Background: Bacterial reaction centers catalyze light-induced transmembrane electron transport using only one of two chemically equivalent pathways.
Results: Multiplexed screening uncovers semirandom mutations that unexpectedly activate the unused pathway by employing ionizable residues.
Conclusion: High throughput mutagenesis approaches reveal structure/function relationships that govern electron transfer efficiency.
Significance: Directed molecular evolution can reveal principles that enable efficient, unidirectional, transmembrane electron transfer for the design of de novo pathways or biomimetic devices.
Keywords: Bioenergetics-Electron Transfer Complex, Molecular Biology, Photosynthesis, Protein Engineering, Protein Evolution, Directed Molecular Evolution, Membrane Protein Purification, Quinone Reduction, Time-resolved Spectroscopy, Transmembrane Electron Transfer
Abstract
Photosynthetic reaction centers convert light energy into chemical energy in a series of transmembrane electron transfer reactions, each with near 100% yield. The structures of reaction centers reveal two symmetry-related branches of cofactors (denoted A and B) that are functionally asymmetric; purple bacterial reaction centers use the A pathway exclusively. Previously, site-specific mutagenesis has yielded reaction centers capable of transmembrane charge separation solely via the B branch cofactors, but the best overall electron transfer yields are still low. In an attempt to better realize the architectural and energetic factors that underlie the directionality and yields of electron transfer, sites within the protein-cofactor complex were targeted in a directed molecular evolution strategy that implements streamlined mutagenesis and high throughput spectroscopic screening. The polycistronic approach enables efficient construction and expression of a large number of variants of a heteroligomeric complex that has two intimately regulated subunits with high sequence similarity, common features of many prokaryotic and eukaryotic transmembrane protein assemblies. The strategy has succeeded in the discovery of several mutant reaction centers with increased efficiency of the B pathway; they carry multiple substitutions that have not been explored or linked using traditional approaches. This work expands our understanding of the structure-function relationships that dictate the efficiency of biological energy-conversion reactions, concepts that will aid the design of bio-inspired assemblies capable of both efficient charge separation and charge stabilization.
Introduction
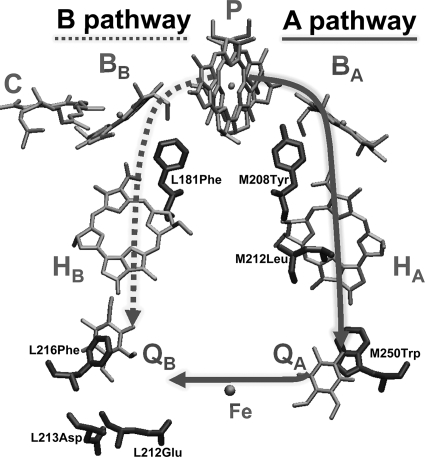
Light energy is converted to chemical energy in a series of extremely efficient electron transfer (ET)4 reactions performed by photosynthetic reaction centers (RCs). The structures of these transmembrane protein-cofactor complexes from photosynthetic bacteria and higher plants reveal an axis of approximate 2-fold symmetry that relates both core protein subunits and functionally asymmetric sets of cofactors (Fig. 1) (1–4). In many photosynthetic bacteria, these complexes are composed of three subunits: the homologous, integral L and M polypeptides and the membrane-tethered H polypeptide. Bacterial cofactors include a bacteriochlorophyll dimer (P) that serves as the primary electron donor, two bacteriochlorophyll monomers (BA and BB), two bacteriopheophytins (HA and HB), and two ubiquinone10 molecules (QA and QB). In type I RCs (e.g. Photosystem I), spectroscopic evidence suggests that both pathways are active in producing reduced quinone via light-induced ET (see e.g. Refs. 5–8). Yet, in type II RCs (e.g. bacterial RCs and Photosystem II), the B pathway is silent, and possibly vestigial; only the A branch cofactors are active.
FIGURE 1.
Electron transfer pathways and positions of substituted amino acids in bacterial RCs. Despite equivalent sets of symmetry-related cofactors, only the A branch is active in native RCs. For visual clarity, all isoprenyl anchoring groups and bulky porphyrin ring substituents were removed from cofactors.
The relative free energies of the charge-separated states are critical in determining the directionality, rates, and yields of ET in the RC. Some strategies to manipulate these factors genetically over the last 20 years (for review, see Ref. 9) include: addition or removal of hydrogen bonds to cofactors (10–15); addition or removal of axial ligands with attendant changes of bacteriopheophytin to bacteriochlorophyll or vice versa (16–27); modification of conserved residues or helical segments that are related by the pseudo-2-fold symmetry axis of the complex (28, 29); and introduction or removal of polar or ionizable residues near the cofactors (12, 13, 16, 17, 30–32). To disable ET from HA to QA or from QA to QB specifically, the occupancy and environments of the quinone binding pockets have been manipulated by mutation (16, 23–25, 33–37).
The highest yields of ET from the excited dimer (P*) to HB (thus producing P+HB−) have been achieved in mutant Rhodobacter (R.) capsulatus RCs (17, 31, 32, 35, 38). In mutant RCs lacking QA, reduction of QB via sole activity of the B branch cofactors has been observed unambiguously (16, 24, 35, 37, 39–41). No cases yet reproduce any of the (essentially) 100% yields characteristic of the native A side ET reactions. Rather, mutant RCs have demonstrated the magnitude of the B pathway inefficiency. The combination of a few site-specific mutations has not managed to modulate the energetics (or electronic factors) of the B branch cofactors to provide for rapid electron transfer. At present, we lack the understanding needed to use rational design to accomplish the drastic leap required to redesign the RC to use the B pathway exclusively and efficiently.
Construction of such a re-engineered RC requires sampling of a much larger number of variants than has been examined previously. Thus, an approach has been adopted that combines semirandom mutagenesis and rapid screening techniques that require minimal sample volumes. Several methods have been streamlined by implementation of an automated liquid handler, and purification of mutant RCs has been semiautomated. A high throughput spectroscopic assay evaluates the competency of candidate RCs for QB reduction via the B side cofactors. This approach has been made feasible by advances in synthetic biology; favorable mutations that have been identified can be fed into directed molecular evolution experiments for additional optimization of the ET pathway. Techniques that have been developed here are likely to be applicable to other multisubunit protein complexes with similar gene architectures.
