Background: Secretins are key components of complex DNA and protein transport machineries.
Results: An unusual secretin ααβαββα fold was identified as a ring-building motif essential for piliation but not for transformation.
Conclusion: Type IV pilus structures are not essential for transformation in T. thermophilus.
Significance: This is the first report of a ring-building domain of a unique secretin complex in T. thermophilus.
Keywords: DNA Transformation, Membrane Proteins, Membrane Transport, Protein Assembly, Protein Complexes, Secretins, Thermophile, Type IV Pili
Abstract
DNA translocators of natural transformation systems are complex systems critical for the uptake of free DNA and provide a powerful mechanism for adaptation to changing environmental conditions. In natural transformation machineries, outer membrane secretins are suggested to form a multimeric pore for the uptake of external DNA. Recently, we reported on a novel structure of the DNA translocator secretin complex, PilQ, in Thermus thermophilus HB27 comprising a stable cone and cup structure and six ring structures with a large central channel. Here, we report on structural and functional analyses of a set of N-terminal PilQ deletion derivatives in T. thermophilus HB27. We identified 136 N-terminal residues exhibiting an unusual ααβαββα fold as a ring-building domain. Deletion of this domain had a dramatic effect on twitching motility, adhesion, and piliation but did not abolish natural transformation. These findings provide clear evidence that the pilus structures of T. thermophilus are not essential for natural transformation. The truncated complex was not affected in inner and outer membrane association, indicating that the 136 N-terminal residues are not essential for membrane targeting. Analyses of complex formation of the truncated PilQ monomers revealed that the region downstream of residue 136 is required for multimerization, and the region downstream of residue 207 is essential for monomer stability. Possible implications of our findings for the mechanism of DNA uptake are discussed.
Introduction
Microbial life has been detected in virtually every environment on earth, ranging from hydrothermal vents, salt saturated alkaline ponds, acidic hot springs, Antarctic ice, dry dessert soils, and the upper atmosphere to animal and plant hosts. To exploit such different environments, microorganisms must have evolved phenotypic traits allowing survival under very different environmental conditions, and acquisition of novel genetic information is suggested to be a major force for bacterial adaptation to changing environments (1–4). Natural transformation in bacteria, which describes the uptake and incorporation of naked DNA, allows the uptake of genetic material from diverse bacterial species and even members of different domains and is perhaps the most versatile mechanism of DNA transfer (2, 3). Natural transformation machineries are complex DNA translocators found in many bacteria, including bacteria thriving under extreme conditions (2, 5–8).
A common feature of the DNA translocators of natural transformation systems is the involvement of proteins that are similar to components of type IV pili (T4P)2 and type II protein secretion systems (8–12). The DNA translocators of Gram-negative bacteria are suggested to be composed of three distinct subassemblies: (i) an inner membrane platform associated with a cytoplasmic AAA ATPase, (ii) a pilin-comprising DNA translocator shaft spanning the periplasm, and (iii) a large, pore-forming outer membrane complex consisting of secretins guiding the translocator shaft through the outer membrane. Despite the broad analyses of DNA translocator proteins, information on the structure and function of DNA translocators is scarce.
To get insights into the structure and function of natural transformation systems, we chose the transformable thermophilic Thermus thermophilus HB27 as a model bacterium for molecular analyses of the DNA translocator. These studies led to a tentative model of the DNA translocation process. DNA is bound to the secretin complex in the outer membrane, which guides the DNA translocator shaft composed of pilins through the outer membrane. The DNA transport might require dynamics of the DNA translocating shaft, which could be powered by the AAA ATPase PilF (6, 13–15). The similarities of the T. thermophilus DNA translocator proteins to components of T4P and the type II protein secretion system raised the question of a functional linkage of both systems, which was unraveled by mutant studies (6, 14, 15). However, the question whether pili themselves are implicated in DNA uptake has not been answered to date.
