Background: Mechanisms of cell death in mucolipidosis type IV, a lysosomal storage disease caused by mutations in gene coding for ion channel TRPML1, are unknown.
Results: Acute TRPML1 knockdown increases apoptosis mediated by cytoplasmic cathepsin B (CatB) and Bax activity.
Conclusion: TRPML1 loss results in Bax- and CatB-dependent apoptosis.
Significance: This shows the first mechanistic link between TRPML1 loss, lysosomal deficiencies, and cell death.
Keywords: Apoptosis, Bax, Cysteine Protease, Lysosomal Storage Disease, TRP Channels
Abstract
Mucolipidosis type IV (MLIV) is a lysosomal storage disease caused by mutations in the gene MCOLN1, which codes for the transient receptor potential family ion channel TRPML1. MLIV has an early onset and is characterized by developmental delays, motor and cognitive deficiencies, gastric abnormalities, retinal degeneration, and corneal cloudiness. The degenerative aspects of MLIV have been attributed to cell death, whose mechanisms remain to be delineated in MLIV and in most other storage diseases. Here we report that an acute siRNA-mediated loss of TRPML1 specifically causes a leak of lysosomal protease cathepsin B (CatB) into the cytoplasm. CatB leak is associated with apoptosis, which can be prevented by CatB inhibition. Inhibition of the proapoptotic protein Bax prevents TRPML1 KD-mediated apoptosis but does not prevent cytosolic release of CatB. This is the first evidence of a mechanistic link between acute TRPML1 loss and cell death.
Introduction
TRPML1 is a member of the transient receptor potential (TRP)3 family of ion channels. Unlike most TRP channels, TRPML1 and its relatives TRPML2 and TRPML3 primarily reside in the membranes of the endocytic pathway (1–6). TRPML1 is localized to later compartments along the endocytic pathway (late endosomes and lysosomes) by dileucine targeting sequences on its N and C termini (5, 6) and has been shown to be an inwardly rectifying cation channel potentiated by low, typically lysosomal, levels of pH (7, 8). TRPML1 activation by phosphatidylinositol 3,5-bisphosphate (9), which is predominantly found in late endosomes and lysosomes, provides a physiological context for its localization suggesting that it is either activated by delivery to the lysosomes or serves as a sensor for phosphatidylinositol 3,5-bisphosphate-rich compartments matching lysosomes and late endosomes for fusion. TRPML1 has been suggested to regulate fusion/fission of vesicles in the endocytic pathway (4, 10–12), and/or some aspect of lysosomal ion homeostasis such as pH, iron, or zinc content (9, 13–15).
TRPML1 down-regulation due to mutations in the gene MCOLN1 results in the rare lysosomal storage disease MLIV (2, 16). The disease is associated with the buildup of storage bodies of largely unknown origin. MLIV has a profound neurodegenerative profile, which, in mouse models has been linked to retinal degeneration and demyelination of corpus callosum, deep layer neocortex, and cerebellar white matter tracts (17, 18). Gastric abnormalities including degeneration of parietal cells and hypochlorhydria have been reported in MLIV patients and model mice (19–22). As with most lysosomal storage diseases, the mechanisms of cell death in MLIV are unclear. We have previously suggested that lysosomal deficiencies in MLIV and, perhaps, other storage diseases, lead to autophagic deficits and buildup of effete mitochondria, which may expose cells to proapoptotic effects of cell stimulation with Ca2+ mobilizing agonists (23). Autophagy deficits have been confirmed in MLIV and several other lysosomal storage models (18, 24–28, 30). Nonetheless, the selectivity of cellular loss in storage diseases remains puzzling. We believe that the key to identifying the cell death pathways in lysosomal storage diseases lies in deconstructing the early events accompanying the loss of TRPML1 or other components of the endocytic pathway. This task is difficult to accomplish in cells cultured from patients due to the possible, and indeed likely, contribution of secondary effects due to chronic accumulation of storage material.
To delineate the early events associated with the loss of TRPML1, we used siRNA-mediated knockdown (KD) to acutely down-regulate TRPML1 in HeLa cells. Knockdown of palmitoyl-protein thioesterase 1 (PPT1), an enzyme mutated in another lysosomal storage disease, infantile neuronal lipofuscinosis, was used as a comparative control (31, 32). We show that TRPML1 loss specifically causes, within 48 h of KD, an increase in the lysosomal protease CatB and the lysosomal membrane protein LAMP-1. These changes are specific to TRPML1 loss and are controlled at a post-transcriptional level. TRPML1 KD also resulted in a cytoplasmic buildup of CatB. Apoptosis is elevated in TRPML1 KD cells and is blocked by inhibition of either CatB or the proapoptotic protein Bax. Inhibition of Bax activity did not prevent CatB release, suggesting that this protein lies downstream of CatB or in a separate apoptotic pathway. These results illustrate, for the first time, the early events leading to cell death in TRPML1-deficient cells.
EXPERIMENTAL PROCEDURES
Cell Culture
HeLa cells were maintained in DMEM (Sigma) supplemented with 7% FBS, 100 μg/ml of penicillin/streptomycin, and 5 μg/ml of plasmocin prophylactic (Invivogen, San Diego, CA). For siRNA KD, antibiotic-free media was used. Antibiotic-free media supplemented with 100 mm sucrose was used for sucrose treatments.
siRNA-mediated KD
siRNA were designed as described previously (13) and custom synthesized as ON-TARGET plus constructs by Dharmacon (Lafayette, CO). The TRPML1 siRNA probe targeting the sequence 5′-CCCACATCCAGGAGTGTAA-3′ in MCOLN1 was used for all TRPML1 KDs. The PPT1 siRNA probe targeting the sequence 5′-GGTACTCACATAAATGCTT-3′ in PPT1 was used for all PPT1 KDs. Control siRNA #1 (Sigma) was used as a negative control. 6-Well plates were transfected using Lipofectamine 2000 (Invitrogen). 7-Day long KDs were maintained by splitting cells every 3 days and retransfecting them in suspension. Transfections were performed as described by the manufacturer's protocol using 300 nm siRNA per well. All KDs were confirmed using SYBR Green-based quantitative real-time RT-PCR and Western blot analysis.
Reverse Transcriptase and Quantitative PCR (qPCR)
RNA was isolated from cells using TRIzol (Invitrogen) according to the manufacturer's protocol. cDNA was synthesized using the GeneAmp RNA PCR system (Applied Biosystems, Carlsbad, CA) with 2 μl of oligo(dT) priming. qPCR was performed using 2 μl of cDNA, 2× SYBR Green (Fermentas, Glen Burnie, MD), and 5 μl of 4 μm primer per 50-μl reaction. The amount of cDNA loaded was normalized to starting RNA concentrations, with a final concentration of 6 ng of RNA loaded per experimental well. Six-point standard curves were generated for each primer using 1:2 dilutions of cDNA and loading 2 μl/well. Dilutions started at 20 ng of starting RNA. The following Quantitect primer assays were used: ACTB (β-actin, QT00095431) and CTSB (CatB, QT00088641). cDNA for the following genes were amplified using the indicated primers (IDT, Coralville, IA); MCOLN1, forward, 5′-TCTTCCAGCACGGAGACAAC-3′ and reverse, 5′-AACTCGTTCTGCAGCAGGAAGC-3′; PPT1, forward, 5′-CCTGTAGATTCGGAGTGGTTTGGATT-3′ and reverse, 5′-CAGGCGGTCCTGTGTGTACA-3′. All primers were designed to span exons, and negative RT controls were tested to ensure amplification of cDNA only. qPCR was performed using the Standard Curve method on the 7300 Real Time System (Applied Biosystems). Reactions were run on the following parameters: 2 min at 50 °C, 10 min at 95 °C, and 40 cycles at 95 °C for 15 s followed by 60 °C for 1 min. All experimental samples were run in triplicate and normalized to a β-actin endogenous control. Microsoft Excel was used to generate standard curves and analyze qPCR results.
