Background: Reversible phosphorylation of the RNA Polymerase II CTD coordinates co-transcriptional recruitment of factors.
Results: Ssu72 is required for erasure of phospho-serine7, and it facilitates Fcp1-mediated phospho-serine2 removal.
Conclusion: Removal of phospho-Ser7 mark plays a key role in the transcription cycle.
Significance: Persistent negative charge at position 7 of the CTD renders cells non-viable, and Ssu72 plays a prominent role in removing phospho-Ser7.
Keywords: CDK (Cyclin-dependent Kinase), Chromatin Immunoprecipitation (ChiP), Phosphatase, Proteomics, RNA Polymerase II, CTD, Fcp1, S7E, Ser7, Ssu72
Abstract
The C-terminal domain (CTD) of the largest subunit of RNA polymerase II (Pol II) serves an important role in coordinating stage-specific recruitment and release of cellular machines during transcription. Dynamic placement and removal of phosphorylation marks on different residues of a repeating heptapeptide (YSPTSPS) of the CTD underlies the engagement of relevant cellular machinery. Whereas sequential placement of phosphorylation marks is well explored, genome-wide engagement of phosphatases that remove these CTD marks is poorly understood. In particular, identifying the enzyme that erases phospho-Ser7 (Ser7-P) marks is especially important, because we find that substituting this residue with a glutamate, a phospho-mimic, is lethal. Our observations implicate Ssu72 as a Ser7-P phosphatase. We report that removal of all phospho-CTD marks during transcription termination is mechanistically coupled. An inability to remove these marks prevents Pol II from terminating efficiently and will likely impede subsequent assembly into the pre-initiation complex.
Introduction
The C-terminal domain (CTD)5 of the largest subunit in RNA polymerase II (Pol II) serves as the primary point of contact for a wide variety of cellular machines involved in RNA biogenesis (1, 2, 3, 4). The CTD consists of the repeating heptapeptide Y1S2P3T4S5P6S7 of which each residue is subject to various post-translational modifications that can be read by the transcriptional machinery. The patterns of modification dictate the association or disassociation of complexes and may therefore define a possible “CTD code” (1, 5–7). During transcription initiation and promoter escape, phosphorylation of Ser-5 (Ser5-P) recruits enzymes required for capping nascent transcripts (8–10). Concurrently, Ser-7 is phosphorylated (Ser7-P), establishing a bivalent mark at both protein-coding (pc) and noncoding (nc) genes (11, 12, 13). Shortly after promoter clearance, Ser5-P is rapidly erased while phosphorylated Ser-2 (Ser2-P) and Ser7-P continue to accrue (14–16). These modifications recruit complexes required for transcription elongation, 3′-end processing, and transcription termination (17–19). Finally, all CTD marks are rapidly erased at the end of transcription, and the hypophosphorylated Pol II is licensed to assemble into the pre-initiation complex and re-initiate transcription (20–22). Taken together, dynamic phosphorylation and de-phosphorylation of the CTD plays a critical role at every stage of transcription.
While much effort has been devoted toward identifying the kinases that place phospho-CTD marks, the removal of these marks has been underexplored. This is surprising because the erasure of stage-specific phospho-CTD marks is critical for RNA biogenesis. The inability to remove these marks is often lethal, and in instances where it is not lethal, it severely impairs cellular growth (23, 24). The importance of erasing phospho-CTD marks is further emphasized by the consequences of disrupting the activity of CTD phosphatases. Among the many CTD phosphatases, three principal enzymes that act at different stages of transcription and on different phospho-CTD marks have been identified and are highly conserved across eukaryotes. Rtr1 de-phosphorylates Ser5-P at the 5′-end of genes, whereas Ssu72 works with the prolyl-isomerase Ess1 to remove the remaining Ser5-P marks at the cleavage and polyadenylation site (CPS) (22, 25–29). Fcp1 is the primary Ser2-P phosphatase, and it also acts primarily at the 3′ end of genes (21). Two of the phosphatases, Fcp1 and Ssu72, are essential; deletion of either phosphatase is lethal (30, 31). Temperature-sensitive mutants of Fcp1 and Ssu72 exhibit termination defects and significant read-through at Nrd1-dependent genes (32–34). Similar read-through at Nrd1-dependent genes has been observed upon deletion of Rtr1 (29). In contrast, inhibition or deletion of CTD kinases is better tolerated. Neither the primary Ser2-P kinase Ctk1/CDK12 nor the mediator-associated Ser5-P kinase Srb10/CDK8 are essential. Additionally, specific chemical inhibition of Kin28/CDK7, the primary promoter-proximal Ser5-P/Ser7-P kinase, does not have a significant influence on transcription (35). Thus, the data suggest that once the CTD marks are placed, erasing those marks during specific stages of the transcription cycle is crucial for cellular fitness and viability.
Despite its importance, there are significant gaps in our understanding of CTD de-phosphorylation. It is unclear if retention of negative charge at Ser-7 has any impact of cellular survival. It is unknown which phosphatase, or combination of phosphatases, removes Ser7-P. Additionally, the interdependency between phosphatases that act on different phospho-CTD marks remains to be explored. The placement of Ser7-P is closely coupled to the phosphorylation of Ser-5 and Ser-2 (11, 12, 14, 36, 37); it is possible the removal of Ser7-P may also be a coupled event. Finally, the relationship between the CTD kinases and phosphatases is not well characterized. Do these enzymes associate in a sequential and non-overlapping manner at specific stages during transcription, or is there simultaneous dynamic placement and removal of phospho-CTD marks during different stages of transcription? Thus, many important questions regarding the de-phosphorylation of the CTD remain to be addressed. Here we demonstrate that retention of negatively charged residues at position 7 is lethal. We also identify Ssu72 as a major Ser7-P phosphatase and reveal genome-wide distributions of Fcp1 and Ssu72. Finally, we observe coupled erasure of all phospho-CTD marks at 3′-ends of genes. Our results provide new insights into the role of sequential and dynamic remodeling of phospho-CTD marks in transcription termination and, more importantly, in the resetting of Pol II so as to permit its assembly in the pre-initiation complex.
EXPERIMENTAL PROCEDURES
Mutant CTD Construction and Plasmid Shuffle
Mutant Rpb1 CTDs were constructed and tested for viability in vivo as previously described (23) with some modifications. Briefly, the CTD repeats were constructed by annealing and ligating oligonucleotides containing mutant or WT codons at position 7: S7A (5′-CCGACTTCACCAGCTTATTCC-3′ and 5′-TCGGGGAATAAGCTGGTGAAG-3′), S7E (5′-CCGACTTCACCAGAATATTCC-3′ and 5′-TCGGGGAATATTCTGGTGAAG-3′), WT (5′-CCGACTTCACCAAGTTATTCC-3′ and 5′-TCGGGGAATAACTTGGTGAAG-3′). WT and mutant Rpb1 CTDs were cloned into pY1 (LEU2), which contains a CTD-less Rpb1 gene. pY1 constructs were transformed into Z26 (MATa his3Δ200 ura3–52 leu2–3,112 rpb1Δ187:: HIS3 GAL+ (pRP112)), which contains a URA3 linked WT Rpb1 gene (38). Single transformants were streaked on synthetic complete media (SC) lacking uracil and leucine, or on SC supplemented with 5-FOA (0.1%) (Toronto Research Chemicals) and incubated at 30 °C for 3 days.