EXPERIMENTAL PROCEDURES
Growth
R. capsulatus strains were cultured on SuperRCVPY (20) medium under semiaerobic, chemoheterotrophic conditions in the dark (165 rpm, silicone sponge closures, 33 °C). Escherichia coli strains were propagated on LB or 2×TY medium (42). Plasmids were selected with kanamycin (30 μg/ml for both organisms). Strains and plasmids used in this study are listed in supplemental Table S1.
Molecular Biology
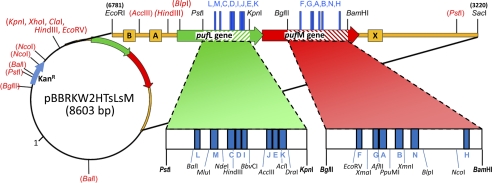
The expression vector used in this study, pBBRKW2HTsLsM (Fig. 2; GenBank Accession Number JN565028), was constructed from puf operon genes derived as an EcoRI-SacI fragment from plasmid pUHTMluBgl:α− (24) that was inserted into a derivative of broad host range cloning vector pBBR1MCS-2 (43), generating pBBRKW2HTBALMX. The expression plasmid was then modified extensively to facilitate the approach described herein. Portions of both the pufL and pufM genes were synthesized (GenScript) to encode silent changes that introduced restriction sites flanking each of the segments chosen for mutagenesis (Fig. 2). To eliminate duplicate restriction sites elsewhere, 11 restriction sites in pBBR1MCS-2 and 4 sites in the puf operon were removed (supplemental Table S2) using either site-directed mutagenesis (QuikChange; Stratagene) or mung bean nuclease. The synthetic segment of pufL was then cloned into pBBRKW2HTBALMX (Table S2) as a PstI-KpnI fragment, and the synthetic pufM segment was cloned using BglII and BamHI, thus creating pBBRKW2HTsLsM. A His7 tag is encoded at the C terminus of pufM. The H32R mutation in the pufA gene prevents assembly of the light-harvesting (LH) I antenna complex (44). Thus, genes expressed from this plasmid in R. capsulatus strain U43 result in the antennaeless, “RC-only” photosynthetic membrane needed for quantitative extraction of RCs by mild detergents (24). A summary of substitutions carried by mutant RCs and the nomenclature used to refer to them is contained in supplemental Table S3.
FIGURE 2.
Engineered expression plasmid for RC mutagenesis. Segments of the L and M genes targeted for mutagenesis (striped regions and Table 1) are flanked by unique restriction sites that facilitate the insertions of oligonucleotide cassettes encoding multiple substitutions. Fifteen restriction sites (parentheses) in the plasmid were removed to enable the cassette-based mutagenesis approach.
Construction of the F(L181)X+FHV, F(L216)X+YFHV and E(L212)A-D(L213)A-F(L216)X+YFHV sets of mutant strains was expedited with cassette-based mutagenesis. The W(M250)V control strain was constructed by site-directed mutagenesis of the synthetic BglII-BamHI fragment of the pufM segment described above. Protocols for phosphorylation and annealing of oligonucleotides, ligations, transformations, and plasmid screening were adapted for use with a liquid handling system (Biomek FX; Beckman Coulter). For each mutant set, a library of oligonucleotides was designed to encode all 20 amino acids at residue X. Oligonucleotides for the F(L181)X+FHV set were flanked by HindIII and BbvCI restriction sites, and the oligonucleotides for the F(L216)X+YFHV and E(L212)A-D(L213)A-F(L216)X+YFHV cassettes were flanked by AccIII and AclI restriction sites (Table 1 and Fig. 2). Oligonucleotides were phosphorylated, annealed, and ligated to pBBRKW2HTsLsM digested with the appropriate restriction enzymes. Candidate plasmids from DH5α transformants were screened using restriction fragment analysis and sequenced. Correct plasmids were used to transform mobilizing donor strain S17-1 for conjugal transfer to R. capsulatus deletion strain U43 (24). To facilitate rapid recovery of expression strains, a 100-μl aliquot of a 1:25,000 dilution in RCV medium (45) was plated directly to SuperRCVPY agar containing kanamycin. After 2 days, R. capsulatus colonies were cultured on SuperRCVPY containing kanamycin for the preparation of archival stocks.
TABLE 1.
Regions targeted for mutagenesis
| Cofactor | Amino acid segment (region) | 5′-Flanking enzyme | 3′-Flanking enzyme |
|---|---|---|---|
| BB | M150–160 (A) | AflII | PpuMI |
| BB | M173–184 (B) | PpuMI | XmnI |
| BB | L168–178 (C) | NdeI | HindIII |
| BB/HB | L179–190 (D) | HindIII | BbvCI |
| HB | L215–220 (E) | AccIII | AclI |
| HB | M124–131 (F) | EcoRV | XmaI |
| HB | M144–151 (G) | XmaI | AflII |
| HB | M271–275 (H) | NcoI | BamHI |
| QB | L186–194 (I) | HindIII | BbvCI |
| QB | L212- 216 (J) | AccIII | AclI |
| QB | L220–232 (K) | AclI | DraI |
| BA | L127–131 (L) | BalI | MluI |
| BA | L146–157 (M) | MluI | NdeI |
| BA | M195–207 (N) | XmnI | BlpI |
Expression Screening
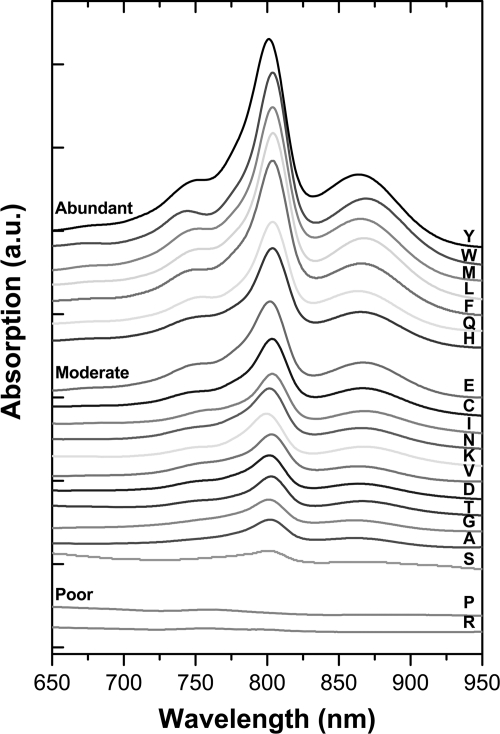
To survey expression levels, crude RCs were purified in tandem according to standard methods (as detailed in supplemental Methods) with the following modifications. Cells from small cultures (80 ml) were lysed using a microfluidizer (Microfluidics, Inc.) and mutant RCs were solubilized with Deriphat 160-C. Following overnight binding to 200 μl of nickel-nitrilotriacetic acid resin (Qiagen), chromatography proceeded in 2-ml, 96-well filter plates. UV-visible spectra of dark-adapted (10 min) samples were recorded (SpectraMax M5e; Molecular Devices). Based on A865 nm, mutant RCs were classified into three expression categories relative to yields of WT RCs: abundant (>25%), moderate (2.5–25%), and poor (<2.5%).