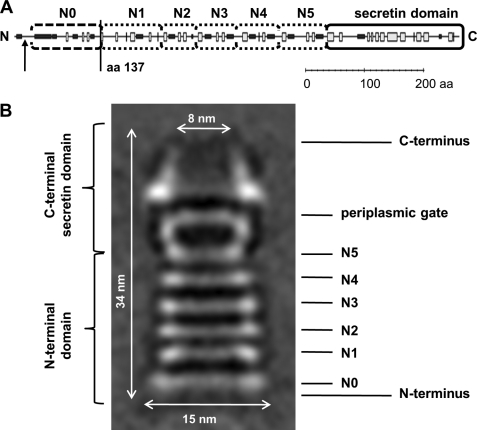
Recently, we reported on the structure and function of a DNA translocator subassembly comprising the secretin PilQ. This secretin forms a much larger complex (∼34-nm length) than known secretins comprising a stable cone and cup structure and six stacked ring structures with a large central channel potentially accommodating a pseudopilus mediating DNA transport across the outer membrane and the periplasmic space (16). Secondary structure analyses suggest that the individual rings are formed by conserved domains of alternating α-helices and β-sheets within the extended N terminus, and we hypothesized that the architecture of the N terminus facilitates the assembly of the six ring systems underneath the cuplike structure.
Here, we used truncated PilQ proteins to identify ring-building motifs and domains essential for complex assembly to gain insights into the role of the modular organization of the extended N terminus in the architecture and assembly of the macromolecular PilQ complex. Deletion of the N-terminal ring-building domain abrogated adhesion, twitching motility, and piliation but had no effect on natural transformation and on the association of the PilQ complex with the inner membrane. Taken together, these results show that pilus structures themselves are not essential for natural transformation in T. thermophilus.
EXPERIMENTAL PROCEDURES
Organisms and Cultivation
All strains used in this study are listed in supplemental Table S1. Escherichia coli DH5α was used as general cloning strain (17) and grown under standard conditions (18). T. thermophilus HB27 was grown at 68 °C on TM+ complex medium containing 8 g of tryptone peptone, 4 g of yeast extract, and 3 g of NaCl/liter, pH 7.5 (19). T. thermophilus mutants were grown on TM+ complex medium containing 5 or 15 μg/ml bleomycin (Bleo) and 20 or 40 μg/ml kanamycin in liquid or solid medium, respectively. For PilQ purification or protein analyses, cells were harvested in the late exponential growth phase.
Twitching Motility and Adhesion Studies
Twitching motility and adhesion studies were performed on minimal medium containing 1% bovine serum albumin (BSA) and 0.01% yeast extract (20). Plates were incubated at 68 °C under humid conditions for 3 days. Cells were stab-inoculated through the solid medium down to the Petri dish. To visualize adhered cells, the solid medium was removed from the Petri dish, and cells were stained by Coomassie Blue (21).
Gradient SDS-PAGE and Immunoblotting
Detection of PilQ proteins was performed by Western blot analyses as described previously (16).
Generation of ΔpilQ::bleo Deletion Mutant
The plasmids and primers are listed in supplemental Tables S1 and S2, respectively. To construct a ΔpilQ::bleo deletion mutant, the ∼1000-bp upstream and ∼1000-bp downstream regions of pilQ was amplified using specific primer pairs (PilOW-for-SalI and PilOW-rev-EcoRV or Chor-for-PstI and Chor-rev-NotI, respectively). The bleomycin cassette was amplified from the E. coli/T. thermophilus shuttle vector pWUR112 using the specific primers Bleo-for-EcoRV and Bleo-rev-PstI. The upstream region (“pilOW”), bleomycin cassette (“bleo”), and downstream region (“chor”) were inserted into pBIISK using restriction sites SalI, EcoRV, PstI, and NotI, respectively, resulting in pBIISK-pilOWbleoChor (supplemental Fig. S1). Linearized pBIISK-pilOWbleoChor was transformed into T. thermophilus HB27, and transformants were selected on TM+/Bleo medium (22). The deletion mutant was verified by Southern blot analysis.
Southern Blot Analyses
Sequence-specific probes were generated using PCR DIG Labeling Mix (Roche Applied Science). Labeling reactions were performed according to the manufacturer's protocol using different primers listed in supplemental Table S2. Three different probes were used for Southern blot hybridization: one against the pilQ gene, which should be deleted; one against the bleomycin cassette; and one against the pilQ downstream region. Southern blot hybridization was performed as described by Ausubel et al. (18), and signals were detected using the chemiluminescent substrate for alkaline phosphatase detection (CSPD) as recommended by the manufacturer (Roche Applied Science).