Chromatin Immunoprecipitation on Microarray Chips (ChIP-chip)
All strains used are described in supplemental Table S1. Chromatin immunoprecipitation (ChIP) was performed as previously described (14) with several modifications. TAP-ChIP was performed using protein-IgG beads whereas the HA-ChIP was done using the 12CA5 antibody (Abcam). For the degron-dependent experiments, cells were grown to an A600 of 1.0 at 25 °C. The culture was then split with half of the cells in culture continuing under previous growth conditions whereas the other half was subjected to non-permissive temperature of 37 °C for one hour before both sets of cultures were crosslinked and harvested. ChIP was performed using an anti-Rpb3 antibody (Neoclone) against Pol II and phospho-CTD specific antibodies against Ser2-P (Bethyl) and Ser7-P (a gift from Dr. Dirk Eick). All ChIP samples were amplified using ligation-mediated PCR and hybridized to high density tiling microarrays from NimbleGen (Roche NimbleGen, Inc.; Madison, WI). ChIP-chip immunoprecipitated (IP) data were mean-scaled against its respective “Input” sample data and then the ratio of scaled IP to the Input was log2 transformed. The data were subjected to computational repeat sequence masking based on probe sequence repetitiveness relative to the sequence composition of the probes on the microarray. A moving average was used to smooth the microarray data, and baseline corrections were applied through comparison between polymerase and transcript data.
Average transcription unit analysis was applied to the data to obtain occupancy profiles normalized for gene length for a set of well isolated protein-coding genes (14). This allowed for alignment of the occupancy patterns for genes of varying lengths. k-means clustering was performed to partition the genes into categories based on their occupancy profiles. These clusters were then manually collapsed into visually distinct representative groups.
Multidimensional Protein Identification Technology (MudPIT) Analysis of Ssu72
Purified protein samples were TCA-precipitated, urea-denatured, reduced, alkylated, digested with LysC/Trypsin enzymes, and then analyzed by LTQ linear ion trap MS equipped with a nano-LC electrospray ionization source (ThermoFisher) coupled with a Quaternary Agilent 1100 series HPLC pump (Agilent Technologies, PaloAlto, CA) as described previously (39) (40). The samples were loaded onto a 3-phase column containing C18 reverse phase particles (Aqua, Phenomenex), followed by strong cation exchange resin (Partisphere SCX, Whatman), and finally a fused silica microcapillary column. The column was placed in line with mass spectrometer and fully automated 12-step MudPIT run was performed as described previously. Each full MS scan (from 400 to 1600 m/z range) was followed by five MS/MS events using data-dependent acquisition, and the top five intense ions of each MS scan subjected to Collision Induced 14 Dissociation (CID). A label-free spectral counting approach named the distributed Normalized Spectral Abundance Factor (dNSAF) was applied for protein quantification and an in-house developed NSAF v7 was used to create the final reports (41). The NSAF v7 calculations take the consideration of protein length and the relative spectral abundance of proteins across various preparations to prevent the redundancy in spectral assignment, and calculate their respective dNSAF values.
Phosphatase Assay
Reactions were performed with ∼5 pmol of GST-CTD (42) phosphorylated with Kin28-TAP (TFIIK), Ctk1-TAP (CTDKI), or activated recombinant MAPK (Millipore) in the presence of 1 mm ATP for 1 h at 30 °C. Residual ATP was removed using gel filtration spin columns as previously described (29). Phosphatase reactions were performed in phosphatase buffer (50 mm Tris-HCl, pH 6.5, 10 mm MgCl2, 20 mm KCl, and 5 mm dithiothreitol) through incubation of recombinant GST-purified Ssu72 with the phosphorylated substrates for 1 h at 30 °C (29). Increasing concentrations of Ssu72 (1.25, 2.5, and 5 pmol) were used for each experiment. Reactions were quenched by the addition of SDS loading buffer and incubation at 98 °C for 5 min prior to analysis of the reaction products by SDS-PAGE and Western blotting. The extent of GST-CTD de-phosphorylation was assessed using antibodies specific for the serine 5-, serine 2-, or serine 7-phosphorylated forms of the CTD as indicated, and quantitation of the reactions was performed using densitometry.
RESULTS
Substituting Serine 7 with the Phospho-mimic Glutamate Is Lethal
Although serine 7 of the CTD hepapeptide is conserved from yeast to humans, its importance has been traditionally overlooked because of mutational studies that eliminated this mark. Mutating serine 7 to alanine (S7A) in the budding yeast Saccharomyces cerevisiae causes growth defects, but the cells are still viable (43). Similar results were reported in fission yeast and mammalian cells (44, 45). Importantly, the inability to erase this mark or remove the negative charge at this position during different stages of transcription has not been explored in yeast. To test the effects of a persistent Ser7-P mark on cellular viability, we constructed a CTD with serine 7 mutated to the phospho-mimic glutamate (S7E). Because the CTD of the largest subunit of Pol II is essential, we used a plasmid shuffle strategy developed by West and Corden (23) to co-express wild type and mutated proteins in the same cells. The wild type Rpb1 is expressed from a plasmid bearing the URA3 auxotrophy marker whereas the mutant Rpb1 is expressed from a second plasmid bearing a different auxotrophy marker. The URA3 gene converts 5-fluoroorotic acid (5-FOA) to a potent toxin and cells bearing this plasmid are unable to grow. Strains that lose the URA3 bearing plasmid during cell division are able to survive in 5-FOA, and under the appropriate selection conditions, these strains retain the plasmid bearing the mutant version of the CTD. The physiological consequences of a given mutation can be monitored in the strains that survive in the presence of 5-FOA. We found that CTD bearing the S7E mutation was unable to grow on plates with 5-FOA, suggesting that the S7E mutation is lethal in yeast (Fig. 1). Several independent transformants were tested (#1–6 in Fig. 1), and while each transformant grew in the appropriate selection media, indicating that both plasmids were present in the strains, none of them grew in the presence of 5-FOA. In contrast, substituting serine 7 with an alanine (S7A) only hindered growth (supplemental Fig. S1). Our results suggest that while the placement of Ser7-P mark aids cellular function, its removal is crucial for survival. Thus, identifying the Ser7-P phosphatase is vital for understanding the regulation and dynamics of this mark and its vital importance in maintaining cellular viability.
FIGURE 1.
Substitution of serine 7 with glutamate is lethal. The URA3+ strain Z26 was transformed with the indicated plasmids (right panel). Plasmid shuffle was performed on synthetic complete plates in the presence (SC +5-FOA) or absence (SC -Leu -Ura) of 5-fluoroorotic acid. Cells transformed with the plasmid that expresses Rpb1 bearing the WT CTD were viable, whereas cells transformed with a deleted CTD (ΔCTD) were inviable. None of the independent transformants bearing the serine 7 to glutamate substitution (S7E CTD clones #1–6) were able to support growth.
