdac3 cooperated to increase sensitivity to Fas-induced apoptosis in U937 transfectants. U937 cells were stably transfected with vectors to overexpress Icsbp, Tel, Hdac3, Icsbp + Tel, Icsbp + Tel + Hdac3, or with empty control vectors (as in Fig. 4). Cells were treated with a Fas-agonist antibody or irrelevant control antibody and apoptosis was determined by annexin V staining and flow cytometry. Results are presented as % of cells staining for annexin V. Results that are not statistically significantly different (p > 0.05 for all comparisons) are indicated by lowercase letters (a and b). Statistically significant differences (p < 0.01 for all comparisons) are indicated by *, **, ***, #, ##, ###, &, or &&. B, knockdown of Tel or Hdac3 expression decreased sensitivity to Fas-induced apoptosis in primary murine myeloid progenitor cells. Primary murine bone marrow cells were harvested from WT mice, transduced with retroviral vectors to express Tel-specific shRNAs, Hdac3-specific shRNAs or scrambled shRNA, as above. Cells were treated with either SLV peptide (to block the Fas/Fap1 interaction) or VLS control peptide. Cells were assayed for apoptosis in response to a Fas-agonist antibody (or control antibody) by staining for annexin V. Differences that are not statistically significantly different (p > 0.2 for all comparisons) are indicated by a lowercase letter (a, b, and c). Statistically significant differences in apoptosis are indicated by *, **, ***, #, or ##.
Interaction between Fap1 and Fas is blocked by a specific tripeptide representing the C-terminal amino acids of Fas (serine-leucine-valine; SLV peptide) (29). In previous studies, we found that Fas resistance of Icsbp−/− primary murine myeloid progenitor cells was reversed by treatment with the SLV peptide (7). In the current studies, we used transduced primary murine bone marrow cells, as described above, to investigated the effect of Tel or Hdac3 knockdown on Fap1-dependent Fas resistance. Cells were assayed for annexin V staining by flow cytometry after treatment with the Fas agonist antibody (or control antibody).
We found that knockdown of either Tel or Hdac3 significantly decreased the % of myeloid progenitor cells that were sensitive to Fas-Ab-induced apoptosis (for comparison to control vector-transduced cells, p = 0.003 and 0.001, respectively, n = 3) (Fig. 5B). To determine the role of Fap1 in Fas resistance, these cells were incubated with SLV peptide (or VLS control peptide). We found that treatment with SLV peptide significantly increased apoptosis in Fas-Ab-treated cells with knockdown of Tel or Hdac3 (p = 0.006 and 0.002 respectively, n = 3) (Fig. 5B). Apoptosis was not significantly different in experiments with VLS control peptide versus no added peptide (p > 0.5, n = 6), so VLS treatment is used as the negative control in these studies.
Tel-PdgfRβ Impairs Repression of PTPN13 Promoter by Icsbp, Tel, and Hdac3
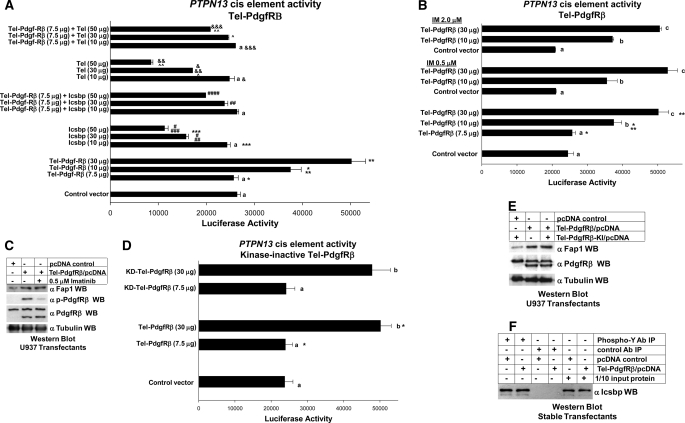
Because binding of Tel to the PTPN13 cis element is essential for assembly of the complex with Icsbp and Hdac3, we hypothesized that the Tel-PdgfRβ fusion protein might influence activity of the PTPN13 cis element. To test this hypothesis, U937 cells were co-transfected with the PTPN13 cis element-containing reporter construct (or reporter vector control) and a vector to express Tel-PdgfRβ (or control expression vector). We found that overexpression of Tel-PdgfRβ significantly increased activity of the PTPN13 cis element in a dose-dependent manner (p = 0.002 and 0.0001 for comparison of transfectants with 10 or 30 μg of Tel-PdgfRβ plasmid with control plasmid transfectants, n = 3) (Fig. 6A).
FIGURE 6.