In trans Complementation of HB27 ΔpilQ::bleo and Generation of N-terminal Truncated pilQ Constructs
Primers used for generating pilQ deletion derivatives are listed in supplemental Table S2. The wild type pilQ gene was amplified using primers PilQ-for-NdeI and PilQ-rev-NotI. The resulting pilQhis fusion was cloned into pDM12 under the control of the Thermus bc1 promotor using restriction sites NdeI and NotI, thereby generating vector pDM12-pilQhis-Q (Fig. 1). pDM12-pilQhis-Q was used as template for generating N-terminal truncated pilQ constructs via the Phusion site-directed mutagenesis (SDM) protocol from Finnzymes. Phosphorylated pDM12PilQ-rev was used as reversed primer in all SDM reactions for amplifying the signal peptide-encoding region of pilQ and the non-coded strand of pDM12-pilQhis-Q. Phosphorylated forward primers pDM12PilQ-for-A, -B, -D, -E, and -F were used for generating 5′ deletions of pilQ by amplifying the encoded strand at defined positions. The template vector pDM12-pilQhis-Q was degraded by DpnI. The amplified fragments were ligated and transformed into E. coli DH5α. Resulting vectors pDM12-pilQΔ25–34his, -pilQΔ25–64his, -pilQΔ25–125his, -pilQΔ25–207his, and -pilQΔ25–262his (supplemental Table S1) were verified by sequencing and transferred into HB27 ΔpilQ::bleo by electroporation (23).
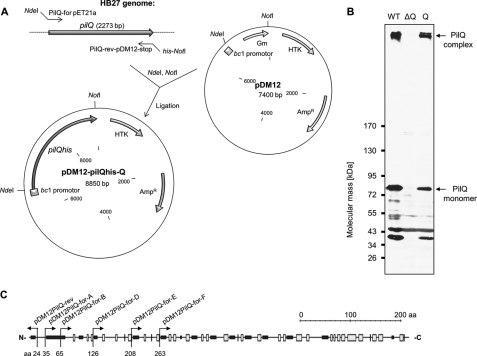
FIGURE 1.
In trans complementation of pilQ deletion mutant HB27 ΔpilQ::bleo. A, the pilQ gene was amplified by PCR and cloned into pDM12 using restriction sites NdeI and NotI. The resulting plasmid pDM12-pilQhis-Q was electroporated into HB27 ΔpilQ::bleo. B, crude extracts of HB27 wild type (lane WT), HB27 ΔpilQ::bleo (lane ΔQ), and HB27 ΔpilQ::bleo carrying pDM12-pilQhis-Q (lane Q) were subjected to 3–12% polyacrylamide gradient SDS-PAGE followed by Western blot analysis. C, scheme of the putative secondary structure of PilQ and corresponding primer binding sites for the generation of N-terminal PilQ deletion derivatives by SDM using pDM12-pilQhis-Q as the template vector. The first maintained amino acid residues after SDM are indicated. Black boxes, α-helical domains; white boxes, β-sheets; aa, amino acid residues.
Purification of Truncated PilQ Complexes
Purification of N-terminal truncated PilQ complexes was performed as described recently (16). Dissociation of PilQ complexes was carried out as described (16).
Mass Spectrometry
A detailed description of all mass spectrometric analyses is available in the supplemental information. In brief, for MALDI-MS analysis, protein solutions were treated with trifluoroacetic acid (TFA), purified, and spotted in dihydroxyacetophenone matrix on ground steel targets. MALDI mass spectra were recorded in a Bruker Autoflex III Smartbeam mass spectrometer after external calibration.
Characterization of SDS-PAGE-separated proteins was performed on reduced and alkylated, trypsin- and chymotrypsin-digested samples prepared by standard mass spectrometry protocols (ProteoExtract digestion kit, Calbiochem). The proteolytic digests were analyzed using LC-MSn (Proxeon easy-nanoHPLC coupled to a Bruker maXis electrospray ionization-quadrupole TOF mass spectrometer as described in the supplemental information).
Electron Microscopy and Image Analysis
PilQ complexes were negatively stained with 1% (w/v) uranyl acetate. Electron micrographs were collected using a Philips CM120 at 120 kV at a magnification of 44,000× on Eastman Kodak Co. SO-163 film, which was digitized on a PhotoScan scanner at a pixel size of 7 μm. Subsequently, adjacent pixels were averaged to yield a pixel size on the specimen of 4.77 Å. Particles were selected using the boxer module from EMAN (24). Image processing was done with Imagic V (25). The images were band pass-filtered and subjected to reference-free alignment and multivariate statistical analysis. The images were classified, and images assigned to the same class were averaged.