Coincidence of Specific Phospho-CTD Marks with Specific Kinases and Phosphatases
As a first step toward understanding the dynamics and regulation of phospho-CTD marks, we examined the distribution of known CTD phosphatases and kinases across the genome. We reasoned that the precise sites of association of these CTD-modifying enzymes would provide insight into the identity of the Ser7-P phosphatase. We examined the occupancy of these factors by performing chromatin immunoprecipitation on tiled microarrays of the yeast genome (ChIP-chip). ChIP was performed against epitope-tagged CTD kinases and phosphatases, and these profiles were aligned against prior phospho-CTD profiles (14). Two representative genes, the nc-gene SNR13 and the pc-gene SED1, were selected because of their canonical phospho-CTD profiles: Ser5-P (red) and Ser7-P (purple) show maximal abundance at the transcription start site (TSS), while on protein coding (pc)-genes Ser2-P (green) levels accrue during early elongation reaching maximal levels by 500 base pairs and are retained at elevated levels until past the 3′-end of SED1 (Fig. 2A). These profiles are consistent across the compilation of snoRNAs less than 100 bp (SNR) and a set of well-isolated pc-genes. Importantly, the positions of the three primary CTD kinases are in remarkable agreement with phospho-CTD patterns (Fig. 2B). Kin28 (maroon) is positioned at the TSS at nc- and pc-genes, consistent with its role as the promoter-proximal Ser5-P and Ser7-P kinase. We find that Bur1 (blue) consistently associates upstream of Ctk1 (light green). The reproducible detection of this subtle difference in binding profiles on a genome scale is significant and consistent with previous single gene studies that showed sequential recruitment of Bur1 by promoter-proximal Ser5-P marks and subsequent Ctk1 recruitment due to Bur1 phosphorylation of Ser-2 (36, 37). This observation was further validated by quantitative ChIP-PCR analysis at the pc-gene GLN1 (supplemental Fig. S2, A and B). Interestingly, while Bur1 and Ctk1 are present at similar levels at pc-genes, Ctk1 is strikingly under-represented at nc-genes while Bur1 occupancy resembles levels observed at pc-genes. These data indicate surprising gene class-specific differences in CTD kinase recruitment, but they do not rule out the possibility that lower levels of Ser2-P on nc-genes may arise from unchecked activity of Ser2-P phosphatases.
FIGURE 2.
Profiles of CTD phosphorylation, kinases, and phosphatases. A, phospho-CTD ChIP-chip profiles at representative single genes (SNR13 and SED1) and across a compilation of short nc genes (SNRs) and a set of well isolated pc genes (data from Tietjen et al., 14). All three phospho-CTD modifications profiles are displayed: Ser2-P (green), Ser5-P (red), and Ser7-P (purple). The black arrow represents the transcription start site (TSS), the red bar the cleavage and polyadenylation site (CPS), and the black boxes the translation boundaries. The x axis displays the distance in bp from the TSS, while the y axis shows the fold enrichment of the immunoprecipitation over input on a log2 scale. B, CTD kinase and phosphatase profiles at nc- and pc-genes. Three CTD kinases (Bur1, blue; Ctk1, light green; Kin28, maroon) and two CTD phosphatases (Fcp1, dark green; Ssu72, orange) were analyzed. The y axis shows the percentage of maximum fold enrichment of the immunoprecipitation over input on a log2 scale.
We focused our efforts on Fcp1 and Ssu72 because studies with an rtr1Δ strain suggested that Rtr1 is not involved in Ser7-P de-phosphorylation (12). Despite their importance, the distribution of these phosphatases across the genome remains unknown. Additionally, clear discrepancies exist between earlier gene-specific ChIP-PCR studies of Ssu72. In one study Ssu72 was found associated both at the 5′- and the 3′-ends of transcription units (46) whereas the other study only detected Ssu72 occupancy at 3′-ends of genes (47). To resolve this discrepancy and explore the sites of action of CTD phosphatases, we performed genome-wide location analysis (ChIP-chip) using HA-tagged Ssu72 (orange) and TAP-tagged Fcp1 (dark green) (Fig. 2B and supplemental Figs. S2C–S3). We observe overlapping peaks for Fcp1 and Ssu72 across SNR13 and other snoRNAs. To our knowledge, this is the first report of Fcp1 association at nc-genes. The strong presence of Fcp1, in combination with the reduced levels of Ctk1 at nc-genes, is likely responsible for the low Ser2-P levels observed across this class of genes. At pc-genes, Fcp1 is detected shortly after Bur1 and Ctk1 associate with the elongating polymerase and it peaks at the CPS after Bur1 and Ctk1 have dissociated. In contrast, we observe a low-abundance Ssu72 peak near the TSS and a much larger peak at the CPS of the gene, but it is relatively depleted within the open reading frame (ORF). The profile of Ssu72 is consistent with earlier studies that found Ssu72 associated with both 5′ and 3′-ends of genes, as opposed to being only at the 3′-end (46, 47). The position of the Ssu72 peaks corresponds with regions of rapidly decreasing Ser7-P, suggesting Ssu72 may be involved in Ser7-P de-phosphorylation.
MudPIT Analysis Suggests That Ssu72 Can Exist by Itself or in Complex with APT
The genome-wide occurrence of Ssu72 at the 5′- (TSS) and 3′-ends (CPS) of genes poses the question of whether Ssu72 is involved with different complexes at these sites. Ssu72 was initially thought to be involved in start site selection with TFIIB, but later studies found Ssu72 associated with components of the Pta1 (APT) complex and the Cleavage and Polyadenylation Factor (CPF) complex (30, 47–49). To characterize Ssu72-interacting proteins, we performed MudPIT analysis using Ssu72 as bait. Our pull-down identified, at low levels, several transcription initiation proteins, including components of the TFIID complex (TAF3 and TAF6) and regulators of the TFIIH complex (MET18) (supplemental Table S2). We also detected the Pol II subunit Rpb2, consistent with previous pull downs of Ssu72 (48). However, the lower spectral abundance (dNSAF) suggests that these proteins are involved in only transient interaction with Ssu72. Surprisingly, in contrast with published results, we did not detect Kin28/TFIIH or TFIIB in complex with Ssu72 (34). We hypothesize that Ssu72 may be not be directly involved in transcription initiation, but rather, Ssu72 may facilitate the transition from initiation to elongation after Kin28/TFIIH dissociates from Pol II during promoter escape. We also observe a high abundance of ribosomal proteins in our MudPIT analysis. While ribosomal proteins often represent contaminants from the protein purification, other CTD remodelers like Ctk1 have been shown to interact with the ribosome and play an important role in translation (50). The intriguing possibility that Ssu72 may be involved in translation remains to be explored.
MudPIT also identified every component of the APT and CPF complexes as Ssu72-interacting partners (Fig. 3A). These components were present in all nine replicates (three technical replicates of three biological samples), indicative of a strong in vivo interaction (Fig. 3, B and C). The interaction of Ssu72 with the APT and CPF complexes is consistent with past reports and its well defined role in termination and 3′-processing of nc-genes and short pc-genes (33, 34, 47, 51). We did not find any member of the Cleavage Factor 1 (CF1) complex in our compilation of Ssu72-associated proteins, even though CF1 is part of the holo-CPF complex. However, this result is consistent with published ChIP-chip traces of the CF1 member Pcf11 (15), which typically peaks immediately after Ssu72 levels begin to drop (supplemental Fig. S4). Interestingly, the dNSAF values for Ssu72 bait were substantially higher than those of the APT (p = 1.04 × 10−5) and CPF (p = 3.33 × 10−6) complex members, especially when compared with pull-downs of Pol II subunits (29). The disproportionately high levels of Ssu72 suggest a possible pool of Ssu72 that is not in complex with APT/CPF (Fig. 3C). It is possible that this form of the phosphatase associates with TFIIB and the promoter-proximal transcriptional machinery and represents the low-abundance ChIP peak at the 5′-ends of pc-genes. However, this possibility remains to be verified.