Tel-PdgfRβ impairs repression of the Icsbp-binding PTPN13 by Icsbp and Tel. A, Tel-PdgfRβ expression increased activity of the PTPN13 cis element and impaired repression by overexpressed Icsbp and Tel. U937 cells were stably transfected with a vector to express Tel-PdgfR (or control expression vector) and co-transfected with a reporter vector with three copies of the Icsbp-binding PTPN13 cis element linked to a minimal promoter (ptpn13L3-p) or control reporter vector, and vectors to express various amounts of Icsbp or Tel (or empty control vector). Activity from the empty reporter vector was <10% of the ptpn13GL3-p, which was subtracted as background. Reporter activity not statistically significant (p > 0.2) is indicated by a lowercase a. Statistically significant differences in reporter activity are indicated by *, **, ***, #, ##, ###, &, &&, &&&, ∧, or ∧∧. B, Tel-PdgfRβ-induced PTPN13 cis element activity is independent of tyrosine kinase activity of the fusion protein (imatinib studies). U937 cells that were stably transfected with a vector to express Tel-PdgfR or control expression vector were co-transfected with a reporter vector with three copies of the Icsbp-binding PTPN13 cis element linked to a minimal promoter (ptpn13GL3-p) or control reporter vector. Some transfectants were treated with the tyrosine kinase inhibitor imatinib (IM). Activity from the empty reporter vector was <10% of the ptpn13GL3-p, which was subtracted as background. Reporter not statistically significantly different (p > 0.2 for all comparisons) is indicated by a lowercase letter (a, b, and c). Statistically significant differences (p < 0.01 for all comparisons) in reporter activity are indicated by * or **. C, Tel-PdgfRβ-induced increase in Fap1 protein expression is not inhibited by imatinib. U937 myeloid cells were stably transfected with a vector to express Tel-PdgfRβ or empty control vector. Some transfectants were treated with imatinib as indicated. Total cell lysates were analyzed for Tel-PdgfRβ tyrosine phosphorylation (activation) by Western blots (WB), which were serially probed with antibodies to Fap1, phospho-PdgfRβ, total PdgfRβ, or tubulin (as a loading control). This blot represents transfectants performed at the same time as one of the reporter gene assays from B. D, Tel-PdgfRβ-induced PTPN13 cis element activity is independent of tyrosine kinase activity of the fusion protein (studies with kinase inactive fusion protein). U937 cells that were stably transfected with a vector to express Tel-PdgfRβ, a kinase inactive form of Tel-PdgfRβ (KD-Tel-PdgfRβ), or control expression vector were co-transfected with a reporter vector with three copies of the Icsbp-binding PTPN13 cis element linked to a minimal promoter (ptpn13GL3-p) or control reporter vector. Reporter activity not statistically significantly different is indicated by a lowercase letter (a, b, and c). Statistically significant differences in reporter activity are indicated by *. E, Tel-PdgfRβ increased Fap1 protein expression in a tyrosine kinase independent manner. U937 myeloid cells were stably transfected with a vector to express Tel-PdgfRβ, KD-Tel-PdgfRβ, or empty control vector. Total cell lysates were analyzed by Western blots, which were serially probed with antibodies to PdgfRβ, Fap1, or tubulin (as a loading control). The anti-PdgfRβ antibodies used in this experiment recognize both endogenous PdgfRβ and the product of the Tel-PdgfRβ transgene. This blot represents transfectants performed at the same time as one of the reporter gene assays from B. F, Tel-PdgfRβ expression does not alter expression or tyrosine phosphorylation of Icsbp in myeloid cell line transfectants. Tel-PdgfRβ stable transfectants were also analyzed for phosphorylation and expression of Icsbp. Cell lysates were immunoprecipitated (under denaturing conditions) with an antibody to phosphotyrosine or irrelevant control antibody. Co-precipitating proteins were separated by SDS-PAGE and Western blots were probed with an antibody to Icsbp. Nonprecipitated lysates were also included on the SDS-PAGE (indicated as 1/10 input protein) as a loading control for Icsbp abundance.
We considered the possibility that Tel-PdgfRβ might prevent Icsbp and/or Tel from binding to and repressing the PTPN13 cis element. To investigate this hypothesis, U937 transfection experiments were performed with the PTPN13 cis element reporter vector (or control), Tel-PdgfRβ expression vector (or control), and a dose titration of Icsbp or Tel expression vector. We found that activity of the PTPN13 cis element was significantly greater in cells co-transfected with vectors to express both Tel-PdgfRβ and Icsbp in comparison to transfectants overexpressing Icsbp only (p = 0.0003 and 0.00001 for comparison of 30 or 50 μg of Icsbp expression vector ± Tel-PdgfRβ, n = 4) (Fig. 6A). Similarly, we found that PTPN13 cis element activity was significantly greater in cells co-transfected with vectors to express both Tel-PdgfRβ and Tel in comparison to transfectants overexpressing Tel only (p < 0.00001 for comparison of 30 or 50 μg of Tel expression vector ± Tel-PdgfRβ, n = 4).
We considered the possibility that endogenous tyrosine kinase activity of Tel-PdgfRβ might be responsible for activation of the PTPN13 cis element. To investigate the role of Tel-PdgfRβ kinase activity on PTPN13 cis element activity directly, we repeated the experiment in the presence of the tyrosine kinase inhibitor imatinib (IM). Although this tyrosine kinase inhibitor was developed to inhibit the Bcr-abl oncogene, it also inhibits Tel-PdgfRβ (32). We found that PTPN13 cis element activity in Tel-PdgfRβ expressing transfectants was not significantly altered by treatment with 0.5 or 2.0 μm doses of IM in comparison to sham treatment (p ≥ 0.2 in three-way analysis of variance for comparison of untreated, low dose and high dose IM-treated Tel-PdgfRβ expressing transfectants, n = 3) (Fig. 6B).
We verified that these doses of IM significantly inhibited Tel-PdgfRβ kinase activity (autophosphorylation) by Western blot of lysate proteins from the transfectants (Fig. 6C). This experiment also demonstrated an equivalent increase in endogenous Fap1 protein in transfectants with Tel-PdgfRβ ± IM. This Western blot was performed with cell lysate proteins from transfectants used to generate reporter gene assay data.