Electron Microscopy of HB27 Cells
Thermus cells were grown for 2 days on TM+ agar. Cells from a 0.7 × 0.5-cm area were resuspended in H2Odest., loaded on a copper grid (400 mesh), and washed three times in H2O. Shadowing of the cells was carried out in a BAF 060 freeze-fracture system (BAL-TEC, Principality of Liechtenstein) with a pressure of 3–4 × 10−7 millibar at 28 °C specimen table temperature in an angle of 25° unidirectional and thickness of 1.5-nm platinum/carbon. Samples were inspected by a transmission electron microscope (EM 208S, FEI Co.) at 80 kV. A 1024 × 1024 slow scan camera (Tietz, TVIPS GmbH, Gauting, Germany) was used for image acquisition.
Transformation Studies
Transformation studies were performed using 5 μg of genomic DNA of a spontaneous streptomycin-resistant HB27 mutant (26).
Cellular Fractionation
Inner and outer membranes were prepared by N-lauroylsarcosine extraction as described previously (16, 27). Purity of the inner membrane fractions was confirmed by Western blot analysis using S-layer protein-specific antibodies.
RESULTS
Residues 126–207 Are Essential for PilQ Complex Formation
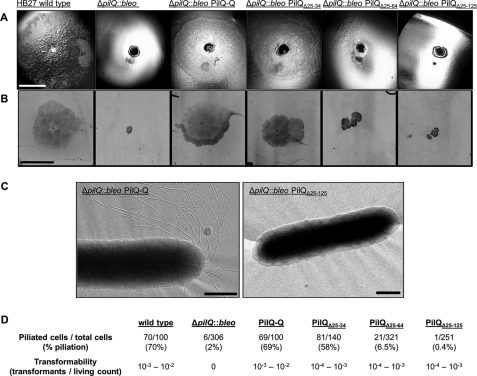
To analyze the structural and functional role of PilQ domains in a pilQ-negative genetic background, a pilQ deletion mutant was generated by marker exchange mutagenesis using a bleomycin resistance marker (supplemental Fig. S1). Electron microscopy analyses revealed that the resulting ΔpilQ::bleo mutant was deficient in piliation. Analyses of the transformation phenotype showed that the ΔpilQ mutant was also deficient in natural transformation. These findings are consistent with our results obtained with a pilQ::kat disruption mutant (15).
To understand the role of distinct domains in structure and function of the PilQ complex, stepwise N-terminal truncations of PilQ were generated via SDM. Therefore, a template vector carrying the complete pilQ gene was generated using the Thermus/E. coli shuttle vector pDM12. To allow expression of the pilQ gene, pilQ with a C-terminal histidine tag was cloned downstream of the constitutively expressed bc1 promotor, resulting in pDM12-pilQhis-Q (Fig. 1A). Expression and functionality of the PilQhis fusion was confirmed by Western blot analysis and transformation studies. As shown in Fig. 1B, a PilQ complex comparable with the PilQ complex in wild type cells was detected in crude extracts of a ΔpilQ::bleo pDM12-pilQhis-Q transformant. Analyses of the transformation frequencies revealed that the PilQ complex comprising PilQhis fusion proteins was fully functional; transformation frequencies of 10−3–10−2 transformants/living count comparable with wild type cells were obtained.
Using SDM, defined parts of pilQ in pDM12-pilQhis-Q were deleted, thereby maintaining the first 24 residues encoding a predicted 22-residue signal peptide sequence and 2 additional residues (Fig. 1C). Resulting vectors and introduced N-terminal deletions are listed in supplemental Table S1. The constructs were electroporated into the HB27 ΔpilQ::bleo deletion mutant, and PilQ production was verified by Western blot analyses (Fig. 2 and Table 1). The three deletion derivatives PilQΔ25–34, PilQΔ25–64, and PilQΔ25–125 assembled into PilQ complexes that migrated faster than the PilQ complex from wild type cells. The detection of these complexes after separation by SDS-PAGE indicates that they are SDS-resistant, which is consistent with our recent finding that the purified PilQ complex from T. thermophilus exhibits SDS resistance (16).
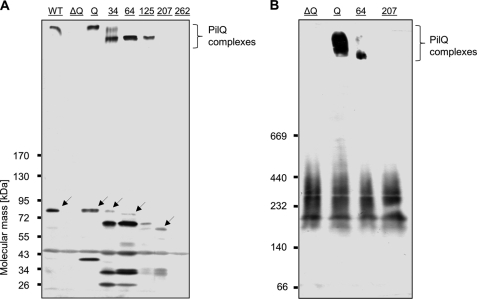
FIGURE 2.