FIGURE 3.
Ssu72, but not co-complex APT phosphatase Glc7, de-phosphorylates Ser7-P. A, identification of Ssu72-associated complexes via MudPIT. Pull-down of TAP-tagged Ssu72 identified every component of the APT complex (orange) and the CPF complex (gray), including the APT phosphatase Glc7 (white and bold). B, distributed normalized spectral abundance factor (dNSAF) of Ssu72-associated proteins from the APT and CPF complex is displayed for three biological replicates. C, dNSAF of Ssu72-associated proteins from the APT and CPF complex averaged across all biological and technical replicates. Ssu72 was present at much greater abundance compared with the average APT (p = 1.04 × 10−5) or CPF (p = 3.33 × 10−6) complex member. D, Ser7-P ChIP-chip profiles upon depletion of degron-tagged (td) Ssu72 (left panel). Replacing it with the catalytically inactive C15S mutant (dashed purple) caused a significant increase in Ser7-P at non-permissive temperature (37 °C) compared with replacement with wild type Ssu72 (solid purple). Ser7-P was elevated at nc- (SNR13 and SNR) and pc-genes (SED1 and protein coding). Ser7-P ChIP-chip was also performed in the Glc7-td strain at permissive (25 °C) and non-permissive temperature (37 °C). Only a small increase at the 5′-end of pc-genes was observed at non-permissive temperatures in a Glc7-td strain (right panel). All Ser7-P traces were normalized to Pol II. E, wild type GST-Ssu72 directly de-phosphorylated a Kin28/TFIIK or MAPK phosphorylated GST-CTD substrate at Ser5-P and Ser7-P in a concentration-dependent manner. Ssu72 was not able to de-phosphorylate Ser2-P on a Ctk1/CTDK-1-phosphorylated GST-CTD substrate. F, histograms represent the quantification of the phosphatase assays from E. The y axis shows the percent of CTD phosphorylation in the sample untreated with phosphatase.
Ssu72 De-phosphorylates Ser7-P While Glc7 Does Not
Glc7, a member of the APT/Ssu72 complex, is a phosphatase involved in a wide variety of cellular processes including the processing of nc-genes (52, 53). Consistent with its association with the APT complex, Glc7 also localizes to the 3′-end of genes (47). Because of the promiscuity of Glc7, a PP2A phosphatase, it is possible that Glc7 also functions as a Ser7-P phosphatase. Therefore, we sought to investigate if Ssu72 and/or Glc7 influenced Ser7-P levels in vivo. We assessed Ser7-P levels across the genome following Ssu72 or Glc7 depletion. Ssu72 and Glc7 were depleted via a degron-tag strategy (Ssu72-td and Glc7-td) (22, 54), where the degron-tagged protein is degraded upon shifting to non-permissive temperatures (37 °C). Because Ssu72 is a part of an essential complex in termination and 3′ processing, we also accounted for any non-enzymatic structural roles Ssu72 may have by using a strain of Ssu72-td that co-expressed either the wild type (WT) Ssu72 or a catalytically inactive Ssu72-C15S (C15S) mutant. Depleting Ssu72 in the C15S mutant (purple dotted line) caused a significant increase in Ser7-P levels compared with WT (purple solid line) (Fig. 3D). Elevated Ser7-P levels were observed at the single genes SNR13 and SED1as well as across the compilation of nc- and pc-genes. The increase in Ser7-P begins at the TSS and was maintained across the ORF and into the CPS. The position of Ser7-P increase correlates closely with the location of the Ssu72 protein, strongly implying that Ssu72 is a Ser7-P phosphatase that acts at both nc- and pc-genes (Fig. 2B). In contrast, the elevated levels of Ser7-P across the transcript were not observed in the Glc7-td strain at non-permissive temperatures. At nc-genes, such as SNR13 a small increase in Ser7-P is observed, but across a collection of short snoRNAs, there is no significant change in Ser7-P levels or profiles. At pc-genes, we see a distinct increase in Ser7-P near the TSS, consistent with the promoter-proximal Ssu72 ChIP peaks. However, the higher Ser7-P levels in Glc7-td were not maintained across the transcript. Therefore, the increase may be due to indirect perturbations of the transcriptional machinery.
Although we observe a significant increase in Ser7-P upon depleting Ssu72, it is possible that Ssu72 is playing an indirect role in vivo. Therefore, we decided to test if Ssu72 can directly remove Ser7-P marks. Recombinant GST-fused WT Ssu72 was affinity purified from Escherichia coli using glutathione beads. Increasing amounts of phosphatase were challenged with a GST-CTD substrate phosphorylated by the Ser5-P/Ser7-P kinase Kin28 or by the nonspecific kinase MAPK. The level of phosphorylation following the phosphatase assay was evaluated via immunoblots with phospho-specific Ser5-P or Ser7-P antibodies. We observe a concentration-dependent decrease in Ser5-P and Ser7-P levels upon addition of the Ssu72 WT (Fig. 3, E and F). The phosphatase activity is specific, as Ssu72 is unable to de-phosphorylate Ser2-P marks placed by Ctk1 (Fig. 3, E and F). Our biochemical analysis supports the conclusion that Ssu72 can directly erase Ser7-P and is also consistent with the altered in vivo profiles of Ser7-P in catalytically inactive Ssu72 strains. Taken together, these results suggest that Ssu72 is a key Ser7-P phosphatase in vivo.
Genomic Distribution of CTD Phosphatases
Previous genome-wide phospho-CTD profiles revealed gene-class specific distributions of CTD marks that corresponded to the different termination mechanisms at these genes (14). To assess the diversity of CTD phosphatase patterns, Ssu72 and Fcp1 profiles across well isolated pc-genes were sorted by unrestrained k-means clustering, which partitions the genes into k number of clusters based on the distinct profiles observed at different genes. We observe four main clusters of profiles for HA-Ssu72 (5′, 5′+3′, CPS, and Post CPS) (Fig. 4A and supplemental Figs. S5 and S6). The vast majority of genes show Ssu72 enriched at the CPS or immediately after the CPS (Post CPS) with a low-abundance peak at the 5′-end. On a genome-wide level, we do not observe a change in the shape of Ssu72 profiles as a function of gene expression or transcript length (Fig. 4B) (55), although the magnitude of Ssu72 occupancy is higher in well-transcribed genes. The widespread recruitment of Ssu72 to 3′-ends of pc-genes is consistent with a role in termination and 3′ RNA processing of most, if not all, Pol II-dependent genes. This expands the role of Ssu72 from only acting at nc-genes and a subset of pc-genes to every Pol II transcript (33, 34, 48, 51).
FIGURE 4.
Genome-wide distribution of Ssu72. A, representative clusters from the k-means clustering analysis (see “Experimental Procedures”) for Ssu72 across well isolated protein-coding genes using average transcription unit analysis. Four profiles emerged from the clustering: 5′-enriched (4 genes), 5′ + 3′ enriched (9), CPS-enriched (31 genes), and post-CPS-enriched (13 genes). The pie chart diagram shows the distribution of genes within each cluster, with the majority of the genes being enriched at the CPS or post CPS. B, Ssu72 profile at genes of various expression levels (left panel) and transcript lengths (right panel). All genes ≥ 320 bp in length were represented in this analysis.