Western Blot Analysis
Cells were solubilized for 10 min at room temperature in a 1× detergent solution (0.5 m EDTA, pH 8.0, 1 m Tris, pH 8.0, 0.4% deoxycholate, 1% Nonidet P-40 substitute) containing protease inhibitor mixture III (Calbiochem, Gibbstown, NJ) and centrifuged at 16,000 × g for 5 min. The supernatant was collected and protein concentrations were determined using a Bradford assay. Protein was incubated at 100 °C for 5 min in sample buffer containing 14% β-mercaptoethanol. Equal amounts of protein were loaded on a 12.5% precast Tris-HCl polyacrylamide gel (Bio-Rad) for each experimental sample. Proteins were transferred to PVDF membrane (Millipore) and blocked in 10% nonfat dry milk for 1 h. The following primary antibodies were used: monoclonal cathepsin B (CA10 clone, Calbiochem, IM27L) at 1:250 dilution, polyclonal PPT1 (Sigma, SAB1400222) at 1:250, polyoclonal lysosomal acid lipase (Abcam, Cambridge, MA, ab73445) at 1:500 dilution, monoclonal LAMP1 (H4A3 clone, Santa Cruz, Santa Cruz, CA) at 1:1,000 dilution, monoclonal cathepsin D (CTD-19 clone, Sigma) and monoclonal HA (HA.C5, Abcam) at 1:500, and monoclonal GAPDH (Millipore) and monoclonal β-actin (AC-15 clone, Abcam) at 1:5,000 dilution. HRP-conjugated goat anti-mouse or anti-rabbit secondary antibodies (Amersham Biosciences) were used at 1:20,000 and 1:1,500 dilutions, respectively. Immunodetection was performed with the Luminata Forte HRP substrate (Millipore). Band densities were measured using ImageJ (Bethesda, MD). For measurement of TRPML1 KD, cells were transfected with siRNA as described above. After 24 h, 2 μg of HA-TRPML1 DNA was transfected into cells using Lipofectamine 2000 (Invitrogen) as described by the manufacturer's protocol. Cells were then immunoblotted for the HA tag 24 h later.
Radioactive Synthesis and Degradation
2 days after siRNA knockdown, cells were starved for 30 min in media lacking cysteine and methionine. Cells were then labeled for 2 h at 37 °C with 100 μCi/ml of Tran35S-label (MP Biomedicals, Solon, OH). Cells were chased with serum-free DMEM containing unlabeled cysteine and methionine. At the indicated time points, cells were lysed using 1× detergent solution and labeled proteins were immunoprecipitated overnight at 4 °C with the CatB monoclonal antibody (CA10 clone, Calbiochem, IM27L). Antigen-antibody complexes were collected using pansorbin cells coated with rabbit anti-mouse antibodies 24 h later. Pansorbin cells were washed 3 times in RIPA buffer, and then resuspended in 20 μl of 4× sample buffer, boiled for 3 min, and pelleted at 16,000 × g for 2 min. Supernatant was run on a 12.5% gel. Dried gels were analyzed using a PhosphorImager and analyzed using Quantity One software (Bio-Rad).
CatB Secretion
2 days after KD, cell media was replaced with 0.5% FBS antibiotic-free media. Media was then concentrated on Millipore 10K Ambion filters. Concentrated protein was subjected to Western blot analysis as described above.
Membrane/Cytosolic Separation
Cells were scraped into Reagent A from the Pierce Lysosomal Extraction Kit and solubilized using 40 strokes of a pre-chilled Dounce homogenizer. An equal volume of Reagent A was added to the lysed cells and nuclear fractions were removed by an 800 × g spin. The cytosolic and membrane fractions were separate using a 60,000 × g spin for 20 min at 4 °C. The supernatant (cytosolic fraction) was concentrated using Millipore 10K Ambion filters (Millipore). The membrane pellet was washed 3 times with PBS and solubilized in 1× detergent solution. GAPDH was used as a loading control for the cytosolic fraction and CatD was used as a loading control for the membrane fraction.
Lysosomal Extraction
Cells were collected and processed using the ThermoScientific Lysosomal Enrichment Kit (Rockford, IL) following the manufacturer's instructions. Lysosomal pellets were solubilized using 1× detergent solution and subjected to Western blot analysis.
Cathepsin B Activity
Cathepsin B activity was analyzed using the EMD Innozyme Cathepsin B detection kit (Calbiochem, CBA001) following the manufacturer's protocol. The kit measures cathepsin B activity using the substrate Z-Arg-Arg AMC (benzyloxycarbonyl-l-arginyl-l-arginine-4-methylcoumaryl-7-amide), which fluoresces when cleaved. Fluorescence was measured using a fluorometer, at an excitation wavelength of 360 nm and an emission wavelength of 440 nm. Pre-cleaved substrates were used to create AMC standard curves and fluorescence in experimental samples was plotted against these curves to determine concentrations (in μm) of cleaved AMC. All values were normalized to the total protein load.
Confocal and Electron Microscopy
Confocal and electron microscopy was performed as previously described (33). Primary monoclonal anti-TFEB antibodies were from Abcam (ab56330). For indirect detection, goat anti-mouse secondary antibodies from Invitrogen were used.
Apoptosis Assay
Cells were prepared and measured using the EnzChek Caspase-3 Assay Kit number 1 (Invitrogen) following the manufacturer's instructions. AMC substrate fluorescence was measured using a fluorometer at an excitation wavelength of 342 nm and an emission wavelength of 441 nm.
Statistical significance was calculated using a one-tailed, unpaired t test with p ≤ 0.05 considered significant. Data are presented as mean ± S.E.
RESULTS
TRPML1 and PPT1 KD Results in Decreased mRNA Levels and a Storage Phenotype
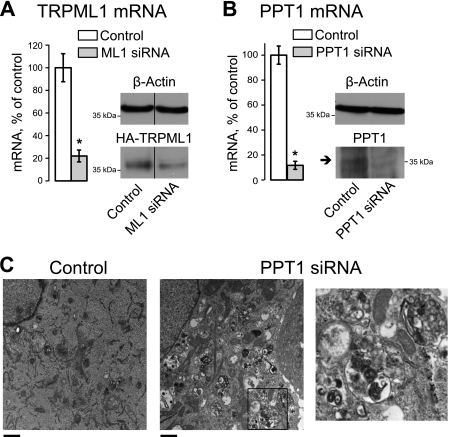
Fig. 1, A and B, show that our siRNA-mediated KD protocol resulted in a significant decrease in TRPML1 or PPT1 mRNA levels (a decrease of 78.0 ± 8.6 and 98.3 ± 3.7%, respectively, p < 0.001, n = 8) 48 h after transfection of HeLa cells. Knockdown was verified using Western blot analysis of heterologously expressed recombinant HA-tagged TRPML1 or endogenous PPT1 (Fig. 1, A and B). Cellular inclusions have been previously shown to accumulate in the same TRPML1 KD model, further confirming that this model represents the early stages of MLIV (13). Electron micrographs of PPT1 KD cells show a large number of cellular inclusions (Fig. 1C). Therefore siRNA-mediated PPT1 KD is an effective model of lysosomal dysfunction.
FIGURE 1.
siRNA-mediated down-regulation of TRPML1 and PPT1 result in decreased mRNA and protein levels and recapitulate a storage phenotype. A, HeLa cells were transfected with TRPML1 siRNA 48 h before collection. Total RNA was isolated and cDNA was synthesized and probed using TRPML1-specific exon-spanning primers. For protein detection, cells were transfected with N-terminal-tagged HA-TRPML1 DNA 24 h after siRNA transfection. Whole cell lysates were collected 24 h later and subjected to Western blot analysis, probing for HA. The 37-kDa band represents the N-terminal HA-tagged cleaved fraction of TRPML1, which is easier to detect than full-length protein. Bands were separated by a lane containing GFP-tagged HA-TRPML1 lysate (denoted by a black line). All protein and mRNA levels were normalized to β-actin. B, PPT1 mRNA and protein levels were measured 48 h after siRNA transfection as described above. Arrow represents mature form of PPT1. C, cells were collected and fixed for EM images 48 h after siRNA transfection. Scale bar represents 1 μm. Right panel represents a zoom-in image of the area on the left panel indicated by the black square. *, p < 0.05, n = 8 for both A and B.