Detection of N-terminal truncated and SDS-stable PilQ complexes. A, crude extracts of HB27 wild type (lane WT), HB27 ΔpilQ::bleo (lane ΔQ), and HB27 ΔpilQ::bleo carrying pDM12-pilQhis-Q (lane Q), -pilQΔ25–34his (lane 34), -pilQΔ25–64his (lane 64), -pilQΔ25–125his (lane 125), -pilQΔ25–207his (lane 207), or -pilQΔ25–262his (lane 262) were separated by 3–12% polyacrylamide gradient SDS-PAGE. PilQ production and SDS-stable complex assembly were verified by Western blot analysis. Arrows indicate expected PilQ derivative monomers. B, to identify potential SDS-unstable PilQ complexes, membranes of the indicated mutants were solubilized with 4% Triton-X-100, and solubilizates were subjected to 3–12% polyacrylamide gradient blue native PAGE followed by Western blot analysis.
TABLE 1.
Monomer and complex formation of PilQ deletion derivatives in Fig. 2
| PilQ construct | Expected molecular mass (processed) | Molecular mass of detected proteins | Complex formation |
|---|---|---|---|
| kDa | kDa | ||
| PilQ-Q | 81.1 | 81.1, 38.5 | + |
| PilQΔ25–34 | 80 | 79.9, 66.2, 42.8, 32.5, 26.9 | + |
| PilQΔ25–64 | 76.9 | 76.5, 66.2, 48.4, 42.8, 33.5, 32.5, 26.9 | + |
| PilQΔ25–125 | 69.9 | 66.2 | + |
| PilQΔ25–207 | 61 | 61.5, 33.5, 32 | − |
| PilQΔ25–262 | 54.9 | − |
The PilQΔ25–34 construct led to the detection of an additional complex that showed almost the same migration behavior as the positive control PilQhis (named PilQ-Q; produced in strain ΔpilQ::bleo pDM12-pilQhis-Q; Fig. 2A). Deletion of residues 25–207 or 25–262 abolished the formation of SDS-resistant PilQ complexes. Taken together, residues 126–207 are essential for the SDS stability or the assembly of SDS-resistant PilQ complexes. To verify the role of these residues in assembly of SDS-unstable complexes, membrane solubilizates of the mutant encoding the deletion derivative PilQΔ25–207, complex-forming mutants, and the negative control HB27 ΔpilQ::bleo were subjected to blue native PAGE followed by Western blot analyses (Fig. 2B). No PilQ complexes were detected in the ΔpilQ::bleo mutant carrying pDM12-pilQΔ25–207his, which suggests that the residues 126–207 are absolutely essential for complex assembly.
Residues 208–262 Are Essential for PilQ Monomer Stability
In addition to the PilQ complexes, mutants carrying pDM12-pilQΔ25–34his, pDM12-pilQΔ25–64his, or pDM12-pilQΔ25–207his produced PilQ proteins of the deduced molecular mass of the truncated PilQ monomers (Fig. 2A, indicated by arrows, and Table 1). Interestingly, the deletion of residues 25–34 and 25–64 led to the detection of only minor amounts of the expected truncated PilQ monomers (Fig. 2A and Table 1), but in both extracts, major amounts of a ∼66.2-kDa PilQ deletion derivative were detected. This indicated that major amounts of the 80- and 76.9-kDa N-terminal deletion derivatives PilQΔ25–34 and PilQΔ25–64, respectively, undergo a further proteolytic cleavage to a ∼66.2-kDa protein. Furthermore, a ∼66.2-kDa protein was also detected in extracts of the mutant carrying pDM12-pilQΔ25–64his instead of the expected ∼69.9-kDa PilQΔ25–125 monomer. Additional smaller proteins that cross-reacted with the specific PilQ antibody but were not detectable in crude extracts of the ΔpilQ::bleo deletion mutant probably represent degradation products of the truncated monomers. In the ΔpilQ::bleo mutant carrying pDM12-pilQΔ25–262his, no PilQ monomers were detected, suggesting that residues 208–262 are essential for monomer stability.
Purification and Characterization of Truncated PilQ Complexes
With respect to a smaller PilQ complex of apparently the same size in mutants carrying pDM12-pilQΔ25–34his, pDM12-pilQΔ25–64his, or pDM12-pilQΔ25–125his, we hypothesized that the smaller PilQ complex comprises the uniformly detected ∼66-kDa truncated PilQ proteins (Fig. 2A). To challenge this hypothesis and to analyze the structural and functional role of residues 25–125, the PilQ complexes of the mutants encoding the truncated deletion derivatives PilQΔ25–64 and PilQΔ25–125 were purified as described recently (16). As shown in supplemental Fig. S2, after gel filtration, homogeneous PilQ complex preparations were obtained. Interestingly, neither the expected ∼76.9-kDa monomer protein for PilQΔ25–64 nor the expected ∼69.9-kDa protein in the case of PilQΔ25–125 could be detected in the purified samples, but the ∼66-kDa protein was detected in both preparations (supplemental Fig. S2, B and D, lane 10). This was further evidence that these ∼66-kDa truncated PilQ proteins are subunits of the purified truncated PilQ complexes.