U937 cells were also co-transfected with the PTPN13 cis element-containing reporter construct (or reporter vector control) and a vector to express a kinase inactive form of Tel-PdgfRβ (KD-Tel-PdgfRβ) versus “wild type” Tel-PdgfRβ (or vector control) (25). We found that activity of the PTPN13 cis element-containing construct was increased equivalently by expression of either Tel-PdgfRβ or KD-Tel-PdgfRβ in this assay (p = 0.8, n = 3) (Fig. 6D). The two forms of Tel-PdgfRβ were equivalently expressed in this experiment, and both forms of Tel-PdgfRβ increased endogenous Fap1 protein expression (Fig. 6E).
Because Icsbp knockdown increases PTPN13 promoter activity (7), we considered the possibility that Tel-PdgfRβ might decrease Icsbp expression, as has been observed for Bcr-abl (14, 22). Also, because tyrosine phosphorylation of Icsbp increases binding affinity for the PTPN13 cis element, we considered the possibility that Tel-PdgfRβ might decrease Icsbp tyrosine phosphorylation. The latter was contraintuitive, but might occur through an indirect mechanism, such as phosphatase activation by Tel-PdgfRβ. We investigated these possibilities by performing phosphotyrosine immunoprecipitation and Western blotting for Icsbp in cells stably expressing Tel-PdgfRβ (or transfected with control vector). We found that Icsbp protein abundance and tyrosine phosphorylation were not influenced by Tel-PdgfRβ expression (Fig. 6F).
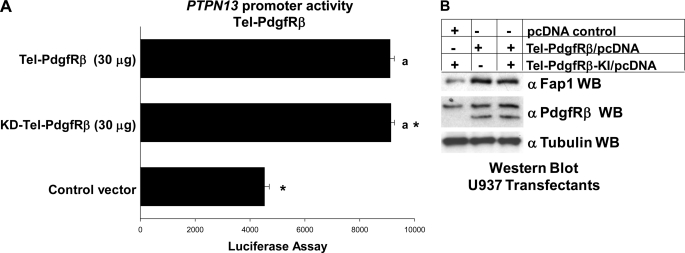
These studies suggested that expression of the Tel-PdgfRβ fusion protein interfered with PTPN13 repression by Icsbp and Tel. However, these studies were performed with an artificial promoter construct with the isolated cis element. Although this assay was specific for influence of these proteins on a common cis element, we performed control studies to assure that Tel-PdgfRβ also influenced the intact PTPN13 promoter. To investigate this, additional transfection experiments were performed using a construct with 670 bp of the PTPN13 5′ flank (or empty reporter vector) and vectors to express Tel-PdgfRβ (30 μg), or a kinase inactive form of Tel-PdgfRβ (30 μg) (or empty expression vector).
These studies demonstrated that both forms of Tel-PdgfRβ were equivalently efficient at activation of the PTPN13 promoter (Fig. 7A). The extent of promoter activation for the two forms of Tel-PdgfRβ was similar to the studies with the isolated cis element (Figs. 6D and 7A). We found similar results in transfection experiments with a 2.0-kb PTPN13 promoter construct (not shown).
FIGURE 7.
Tel-PdgfRβ increased activity of the PTPN13 promoter. A, Tel-PdgfRβ increased PTPN13 promoter activity in a kinase-independent manner. U937 cells that were stably transfected with a vector to express Tel-PdgfRβ, a kinase inactive form of Tel-PdgRβ (KD-Tel-PdgfRβ), or control expression vector were co-transfected with a reporter vector with 670 bp of the PTPN13 promoter (ptpn13GL3) or control reporter vector. Activity from the empty reporter vector was <10% of the ptpn13GL3, which was subtracted as background. Differences in reporter activity not statistically significantly different (p > 0.6) are indicated by a lowercase a. Statistically significant differences (p < 0.0001) in reporter activity are indicated by *. B, Tel-PdgfRβ increased Fap1 protein expression in a tyrosine kinase-independent manner. U937 myeloid cells were stably transfected with a vector to express Tel-PdgfRβ, KD-Tel-PdgfRβ, or empty control vector. Total cell lysates were analyzed by Western blots (WB), which were serially probed with antibodies to PdgfRβ, Fap1, or tubulin (as a loading control). The anti-PdgfRβ antibodies used in this experiment recognize both endogenous PdgfRβ and the product of the Tel-PdgfRβ transgene. This blot represents transfectants performed at the same time as one of the reporter gene assays from A.
In control experiments, we found that expression of either Tel-PdgfRβ or kinase-inactive Tel-PdgfRβ was equivalent and each increased expression of the endogenous Fap1 protein (Fig. 7B). These experiments were performed with cell lysate proteins from reporter gene assays from Fig. 7A. We found that activity of empty reporter vector (pGL3) was not influenced by expression of either form of Tel-PdgfRβ and was subtracted as background.
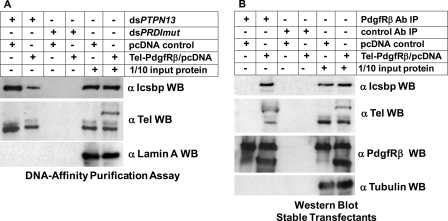
Tel-PdgfRβ Impairs Binding of Tel and Icsbp to PTPN13 Promoter Cis Element
These studies led to the hypothesis that Tel-PdgfRβ might prevent binding of Icsbp, Tel, or both to the PTPN13 cis element. To investigate this possibility, we performed DNA-affinity purification assays, similar to studies above, with cells stably transfected with a Tel-PdgfRβ expression vector (or control vector). Nuclear proteins from these transfectants were incubated with a biotin-labeled, double-stranded oligonucleotide probe with either the PTPN13 cis element or mutant, nonbinding control probe. Proteins that bound to the probes were purified by affinity to avidin beads, and Icsbp or Tel were identified by Western blot. We found that expression of Tel-PdgfRβ significantly decreased binding of both Icsbp and Tel to the PTPN13 cis element probe (Fig. 8A). The Tel antibody used in these experiments recognizes both endogenous Tel and the Tel-PdgfRβ fusion protein. Therefore, this experiment also demonstrated that the fusion protein did not bind to the PTPN13 cis element probe in vitro.