Semiautomated RC Purification
Mutant strains that expressed RCs abundantly or moderately were grown in 2–6-liter volumes depending on the expression level. RCs were purified via semiautomated methods using ÄKTA-FPLCs (46).
RC Functionality Screening
Assaying mutant RCs for formation of P+QB− via B branch ET was accomplished with a home-built, time-resolved spectrometer designed for 100–200-μl samples arrayed in clear-bottom 96-well plates (BD Falcon). Rapid data collection and reproducibility were facilitated via use of a computer-controlled translation stage (Aerotech) to position the sample well under collinear, vertical excitation and probing (measuring) beams. RCs were excited with a single subsaturating, 5-ns, 532-nm flash from a Nd:YAG laser. The magnitude and decay of bleaching of the ground state absorbance of P were probed by a stable, continuous-wave, 850-nm laser diode (Melles-Griot) attenuated with neutral density filters so that no discernible actinic effects were observed. For added sample protection, the probe light was incident on the sample only during data collection and was otherwise blocked with a computer-controlled shutter. Following passage through the sample, the probe light was isolated from the collinearized pump-probe beams by dichroic and 850-nm interference filters. The isolated probe light was collected onto a near-infrared-sensitive photosensor module (Hamamatsu H-5784-20), whose output was recorded on a digital oscilloscope. The apparatus is capable of measuring transient spectroscopic signals spanning 50 μs to several minutes. For 100-μl volume, the effective measuring pathlength was 2 mm; sample concentrations were adjusted such that A865 nm was 0.45–0.55. Signal intensities were normalized to the same ground state absorbance of the P band during data analysis. Because RCs are QA-less (except for wild type) due to the W(M250)V mutation described below, the initial amplitude of each kinetic trace is proportional to the yield of ET to QB solely via the B branch, as assayed relative to control RCs on the same plate.
Further characterization of primary electron transfer events in select mutant RCs utilized a transient absorption apparatus having 130-fs excitation and white light probe flashes operating at 10 Hz (described previously in Refs. 31, 38). RCs were held at ∼10 °C in an ice-cooled reservoir and flowed through a 2-mm pathlength cell.
RESULTS
A system to construct and express mutants of the L and M genes of the polycistronic, oxygen-regulated puf operon of R. capsulatus was developed previously (47–49). The expression host strain, U43, does not synthesize any light-harvesting antennae or RC complexes of the photosynthetic apparatus (i.e. LHI− LHII− RC−). The original broad host range expression plasmid cannot be used for this type of study because its complete sequence has not been determined and its large size (32 kb) hinders its application to an approach based upon restriction enzyme cloning of mutated cassettes. Thus, pBBR1MCS-2 (5.1 kb), whose sequence is known (43), was selected as the vector for the expression plasmid and modified as described above. Yields of RCs purified from strains carrying the newly engineered expression plasmid were found to be 2-fold greater than those obtained from plasmid pUHTMluBgl:α−.
The initial mutagenesis template was FHV. The F mutation substitutes Phe for the native Tyr at M208 near BA and diminishes ET to the A side. The H mutation substitutes His for Leu at M212 near HA and results in incorporation of a bacteriochlorophyll in the HA site. Because this cofactor is more difficult to reduce, the A pathway is disabled further. This mutation also removes HA absorption from the 510–550 nm spectral region, allowing quantitative assessment of HB reduction via ultrafast (picosecond) spectroscopic measurements. The V mutation, which substitutes Val for the conserved Trp at M250, prevents QA binding and, thus, prevents ET to QB via QA. In other words, this mutation ensures that electrons can reach QB only via the B side cofactors.
M208Tyr is related via the C2 symmetry axis to residue L181Phe near BB. This conserved asymmetric pair was the target of some of the earliest mutagenesis studies of the purple bacterial RC. The swap of Tyr for the native Phe at L181 and Phe for Tyr at M208 reverses the asymmetry and enhances ET to HB while diminishing ET to the A side. When the F(L181)Y mutation is added to the FHV template, the result is the YFHV RC, which serves as another control (in addition to the wild type that results in quantitative reduction of QB via A branch activity) for the ms screening assay because it displays ∼30% yield of P+HB− and ∼15% yield of P+QB− (31, 35, 39, 40, 50).
A large number of residues within 5–14 amino acid segments near the cofactors have been identified as mutagenesis targets (Table 1 and Fig. 2). At the outset, regions proximal to BB, HB, and QB were targeted to further enable (compared with YFHV) the B side pathway. For all of the mutant strains reported here, protein yields from larger scale cultures and automated purification procedures correlated well with yields determined from the small scale expression screening. The use of Deriphat 160C for RC solubilization and purification ensured full occupancy of QB in the native binding site (24).