For further experimental verification of the PilQ derivative complex composition, the purified PilQ complexes were dissociated into their monomers by treatment with hot phenol and analyzed by SDS-PAGE followed by Western blot analyses using the wild type PilQ as a control. As shown in Fig. 3A, the dissociation of these PilQ derivative complexes led to the detection of only a ∼66-kDa PilQ protein. For a more concise determination of the molecular mass of the truncated PilQ monomers, the PilQ preparations were submitted to MALDI-TOF mass spectrometric analysis. Although the wild type PilQ showed the expected molecular mass of the processed protein (80.3 kDa), the PilQ deletion variants displayed molecular masses of 68.7 and 68.9 kDa (Fig. 3B), suggesting a truncation between residues 134 and 138.
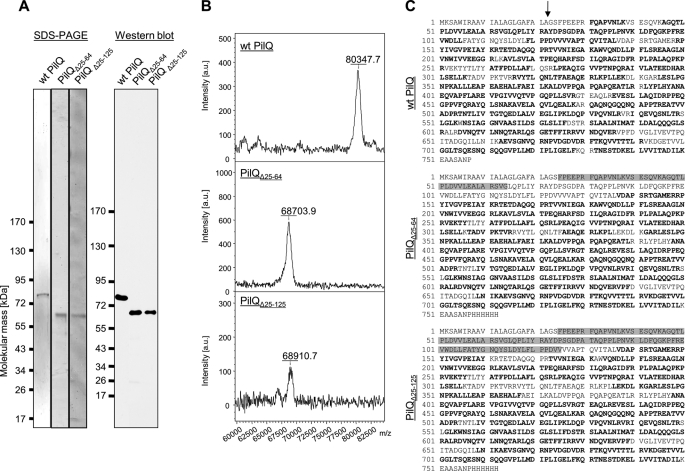
FIGURE 3.
Identification of molecular mass of PilQ deletion derivatives by MALDI-MS. A, purified PilQ, PilQΔ25–64, and PilQΔ25–125 complexes were separated by blue native PAGE, electroeluted, and treated with hot phenol. These samples were subjected to SDS-PAGE and verified by immunoblotting using PilQ antibodies. B, purified PilQ, PilQΔ25–64, and PilQΔ25–125 complexes were dissociated using TFA and purified using ZipTips. The molecular mass of the monomers was analyzed by MALDI-MS. C, peptide mass fingerprinting was performed to characterize the sequence of PilQ, PilQΔ25–64, and PilQΔ25–125 monomers. Samples were prepared from SDS-polyacrylamide gels, digested using trypsin/chymotrypsin, and analyzed in a nano-HPLC-coupled electrospray ionization-quadrupole TOF mass spectrometer with data-dependent tandem MS spectrum acquisition. Matched peptides are shown in bold letters. Gray boxes, residues deleted by SDM; arrow, predicted signal peptide cleavage site. a.u., arbitrary units.
To obtain further information on the exact position of the processing site, both wild type PilQ and the two deletion variants were subjected to peptide mass fingerprinting (Fig. 3C). Although multiple peptides in the N-terminal region were detected in the sample containing the wild type PilQ, both deletion variants only afforded sequence coverage starting at residue 137. Truncation at this position would yield a protein with a calculated molecular mass of 68.78 kDa, a value that is in excellent agreement with the data obtained in the MALDI-MS experiments.
Taken together, these results suggest that the PilQ deletion derivatives are both processed after position 136, resulting in identical proteins with a molecular mass of ∼68.8 kDa. Due to the relatively weak signal intensities in the MALDI-MS experiments and the limitations of proteolytic digests, the truncation site can only be estimated with an accuracy of several amino acids. Hereafter this protein is labeled PilQΔ136.