FIGURE 8.
Tel-PdgfRβ interacted with Tel and Icsbp and decreased binding of these proteins to the PTPN13 cis element. A, Tel-PdgfRβ expression in myeloid cell lines decreased binding of Tel and Icsbp to the PTPN13 cis element in vitro. DNA affinity purification assays were performed with KG1 nuclear proteins stably expressing Tel-PdgfRβ (or with empty control vector), and biotin-labeled, double-stranded oligonucleotide probes representing the Icsbp/Tel binding site in the PTPN13 promoter (dsPTPN13) or a non-Icsbp binding mutant sequence (dsPRDImut). Affinity purified proteins were separated by SDS-PAGE and Western blots were probed with antibodies to Icsbp and Tel (using an antibody that recognizes both wild type Tel and the Tel-PdgfRβ fusion protein). Nonprecipitated proteins were also included on the SDS-PAGE (indicated as 1/10 input protein) and Western blots were probed with a Lamin A antibody as a loading control. B, Tel and Icsbp co-immunoprecipitated with Tel-PdgfRβ from myeloid cells. Total cell lysates from these KG1 stable transfectants were immunoprecipitated with an antibody to PdgfRβ (or with irrelevant antibody control). Immunoprecipitates were separated by SDS-PAGE and Western blots were serially probed with antibodies to Icsbp, Tel, or PdgfRβ. The Tel and PdgfRβ antibodies used in these experiments recognize the endogenous protein and the Tel-PdgfRβ fusion proteins. Nonprecipitated proteins were included on the SDS-PAGE (indicated as 1/10 input protein) and Western blots were also probed with tubulin antibody as a loading control.
Tel-PdgfRβ interacts with wild type Tel through the basic helix loop helix domains in the two proteins (23). Therefore, we hypothesized that the fusion protein might bind to wild type Tel and make it unavailable for interaction with the PTPN13 cis element. To investigate this possibility, we determined if endogenous Tel interacted with Tel-PdgfRβ using the transfectants described above. We found that an anti-PdgfRβ antibody successfully co-immunoprecipitated endogenous Tel in experiments with cells expressing Tel-PdgfRβ expressing cells, but not control cells (Fig. 8B).
Because Icsbp binding to the PTPN13 cis element required pre-binding of Tel, these results suggested that decreased availability of Tel in Tel-PdgfRβ expressing cells might decrease Icsbp interaction with this cis element. Also, because Tel interacts with Irf proteins, it was possible that expression of Tel-PdgfRβ might deplete available Icsbp in the transfectants; either by direct interaction between Icsbp and the fusion protein, or indirectly through interaction with Tel bound to Tel-PdgfRβ. Therefore, we also investigated Icsbp co-immunoprecipitation with Tel-PdgfRβ using these transfectants. We found significant co-precipitation of Icsbp with PdgfRβ in experiments with cells expressing the fusion protein, but not with endogenous PdgfRβ in control cells (Fig. 8B).
Tel-PdgfRβ Increases Fap1 Expression and Impairs Fas-induced Apoptosis
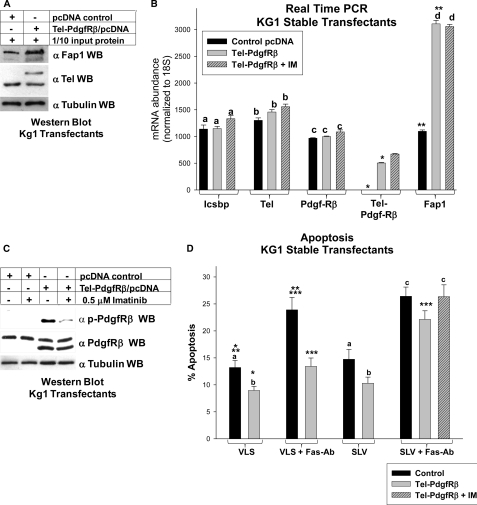
Based on these results, we investigated the effect of Tel-PdgfRβ on Fap1 protein and mRNA using stable transfectants of the Kg1 myeloid line. We found increased expression of the Fap1 protein in Tel-PdgfRβ expressing cells in comparison to control cells by Western blot (Fig. 9A). We also investigated Fap1 mRNA expression in these transfectants. Consistent with protein expression results, we found significantly more Fap1 mRNA in Tel-PdgfRβ expressing transfectants in comparison to control transfectants (p < 0.00001, n = 3) (Fig. 9B). This increase in Fap1 mRNA expression was not altered by treatment of the transfectants with IM (p = 0.8, n = 3 for Tel-PdgfRβ expressing transfectants with versus without IM) (Fig. 9B). These experiments were performed at an IM dose that inhibited Tel-PdgfRβ activity (determined by autophosphorylation; Fig. 9C).
FIGURE 9.