CatB Buildup in TRPML1 KD Cells
CatB is a ubiquitous lysosomal protease, which has previously been implicated in cell death upon lysosomal permeabilization. It has been shown that CatB released into the cytoplasm is active and promotes cleavage and activation of caspases involved in apoptosis (34–38). CatB is processed through the ER and Golgi and enters the endocytic pathway as an inactive zymogen (pro-CatB), which is cleaved by active lysosomal enzymes as well as by autocatalysis into a mature single peptide form of about 35 kDa and into dipeptide forms with a heavy chain of 25–29 kDa (39–41).
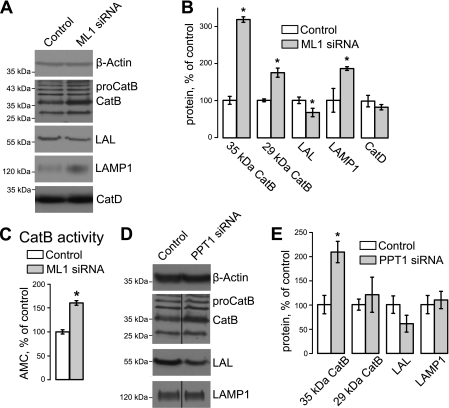
To determine whether TRPML1 KD had an effect on CatB levels, we performed Western blot analysis on control and TRPML KD HeLa cells using CatB antibodies. Cell lysates were collected 48 h after siRNA transfection. All protein levels were normalized to β-actin loading controls. Fig. 2, A and B, show that TRPML1 KD results in a robust increase of mature CatB levels, whereas pro-CatB levels are unaltered. Mature CatB levels increased by 213.3 ± 9.2% for the 35-kDa form and by 81.0 ± 16.5% for the 29-kDa form (p < 0.01, n = 4 each) in TRPML1 KD cells.
FIGURE 2.
TRPML1 KD and PPT1 KD specifically alter steady state protein levels of CatB, LAL, and LAMP-1. A and B, representative Western blot (A) and blot density analysis (B) of HeLa whole cell lysates 48 h after siRNA transfection show increased mature CatB and LAMP-1 levels, and decreased LAL levels in TRPML1 KD cells. The CatB label points to the 35-kDa mature form of CatB; the 29-kDa heavy chain can be seen below. CatD levels did not seem to change in TRPML1 KD cells. C, CatB activity was measured using the EMD Innozyme Cathepsin B detection kit. Precleaved AMC substrates were used to create a 6-point standard curve, micromolar AMC were determined using this curve. Values were normalized to total protein load. *, p < 0.05, n = 5. D and E, representative Western blot (D) and blot density analysis (E) of PPT1 KD cells shows an increase in pro-CatB not mature CatB, LAL, or LAMP-1. Lanes containing control and PPT1 KD samples were not run in continuous lanes, which is denoted by the dividing line in the CatB and LAMP-1 blots. *, p < 0.05, n ≥ 3.
In contrast to CatB, another lumenal protein lysosomal acid lipase (LAL) was decreased in TRPML1 KD cells (Fig. 2, A and B; 32.5 ± 13.3% decrease, p < 0.05, n = 7), whereas the levels of lysosomal membrane protein LAMP-1 were increased (72.4 ± 11.7% increase, p < 0.01, n = 3). The changes in LAMP1 are consistent with previous studies showing that levels of specific lysosomal markers are elevated after TRPML1 loss (18, 30). CatD levels did not change in TRPML1 KD cells (Fig. 2, A and B) indicating that the effect of TRPML1 KD on CatB is specific.
The physiological significance of CatB up-regulation in TRPML1 KD HeLa cells was evaluated using the Innozyne CatB Activity Assay. CatB activity was up-regulated by 60.4 ± 15.6% (p < 0.05, n = 5) in TRPML1 KD whole cell lysates (Fig. 2C), confirming the specificity of our Western blot results and demonstrating that the mature, active form of CatB is up-regulated as a consequence of TRPML1 loss.
As a comparison, we analyzed LAMP1, CatB, and LAL levels in PPT1 KD to determine whether the changes we observed were specific to TRPML1 KD. There was no significant change in LAL and LAMP-1 levels (n = 3, p = 0.4 and 0.1, respectively) in PPT1 KD cells (Fig. 2, D and E). We did observe a 107.8 ± 21.9% (p < 0.05, n = 3) increase in the 35-kDa CatB levels, however, there was no significant increase in mature, 29-kDa CatB levels in PPT1 KD cells. These results suggest that PPT1 KD affects the maturation of the inactive CatB zymogen. Therefore, changes in lysosomal protein levels are specific for individual lysosomal storage disease pathways.
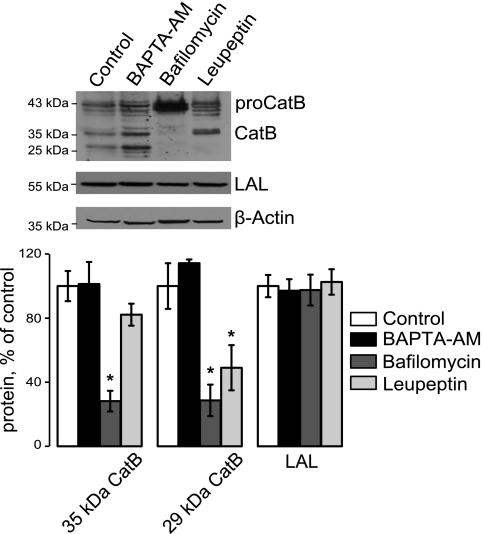
To confirm that the changes we observe in TRPML1 KD cells are specific and not caused by general endocytic inhibition, HeLa cells were treated for 48 h with 10 μm BAPTA-AM, a Ca2+ chelator that inhibits fission/fusion events and results in a membrane trafficking defect (13); 10 nm bafilomycin A1, an inhibitor of the lysosomal H+ ATPase pump that blocks the acidification of lysosomes (42); or 10 μm leupeptin, a serine/cysteine protease inhibitor that prevents degradation of protein substrates in the lysosome (43, 44). These inhibitors did not alter LAL levels and did not result in CatB increase (Fig. 3), suggesting that these changes are TRPML1 KD specific.
FIGURE 3.
Increased levels of CatB and decreased LAL levels are TRPML1 specific and not due to general endocytic disruption. HeLa cells were treated with the endocytic inhibitors BAPTA-AM (10 μm), bafilomycin A1 (10 nm), and leupeptin (10 μm) for 48 h. These inhibitors did not affect LAL levels and did not lead to an increase in CatB. Therefore, general endocytic disruption does not cause protein changes similar to that of TRPML1 and PPT1 KD (*, p < 0.05, n ≥ 5).
As shown before, and in agreement with the requirement for active lysosomal machinery for pro-CatB processing to mature CatB (39–41), bafilomycin A1 treatment resulted in a buildup of the 43-kDa pro-CatB and disappearance of mature CatB (Fig. 3). Leupeptin treatment prevented the autocatalytic conversion of 34-kDa monopeptide into the 29-kDa form, also in agreement with the previously published data (41). These results confirm the molecular identity of mature and pro-CatB in our system.