N-terminal 25–136 Residues Form Lowest N-terminal Ring Domain of PilQ Complex
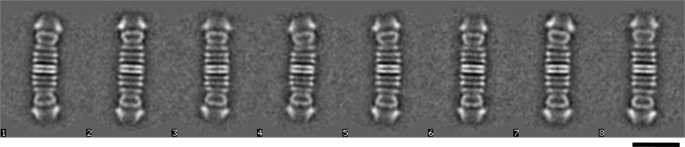
The purified truncated PilQΔ25–64 and PilQΔ25–125 complexes both comprising N-terminal truncated 68.8-kDa PilQ monomers (PilQΔ136) were negatively stained and studied by electron microscopy. The two samples looked identical and contained exclusively very long complexes (∼61 nm), which are clearly recognizable as dimers of the PilQ complex with the characteristically shaped secretin domains on the outside, indicating a total length of ∼30.5 nm for one complex. This was confirmed by image analysis of the PilQΔ25–64 sample (Fig. 4). Each complex comprises five distinct rings followed by a characteristic cone and cup structure that represents the conserved C-terminal secretin domain that was also detected in the structure of the wild type PilQ complex that we reported recently (16). In contrast to the truncated PilQ complexes, the ∼34-nm-long wild type PilQ complex comprises six distinct ring domains underneath the cup structure (see Fig. 7). Taken together, these results provide clear evidence that the N-terminal domain spanning residues 25–136 is the ring-building domain of the first ring structure of PilQ.
FIGURE 4.
Electron microscopy analysis of PilQΔ25–64. Class averages after multivariate statistical analysis and classification of 800 particles reveal dimers of a truncated PilQ complex. Each class contains ∼65 images. Scale bar, 25 nm. 1–8, eight representative images of class averages.
FIGURE 7.
Secondary structure and subdomain prediction of T. thermophilus PilQ. The PredictProtein server was used to predict potentially α-helical (black boxes) and β-sheet (white boxes) regions within PilQ (A). The putative signal peptide processing site is indicated by an arrow, and the determined truncation site of the PilQΔ136 complex is displayed. Rounded boxes indicate putative N0, N1, N2, N3, N4, and N5 subdomains and the secretin domain. Subdomains are indicated in the PilQ complex structure (B). aa, amino acid(s).
N-terminal Ring Structure Is Essential for Piliation but Not for Natural Transformation
To analyze the role of the N-terminal ring structure of the PilQ complex in natural transformation, the transformation phenotypes of HB27 ΔpilQ::bleo mutants carrying vector pDM12-pilQΔ25–34his, pDM12-pilQΔ25–64his, or pDM12-pilQΔ25–125his were determined. The three mutants producing PilQΔ136 complexes devoid of the first N-terminal ring exhibited transformation frequencies of 10−4–10−3. This provides clear evidence that the N-terminal ring is not essential for transformation.
HB27 produces T4P, and the secretin PilQ was shown to be essential for biogenesis of T4P (15). T4P are homopolymeric structures on the surface of many Gram-negative bacteria that mediate adhesion and bacterial translocation over moist surfaces occurring by extension, tethering, and then retraction of polar T4P. At the macroscopic level, twitching motility can be manifested by the formation of flat spreading colonies with a characteristic rough appearance on agar surfaces or even at the interface of solid medium and the Petri dish. The latter can be assayed by a stab assay (58). In numerous Gram-negative bacteria, T4P have been shown to mediate adhesion and twitching motility. But so far, this has not been reported for T. thermophilus. By performing a stab assay, we could show for the first time that HB27 exhibits both twitching motility and adhesion (Fig. 5, A and B). The ΔpilQ::bleo deletion mutant was deficient in adhesion and twitching motility, which together with the piliation defect, leads to the conclusion that T. thermophilus pili are essential for both functions.
FIGURE 5.
Colony morphology and cell adhesion on solid surfaces of T. thermophilus and HB27 ΔpilQ::bleo mutants. A, cells of T. thermophilus HB27 wild type, HB27 ΔpilQ::bleo, and HB27 ΔpilQ::bleo carrying pDM12-pilQhis-Q (PilQ-Q), -pilQΔ25–34his (PilQΔ25–34), -pilQΔ25–64his (PilQΔ25–64), or -pilQΔ25–125his (PilQΔ25–125) were stab-inoculated on minimal medium plates containing 1% BSA and incubated for 3 days at 68 °C under humid conditions. Colony morphology was documented under a binocular microscope. Scale bar, 5 mm. B, for analyzing cell adhesion and twitching motility, medium was removed from the Petri dish, and adhered cells were visualized by Coomassie staining. Scale bar, 10 mm. C, electron micrographs of piliated HB27 and non-piliated HB27 mutant cells. Scale bars, 0.5 μm. D, statistical analyses of piliation of the indicated complementation mutant cells by electron microscopy.