Twenty F(L181)X+FHV constructs comprised the initial mutant set. Residue L181 (segment D, near BB) was chosen because of the known influence of this site on initial B side charge separation as indicated above. Eighteen of the mutant RCs were expressed moderately or abundantly (Fig. 3). None of the substitutions at L181 resulted in pronounced effects on the RC ground state absorption spectra except for the KFHV mutant, with K at L181 previously having been shown to provide a sixth ligand to BB (11).
FIGURE 3.
Expression yields of mutant RCs. UV-visible spectra of RCs purified from the F(L181)X+FHV set of mutant RCs indicate three expression categories. The residue present at the L181 site is designated with the single-letter code.
Additional rounds of mutagenesis focused on residues near QB in attempts to increase the low (∼40%) yield of ET from HB to QB (24, 35, 39). Saturation mutagenesis was performed at conserved residue L216Phe (segment J) because its C2-symmetry-related counterpart, M250Trp, is essential for rapid ET from HA to QA (33) and is important for QA binding. The F(L216)X substitutions were coupled with YFHV. Additionally, because previous work showed that replacement of the native Glu and Asp at L212 and L213, respectively, with alanines stabilizes P+QB− (51), the F(L216)X variants were coupled with the E(L212)A-D(L213)A+YFHV (AA+YFHV) substitutions in another set of mutants. These two rounds of mutagenesis yielded 40 additional strains with 36 expressing RCs at abundant or moderate levels, 19 F(L216)X+YFHV mutants and 17 AA+F(L216)X+YFHV mutants (supplemental Table S4). Only the D, H, and K substitutions failed to produce RCs in the AA+F(L216)X+YFHV background.
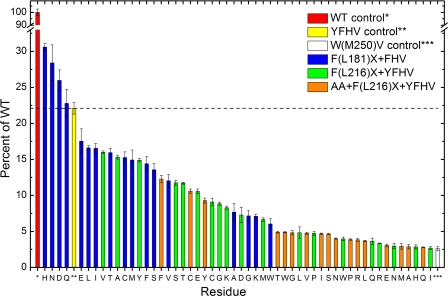
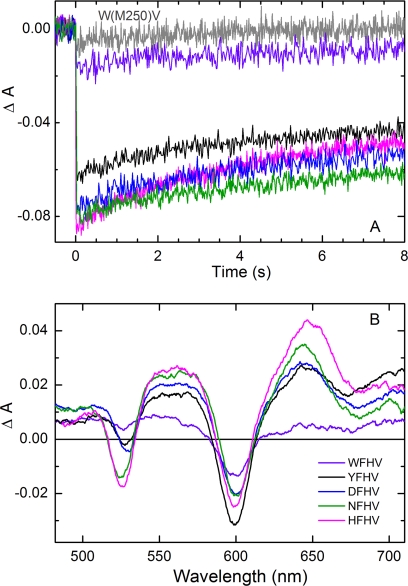
All 54 of the mutant RCs studied here gave light-induced formation of P+QB− via the B side cofactors in the ms transient absorption assay. The relative amounts of P+QB− formed are plotted in Fig. 4. Of particular note are the F(L181)H+FHV (HFHV), F(L181)N+FHV (NFHV), and F(L181)D+FHV (DFHV) mutant RCs, which produce P+QB− signals that are 20–30% larger than that of the YFHV control. The A, G, K, and W mutations produce RCs with the poorest yields of P+QB− in the F(L181)X+FHV series. Fig. 5A shows kinetic traces derived from the HFHV, NFHV, DFHV, and WFHV RCs along with the YFHV and W(M250)V-only control RCs from the ms screening assay. The profiles, displaying decay time constants ≥10 s, are characteristic of P+QB− charge recombination when this state is formed via activity of the B branch cofactors; in these RCs, decay of this state via routes involving QA is prohibited by the W(M250)V mutation (24). Thus, the initial amplitudes of the kinetic traces in Fig. 5A are measures of the (relative) yields of P+QB− formed solely via B branch activity; this value is plotted for all of the mutant RCs in Fig. 4.
FIGURE 4.
B branch quinone reduction in mutant RCs. Yields of B side P+QB− formation from time-resolved assays of mutant RCs relative to signals from wild-type RCs are shown. Signals from the YFHV (yellow) and W(M250)V (white) RCs serve as benchmark controls. Error bars reflect variability in replicate spectroscopic screening measurements.
FIGURE 5.
Transient light-induced optical signals indicative of B branch activity in mutant RCs. A, P-bleaching amplitudes and decay profiles for state P+QB− observed in the ms screening assay. The gray trace is for the W(M250)V mutant RC; the other traces are as indicated in the legend in B. Each trace represents the average of seven data acquisitions (single excitation flash events). Signal amplitudes were corrected by factors of ≤1.2 to normalize the sample concentrations. B, comparison of transient absorption spectra acquired using 130-fs excitation flashes at 850 nm in ultrafast studies of the primary photochemistry. RC concentrations and laser excitation conditions were matched. The spectra shown were acquired at 1 ns after the flash to minimize the contribution of shorter lived states. The relative yields of P+HB− (determined by the magnitude of bleaching of the band of HB at 525–528 nm) agree well with those measured for the P+QB− yields in the ms screening assay (A).
The results for the HFHV, NFHV, DFHV, and WFHV mutant RCs were confirmed by ultrafast transient absorption measurements (Fig. 5B). These spectra show the magnitudes of bleaching of HB at ∼528 nm, reflecting the relative yields of ET from P* to HB and confirm an ordering of H>N>D>Y≫W, the same ordering as determined in the P+QB− yield assay. Additional ultrafast measurements are under way and will be reported elsewhere.
Expression yields for the F(L216)X+YFHV and AA+F(L216)X+YFHV sets of mutant RCs were quite high, especially for the latter set given the number of mutations introduced into the RC. However, yields of P+QB− in these mutant RCs did not exceed that of the template YFHV. In general, there is less ET to QB in RCs containing the E(L212)A-D(L213)A substitutions than in mutant RCs carrying the native residues at these sites. The relative rank order of the residues in each set is different (see, e.g. the effect of a Val substitution in each set), suggesting complex interplay of effects introduced by substitution of the small, nonpolar alanines for the larger, ionizable Glu and Asp.