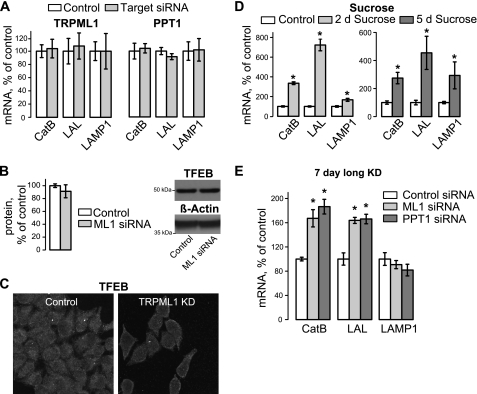
CTSB, the gene coding for CatB, is a member of the CLEAR degradative gene network regulated by the transcription factor TFEB, as are genes coding for LAL and LAMP-1 (45). Although the CLEAR network was shown to coordinate changes in lysosomal genes, input signals regulating it are currently unclear. In our system, there was no significant change in CatB, LAL, and LAMP1 mRNA levels in TRPML1 and PPT1 KD cells compared with control cells within 48 h of KD (Fig. 4A). Values were normalized to β-actin mRNA levels. Western blotting and confocal microscopy show no change in TFEB protein levels or localization in TRPML1 KD cells (Fig. 4, B and C). Therefore, the changes we observe in CatB, LAL, and LAMP1 occur at a post-transcriptional level.
FIGURE 4.
TFEB-regulated lysosomal genes and TFEB levels are unaltered in acute TRPML1 and PPT1 KD cells. A, mRNA levels were measured in TRPML1 and PPT1 KD cells using qRT-PCR using CatB, LAL, and LAMP-1 specific primers (n ≥ 3). No significant change was seen in mRNA levels under either condition. B, TFEB protein levels were measured by Western blot analysis in TRPML1 KD cells. No difference was seen. C, TFEB cellular localization was analyzed by confocal microscopy. No change in localization occurs after 48 h of TRPML1 KD (n = 3). D, qRT-PCR analysis of CatB, LAL, and LAMP1 mRNA in HeLa cells exposed to 100 mm sucrose for 2 or 5 days to induce general inhibition of the lysosomal function as described before. E, qRT-PCR analysis of the same genes in cells after a 7-day KD of TRPML1 or PPT1. β-Actin was used as a loading control (*, p < 0.05, n = 3).
To confirm that the TFEB gene network reports the functional status of lysosomes in our experimental system, we used protocols previously used to establish this paradigm. First, incubation of cells with 100 mm sucrose, previously shown to disrupt lysosomal traffic and stimulate the TFEB network (45), was performed and the transcriptional levels of CatB, LAL, and LAMP1 were analyzed using qRT-PCR. Fig. 4D shows that 2- and 5-day sucrose treatments induce an increase in levels of these TFEB-regulated genes, consistent with previous studies.
Prolonged (7 days) TRPML1 KD in HeLa cells is associated with an increased buildup of storage bodies (33). We reasoned that if the TFEB network reports the general disruption of the lysosomal function and/or buildup of storage bodies, then the genes belonging to the TFEB network would be up-regulated after prolonged TRPML1 KD. Fig. 4E shows that 7-day KD of TRPML1 or PPT1 results in up-regulation of CatB and LAL mRNA (CatB levels, respectively, 67.3 ± 14.2 and 86.5 ± 12.1%; LAL levels, respectively, 63.7 ± 5.0 and 66.1 ± 7.8%, n = 3, p < 0.01). Because both genes belong to the TFEB network, we conclude that TFEB activation most likely depends on general lysosomal cues such as storage bodies. It is interesting to note that LAMP1 mRNA was unaltered after a 7-day KD, suggesting that additional cues are necessary to change expression of this gene. The elevated level of LAMP1 expression in our sucrose treatment appears to be time dependent (Fig. 4D), suggesting that increased levels of lysosomal dysfunction may be required for increased LAMP1 gene expression. These results suggest that the biochemical events leading to CatB up-regulation precede the critical juncture in lysosomal dysfunction that is necessary for activation of the CLEAR network.
Taken together, the results described above show up-regulation of pro-CatB and mature CatB shortly after TRPML1 KD. The buildup of pro-CatB in both TRPML1 and PPT1 KD cells can be explained by general lysosomal malfunction leading to delayed processing of CatB. However, up-regulation of the mature form of CatB appears to be TRPML1 specific and, under the conditions of acute TRPML1 KD, it is not caused by changes in gene expression.
CatB Release in TRPML1 KD Cells
Lysosomal proteases have been implicated in cell death via the apoptosis program due to their release into the cytoplasm and activation of apoptotic caspases (38, 46–48). Several mechanisms of CatB release have been proposed. Some studies suggest general loss of lysosomal integrity, whereas in some studies lysosomes seem to remain intact during the CatB rise in the cytoplasm, suggesting a specific mechanism of CatB release. Because MLIV has a neurodegenerative profile, ostensibly due to neuronal loss, CatB changes in TRPML1-deficient cells may shed new light on cell death caused by MLIV. To elucidate the mechanism of the CatB increase as well as the cellular localization of this protein, we focused on the handling of CatB by TRPML1 KD cells.
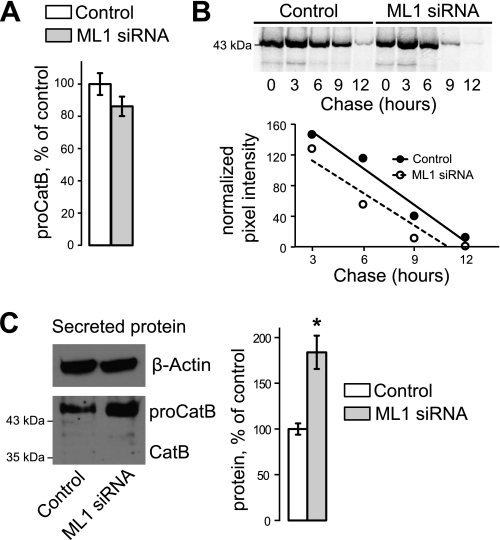
To determine whether TRPML1 KD induced CatB up-regulation is caused by increased protein synthesis or decreased degradation, cells were pulsed with 35S-labeled cysteine-methionine for 2 h, then chased with serum-free media for 0, 3, 6, 9, and 12 h. Whole cell lysates were collected at each time point and 35S-labeled CatB was immunoprecipitated, and visualized on a SDS-PAGE gel. A small aliquot of lysate was collected and subjected to Western blot analysis to measure β-actin levels; all values were normalized to β-actin. There were no appreciable differences in pro-CatB synthesis, as measured at the 0-h time point (Fig. 5A). To ensure that CatB synthesis was similar during the 2-h pulse interval, cell lysates were collected, immunoprecipitated with CatB antibody, and levels were quantitated after 30, 60, 90, or 120 min of labeling (supplemental Fig. S1). In TRPML1 KD cells, the rate of 35S-labeled pro-CatB accumulation during the labeling period was slightly lower than in control cells. Together, these results show that the synthesis of pro-CatB is not increased in TRPML1 KD cells. Additionally, there was little difference between CatB degradation rates in control and TRPML1 KD cells (Fig. 5B).
FIGURE 5.
Analysis of CatB handling shows no change in synthesis or degradation, whereas secreted levels of pro-CatB are increased. A, synthesis of 35S-labeled CatB was measured beginning immediately after a 2-h labeling period. Levels were normalized to β-actin (n = 3). B, degradation rates of CatB were measured over a 12-h time course. 35S-Labeled pro-CatB levels were normalized to β-actin and rates of degradation were determined using a linear fit curve. C, levels of CatB secretion were measured by Western blot analysis. Higher levels of pro-CatB are detected in the media; mature CatB is undetectable (*, p < 0.05, n = 3).
Proteins in the medium were concentrated and subjected to Western blot analysis to measure CatB secretion levels. There was an 83.8 ± 18.2% (p < 0.01, n = 3) increase in extracellular pro-CatB in TRPML1 KD cells, normalized to cellular β-actin (Fig. 5C). Secretion of pro-CatB has been observed previously and suggests that extracellular CatB originates from pre-lysosomal compartments (40, 41). Increased pro-CatB secretion in TRPML1 KD cells may account for at least some of the decrease in pro-CatB synthesis in TRPML1 KD cells noted above.