Remarkably, four clusters of profiles of the Ser2-P phosphatase Fcp1 (Uniform, 5′, CPS, Post CPS) emerged from k-means clustering (Fig. 5A and supplemental Figs. S7 and S8). Importantly, genes with higher levels of Fcp1 within the ORF (Uniform profile) are generally longer genes that are highly expressed (Fig. 5B). Unlike Ssu72, whose promoter-proximal peak is offset from the Kin28 kinase, there is significant overlap between the uniform Fcp1 profile and its kinase counterparts Bur1 and Ctk1. Importantly, three of the k-means clustered profiles (Uniform, CPS, Post CPS) display maximal occupancy at the CPS. These Fcp1 maxima occur after Bur1 and Ctk1 have dissociated, which provides a window of opportunity for Fcp1 to completely strip the remaining Ser2-P marks off Pol II. Interestingly, the Fcp1 peaks at the CPS overlap with the 3′ Ssu72 peak. Previous studies have demonstrated that the placement of phospho-CTD marks is coupled (14, 36, 37); however, the interdependencies between CTD phosphatases and removal of the CTD marks come into focus from our genome-wide occupancy profiles.
FIGURE 5.
Genome-wide distribution of Fcp1 and the coupled de-phosphorylation of the CTD. A, four distinct profiles were visualized upon k-means clustering for Fcp1-TAP: uniformly enriched (16 genes), 5′-enriched (11), CPS- enriched (25), and post-CPS-enriched (8). B, Fcp1-TAP profile at genes of various expression levels (left panel) and transcript lengths (right panel). All genes ≥ 320 bp in length were represented in this analysis. C, Ser2-P ChIP-chip profiles upon depletion of Ssu72-td with either the WT (solid green) or C15S mutant (dotted green) as replacement. The Ser2-P traces were normalized to Pol II. The position of Fcp1 (brown) overlays the Ser2-P profiles. Single gene traces and gene compilations for non-coding and protein-coding genes are displayed.
CTD De-phosphorylation Is Coupled at the CPS
The overlap between Ssu72 and Fcp1 peaks prompted us to investigate whether de-phosphorylation of the different CTD marks is coupled. We examined genome-wide Ser2-P profiles following degron-dependent depletion of Ssu72-td. We observed a strong increase in Ser2-P levels at nc-genes in strains bearing the catalytically inactive Ssu72-C15S (dotted green trace) as compared with strains with Ssu72-WT (solid green) (Fig. 5C). The rise in Ser2-P levels is apparent even after normalization to Pol II. This increase is present across the transcribed region of the SNR13 gene as well as across a compilation of short snoRNAs. At pc-genes, we observe an increase in Ser2-P primarily at the 3′-end of genes near the CPS, but we detect no difference in Ser2-P levels at the TSS and across the ORF of well isolated pc-genes. In contrast to the change in Ser2-P patterns, the increase in Ser7-P upon Ssu72 depletion is preserved across the transcribed region rather than being restricted only to the CPS. This observation suggests that rather than directly removing Ser2-P, Ssu72 may have an indirect effect on Ser2-P erasure, perhaps through the Ser2-P phosphatase Fcp1. Indeed, we observe a striking overlap in the peak position of Fcp1 (brown) around the CPS with the 3′ increase in Ser2-P. In contrast, Fcp1 does not overlap with Ssu72 at the TSS or within the transcribed region of well isolated pc-genes, which could explain why there is no increase in Ser2-P at these sites. Importantly, Ser2-P levels do not change within the ORF where Ssu72 is diminished. Our results suggest Ssu72 activity may be important for Fcp1 function, thereby coupling the de-phosphorylation of Ser2-P to the removal of Ser7-P and Ser5-P.
Inactivation of Ssu72 Triggers Transcription Read-through and May Impede Re-initiation
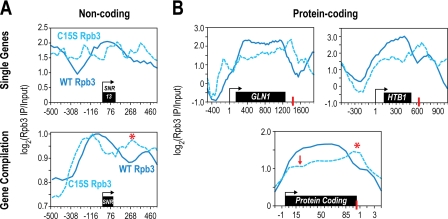
Phosphorylation of the CTD is intricately involved in the recruitment of factors important in transcription termination and processing. Therefore, we investigated whether the perturbation of phospho-CTD marks from the inactivation of Ssu72 influences transcription termination. We observed elevated levels of Pol II in the Ssu72 C15S strain at the 3′-end of several nc-genes, including SNR13 (Fig. 6A). Elevated Pol II at the 3′-end is often indicative of transcription read-through, which would be consistent with previous temperature-sensitive mutants of Ssu72 (33, 34, 56). To our surprise, we also observe increased Pol II at the 3′-end of most, if not all, protein-coding genes (Fig. 6B). Read-through at pc-genes in Ssu72 mutants has been previously attributed to terminator read-through of an upstream non-coding transcript (28, 34). However, we observe persistent Pol II at the CPS of pc-genes GLN1, HTB1, and AGP1, which are devoid of upstream nc-genes or cryptic unstable transcripts (CUTs) (Fig. 6B and supplemental Fig. S9). Our results strongly suggest that Ssu72 is playing a significant role in the termination of all pc-genes, and this function is not dependent on read through of upstream transcription of nc-genes. Intriguingly, Pol II levels at the TSS of pc-genes decrease in the Ssu72 C15S mutant strains as compared with those with Ssu72 WT. The lower levels of Pol II within genes suggest that there is a defect in transcription initiation by Pol II. These results are consistent with the inference that inactivating Ssu72 prevents the de-phosphorylation of the CTD, which then impedes Pol II assembly into the promoter-bound preinitiation complex, thereby decreasing the reinitiation of the next round of transcription (Fig. 7).
FIGURE 6.
Pol II read-through at non-coding and protein-coding genes. A, Pol II ChIP-chip profiles upon depletion of Ssu72-td in strains bearing the WT (solid blue) or catalytically inactive C15S mutant (dashed blue). Pol II was elevated at the 3′-end of SNR13 and a compilation of short SNRs, indicative of read-through (star). B, Pol II levels remain high beyond the CPS of several protein-coding genes (GLN1 and HTB1) and across a compilation of widely distributed protein coding genes that are well separated from other transcribed regions (red asterisk). However, Pol II levels are reproducibly lower during early elongation of protein coding genes (see WT (red arrow) compared with C15S mutant), suggesting that the inability to erase phospho-CTD marks during termination compromises subsequent rounds to transcription.
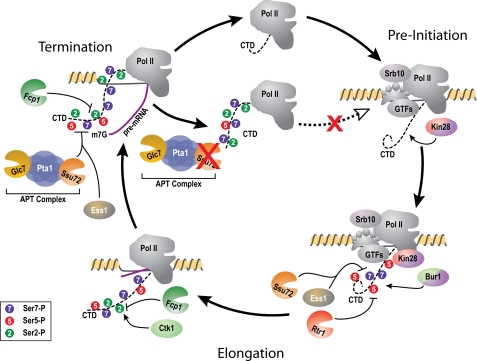
FIGURE 7.