DISCUSSION
Directionality of ET in type II RCs is a conundrum that has existed ever since the 1985 structure of the bacterial RC (3) revealed two branches of cofactors related by an axis of approximate C2 symmetry whereas spectroscopic measurements demonstrated activity of only one of them. The free energies of the charge-separated states on the A pathway (P+BA−, P+HA−, and P+QA−) are successively downhill from each other and from P*, facilitating rapid forward ET. In contrast, B branch inactivity presumably stems in large measure from P+BB− being somewhat higher in free energy than P*, resulting in much slower, noncompetitive, B side charge separation. Differences between the electronic couplings of P* with BA and BB may also contribute to unidirectional A side ET. The rate constant for P* → P+HB− on the native B pathway has been measured to be ∼(100 ps)−1 (17, 31), approximately 30-fold slower than charge separation on the A side (3–4 ps for P* → P+HA−), suggesting that yields of B side ET in wild-type RCs should be ∼3%.
Although significant yields of P+HB− formation have been reported in some R. capsulatus mutants (17, 31, 32, 35, 38), the largest rate constant for P* → P+HB− is ∼(40 ps)−1, obtained in the YFH mutant RC (31). The best efforts by several laboratories over many years have thus achieved approximately only a 2-fold increase in the rate of ET to HB compared with the wild-type value. This enhanced rate is still a factor of 10 slower than ET to the A side. Interestingly, rapid initial charge separation to the B side has been reported in a mutant where a bacteriopheophytin substitutes for BB. Whereas ET from P* to this new cofactor takes place in ∼10 ps, ET onward to HB does not occur (18), creating another, different, bottleneck in the quest for efficient use of the B side cofactors for transmembrane charge separation. Similar issues occur at the final step of ET from HB to QB, which has a yield of only ∼40% as mentioned above. The rate of this ET reaction is ∼(4 ns)−1, whereas the rate of competing charge recombination of P+HB− is ∼(3 ns)−1 (39). For comparison, the rates of the corresponding reactions on the A side are (200 ps)−1 and ∼(20 ns)−1.
To sample a larger number of variants and remove much of the prejudice inherent in rational design schemes, a directed molecular evolution approach has been adopted for engineering of the RC to enable efficient B branch ET. Directed molecular evolution has been applied to the discovery of new functions, increased expression, increased stability, and enhanced activity for many proteins (for review, see Refs. 52, 53) but has had relatively limited application to membrane proteins (e.g. Refs. 54–57). The present study merges principles of both rational design and directed molecular evolution to generate versions of the heterotrimeric, protein-cofactor RC complex that have acquired increased ability to utilize a vestigial ET pathway for transmembrane charge separation.
The mutagenesis employed here is best termed “semirandom” and relies on oligonucleotide cassettes rather than error-prone PCR or fragment shuffling. The rationale for this approach is based on knowledge of the structure, awareness of the substitutions that have been tried before, the concern that massively random mutagenesis will produce RCs with assembly and/or stability problems, the difficulties imposed by the high degree of sequence similarity between the pufL and pufM genes (especially in highly conserved regions that surround the cofactors), and the fact that their coding regions overlap (Fig. 2). Because this approach has worked well here for these genes embedded in the polycistronic puf operon, it is likely that the chosen mutagenesis strategy could be applied to many gene clusters encoding transmembrane complexes where components share a high degree of sequence similarity, a common occurrence in sequence data emerging from representatives of all kingdoms.
Directional cloning of cassettes required significant downsizing and engineering of the broad host range expression vector and was enabled by synthetic biology. Surprisingly, the yield of RCs expressed from the new, partially synthetic platform vector was 2-fold greater than the yield obtained from the original expression plasmid. This result may be due to differences in plasmid copy number and/or unexpected changes in (i) transcript stability, (ii) codon preferences, or (iii) modification of regulatory elements that occurred as a result of the engineering that was required.
In the initial round of saturation mutagenesis at the L181 site, surprises were encountered when yields of B side ET were found to be highest in RCs carrying His, Asn, Asp, and Gln mutations. Numerous RCs carrying single mutations at this site have been reported previously (9, 11, 58), but none of these studies had ever suggested that any of the above residues would match or exceed a Tyr at L181 in enhancing B branch activity or that the functional consequences of a Glu substitution would be similar to those conferred by a Tyr substitution (Fig. 4). In fact, considering that placement of an acidic residue close to BA in the G(M201)D RC disfavors ET to the A side (12), it was surprising that an Asp substitution at L181 near BB had the opposite effect, enabling more B branch activity. Similar effects observed for the polarizable His, Asn, Asp, and Gln residues at L181 cannot all be attributable to a common fundamental mechanism, but all of these substitutions will influence, rather unpredictably, the dielectric environment of the cofactors involved in the initial electron transfer reactions. The close parallel effects on B side ET for the Asn/Asp and Gln/Glu pairs, with the smaller Asn/Asp substitutions producing RCs with larger yields of B side ET, are noteworthy. In general, an assortment of uncharged substitutions at the L181 site is tolerated with neutral functional consequences; an additional surprise is that substitutions of both small (Gly, Ala) and large (Trp) residues produce nearly equivalent yields of P+QB−.
Although substitutions at the L216 site (regardless of whether they are accompanied by the Ala substitutions at L212–213) do not increase the amount of reduced quinone, trends are observed, some of which can be related to amino acid properties. In general, large residues at the L216 site result in lower yields of reduced quinone. RCs in which ionizable residues are substituted at the L216 site, when in combination with the Ala substitutions at L212–213, either fail to assemble or perform poorly in the functional assay. It is possible that substitutions within this region of the QB binding site affect its occupancy. A disappointment is that the Trp substitution at this site does not result in increased ET from HB to QB whereas the conserved Trp residue at the symmetry-related M250 site is critical for efficient ET from HA to QA in the wild-type RC (33). These results suggest that targeting of residues based on their relationship to the C2 symmetry axis in this region of the RC is less productive and emphasize that a more random, evolutionary approach will be required to enhance secondary electron transfer reactions of the B pathway.