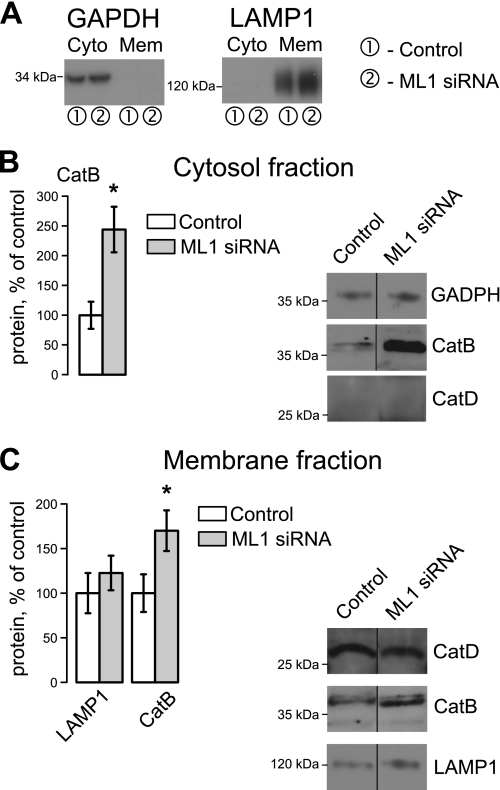
Cytosolic release of mature CatB in response to cell stress has been observed in several reports. Previous studies demonstrated CatB localization through the expression of GFP-CatB, whereas others established endogenous CatB localization using fractionation assays (38, 49–51). Despite numerous attempts we were unable to clearly visualize endogenous CatB in HeLa cells using confocal microscopy and CatB antibodies. We therefore established CatB localization using a cytosolic/membrane fractionation protocol. Fractions from control and TRPML1 KD cells were isolated and CatB, LAMP-1, GAPDH, and CatD were compared in these fractions. GAPDH was detected only in the cytosolic fractions, whereas LAMP-1 was only detected in the membrane fraction (Fig. 6A), confirming specificity of our preparation. CatD, whose levels remain unaltered in TRPML1 KD cells, was used to normalize membrane levels of CatB and LAMP-1. GADPH was used to normalize CatB in the cytosolic fractions. Our data show a significant increase of a mature, 35-kDa form of CatB in both fractions (Fig. 6, B and C) and increased LAMP-1 levels in membrane fractions (Fig. 6C), consistent with our whole cell lysate analysis. Interestingly, cytosolic CatB levels increased to 144.1 ± 38.2% (p < 0.05, n = 3) of control levels in TRPML1 KD cells, suggesting that a large amount of CatB is released into the cytosol. CatD was not observed in the cytosolic fraction, confirming that the release of CatB is specific and not due to general lysosomal permeabilization (Fig. 6B). Our results indicate that TRPML1 KD leads to the selective release of CatB into the cytoplasm.
FIGURE 6.
TRPML1 KD results in high levels of cytosolic CatB. A, cytosolic and membrane fractions were isolated from HeLa lysates through differential centrifugation 48 h at TRPML1 KD and identified using GAPDH and LAMP-1 antibodies. B, CatB levels were measured in cytoplasmic fractions by Western blot analysis and normalized to GAPDH. C, membrane fractions show increases in mature CatB and LAMP-1 levels, consistent with whole cell lysate analyses. Due to changes in LAMP-1 levels, CatD was used as a loading control for both CatB and LAMP-1 quantification (*, p < 0.05, n = 3).
CatB and Apoptosis in TRPML1-deficient Cells
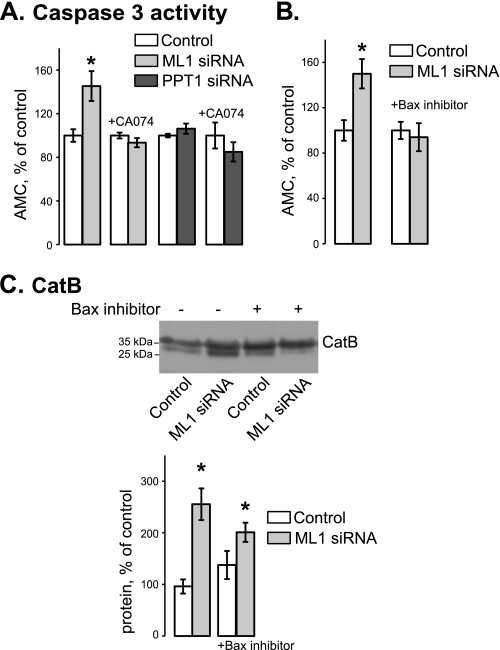
Leakage of CatB and other lysosomal proteases into the cytosol has been shown to lead to apoptosis (52–56). This phenomenon seems to take place in our system as well. Apoptotic levels were tested in control, TRPML1 KD, and PPT1 KD cells using a caspase 3 fluorogenic substrate assay. Apoptosis was significantly up-regulated in TRPML1, but not in PPT1 KD cells (Fig. 7A). In TRPML1 KD cells, caspase 3 activity averaged 162.9 ± 9.3% of its value in control cells, whereas in PPT1 KD cells it averaged 107.31 ± 4.7% of control values (n = 3 for each set of conditions; p < 0.01 for TRPML1 KD cells). 24 h incubation of control and TRPML1 KD cells with the cell-permeable CatB inhibitor Ca-074Me (2.5 μm) resulted in decreased apoptosis in TRPML1 KD cells as compared with untreated control (67.9 ± 7.2%, p < 0.01, n = 3). Ca-074Me did not alter apoptosis in control-treated cells as seen in Fig. 7A. This suggests that apoptosis in TRPML1 KD cells is CatB dependent and that apoptosis is an early specific consequence of TRPML1 loss.
FIGURE 7.
TRPML1 KD results in cell death via Bax- and CatB-dependent mechanism. A, apoptosis was measured using the EnzChek caspase-3 apoptotic assay 48 h after TRPML1 and PPT1 KD. Ca-074Me (2.5 μm) was added 24 h after KD (*, p < 0.05, n = 3). B, caspase-3 activity was measured in TRPML1 KD cells 48 h after KD in the presence and absence of Bax inhibiting peptide (100 μm, added 24 h after KD) (*, p < 0.05, n ≥ 3). C, Western blot example and statistical analysis of mature CatB in 48-h TRPML1 KD cells treated with Bax inhibiting peptide (100 μm, added 24 h after KD) (*, p < 0.05, n ≥ 3).
The proapoptotic protein Bax has been implicated in CatB release in several systems (34, 38, 57, 58). To test whether Bax mediated apoptosis in TRPML1 KD cells and contributed to CatB release, we treated cells 24 h after siRNA transfection with a Bax inhibiting peptide (100 μm, Calbiochem) and collected samples 24 h later. Fig. 7B shows that Bax inhibition prevented apoptosis in TRPML1 KD cells without significantly affecting apoptosis in control cells. TRPML1 KD cells treated with the inhibitor still had elevated cytoplasmic CatB levels compared with both untreated and Bax inhibited control cells as seen in Fig. 7C (n = 4, p < 0.01, p = 0.05, respectively). These data suggest that CatB release lies upstream of Bax activation or in a separate apoptotic pathway. Further studies will focus on elucidating these pathways and determining the sequence of events that leads to Bax activation, caspase-3 activation, and apoptosis in these cells.
DISCUSSION
The present study shows that shortly after TRPML1 KD in HeLa cells, the lysosomal protease CatB is translocated into the cytoplasm where it triggers cell death through apoptosis. Apoptosis is mediated by CatB and Bax; whether these proteins act in the same or parallel pathways will need to be determined. These data are in line with some of the previous reports on lysosomal toxicity (34–38, 52–58). They do, however, raise several important questions relevant to lysosomal storage diseases and lysosomal function.