Dynamic and sequential remodeling of RNA Polymerase II. Pol II arrives at the pre-initiation complex (PIC) with its CTD in a hypophosphorylated state. During transcription initiation and promoter escape, Kin28 and Srb10 phosphorylate Ser-5 residue of the repeating CTD heptad. Rtr1 and possibly promoter-proximal Ssu72 remove this mark during promoter clearance. Ser-7 is phosphorylated by Kin28 and is de-phosphorylated by Ssu72. Bur1, which places Ser2-P marks to prime the CTD for the association of the primary Ser2 kinase Ctk1, also places additional Ser7-P marks on the elongating Pol II. At the end of transcription, APT-associated Ssu72 removes the remaining Ser5-P and Ser7-P marks with the help of the prolyl-isomerase Ess1. The de-phosphorylation of Ser5-P and Ser7-P allows Fcp1 to remove Ser2-P, returning the CTD to a hypophosphorylated state and licensing it for re-initiation. If Ssu72 is unable to de-phosphorylate Ser5-P and Ser7-P, the CTD remains in a hyperphosphorylated state and is unable to effectively terminate transcription or assemble into the pre-initiation complex leading to cellular lethality.
DISCUSSION
In this study, we demonstrate that the essential phosphatase Ssu72 de-phosphorylates Ser7-P on the CTD of RNA polymerase II. We find that depletion of Ssu72 in vivo results in an increase in Ser7-P levels at nc- and pc-genes. Furthermore, the site of increase in Ser7-P correlates exceedingly well with the genomic association profiles of Ssu72. A particularly important finding is that substituting Ser-7 with a glutamate, a phospho-mimic, is lethal. In other words, erasure of the Ser7-P mark is crucial for cellular viability. These results are consistent with studies in mammalian cells where the inability to place phosphoserine marks on alanine substituted (S7A) CTD specifically hinders processing of snRNA genes (45) whereas glutamate substitutions (S7E) lead to rapid cellular inviability (24). The S7E mutants displayed defective transcription and lowered 3′-end processing of transcripts (45). Our results strongly suggest that Ssu72 erases Ser7-P marks on Pol II immediately after cleavage and polyadenylation and resets the terminating Pol II to a hypo-phosphorylated state. While the identification of Ssu72 does not rule out the existence of other Ser7-P phosphatases, it is clear that the erasure of phospho-CTD marks is important for licensing Pol II to assemble into the promoter-bound pre-initiation complex to start the next round of transcription. Inactivation of Ssu72 leads to persistent Ser7-P marks that impede Pol II recruitment to PIC, thereby blocking transcription initiation and resulting in cellular lethality (Fig. 7).
Ssu72 activity is also crucial for transcription-coupled 3′ processing and termination of pc-genes (26, 33, 51). At pc-genes, these processes are facilitated through the cooperative binding of Pcf11 and Rtt103 to Ser2-P marks (56–58). In vitro assays have demonstrated that other CTD marks reduce the association of Pcf11/Rtt103 with Ser2-P. It is possible that Pcf11/Rtt103 requires de-phosphorylation of Ser5-P and Ser7-P by Ssu72 in order to associate with the 3′ elongation complex. Reinforcing this idea, Pcf11 peaks immediately after Ssu72 at pc-genes, and the position of Pcf11 corresponds with the decline in Ser7-P. When Ssu72 is depleted, we observe elevated levels of Pol II extending past the CPS of several pc-genes, which is often indicative of read-through and processing/termination defects. Taken together, the removal of Ser7-P by Ssu72 may be essential for proper transcription processing, termination, and re-initiation.
While it is tempting to assume that Ssu72 at the TSS is involved in CTD de-phosphorylation during PIC assembly, Ssu72 actually peaks after Kin28/TFIIH, the last general transcription factor to arrive during initiation (59). Therefore, it is likely that the Ssu72 serves a different function at this site. From our mass spectrometric MudPIT analysis, we found that the levels of Ssu72 identified were significantly higher compared with other interacting proteins in the APT complex. Although it is possible that the Ssu72 is present in the APT complex at a higher average stoichiometry, the high bait values might reflect a pool of cellular Ssu72 that is not in the APT complex. Such pool of Ssu72 might localize to the TSS and facilitate the transition between CTD modification states. At the promoter-proximal site, Ssu72 abundance increases as Kin28 levels rapidly decline. Interestingly, we did not find Kin28 associated with Ssu72 in our pull downs, suggesting they may interact transiently and likely interact with the CTD substrate sequentially. The subsequent decline of Ssu72 levels in the promoter-proximal region coincides with rising Bur1 and Ctk1 levels. In contrast to the offset in Kin28-Ssu72 profiles, there is significant overlap between the Ser2-P kinases Bur1/Ctk1 and the Ser2-P phosphatase Fcp1, especially at highly expressed genes. It is possible that such association of kinase-phosphatase function lead to dynamic remodeling of Ser2-P marks on the elongating polymerase, thereby priming Pol II for rapid monitoring of signals that favor a switch between elongation and termination. Indeed, competition between cyclin-dependent kinases and protein phosphatases for the same substrate binding site has been proposed as a regulation mechanism for the retinoblastoma tumor suppressor protein (60). The elevated Fcp1 levels across the transcribed region may be important to control Ser2-P levels, as the occupancy of Ser2-P kinases Bur1 and Ctk1 increase at highly expressed genes (supplemental Fig. S10). The interplay of CTD kinases and phosphatases may greatly influence the association and dissociation of elongation and termination factors. Intriguingly, the two peaks of Ssu72 correspond with two transition states in the transcription cycle. The promoter-proximal Ssu72 peak coincides with the association of a broad range of transcription elongation factors, while the 3′ CPS-proximal Ssu72 peak occurs as many of these factors are dissociating (15). Ssu72 activity at these sites likely plays an important role in facilitating these key transitions.
Traditionally, Ssu72 has been described as a Ser5-P phosphatase (22), and several laboratories have recently published crystal structures of Ssu72 bound to a Ser5-P CTD substrate (26, 27). Additionally, Ssu72 was originally described as a tyrosine phosphatase (61). It is possible that the Ssu72 active site may be flexible enough able to support a variety of CTD modifications. The identification of Ssu72 as a Ser7-P phosphatase emphasizes the tight coupling between the Ser5-P and Ser7-P marks. Thus, these two CTD marks share a common kinase, Kin28, and a common phosphatase, Ssu72. The placement of Ser7-P is coupled to the phosphorylation of Ser5-P: no Ser7-P is detected on CTD bearing S5A mutations (24). Although it is unclear whether Ser5-P needs to be removed prior to Ser7-P de-phosphorylation, our results suggest that Ser5-P and Ser7-P erasure is coupled to Ser2-P removal during transcription termination. This is consistent with reported genetic interactions between Ssu72 and Fcp1 (34). In essence, we report that Ssu72 plays a central role in removal of Ser7-P and that the erasure of all phospho-CTD marks at 3′-ends is intricately coupled. The efficient removal of these marks is critical for 3′-processing and termination of Pol II-dependent transcripts. Furthermore, erasure of phospho-CTD marks would license Pol II that is just released after a cycle of transcription for re-association with the pre-initiation complex and progression through the next round of transcription.
Supplementary Material
Acknowledgments
We thank Michael Hampsey, Claire Moore, and Jack Greenblatt for sharing the degron-tagged Ssu72, degron-tagged Glc7, and Ssu72-TAP strains, respectively. We thank Dirk Eick for generously sharing his anti-CTD antibodies.
This work was supported, in whole or in part, by National Institutes of Health NHGRI Training Grant to the Genomic Sciences Training Program (5T32HG002760) (to J. B. R.). This work was also supported by NSF (MCB 07147), W. M. Keck, Shaw Scholar, and Vilas Associate awards (to A. Z. A.).

This article contains supplemental Figs. S1–S10 and Table S1.