Overall, the results reported herein validate the experimental strategy and underscore the plasticity of the RC in accommodating substitutions that result in a range of B branch activity. A subset of the new mutant RCs displays increased quinone reduction by the exclusive use of B branch cofactors. Unexpected, productive combinations of amino acid substitutions that previously have not been discovered by rational design were revealed by the approach described herein. Realization of these mutant RCs was enabled by miniaturization at two screening steps (expression testing and spectroscopic assays) and automation of protein purification and time-resolved spectroscopy. A system is now in place whereby mutant RCs with altered ET functionalities can be identified rapidly as candidates for more exhaustive functional characterization and iterative engineering to produce next generation RCs with desired ET capabilities, even those that are unimagined by Nature.
Supplementary Material
Acknowledgments
We thank Craig Luehr for optimization of scripts for automated protein purification and Elizabeth Landorf for assistance with adaptation of molecular biology methods to higher throughput on the liquid-handling robot.
This work was supported by Grant DE-FG-02-09ER16116 from the United States Department of Energy Office of Basic Energy Sciences, Division of Chemical Sciences, Geosciences, and Biosciences (to C. K., D. H., P. D. L., and D. K. H.). Argonne, a United States Department of Energy Office of Science laboratory, is operated under Contract DE-AC02-06CH11357.

This article contains supplemental Tables S1–S4 and Methods.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s) JN565028.
- ET
- electron transfer
- B
- bacteriochlorophyll
- H
- bacteriopheophytin
- LH
- light harvesting
- Q
- quinone
- P
- primary donor
- RC
- reaction center
- YFHV
- F(L181)Y-Y(M208)F-L(M212)H-W(M250)V
- KFHV
- F(L181)K+FHV
- AA+YFHV
- E(L212)A-D(L213)A+YFHV
- HFHV
- F(L181)H+FHV
- NFHV
- F(L181)N+FHV
- DFHV
- F(L181)D+FHV
- WFHV
- F(181)W+FHV.
REFERENCES
- 1. Allen J. P., Feher G., Yeates T. O., Komiya H., Rees D. C. (1987) Structure of the reaction center from Rhodobacter sphaeroides R-26: the cofactors. Proc. Natl. Acad. Sci. U.S.A. 84, 5730–5734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chang C. H., el-Kabbani O., Tiede D., Norris J., Schiffer M. (1991) Structure of the membrane-bound protein photosynthetic reaction center from Rhodobacter sphaeroides. Biochemistry 30, 5352–5360 [DOI] [PubMed] [Google Scholar]
- 3. Deisenhofer J., Epp O., Miki K., Huber R., Michel H. (1985) Structure of the protein subunits in the photosynthetic reaction center from Rhodopseudomonas viridis at 3 Å resolution. Nature 318, 618–624 [DOI] [PubMed] [Google Scholar]
- 4. Ermler U., Fritzsch G., Buchanan S. K., Michel H. (1994) Structure of the photosynthetic reaction centre from Rhodobacter sphaeroides at 2.65 A resolution: cofactors and protein-cofactor interactions. Structure 2, 925–936 [DOI] [PubMed] [Google Scholar]
- 5. Dashdorj N., Xu W., Cohen R. O., Golbeck J. H., Savikhin S. (2005) Asymmetric electron transfer in cyanobacterial photosystem I: charge separation and secondary electron transfer dynamics of mutations near the primary electron acceptor A0. Biophys. J. 88, 1238–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Guergova-Kuras M., Boudreaux B., Joliot A., Joliot P., Redding K. (2001) Evidence for two active branches for electron transfer in photosystem I. Proc. Natl. Acad. Sci. U.S.A. 98, 4437–4442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Joliot P., Joliot A. (1999) In vivo analysis of the electron transfer within photosystem I: are the two phylloquinones involved? Biochemistry 38, 11130–11136 [DOI] [PubMed] [Google Scholar]
- 8. Ramesh V. M., Gibasiewicz K., Lin S., Bingham S. E., Webber A. N. (2004) Bidirectional electron transfer in photosystem I: accumulation of A0- in A-side or B-side mutants of the axial ligand to chlorophyll A0. Biochemistry 43, 1369–1375 [DOI] [PubMed] [Google Scholar]
- 9. Wakeham M. C., Jones M. R. (2005) Rewiring photosynthesis: engineering wrong-way electron transfer in the purple bacterial reaction centre. Biochem. Soc. Trans. 33, 851–857 [DOI] [PubMed] [Google Scholar]
- 10. Bylina E. J., Kirmaier C., McDowell L., Holten D., Youvan D. C. (1988) Influence of an amino-acid residue on the optical properties and electron transfer dynamics of a photosynthetic reaction centre complex. Nature 336, 182–184 [Google Scholar]
- 11. DiMagno T. L., PD, Reddy N. R., Small G. J., Norris J. R., Schiffer M., Hanson D. K. (1998) Protein-chromophore interactions: spectral shifts report the consequences of mutations in the bacterial photosynthetic reaction center. Spectrochim. Acta Part A 54, 1247–1267 [Google Scholar]
- 12. Heller B. A., Holten D., Kirmaier C. (1996) Effects of Asp residues near the L-side pigments in bacterial reaction centers. Biochemistry 35, 15418–15427 [DOI] [PubMed] [Google Scholar]
- 13. Katilius E., Babendure J. L., Katiliene Z., Lin S., Taguchi A. K. W., Woodbury N. W. (2003) Manipulations of the B-side charge-separated states' energetics in the Rhodobacter sphaeroides reaction center. J. Phys. Chem. 107, 12029–12034 [Google Scholar]