Cell death clearly underlies the key degenerative aspects of lysosomal storage diseases. Its mechanisms in lysosomal storage disorders, however, are not well understood. Some of the current models, including suppressed flow of key substrates or accumulation of effete organelles due to autophagy block, have been discussed (25, 59, 60). Here we show that in addition to the “supply side” issues in lysosomal storage diseases, an active and early CatB release mechanism specifically caused by TRPML1 loss takes place soon after TRPML1 KD. Relative contribution of each of these mechanisms in cell death in specific tissues remains to be established. It is, however, likely that if the propensity for CatB release varies between different tissues, then the CatB release mechanism may explain tissue-specific cell death in lysosomal storage diseases.
Several aspects of CatB behavior in the TRPML1 KD system await clarification. First, the mechanism of CatB buildup in TRPML1 KD cells is not completely clear. Although our labeling experiments (Fig. 5, A and B, and supplemental Fig. S1) do not directly support increased synthesis or decreased degradation of pro-CatB in TRPML1 KD cells, the increased secretion of pro-CatB (Fig. 5C), may be a complicating factor in these assays. Therefore, some possibility that pro-CatB synthesis is up-regulated in TRPML1 KD cells still exists. On the other hand, the majority of CatB that accumulates in TRPML1 KD cells is in the mature form (Fig. 2A), arguing that this increase takes place at the lysosomal, or post-lysosomal stage. The most pronounced increase in CatB in TRPML1 KD cells is in the cytoplasm, as the cytoplasmic CatB rose by almost 150% over control compared with just over 60% for the membrane fraction (Fig. 6). CatB leaking from the lysosomes into the cytoplasm is likely to have a longer lifespan, as it is not accessible to the lysosomal degradation machinery. It is tempting to speculate that this leakage, taking place over the 2-day KD period, is the reason for measurable CatB buildup in TRPML1 KD cells.
The mechanism of CatB release in TRPML1 KD cells remains to be identified. Interestingly, a recent paper identified an increase of CatB in a mouse model of Niemann Pick disease, a lysosomal storage disorder, and found that CatB inhibition decreased liver fibrosis in vivo. This study is an interesting link to our data, suggesting that lysosomal storage defects may result in CatB-mediated cellular changes (61). Furthermore, cytosolic CatB release has been implicated in many instances of cellular toxicity, including those caused by cytoplasmic/mitochondrial and by lysosomal events (52–56). Dissociation of lysosomal membrane by lysosomotropic agents has been proposed as one explanation for this phenomenon. It is important to note that nonselective physical damage to the lysosomal membrane is usually associated with a general loss of lysosomal integrity and release of a number of lysosomal components, including lysosomal dyes, into the cytoplasm. This has not been observed in our system; indeed the lumenal lysosomal component CatD does not appear to change in TRPML1 KD cells (Fig. 2). Furthermore, we do not detect any measurable loss of lysosomal integrity using lysosomal marker Lysotracker (not shown). These observations are in line with previous findings of CatB translocation into cytoplasm without detectable loss of lysosomal integrity. An alternative mechanism of CatB release was suggested to involve Bax. Bax is involved in permeability pore transition in mitochondria, which triggers apoptosis; however, several studies clearly implicated it in lysosomal permeabilization by direct interaction with lysosomes or with another nonmitochondrial membrane fraction (47, 29, 22, 62, 63). The involvement of Bax in lysosomal permeabilization makes it possible that a selective mechanism is responsible for CatB release associated with apoptosis, given that CatB release in our system is fairly selective and does not seem to induce gross dissociation of lysosomal membrane. Although Bax does not seem to be the driving factor behind CatB release in our system, there are other candidates that may play such a role, such as caspase-8 (51).
It is important to remember that Bax has been implicated as a pro-apoptotic signal. The relationship between TRPML1 KD, CatB release, apoptosis, and Bax is a subject of future investigations. One possible mechanism could involve CatB release followed by Bid/Bax activation, cytochrome c release, mitochondrial permeabilization, and caspase cleavage. Alternatively, CatB release could work in a parallel or supplemental apoptotic pathway, independent of the Bax-mediated pathway. Elucidating these systems and the connection between the apoptotic pathways will be the subject of future work.
Supplementary Material
Acknowledgements
We thank the Center for Biologic Imaging at the University of Pittsburgh and Tom Harper in the Dept. of Biological Sciences, University of Pittsburgh, for invaluable help with electron microscopy. We thank Dr. Gustavo Maegawa for fruitful discussion.
This work was supported, in whole or in part, by National Institutes of Health Grants HD058577 and ES01678 (to K. K.).

This article contains supplemental Fig. S1.
- TRP
- transient receptor potential
- CatB
- cathepsin B
- KD
- knockdown
- LAL
- lysosomal acid lipase
- MLIV
- mucolipiodosis type IV
- PPT1
- palmitoyl-protein thioesterase 1
- TRPML1
- transient receptor potential channel mucolipin-1
- BAPTA
- 1,2-bis(2-aminophenoxyl)ethane-N,N,N′,N′-tetraacetic acid
- qPCR
- quantitative PCR
- CatB
- cathepsin B
- Z
- benzyloxycarbonyl
- AMC
- arginine-4-methylcoumaryl-7-amide.
REFERENCES
- 1. Kiselyov K., Chen J., Rbaibi Y., Oberdick D., Tjon-Kon-Sang S., Shcheynikov N., Muallem S., Soyombo A. (2005) TRP-ML1 is a lysosomal monovalent cation channel that undergoes proteolytic cleavage. J. Biol. Chem. 280, 43218–43223 [DOI] [PubMed] [Google Scholar]
- 2. Sun M., Goldin E., Stahl S., Falardeau J. L., Kennedy J. C., Acierno J. S., Jr., Bove C., Kaneski C. R., Nagle J., Bromley M. C., Colman M., Schiffmann R., Slaugenhaupt S. A. (2000) Mucolipidosis type IV is caused by mutations in a gene encoding a novel transient receptor potential channel. Hum. Mol. Genet. 9, 2471–2478 [DOI] [PubMed] [Google Scholar]
- 3. LaPlante J. M., Falardeau J., Sun M., Kanazirska M., Brown E. M., Slaugenhaupt S. A., Vassilev P. M. (2002) Identification and characterization of the single channel function of human mucolipin-1 implicated in mucolipidosis type IV, a disorder affecting the lysosomal pathway. FEBS Lett. 532, 183–187 [DOI] [PubMed] [Google Scholar]
- 4. Hersh B. M., Hartwieg E., Horvitz H. R. (2002) The Caenorhabditis elegans mucolipin-like gene cup-5 is essential for viability and regulates lysosomes in multiple cell types. Proc. Natl. Acad. Sci. U.S.A. 99, 4355–4360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vergarajauregui S., Puertollano R. (2006) Two di-leucine motifs regulate trafficking of mucolipin-1 to lysosomes. Traffic 7, 337–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Miedel M. T., Weixel K. M., Bruns J. R., Traub L. M., Weisz O. A. (2006) Post-translational cleavage and adaptor protein complex-dependent trafficking of mucolipin-1. J. Biol. Chem. 281, 12751–12759 [DOI] [PubMed] [Google Scholar]