Tel-PdgfRβ increased Fap1 expression and induced Fas resistance in myeloid cells. A, Tel-PdgfRβ increased Fap1 protein expression in KG1 myeloid cells. KG1 cells were stably transfected with a vector to express Tel-PdgfRβ or control vector and analyzed for Fap1 protein expression. Western blots of total cell lysates were serially probed with antibodies to Fap1, Tel (using an antibody which identifies endogenous Tel and Tel-PdgfRβ), or tubulin (as a loading control). C, Tel-PdgfRβ increased Fap1 mRNA expression in KG1 myeloid cells. The KG1 stable transfectants described above were also analyzed for Fap1 mRNA expression by real time PCR. Expression of Icsbp, endogenous Tel, endogenous PdgfRβ, and Tel-PdgfRβ were also determined. For these experiments, Tel- and PdgfRβ-specific primer sets were used, which do not recognize the Tel-PdgfRβ fusion transcript, and a Tel-PdgfRβ primer set was used, which only recognizes the fusion transcript. Nonstatistically significant differences in mRNA expression (p > 0.05 for all comparisons) are indicated by lowercase letters (a, b, c, and d). Statistically significant differences (p < 0.001 for all comparisons) in expression are indicated by * or **. B, treatment with imatinib impaired Tel-PdgfRβ autoactivation in KG1 transfectants. KG1 myeloid cells were stably transfected with a vector to express the Tel-PdgfRβ fusion protein or empty control vector. Untreated cells were compared with cells treated with imatinib (IM) as indicated. Total cell lysates were analyzed for Tel-PdgfRβ tyrosine phosphorylation (activation) by Western blots, which were serially probed with antibodies to phospho-PdgfRβ, total PdgfRβ, or tubulin (as a loading control). The anti-PdgfRβ antibodies used in this experiment recognize both endogenous PdgfRβ and the product of the Tel-PdgfRβ transgene. C, Tel-PdgfRβ induced Fap1-dependent Fas resistance in KG1 myeloid cells. KG1 stable transfectants with Tel-PdgfRβ or control vector were analyzed for Fas-induced apoptosis. Cells were treated with SLV (Fas/Fap1 blocking peptide) or VLS control peptide, and Fas-agonist antibody or irrelevant control antibody. Some cells were additionally treated with imatinib. Apoptosis was determined by annexin V staining and flow cytometry, and expressed as % apoptotic cells. Differences in apoptosis not statistically significantly different (p > 0.05 for all comparisons) are indicated by lowercase letters (a, b, and c). Statistically significant differences (p < 0.001 for all comparisons) are indicated by *, **, or ***.
Expression of Icsbp and Tel mRNA was not significantly different in Tel-PdgfRβ expressing transfectants in comparison to control transfectants (p = 0.2 and 0.4, n = 3, respectively) (Fig. 9B). Expression of the fusion protein did not significantly alter expression of endogenous PdgfRβ mRNA in comparison to control vector transfectants (p = 0.05, n = 3) (Fig. 9B).
We anticipated that increased Fap1 expression in Tel-PdgfRβ expressing cells would impair Fas-induced apoptosis. To investigate this, stable transfectants overexpressing Tel-PdgfRβ (or vector control cells) were treated with Fas agonist antibody and SLV peptide or VLS control. Apoptosis was determined by flow cytometry, as above. We found that Fas-antibody treatment significantly increased apoptosis in control Kg1 cells (p = 0.002, n = 4 for % apoptotic cells with versus without Fas-Ab) (Fig. 9D), as in our previous studies (7). Fas-antibody treatment induced significantly less apoptosis in Tel-PdgfRβ expressing cells in comparison to control cells (p = 0.003, n = 4). However, apoptosis in Tel-PdgfRβ expressing cells treated with SLV peptide and Fas-antibody was significantly greater than Tel-PdgfRβ expressing cells treated with Fas-antibody alone (p = 0.001, n = 4) (Fig. 9D). The % of apoptosis for Tel-PdgfRβ expressing cells treated with Fas-Ab + SLV peptide was not significantly different with versus without tyrosine kinase inhibition with IM (p = 0.3, n = 4) (Fig. 9D).
Neither SLV nor VLS peptide alone induced significant apoptosis in Tel-PdgfRβ expressing cells or control cells in the absence of the Fas-agonist Ab, consistent with our previous studies (7).
DISCUSSION
In this study, we determined that repression of PTPN13 transcription by Icsbp involves formation of a multiprotein complex with Tel and Hdac3. We also found that pre-binding of Tel to the cis element is necessary for Icsbp binding, and histone deacetylase activity is necessary for PTPN13 repression. Additionally, we determined that knockdown of Tel or Hdac3 increases PTPN13 transcription, Fap1 expression, and Fap1-dependent Fas resistance in myeloid cells. These studies therefore identify a novel function for Tel; regulation of Fas sensitivity during myelopoiesis. We also found that expression of the leukemia-associated Tel-PdgfRβ oncoprotein increases Fap1 expression and Fas resistance. Importantly, this effect did not require tyrosine kinase activity of the fusion protein. Instead, our studies suggest that interaction with Tel-PdgfRβ results in decreased availability of Tel and Icsbp for binding to the PTPN13 cis element. This is a novel activity for Tel-PdgfRβ and a nontyrosine kinase-related mechanism by which this oncoprotein may contribute to leukemogenesis.