- 14. Williams J. C., Alden R. G., Murchison H. A., Peloquin J. M., Woodbury N. W., Allen J. P. (1992) Effects of mutations near the bacteriochlorophylls in reaction centers from Rhodobacter sphaeroides. Biochemistry 31, 11029–11037 [DOI] [PubMed] [Google Scholar]
- 15. Allen J. P., Williams J. C. (1995) Relationship between the oxidation potential of the bacteriochlorophyll dimer and electron transfer in photosynthetic reaction centers. J. Bioenerg. Biomembr. 27, 275–283 [DOI] [PubMed] [Google Scholar]
- 16. de Boer A. L., Neerken S., de Wijn R., Permentier H. P., Gast P., Vijgenboom E., Hoff A. J. (2002) B-branch electron transfer in reaction centers of Rhodobacter sphaeroides assessed with site-directed mutagenesis. Photosynth. Res. 71, 221–239 [DOI] [PubMed] [Google Scholar]
- 17. Heller B. A., Holten D., Kirmaier C. (1995) Control of electron transfer between the L- and M-sides of photosynthetic reaction centers. Science 269, 940–945 [DOI] [PubMed] [Google Scholar]
- 18. Katilius E., Turanchik T., Lin S., Taguchi A. K. W., Woodbury N. W. (1999) B-side electron transfer in a Rhodobacter sphaeroides reaction center mutant in which the B-side monomer bacteriochlorophyll is replaced with bacteriopheophytin. J. Phys. Chem. B 103, 7386–7389 [Google Scholar]
- 19. Kirmaier C., Gaul D., DeBey R., Holten D., Schenck C. C. (1991) Charge separation in a reaction center incorporating bacteriochlorophyll for photoactive bacteriopheophytin. Science 251, 922–927 [DOI] [PubMed] [Google Scholar]
- 20. Kirmaier C., Laible P. D., Czarnecki K., Hata A. N., Hanson D. K., Bocian D. F., Holten D. (2002) Comparison of M-side electron transfer in R. sphaeroides and R. capsulatus reaction centers. J. Phys. Chem. B 106, 1799–1808 [Google Scholar]
- 21. McDowell L. M., Kirmaier C., Holten D. (1990) Charge transfer and charge resonance states of the primary electron donor in wild-type and mutant bacterial reaction centers. Biochim. Biophys. Acta 1020, 239–246 [Google Scholar]
- 22. Watson A. J., Fyfe P. K., Frolov D., Wakeham M. C., Nabedryk E., van Grondelle R., Breton J., Jones M. R. (2005) Replacement or exclusion of the B-branch bacteriopheophytin in the purple bacterial reaction centre: the H(B) cofactor is not required for assembly or core function of the Rhodobacter sphaeroides complex. Biochim. Biophys. Acta 1710, 34–46 [DOI] [PubMed] [Google Scholar]
- 23. de Boer A. L., Neerken S., de Wijn R., Permentier H. P., Gast P., Vijgenboom E., Hoff A. J. (2002) High yield of B-branch electron transfer in a quadruple reaction center mutant of the photosynthetic bacterium Rhodobacter sphaeroides. Biochemistry 41, 3081–3088 [DOI] [PubMed] [Google Scholar]
- 24. Laible P. D., Kirmaier C., Udawatte C. S., Hofman S. J., Holten D., Hanson D. K. (2003) Quinone reduction via secondary B-branch electron transfer in mutant bacterial reaction centers. Biochemistry 42, 1718–1730 [DOI] [PubMed] [Google Scholar]
- 25. Wakeham M. C., Frolov D., Fyfe P. K., van Grondelle R., Jones M. R. (2003) Acquisition of photosynthetic capacity by a reaction centre that lacks the QA ubiquinone: possible insights into the evolution of reaction centres? Biochim. Biophys. Acta 1607, 53–63 [DOI] [PubMed] [Google Scholar]
- 26. Kirmaier C., Holten D., Bylina E. J., Youvan D. C. (1988) Electron transfer in a genetically modified bacterial reaction center containing a heterodimer. Proc. Natl. Acad. Sci. U.S.A. 85, 7562–7566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Heller B. A., Holten D., Kirmaier C. (1995) Characterization of bacterial reaction centers having mutations of aromatic residues in the binding site of the bacteriopheophytin intermediary electron carrier. Biochemistry 34, 5294–5302 [DOI] [PubMed] [Google Scholar]
- 28. Robles S. J., Breton J., Youvan D. C. (1990) Partial symmetrization of the photosynthetic reaction center. Science 248, 1402–1405 [DOI] [PubMed] [Google Scholar]
- 29. Taguchi A. K., Stocker J. W., Alden R. G., Causgrove T. P., Peloquin J. M., Boxer S. G., Woodbury N. W. (1992) Biochemical characterization and electron-transfer reactions of sym1, a Rhodobacter capsulatus reaction center symmetry mutant which affects the initial electron donor. Biochemistry 31, 10345–10355 [DOI] [PubMed] [Google Scholar]
- 30. Haffa A. L., Lin S., Lobrutto R., Williams J. C., Taguchi A. K., Allen J. P., Woodbury N. W. (2005) Environmental control of primary photochemistry in a mutant bacterial reaction center. J. Phys. Chem. B 109, 19923–19928 [DOI] [PubMed] [Google Scholar]
- 31. Kirmaier C., He C., Holten D. (2001) Manipulating the direction of electron transfer in the bacterial reaction center by swapping Phe for Tyr near BChl(M) (L181) and Tyr for Phe near BChl(L) (M208). Biochemistry 40, 12132–12139 [DOI] [PubMed] [Google Scholar]
- 32. Kirmaier C., Weems D., Holten D. (1999) M-side electron transfer in reaction center mutants with a lysine near the nonphotoactive bacteriochlorophyll. Biochemistry 38, 11516–11530 [DOI] [PubMed] [Google Scholar]
- 33. Coleman W. J., Bylina E. J., Aumeier W., Siegl J., Eberl U., Heckmann R., Ogrodnik A., Michel-Beyerle M. E., Youvan D. C. (1990) in Structure and Function of Bacterial Photosynthetic Reaction Centers (Michel-Beyerle M. E., ed) pp. 273–281, Springer-Verlag, New York [Google Scholar]