- 7. Zhang F., Jin S., Yi F., Li P. L. (2009) TRP-ML1 functions as a lysosomal NAADP-sensitive Ca2+ release channel in coronary arterial myocytes. J. Cell. Mol. Med. 13, 3174–3185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xu H., Delling M., Li L., Dong X., Clapham D. E. (2007) Activating mutation in a mucolipin transient receptor potential channel leads to melanocyte loss in varitint-waddler mice. Proc. Natl. Acad. Sci. U.S.A. 104, 18321–18326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dong X. P., Shen D., Wang X., Dawson T., Li X., Zhang Q., Cheng X., Zhang Y., Weisman L. S., Delling M., Xu H. (2010) PI(3,5)P(2) controls membrane trafficking by direct activation of mucolipin Ca2+ release channels in the endolysosome. Nat. Commun. 1, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pryor P. R., Reimann F., Gribble F. M., Luzio J. P. (2006) Mucolipin-1 is a lysosomal membrane protein required for intracellular lactosylceramide traffic. Traffic 7, 1388–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. LaPlante J. M., Sun M., Falardeau J., Dai D., Brown E. M., Slaugenhaupt S. A., Vassilev P. M. (2006) Lysosomal exocytosis is impaired in mucolipidosis type IV. Mol. Genet. Metab. 89, 339–348 [DOI] [PubMed] [Google Scholar]
- 12. LaPlante J. M., Ye C. P., Quinn S. J., Goldin E., Brown E. M., Slaugenhaupt S. A., Vassilev P. M. (2004) Functional links between mucolipin-1 and Ca2+-dependent membrane trafficking in mucolipidosis IV. Biochem. Biophys. Res. Commun. 322, 1384–1391 [DOI] [PubMed] [Google Scholar]
- 13. Miedel M. T., Rbaibi Y., Guerriero C. J., Colletti G., Weixel K. M., Weisz O. A., Kiselyov K. (2008) Membrane traffic and turnover in TRP-ML1-deficient cells. A revised model for mucolipidosis type IV pathogenesis. J. Exp. Med. 205, 1477–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Soyombo A. A., Tjon-Kon-Sang S., Rbaibi Y., Bashllari E., Bisceglia J., Muallem S., Kiselyov K. (2006) TRP-ML1 regulates lysosomal pH and acidic lysosomal lipid hydrolytic activity. J. Biol. Chem. 281, 7294–7301 [DOI] [PubMed] [Google Scholar]
- 15. Eichelsdoerfer J. L., Evans J. A., Slaugenhaupt S. A., Cuajungco M. P. (2010) Zinc dyshomeostasis is linked with the loss of mucolipidosis IV-associated TRPML1 ion channel. J. Biol. Chem. 285, 34304–34308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bassi M. T., Manzoni M., Monti E., Pizzo M. T., Ballabio A., Borsani G. (2000) Cloning of the gene encoding a novel integral membrane protein, mucolipidin-and identification of the two major founder mutations causing mucolipidosis type IV. Am. J. Hum. Genet. 67, 1110–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Micsenyi M. C., Dobrenis K., Stephney G., Pickel J., Vanier M. T., Slaugenhaupt S. A., Walkley S. U. (2009) Neuropathology of the Mcoln1(−/−) knockout mouse model of mucolipidosis type IV. J. Neuropathol. Exp. Neurol. 68, 125–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Venugopal B., Browning M. F., Curcio-Morelli C., Varro A., Michaud N., Nanthakumar N., Walkley S. U., Pickel J., Slaugenhaupt S. A. (2007) Neurologic, gastric, and opthalmologic pathologies in a murine model of mucolipidosis type IV. Am. J. Hum. Genet. 81, 1070–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Frei K. P., Patronas N. J., Crutchfield K. E., Altarescu G., Schiffmann R. (1998) Mucolipidosis type IV. Characteristic MRI findings. Neurology 51, 565–569 [DOI] [PubMed] [Google Scholar]
- 20. Schiffmann R., Dwyer N. K., Lubensky I. A., Tsokos M., Sutliff V. E., Latimer J. S., Frei K. P., Brady R. O., Barton N. W., Blanchette-Mackie E. J., Goldin E. (1998) Constitutive achlorhydria in mucolipidosis type IV. Proc. Natl. Acad. Sci. U.S.A. 95, 1207–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Siegel H., Frei K., Greenfield J., Schiffmann R., Sato S. (1998) Electroencephalographic findings in patients with mucolipidosis type IV. Electroencephalogr. Clin. Neurophysiol. 106, 400–403 [DOI] [PubMed] [Google Scholar]
- 22. Chandra M., Zhou H., Li Q., Muallem S., Hofmann S. L., Soyombo A. A. (2011) A role for the Ca2+ channel TRPML1 in gastric acid secretion, based on analysis of knockout mice. Gastroenterology 140, 857–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jennings J. J., Jr., Zhu J. H., Rbaibi Y., Luo X., Chu C. T., Kiselyov K. (2006) Mitochondrial aberrations in mucolipidosis type IV. J. Biol. Chem. 281, 39041–39050 [DOI] [PubMed] [Google Scholar]
- 24. Settembre C., Fraldi A., Jahreiss L., Spampanato C., Venturi C., Medina D., de Pablo R., Tacchetti C., Rubinsztein D. C., Ballabio A. (2008) A block of autophagy in lysosomal storage disorders. Hum. Mol. Genet. 17, 119–129 [DOI] [PubMed] [Google Scholar]
- 25. Settembre C., Fraldi A., Rubinsztein D. C., Ballabio A. (2008) Lysosomal storage diseases as disorders of autophagy. Autophagy 4, 113–114 [DOI] [PubMed] [Google Scholar]
- 26. Takamura A., Higaki K., Kajimaki K., Otsuka S., Ninomiya H., Matsuda J., Ohno K., Suzuki Y., Nanba E. (2008) Enhanced autophagy and mitochondrial aberrations in murine G(M1)-gangliosidosis. Biochem. Biophys. Res. Commun. 367, 616–622 [DOI] [PubMed] [Google Scholar]
- 27. Vergarajauregui S., Connelly P. S., Daniels M. P., Puertollano R. (2008) Autophagic dysfunction in mucolipidosis type IV patients. Hum. Mol. Genet. 17, 2723–2737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ballabio A., Gieselmann V. (2009) Lysosomal disorders. From storage to cellular damage. Biochim. Biophys. Acta 1793, 684–696 [DOI] [PubMed] [Google Scholar]
- 29. Polster B. M., Fiskum G. (2004) Mitochondrial mechanisms of neural cell apoptosis. J. Neurochem. 90, 1281–1289 [DOI] [PubMed] [Google Scholar]
- 30. Curcio-Morelli C., Charles F. A., Micsenyi M. C., Cao Y., Venugopal B., Browning M. F., Dobrenis K., Cotman S. L., Walkley S. U., Slaugenhaupt S. A. (2010) Macroautophagy is defective in mucolipin-1-deficient mouse neurons. Neurobiol. Dis. 40, 370–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vesa J., Hellsten E., Verkruyse L. A., Camp L. A., Rapola J., Santavuori P., Hofmann S. L., Peltonen L. (1995) Mutations in the palmitoyl protein thioesterase gene causing infantile neuronal ceroid lipofuscinosis. Nature 376, 584–587 [DOI] [PubMed] [Google Scholar]
- 32. Weimer J. M., Kriscenski-Perry E., Elshatory Y., Pearce D. A. (2002) The neuronal ceroid lipofuscinoses. Mutations in different proteins result in similar disease. NeuroMolecular Med. 1, 111–124 [DOI] [PubMed] [Google Scholar]
- 33. Shimizu S., Kanaseki T., Mizushima N., Mizuta T., Arakawa-Kobayashi S., Thompson C. B., Tsujimoto Y. (2004) Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat. Cell Biol. 6, 1221–1228 [DOI] [PubMed] [Google Scholar]