Icsbp is an interferon regulatory transcription factor that functions as a “tumor suppressor” for myeloid leukemia. Identification of Icsbp target genes has clarified some of the molecular mechanisms involved in this activity. The first identified Icsbp target genes encoded phagocyte-specific proteins, including gp91PHOX, p67PHOX, Toll-like receptor 4, and IL18 (15, 16, 33, 34). Icsbp activates transcription of these genes during myelopoiesis, resulting in acquisition of phagocyte functional competence. For some of these genes, transcriptional activation was shown to involve formation of a multiprotein complex with Icsbp, Pu.1 (an ets protein), Irf1, and the CREB-binding protein (CBP) (8). Assembly of the activation complex involves pre-binding of the ets protein, as we found for PTPN13 in the current studies. These protein-protein interactions are mediated by signal-dependent phosphorylation of Pu.1 and the Irf proteins.
Icsbp target genes involved in regulating cell proliferation were also identified, including genes encoding Neurofibromin 1 (Nf1; a RasGap) and growth arrest-specific protein 2 (Gas2; a calpain inhibitor) (18, 22). Icsbp activates NF1 transcription by forming a multiprotein complex with Pu.1 and Irf2, similar to the mechanism for activation of phagocyte effector genes (18). In cytokine-stimulated myeloid progenitor cells, Icsbp-induced Nf1 expression terminates Ras-related proliferative signals transduced by cytokine receptors (9). In contrast, Icsbp represses GAS2 transcription by participating in a complex with Tel and Hdac3. Decreased Gas2 expression increases calpain-mediated degradation of β-catenin protein (22). Therefore, decreased Icsbp activity results in β-catenin-dependent cell proliferation (22).
These studies suggest that, during normal myelopoiesis, Icsbp is involved in activation of phagocyte-specific genes, proliferation arrest, and increased sensitivity to Fas-induced apoptosis. Conversely, decreased Icsbp expression, which is common in CML and acute myeloid leukemia, results in differentiation block, dysregulation of cytokine-stimulated proliferation, and Fas resistance. Identification of these Icsbp target genes suggested the existence of an “ets/Irf code” or gene regulatory network involved in myelopoiesis. These studies additionally suggest the hypothesis that the ets partner for Icsbp determines whether the Icsbp complex will activate (with Pu.1) or repress (with Tel) transcription. Identification and characterization of additional target genes will determine whether this hypothesis is correct.
Studies in our laboratory and others indicate that the level of Fap1 expression influences Fas resistance in leukemia and other malignancies. Our current studies suggest that increased Fap1 expression would be anticipated in CMMoL with TEL/PDGFRB (due to t(5;12)), although this will require confirmation. CMMoL with this translocation is not common, and gene expression profiles for such patients are not found in publicly available databases. If increased Fap1 expression is associated with Fas resistance in CMMoL subjects, our studies suggest that this would not be amenable to treatment with IM or other tyrosine kinase inhibitors. This may provide a partial explanation for the inferior therapeutic response of CMMoL to IM in comparison to CML.
Previous studies of Tel-PdgfRβ focused on tyrosine kinase activity of the oncogene. These studies demonstrated that this activity dysregulates proliferation in CMMoL cells. Studies of apoptosis in cells expressing Tel-PdgfRβ have not been as clear. One published report indicates that apoptosis is increased in BaF3 cells expressing Tel-PdgfRβ (17). Comparison to our results suggests that the cellular milieu (i.e. lymphoid versus myeloid) may be an important determination of some consequences of Tel-PdgfRβ expression. We found that expression of Tel-PdgfRβ in the Fas-sensitive KG1 myeloid progenitor cell line results in Fas resistance that is overcome by blocking Fap1-Fas interaction. We found that Fap1 expression is not altered by IM treatment of Tel-PdgfRβ expressing cells, nor did IM treatment further influence Fas sensitivity of SLV peptide-treated Tel-PdgfRβ expressing cells.
Our studies identify a mechanism for Fas resistance in Tel-PdgfRβ expressing cells that is independent of tyrosine kinase activity, but dependent on protein-protein interactions with the fusion protein. Our results suggested that Tel-Tel-PdgfRβ interaction prevents Tel from participating in normal gene regulatory activities. Our studies also suggest that Icsbp-Tel-PdgfRβ interaction occurs, perhaps involving a heterotrimer of Tel, Icsbp, and Tel-PdgfRβ. The exact interactions will be of interest for potential targeting of this molecular mechanism in CMMoL.
These studies suggest that inhibition of the Tel-PdgfRβ tyrosine kinase activity may leave some leukemia promoting activities unperturbed. This has implications for understanding CMMoL with this translocation and suggests multifunctionality for this oncoprotein.
Footnotes
- Fap1
- Fas-associated phosphatase 1
- TSA
- trichostatin A
- Hdac3
- histone deacetylase 3
- PTP
- protein-tyrosine phosphatase
- CML
- chronic myeloid leukemia
- Icsbp
- interferon consensus sequence binding protein
- Irf8
- interferon regulatory factor 8
- PdgfRβ
- platelet-derived growth factor receptor β
- CMMoL
- chronic myelomonocytic leukemia
- IM
- imatinib
- SCF
- stem cell factor.