- 34. Coleman W. J., Youvan D. C. (1993) Atavistic reaction centre. Nature 366, 517–518 [DOI] [PubMed] [Google Scholar]
- 35. Kirmaier C., Laible P. D., Hanson D. K., Holten D. (2003) B-side charge separation in bacterial photosynthetic reaction centers: nanosecond time scale electron transfer from HB− to QB. Biochemistry 42, 2016–2024 [DOI] [PubMed] [Google Scholar]
- 36. Li J., Coleman W. J., Youvan D. C., Gunner M. R. (2000) Characterization of a symmetrized mutant RC with 42 residues from the QA site replacing residues in the QB site. Photosynth. Res. 64, 41–52 [DOI] [PubMed] [Google Scholar]
- 37. Paddock M. L., Chang C., Xu Q., Abresch E. C., Axelrod H. L., Feher G., Okamura M. Y. (2005) Quinone (QB) reduction by B-branch electron transfer in mutant bacterial reaction centers from Rhodobacter sphaeroides: quantum efficiency and x-ray structure. Biochemistry 44, 6920–6928 [DOI] [PubMed] [Google Scholar]
- 38. Chuang J. I., Boxer S. G., Holten D., Kirmaier C. (2006) High yield of M-side electron transfer in mutants of Rhodobacter capsulatus reaction centers lacking the L-side bacteriopheophytin. Biochemistry 45, 3845–3851 [DOI] [PubMed] [Google Scholar]
- 39. Kee H. L., Laible P. D., Bautista J. A., Hanson D. K., Holten D., Kirmaier C. (2006) Determination of the rate and yield of B-side quinone reduction in Rhodobacter capsulatus reaction centers. Biochemistry 45, 7314–7322 [DOI] [PubMed] [Google Scholar]
- 40. Laible P. D., Kirmaier C. K., Holten D., Tiede D. M., Schiffer M., Hanson D. K. (1998) in Photosynthesis: Mechanisms and Effects (Garab G., ed) pp. 849–852, Kluwer, Dordrecht [Google Scholar]
- 41. Wakeham M. C., Goodwin M. G., McKibbin C., Jones M. R. (2003) Photoaccumulation of the P+QB− radical pair state in purple bacterial reaction centres that lack the QA ubiquinone. FEBS Lett. 540, 234–240 [DOI] [PubMed] [Google Scholar]
- 42. Sambrook J., Fritsch E., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 43. Kovach M. E., Elzer P. H., Hill D. S., Robertson G. T., Farris M. A., Roop R. M., 2nd, Peterson K. M. (1995) Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166, 175–176 [DOI] [PubMed] [Google Scholar]
- 44. Bylina E. J., Robles S. J., Youvan D. C. (1988) Directed mutations affecting the putative bacteriochlorophyll-binding sites in the light-harvesting I antenna of Rhodobacter capsulatus. Isr. J. Chem. 28, 73–78 [Google Scholar]
- 45. Weaver P. F., Wall J. D., Gest H. (1975) Characterization of Rhodopseudomonas capsulata. Arch. Microbiol. 105, 207–216 [DOI] [PubMed] [Google Scholar]
- 46. Kirmaier C., Bautista J. A., Laible P. D., Hanson D. K., Holten D. (2005) Probing the contribution of electronic coupling to the directionality of electron transfer in photosynthetic reaction centers. J. Phys. Chem. B 109, 24160–24172 [DOI] [PubMed] [Google Scholar]
- 47. Bylina E. J., Ismail S., Youvan D. C. (1986) Plasmid pU29, a vehicle for mutagenesis of the photosynthetic puf operon in Rhodopseudomonas capsulata. Plasmid 16, 175–181 [DOI] [PubMed] [Google Scholar]
- 48. Bylina E. J., Jovine R. V. M., Youvan D. C. (1989) A genetic system for rapidly assessing herbicides that compete for the quinone binding site of photosynthetic reaction centers. Bio/Technology 7, 69–74 [Google Scholar]
- 49. Youvan D. C., Ismail S., Bylina E. J. (1985) Gene 33, 19–30 [DOI] [PubMed] [Google Scholar]
- 50. Kirmaier C., Laible P. D., Hindin E., Hanson D. K., Holten D. (2003) Chem. Phys. 294, 305–318 [Google Scholar]
- 51. Hanson D. K., Baciou L., Tiede D. M., Nance S. L., Schiffer M., Sebban P. (1992) In bacterial reaction centers protons can diffuse to the secondary quinone by alternative pathways. Biochim. Biophys. Acta 1102, 260–265 [DOI] [PubMed] [Google Scholar]
- 52. Yuan L., Kurek I., English J., Keenan R. (2005) Laboratory-directed protein evolution. Microbiol. Mol. Biol. Rev. 69, 373–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jäckel C., Kast P., Hilvert D. (2008) Protein design by directed evolution. Annu. Rev. Biophys. 37, 153–173 [DOI] [PubMed] [Google Scholar]
- 54. Bokma E., Koronakis E., Lobedanz S., Hughes C., Koronakis V. (2006) Directed evolution of a bacterial efflux pump: adaptation of the E. coli TolC exit duct to the Pseudomonas MexAB translocase. FEBS Lett. 580, 5339–5343 [DOI] [PubMed] [Google Scholar]
- 55. Dong S., Rogan S. C., Roth B. L. (2010) Directed molecular evolution of DREADDs: A generic approach to creating next-generation RASSLs. Nat. Protocols 5, 561–573 [DOI] [PubMed] [Google Scholar]
- 56. Sarkar C. A., Dodevski I., Kenig M., Dudli S., Mohr A., Hermans E., Plückthun A. (2008) Directed evolution of a G protein-coupled receptor for expression, stability, and binding selectivity. Proc. Natl. Acad. Sci. U.S.A. 105, 14808–14813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wise K. J., Gillespie N. B., Stuart J. A., Krebs M. P., Birge R. R. (2002) Optimization of bacteriorhodopsin for bioelectronic devices. Trends Biotechnol. 20, 387–394 [DOI] [PubMed] [Google Scholar]
- 58. Jia Y., DiMagno T. J., Chan C. K., Wang Z., Du M., Hanson D. K., Schiffer M., Norris J. R., Fleming G. R., Popov M. S. (1993) Primary charge separation in mutant reaction centers of Rhodobacter capsulatus. J. Phys. Chem. 97, 13180–13191 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.