- CTD
- C-terminal domain
- Pol II
- RNA polymerase II
- CID
- collision-induced 14 dissociation
- dNSAF
- distributed Normalized Spectral Abundance Factor
- 5-FOA
- 5-fluoroorotic acid
- pc
- protein coding
- nc
- noncoding
- MudPIT
- Multidimensional Protein Identification Technology
- CF
- cleavage factor
- CPF
- core cleavage and polyadenylation factor
- APT
- associated with Pta1.
REFERENCES
- 1. Phatnani H. P., Greenleaf A. L. (2006) Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 20, 2922–2936 [DOI] [PubMed] [Google Scholar]
- 2. Buratowski S. (2009) Progression through the RNA polymerase II CTD cycle. Mol. Cell 36, 541–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Richard P., Manley J. L. (2009) Transcription termination by nuclear RNA polymerases. Genes Dev. 23, 1247–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Perales R., Bentley D. (2009) “Cotranscriptionality”: the transcription elongation complex as a nexus for nuclear transactions. Mol. Cell 36, 178–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Buratowski S. (2003) The CTD code. Nat. Struct. Biol. 10, 679–680 [DOI] [PubMed] [Google Scholar]
- 6. Corden J. L. (2007) Transcription. Seven ups the code. Science 318, 1735–1736 [DOI] [PubMed] [Google Scholar]
- 7. Sims R. J., III, Belotserkovskaya R., Reinberg D. (2004) Elongation by RNA polymerase II: the short and long of it. Genes Dev. 18, 2437–2468 [DOI] [PubMed] [Google Scholar]
- 8. Komarnitsky P., Cho E. J., Buratowski S. (2000) Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14, 2452–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schroeder S. C., Schwer B., Shuman S., Bentley D. (2000) Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 14, 2435–2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ghosh A., Shuman S., Lima C. D. (2011) Structural insights to how mammalian capping enzyme reads the CTD code. Mol. Cell 43, 299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Akhtar M. S., Heidemann M., Tietjen J. R., Zhang D. W., Chapman R. D., Eick D., Ansari A. Z. (2009) TFIIH kinase places bivalent marks on the carboxy-terminal domain of RNA polymerase II. Mol. Cell 34, 387–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kim M., Suh H., Cho E. J., Buratowski S. (2009) Phosphorylation of the yeast Rpb1 C-terminal domain at serines 2, 5, and 7. J. Biol. Chem. 284, 26421–26426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Glover-Cutter K., Larochelle S., Erickson B., Zhang C., Shokat K., Fisher R. P., Bentley D. L. (2009) TFIIH-associated Cdk7 kinase functions in phosphorylation of C-terminal domain Ser7 residues, promoter-proximal pausing, and termination by RNA polymerase II. Mol. Cell. Biol. 29, 5455–5464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tietjen J. R., Zhang D. W., Rodríguez-Molina J. B., White B. E., Akhtar M. S., Heidemann M., Li X., Chapman R. D., Shokat K., Keles S. (2010) Chemical-genomic dissection of the CTD code. Nat. Struct. Mol. Biol. 17, 1154–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mayer A., Lidschreiber M., Siebert M., Leike K., Söding J., Cramer P. (2010) Uniform transitions of the general RNA polymerase II transcription complex. Nat. Struct. Mol. Biol. 17, 1272–1278 [DOI] [PubMed] [Google Scholar]
- 16. Kim H., Erickson B., Luo W., Seward D., Graber J. H., Pollock D. D., Megee P. C., Bentley D. L. (2010) Gene-specific RNA polymerase II phosphorylation and the CTD code. Nat. Struct. Mol. Biol. 17, 1279–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ahn S. H., Kim M., Buratowski S. (2004) Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3' end processing. Mol. Cell 13, 67–76 [DOI] [PubMed] [Google Scholar]
- 18. Licatalosi D. D., Geiger G., Minet M., Schroeder S., Cilli K., McNeil J. B., Bentley D. L. (2002) Functional interaction of yeast pre-mRNA 3′-end processing factors with RNA polymerase II. Mol. Cell 9, 1101–1111 [DOI] [PubMed] [Google Scholar]
- 19. Proudfoot N. J., Furger A., Dye M. J. (2002) Integrating mRNA processing with transcription. Cell 108, 501–512 [DOI] [PubMed] [Google Scholar]
- 20. Svejstrup J. Q., Li Y., Fellows J., Gnatt A., Bjorklund S., Kornberg R. D. (1997) Evidence for a mediator cycle at the initiation of transcription. Proc. Natl. Acad. Sci. 94, 6075–6078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cho E. J., Kobor M. S., Kim M., Greenblatt J., Buratowski S. (2001) Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes Dev. 15, 3319–3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Krishnamurthy S., He X., Reyes-Reyes M., Moore C., Hampsey M. (2004) Ssu72 Is an RNA polymerase II CTD phosphatase. Mol. Cell 14, 387–394 [DOI] [PubMed] [Google Scholar]
- 23. West M. L., Corden J. L. (1995) Construction and analysis of yeast RNA polymerase II CTD deletion and substitution mutations. Genetics 140, 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chapman R. D., Heidemann M., Albert T. K., Mailhammer R., Flatley A., Meisterernst M., Kremmer E., Eick D. (2007) Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science 318, 1780–1782 [DOI] [PubMed] [Google Scholar]
- 25. Zhang Y., Zhang M., Zhang Y. (2011) Crystal structure of Ssu72, an essential eukaryotic phosphatase specific for the C-terminal domain of RNA polymerase II, in complex with a transition state analogue. Biochem. J. 434, 435–444 [DOI] [PubMed] [Google Scholar]
- 26. Xiang K., Nagaike T., Xiang S., Kilic T., Beh M. M., Manley J. L., Tong L. (2010) Crystal structure of the human symplekin-Ssu72-CTD phosphopeptide complex. Nature 467, 729–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Werner-Allen J. W., Lee C. J., Liu P., Nicely N. I., Wang S., Greenleaf A. L., Zhou P. (2011) cis-Proline-mediated Ser(P)5 dephosphorylation by the RNA polymerase II C-terminal domain phosphatase Ssu72. J. Biol. Chem. 286, 5717–5726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Singh N., Ma Z., Gemmill T., Wu X., Defiglio H., Rossettini A., Rabeler C., Beane O., Morse R. H., Palumbo M. J., Hanes S. D. (2009) The Ess1 prolyl isomerase is required for transcription termination of small noncoding RNAs via the Nrd1 pathway. Mol. Cell 36, 255–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mosley A. L., Pattenden S. G., Carey M., Venkatesh S., Gilmore J. M., Florens L., Workman J. L., Washburn M. P. (2009) Rtr1 is a CTD phosphatase that regulates RNA polymerase II during the transition from serine 5 to serine 2 phosphorylation. Mol. Cell 34, 168–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sun Z. W., Hampsey M. (1996) Synthetic enhancement of a TFIIB defect by a mutation in SSU72, an essential yeast gene encoding a novel protein that affects transcription start site selection in vivo. Mol. Cell. Biol. 16, 1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kobor M. S., Archambault J., Lester W., Holstege F. C., Gileadi O., Jansma D. B., Jennings E. G., Kouyoumdjian F., Davidson A. R., Young R. A., Greenblatt J. (1999) An unusual eukaryotic protein phosphatase required for transcription by RNA polymerase II and CTD dephosphorylation in S. cerevisiae. Mol. Cell 4, 55–62 [DOI] [PubMed] [Google Scholar]