REFERENCES
- 1. Inazawa J., Ariyama T., Abe T., Druck T., Ohta M., Huebner K., Yanagisawa J., Reed J. C., Sato T. (1996) PTPN13, a Fas-associated protein-tyrosine phosphatase, is located on the long arm of chromosome 4 at band q21.3. Genomics 31, 240–242 [DOI] [PubMed] [Google Scholar]
- 2. Sato T., Irie S., Kitada S., Reed J. C. (1995) FAP-1. A protein-tyrosine phosphatase that associates with Fas. Science 268, 411–415 [DOI] [PubMed] [Google Scholar]
- 3. Ivanov V. N., Lopez Bergami P., Maulit G., Sato T. A., Sassoon D., Ronai Z. (2003) FAP-1 association with Fas (Apo-1) inhibits Fas expression on the cell surface. Mol. Cell. Biol. 23, 3623–3635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Komada Y., Inaba H., Zhou Y. W., Zhang X. L., Tanaka S., Azuma E., Sakurai M. (1997) mRNA expression of Fas receptor (CD95)-associated proteins (Fas-associated phosphatase-1/FAP-1, Fas-associating protein with death domain/FADD, and receptor-interacting protein/RIP) in human leukaemia/lymphoma cell lines. Br. J. Haematol. 99, 325–330 [DOI] [PubMed] [Google Scholar]
- 5. Otten H. G., van Ginkel W. G., Hagenbeek A., Petersen E. J. (2004) Prevalence and clinical significance of resistance to perforin- and FAS-mediated cell death in leukemia. Leukemia 18, 1401–1405 [DOI] [PubMed] [Google Scholar]
- 6. Michor F., Hughes T. P., Iwasa Y., Branford S., Shah N. P., Sawyers C. L., Nowak M. A. (2005) Dynamics of chronic myeloid leukemia. Nature 435, 1267–1270 [DOI] [PubMed] [Google Scholar]
- 7. Huang W., Zhu C., Wang H., Horvath E., Eklund E. A. (2008) The interferon consensus sequence-binding protein (ICSBP/IRF8) represses PTPN13 gene transcription in differentiating myeloid cells. J. Biol. Chem. 283, 7921–7935 [DOI] [PubMed] [Google Scholar]
- 8. Kautz B., Kakar R., David E., Eklund E. A. (2001) SHP1 protein-tyrosine phosphatase inhibits gp91PHOX and p67PHOX expression by inhibiting interaction of PU.1, IRF1, interferon consensus sequence-binding protein, and CREB-binding protein with homologous cis elements in the CYBB and NCF2 genes. J. Biol. Chem. 276, 37868–37878 [DOI] [PubMed] [Google Scholar]
- 9. Huang W., Saberwal G., Horvath E., Zhu C., Lindsey S., Eklund E. A. (2006) Leukemia-associated, constitutively active mutants of SHP2 protein-tyrosine phosphatase inhibit NF1 transcriptional activation by the interferon consensus sequence binding protein. Mol. Cell. Biol. 26, 6311–6332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schmidt M., Nagel S., Proba J., Thiede C., Ritter M., Waring J. F., Rosenbauer F., Huhn D., Wittig B., Horak I., Neubauer A. (1998) Lack of interferon consensus sequence binding protein (ICSBP) transcripts in human myeloid leukemias. Blood 91, 22–29 [PubMed] [Google Scholar]
- 11. Schmidt M., Hochhaus A., Nitsche A., Hehlmann R., Neubauer A. (2001) Expression of nuclear transcription factor interferon consensus sequence binding protein in chronic myeloid leukemia correlates with pretreatment risk features and cytogenetic response to interferon-α. Blood 97, 3648–3650 [DOI] [PubMed] [Google Scholar]
- 12. Qian Z., Fernald A. A., Godley L. A., Larson R. A., Le Beau M. M. (2002) Expression profiling of CD34+ hematopoietic stem/progenitor cells reveals distinct subtypes of therapy-related acute myeloid leukemia. Proc. Natl. Acad. Sci. U.S.A. 99, 14925–14930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Holtschke T., Löhler J., Kanno Y., Fehr T., Giese N., Rosenbauer F., Lou J., Knobeloch K. P., Gabriele L., Waring J. F., Bachmann M. F., Zinkernagel R. M., Morse H. C., 3rd, Ozato K., Horak I. (1996) Immunodeficiency and chronic myelogenous leukemia-like syndrome in mice with a targeted mutation of the ICSBP gene. Cell 87, 307–317 [DOI] [PubMed] [Google Scholar]
- 14. Hao S. X., Ren R. (2000) Expression of interferon consensus sequence binding protein (ICSBP) is down-regulated in Bcr-Abl-induced murine chronic myelogenous leukemia-like disease, and forced coexpression of ICSBP inhibits Bcr-Abl-induced myeloproliferative disorder. Mol. Cell. Biol. 20, 1149–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Eklund E. A., Jalava A., Kakar R. (1998) PU.1, interferon regulatory factor 1, and interferon consensus sequence-binding protein cooperate to increase gp91phox expression. J. Biol. Chem. 273, 13957–13965 [DOI] [PubMed] [Google Scholar]
- 16. Eklund E. A., Kakar R. (1999) Recruitment of CREB-binding protein by PU.1, IFN-regulatory factor-1, and the IFN consensus sequence-binding protein is necessary for IFN-γ-induced p67phox and gp91phox expression. J. Immunol. 163, 6095–6105 [PubMed] [Google Scholar]
- 17. Atfi A., Prunier C., Mazars A., Défachelles A. S., Cayre Y., Gespach C., Bourgeade M. F. (1999) The oncogenic TEL/PDGFRβ fusion protein induces cell death through JNK/SAPK pathway. Oncogene 18, 3878–3885 [DOI] [PubMed] [Google Scholar]