- 34. Luo C. L., Chen X. P., Yang R., Sun Y. X., Li Q. Q., Bao H. J., Cao Q. Q., Ni H., Qin Z. H., Tao L. Y. (2010) Cathepsin B contributes to traumatic brain injury-induced cell death through a mitochondria-mediated apoptotic pathway. J. Neurosci. Res. 88, 2847–2858 [DOI] [PubMed] [Google Scholar]
- 35. Cheriyath V., Kuhns M. A., Kalaycio M. E., Borden E. C. (2011) Potentiation of apoptosis by histone deacetylase inhibitors and doxorubicin combination: cytoplasmic cathepsin B as a mediator of apoptosis in multiple myeloma. Br. J. Cancer 104, 957–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rommelaere G., Michel S., Mercy L., Fattaccioli A., Demazy C., Ninane N., Houbion A., Renard P., Arnould T. (2011) Hypersensitivity of mtDNA-depleted cells to staurosporine-induced apoptosis: roles of Bcl-2 downregulation and cathepsin B. Am. J. Physiol. Cell Physiol. 300, C1090–C1106 [DOI] [PubMed] [Google Scholar]
- 37. Michallet M. C., Saltel F., Preville X., Flacher M., Revillard J. P., Genestier L. (2003) Cathepsin B-dependent apoptosis triggered by antithymocyte globulins. A novel mechanism of T-cell depletion. Blood 102, 3719–3726 [DOI] [PubMed] [Google Scholar]
- 38. Oberle C., Huai J., Reinheckel T., Tacke M., Rassner M., Ekert P. G., Buellesbach J., Borner C. (2010) Lysosomal membrane permeabilization and cathepsin release is a Bax/Bak-dependent, amplifying event of apoptosis in fibroblasts and monocytes. Cell Death Differ. 17, 1167–1178 [DOI] [PubMed] [Google Scholar]
- 39. Mach L., Schwihla H., Stüwe K., Rowan A. D., Mort J. S., Glössl J. (1993) Activation of procathepsin B in human hepatoma cells. The conversion into the mature enzyme relies on the action of cathepsin B itself. Biochem. J. 293, 437–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mach L., Stüwe K., Hagen A., Ballaun C., Glössl J. (1992) Proteolytic processing and glycosylation of cathepsin B. The role of the primary structure of the latent precursor and of the carbohydrate moiety for cell type-specific molecular forms of the enzyme. Biochem. J. 282, 577–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hanewinkel H., Glössl J., Kresse H. (1987) Biosynthesis of cathepsin B in cultured normal and I-cell fibroblasts. J. Biol. Chem. 262, 12351–12355 [PubMed] [Google Scholar]
- 42. Tapper H., Sundler R. (1995) Bafilomycin A1 inhibits lysosomal, phagosomal, and plasma membrane H(+)-ATPase and induces lysosomal enzyme secretion in macrophages. J. Cell. Physiol. 163, 137–144 [DOI] [PubMed] [Google Scholar]
- 43. Seglen P. O., Gordon P. B., Grinde B., Solheim A., Kovács A. L., Poli A. (1981) Inhibitors and pathways of hepatocytic protein degradation. Acta Biol. Med. Ger. 40, 1587–1598 [PubMed] [Google Scholar]
- 44. Scornik O. A. (1984) Effects of inhibitors of protein degradation on the rate of protein synthesis in Chinese hamster ovary cells. J. Cell. Physiol. 121, 257–262 [DOI] [PubMed] [Google Scholar]
- 45. Sardiello M., Palmieri M., di Ronza A., Medina D. L., Valenza M., Gennarino V. A., Di Malta C., Donaudy F., Embrione V., Polishchuk R. S., Banfi S., Parenti G., Cattaneo E., Ballabio A. (2009) A gene network regulating lysosomal biogenesis and function. Science 325, 473–477 [DOI] [PubMed] [Google Scholar]
- 46. Terman A., Kurz T., Gustafsson B., Brunk U. T. (2006) Lysosomal labilization. IUBMB Life 58, 531–539 [DOI] [PubMed] [Google Scholar]
- 47. Boya P., Gonzalez-Polo R. A., Poncet D., Andreau K., Vieira H. L., Roumier T., Perfettini J. L., Kroemer G. (2003) Mitochondrial membrane permeabilization is a critical step of lysosome-initiated apoptosis induced by hydroxychloroquine. Oncogene 22, 3927–3936 [DOI] [PubMed] [Google Scholar]
- 48. Ferri K. F., Kroemer G. (2001) Organelle-specific initiation of cell death pathways. Nat. Cell Biol. 3, E255–263 [DOI] [PubMed] [Google Scholar]
- 49. Yeung B. H., Huang D.C., Sinicrope F.A. (2006) PS-341 (bortezomib) induces lysosomal cathepsin B release and a caspase-2-dependent mitochondrial permeabilization and apoptosis in human pancreatic cancer cells. J. Biol. Chem. 281, 11923–11932 [DOI] [PubMed] [Google Scholar]
- 50. Werneburg N. W., Guicciardi M. E., Bronk S. F., Gores G. J. (2002) Tumor necrosis factor-alpha-associated lysosomal permeabilization is cathepsin B dependent. Am. J. Physiol. Gastrointestinal Liver Physiol. 283, 947–956 [DOI] [PubMed] [Google Scholar]
- 51. Guicciardi M. E., Deussing J., Miyoshi H., Bronk S. F., Svingen P. A., Peters C., Kaufmann S. H., Gores G. J. (2000) Cathepsin B contributes to TNF-α-mediated hepatocyte apoptosis by promoting mitochondrial release of cytochrome c. J. Clin. Invest. 106, 1127–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Uchiyama Y. (2001) Autophagic cell death and its execution by lysosomal cathepsins. Arch. Histol. Cytol. 64, 233–246 [DOI] [PubMed] [Google Scholar]
- 53. Krepela E. (2001) Cysteine proteinases in tumor cell growth and apoptosis. Neoplasma 48, 332–349 [PubMed] [Google Scholar]
- 54. Bursch W. (2001) The autophagosomal-lysosomal compartment in programmed cell death. Cell Death Differ. 8, 569–581 [DOI] [PubMed] [Google Scholar]
- 55. Yuan X. M., Li W., Brunk U. T., Dalen H., Chang Y. H., Sevanian A. (2000) Lysosomal destabilization during macrophage damage induced by cholesterol oxidation products. Free Radic. Biol. Med. 28, 208–218 [DOI] [PubMed] [Google Scholar]
- 56. Isahara K., Ohsawa Y., Kanamori S., Shibata M., Waguri S., Sato N., Gotow T., Watanabe T., Momoi T., Urase K., Kominami E., Uchiyama Y. (1999) Regulation of a novel pathway for cell death by lysosomal aspartic and cysteine proteinases. Neuroscience 91, 233–249 [DOI] [PubMed] [Google Scholar]
- 57. Liu L., Zhang Z., Xing D. (2011) Cell death via mitochondrial apoptotic pathway due to activation of Bax by lysosomal photodamage. Free Radic. Biol. Med. 51, 53–68 [DOI] [PubMed] [Google Scholar]
- 58. Sun L., Zhao Y., Li X., Yuan H., Cheng A., Lou H. (2010) A lysosomal-mitochondrial death pathway is induced by solamargine in human K562 leukemia cells. Toxicol. In Vitro 24, 1504–1511 [DOI] [PubMed] [Google Scholar]
- 59. Ruivo R., Anne C., Sagné C., Gasnier B. (2009) Molecular and cellular basis of lysosomal transmembrane protein dysfunction. Biochim. Biophys. Acta 1793, 636–649 [DOI] [PubMed] [Google Scholar]
- 60. Kiselyov K., Jennigs J. J., Jr., Rbaibi Y., Chu C. T. (2007) Autophagy, mitochondria, and cell death in lysosomal storage diseases. Autophagy 3, 259–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Moles A., Tarrats N., Fernandez-Checa J. C., Mari M. (2012) Cathepsin B overexpression due to acid sphingomyelinase ablation promotes liver fibrosis in Niemann-Pick disease. J. Biol. Chem. 287, 1178–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cregan S. P., Fortin A., MacLaurin J. G., Callaghan S. M., Cecconi F., Yu S. W., Dawson T. M., Dawson V. L., Park D. S., Kroemer G., Slack R. S. (2002) Apoptosis-inducing factor is involved in the regulation of caspase-independent neuronal cell death. J. Cell Biol. 158, 507–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Orrenius S. (2004) Mitochondrial regulation of apoptotic cell death. Toxicol Lett. 149, 19–23 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.