- 32. Gudipati R. K., Villa T., Boulay J., Libri D. (2008) Phosphorylation of the RNA polymerase II C-terminal domain dictates transcription termination choice. Nat. Struct. Mol. Biol. 15, 786–794 [DOI] [PubMed] [Google Scholar]
- 33. Steinmetz E. J., Brow D. A. (2003) Ssu72 protein mediates both poly(A)-coupled and poly(A)-independent termination of RNA polymerase II transcription. Mol. Cell. Biol. 23, 6339–6349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ganem C., Devaux F., Torchet C., Jacq C., Quevillon-Cheruel S., Labesse G., Facca C., Faye G. (2003) Ssu72 is a phosphatase essential for transcription termination of snoRNAs and specific mRNAs in yeast. EMBO J. 22, 1588–1598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kanin E. I., Kipp R. T., Kung C., Slattery M., Viale A., Hahn S., Shokat K. M., Ansari A. Z. (2007) Chemical inhibition of the TFIIH-associated kinase Cdk7/Kin28 does not impair global mRNA synthesis. Proc. Natl. Acad. Sci. U.S.A. 104, 5812–5817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Qiu H., Hu C., Hinnebusch A. (2009) Phosphorylation of the Pol II CTD by KIN28 enhances BUR1/BUR2 recruitment and Ser2 CTD phosphorylation near promoters. Mol. Cell 33, 752–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Viladevall L., St. Amour C. V., Rosebrock A., Schneider S., Zhang C., Allen J. J., Shokat K. M., Schwer B., Leatherwood J. K., Fisher R. P. (2009) TFIIH and P-TEFb coordinate transcription with capping enzyme recruitment at specific genes in fission yeast. Mol. Cell 33, 738–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nonet M., Sweetser D., Young R. A. (1987) Functional redundancy and structural polymorphism in the large subunit of RNA polymerase II. Cell 50, 909–915 [DOI] [PubMed] [Google Scholar]
- 39. Florens L., Washburn M. P. (2006) Proteomic analysis by multidimensional protein identification technology. Methods Mol. Biol. 328, 159–175 [DOI] [PubMed] [Google Scholar]
- 40. Mosley A. L., Sardiu M. E., Pattenden S. G., Workman J. L., Florens L., Washburn M. P. (2011) Highly reproducible label free quantitative proteomic analysis of RNA polymerase complexes. Mol. Cell. Proteomics 10, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang Y., Wen Z., Washburn M. P., Florens L. (2010) Refinements to label free proteome quantitation: how to deal with peptides shared by multiple proteins. Anal. Chem. 82, 2272–2281 [DOI] [PubMed] [Google Scholar]
- 42. Thompson C. M., Koleske A. J., Chao D. M., Young R. A. (1993) A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell 73, 1361–1375 [DOI] [PubMed] [Google Scholar]
- 43. Stiller J. W., Mcconaughy B. L., Hall B. D. (2000) Evolutionary complementation for polymerase II CTD function. Yeast 16, 57–64 [DOI] [PubMed] [Google Scholar]
- 44. Schwer B., Shuman S. (2011) Deciphering the RNA polymerase II CTD code in fission yeast. Mol. Cell 43, 311–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Egloff S., O'Reilly D., Chapman R. D., Taylor A., Tanzhaus K., Pitts L., Eick D., Murphy S. (2007) Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science 318, 1777–1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ansari A., Hampsey M. (2005) A role for the CPF 3′-end processing machinery in RNAP II-dependent gene looping. Genes Dev. 19, 2969–2978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nedea E., He X., Kim M., Pootoolal J., Zhong G., Canadien V., Hughes T., Buratowski S., Moore C. L., Greenblatt J. (2003) Organization and function of APT, a subcomplex of the yeast cleavage and polyadenylation factor involved in the formation of mRNA and small nucleolar RNA 3′-ends. J. Biol. Chem. 278, 33000–33010 [DOI] [PubMed] [Google Scholar]
- 48. Dichtl B., Blank D., Ohnacker M., Friedlein A., Roeder D., Langen H., Keller W. (2002) A role for SSU72 in balancing RNA polymerase II transcription elongation and termination. Mol. Cell 10, 1139–1150 [DOI] [PubMed] [Google Scholar]
- 49. Gavin A. C., Bösche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J. M., Michon A. M., Cruciat C. M. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415, 141–147 [DOI] [PubMed] [Google Scholar]
- 50. Röther S., Strässer K. (2007) The RNA polymerase II CTD kinase Ctk1 functions in translation elongation. Genes Dev. 21, 1409–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. He X., Khan A. U., Cheng H., Pappas D. L., Hampsey M., Moore C. L. (2003) Functional interactions between the transcription and mRNA 3′-end processing machineries mediated by Ssu72 and Sub1. Genes Dev. 17, 1030–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nedea E., Nalbant D., Xia D., Theoharis N. T., Suter B., Richardson C. J., Tatchell K., Kislinger T., Greenblatt J. F., Nagy P. L. (2008) The Glc7 phosphatase subunit of the cleavage and polyadenylation factor is essential for transcription termination on snoRNA genes. Mol. Cell 29, 577–587 [DOI] [PubMed] [Google Scholar]
- 53. Cannon J. F. (2010) Function of protein phosphatase-1, Glc7, in Saccharomyces cerevisiae. Advances Appl. Microbiol. 73, 27–59 [DOI] [PubMed] [Google Scholar]
- 54. He X., Moore C. (2005) Regulation of yeast mRNA 3′-end processing by phosphorylation. Mol. Cell 19, 619–629 [DOI] [PubMed] [Google Scholar]
- 55. Holstege F. C., Jennings E. G., Wyrick J. J., Lee T. I., Hengartner C. J., Green M. R., Golub T. R., Lander E. S., Young R. A. (1998) Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95, 717–728 [DOI] [PubMed] [Google Scholar]
- 56. Kim M., Vasiljeva L., Rando O. J., Zhelkovsky A., Moore C., Buratowski S. (2006) Distinct pathways for snoRNA and mRNA termination. Mol. Cell 24, 723–734 [DOI] [PubMed] [Google Scholar]
- 57. Meinhart A. (2004) Recognition of RNA polymerase II carboxy-terminal domain by 3′-RNA-processing factors. Nature 430, 223–226 [DOI] [PubMed] [Google Scholar]
- 58. Lunde B. M., Reichow S. L., Kim M., Suh H., Leeper T. C., Yang F., Mutschler H., Buratowski S., Meinhart A., Varani G. (2010) Cooperative interaction of transcription termination factors with the RNA polymerase II C-terminal domain. Nat. Struct. Mol. Biol. 17, 1195–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Maxon M. E., Goodrich J. A., Tjian R. (1994) Transcription factor IIE binds preferentially to RNA polymerase IIa and recruits TFIIH: a model for promoter clearance. Genes Dev. 8, 515–524 [DOI] [PubMed] [Google Scholar]
- 60. Hirschi A., Cecchini M., Steinhardt R. C., Schamber M. R., Dick F. A., Rubin S. M. (2010) An overlapping kinase and phosphatase docking site regulates activity of the retinoblastoma protein. Nat. Struct. Mol. Biol. 17, 1051–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Meinhart A., Silberzahn T., Cramer P. (2003) The mRNA transcription/processing factor Ssu72 is a potential tyrosine phosphatase. J. Biol. Chem. 278, 15917–15921 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.