- 18. Zhu C., Saberwal G., Lu Y., Platanias L. C., Eklund E. A. (2004) The interferon consensus sequence-binding protein activates transcription of the gene encoding neurofibromin 1. J. Biol. Chem. 279, 50874–50885 [DOI] [PubMed] [Google Scholar]
- 19. Huang W., Horvath E., Eklund E. A. (2007) PU.1, interferon regulatory factor (IRF) 2, and the interferon consensus sequence-binding protein (ICSBP/IRF8) cooperate to activate NF1 transcription in differentiating myeloid cells. J. Biol. Chem. 282, 6629–6643 [DOI] [PubMed] [Google Scholar]
- 20. Golub T. R., Barker G. F., Lovett M., Gilliland D. G. (1994) Fusion of PDGF receptor beta to a novel ets-like gene, tel, in chronic myelomonocytic leukemia with t(5;12) chromosomal translocation. Cell 77, 307–316 [DOI] [PubMed] [Google Scholar]
- 21. Kuwata T., Gongora C., Kanno Y., Sakaguchi K., Tamura T., Kanno T., Basrur V., Martinez R., Appella E., Golub T., Ozato K. (2002) γ-Interferon triggers interaction between ICSBP (IRF-8) and TEL, recruiting the histone deacetylase HDAC3 to the interferon-responsive element. Mol. Cell. Biol. 22, 7439–7448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huang W., Zhou W., Saberwal G., Konieczna I., Horvath E., Katsoulidis E., Platanias L. C., Eklund E. A. (2010) Interferon consensus sequence binding protein (ICSBP) decreases β-catenin activity in myeloid cells by repressing GAS2 transcription. Mol. Cell. Biol. 30, 4575–4594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jousset C., Carron C., Boureux A., Quang C. T., Oury C., Dusanter-Fourt I., Charon M., Levin J., Bernard O., Ghysdael J. (1997) A domain of TEL conserved in a subset of ETS proteins defines a specific oligomerization interface essential to the mitogenic properties of the TEL-PDGFRβ oncoprotein. EMBO J. 16, 69–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sharf R., Meraro D., Azriel A., Thornton A. M., Ozato K., Petricoin E. F., Larner A. C., Schaper F., Hauser H., Levi B. Z. (1997) Phosphorylation events modulate the ability of interferon consensus sequence binding protein to interact with interferon regulatory factors and to bind DNA. J. Biol. Chem. 272, 9785–9792 [DOI] [PubMed] [Google Scholar]
- 25. Toffalini F., Kallin A., Vandenberghe P., Pierre P., Michaux L., Cools J., Demoulin J. B. (2009) The fusion proteins TEL-PDGFRβ and FIP1L1-PDGFRα escape ubiquitination and degradation. Haematologica 94, 1085–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Larrick J. W., Fischer D. G., Anderson S. J., Koren H. S. (1980) Characterization of a human macrophage-like cell line stimulated in vitro. A model of macrophage functions. J. Immunol. 125, 6–12 [PubMed] [Google Scholar]
- 27. Oberley M. J., Farnham P. J. (2003) Probing chromatin immunoprecipitates with CpG-island microarrays to identify genomic sites occupied by DNA-binding proteins. Methods Enzymol. 371, 577–596 [DOI] [PubMed] [Google Scholar]
- 28. Garrone P., Neidhardt E. M., Garcia E., Galibert L., van Kooten C., Banchereau J. (1995) Fas ligation induces apoptosis of CD40-activated human B lymphocytes. J. Exp. Med. 182, 1265–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yanagisawa J., Takahashi M., Kanki H., Yano-Yanagisawa H., Tazunoki T., Sawa E., Nishitoba T., Kamishohara M., Kobayashi E., Kataoka S., Sato T. (1997) The molecular interaction of Fas and FAP-1. A tripeptide blocker of human Fas interaction with FAP-1 promotes Fas-induced apoptosis. J. Biol. Chem. 272, 8539–8545 [DOI] [PubMed] [Google Scholar]
- 30. Tamura T., Thotakura P., Tanaka T. S., Ko M. S., Ozato K. (2005) Identification of target genes and a unique cis element regulated by IRF-8 in developing macrophages. Blood 106, 1938–1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang H., Lindsey S., Konieczna I., Bei L., Horvath E., Huang W., Saberwal G., Eklund E. A. (2009) Constitutively active SHP2 cooperates with HoxA10 overexpression to induce acute myeloid leukemia. J. Biol. Chem. 284, 2549–2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Carroll M., Ohno-Jones S., Tamura S., Buchdunger E., Zimmermann J., Lydon N. B., Gilliland D. G., Druker B. J. (1997)) CGP 57148, a tyrosine kinase inhibitor, inhibits the growth of cells expressing BCR-ABL, TEL-ABL, and TEL-PDGFR fusion proteins. Blood 90, 4947–4952 [PubMed] [Google Scholar]
- 33. Rehli M., Poltorak A., Schwarzfischer L., Krause S. W., Andreesen R., Beutler B. (2000) PU.1 and interferon consensus sequence-binding protein regulate the myeloid expression of the human Toll-like receptor 4 gene. J. Biol. Chem. 275, 9773–97781 [DOI] [PubMed] [Google Scholar]
- 34. Kim Y. M., Im J. Y., Han S. H., Kang H. S., Choi I. (2000) IFN-γ up-regulates IL-18 gene expression via IFN consensus sequence-binding protein and activator protein-1 elements in macrophages. J. Immunol. 165, 3198–3205 [DOI] [PubMed] [Google Scholar]