The mutant ΔpilQ::bleo carrying vector pDM12-pilQΔ25–64his or pDM12-pilQΔ25–125his forming the PilQΔ136 complex was still deficient in adhesion and twitching motility (Fig. 5, A and B). This leads to the conclusion that the lowest N-terminal ring is essential for these pilus functions. To analyze the role of the N-terminal ring structure in the presence of pili, the piliation of HB27 ΔpilQ::bleo mutants carrying vector pDM12-pilQΔ25–34his, pDM12-pilQΔ25–64his, or pDM12-pilQΔ25–125his was analyzed by electron microscopy. These studies revealed that mutants only producing PilQΔ136 complexes were dramatically reduced in pilus structures (Fig. 5, C and D). These results provide clear evidence that the N-terminal 136 residues of PilQ forming the first ring structure of the PilQ complex are essential for pilus assembly at the cell surface.
Transformation studies of the mutants carrying vector pDM12-pilQΔ25–34his, pDM12-pilQΔ25–64his, or pDM12-pilQΔ25–125his revealed that these mutants were only 10-fold reduced in transformation frequencies (Fig. 5D). Taken together, the transformability of the mutants producing the PilQΔ136 complex together with the piliation defect clearly demonstrate that type IV pilus fibers are not essential for natural transformation in T. thermophilus HB27.
N-terminal Ring of PilQ Is Not Essential for Inner Membrane (IM) Association of PilQ Complex
Recently, we have shown that the wild type PilQ complex was mainly associated with the outer membrane but was also detectable in IM fractions (16). To analyze the role of the N-terminal ring of the PilQ complex in IM association, inner and outer membrane preparations of a HB27 mutant strain producing the PilQΔ136 complex were analyzed by immunoblotting. As seen in Fig. 6, the PilQΔ136 complex was insoluble in N-lauroylsarcosine, indicating an outer membrane localization, whereas minor amounts of PilQΔ136 complexes were also found to be Sarkosyl-soluble. Taken together, these results lead to the conclusion that the lowest N-terminal ring of the PilQ complex is not essential for an inner membrane association of the PilQ complex.
FIGURE 6.
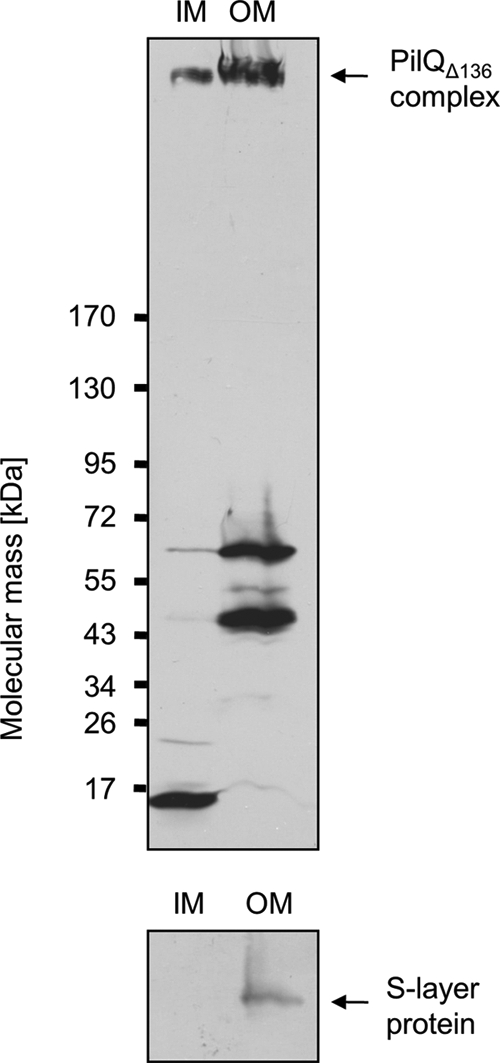
Subcellular localization of PilQΔ136 complex. Total membranes of HB27 ΔpilQ::bleo carrying pDM12-pilQΔ25–64his were separated into IM and outer membrane (OM) fractions. The fractions were subjected to SDS-PAGE in a 3–12% polyacrylamide gradient gel followed by immunoblotting using PilQ antibodies. The PilQ complex is indicated by an arrow. The purity of IM fractions was verified by Western blot analysis using S-layer protein-specific antibodies.
DISCUSSION
Secretins comprise a family of large, homomultimeric, and dynamic protein channels that are localized in the outer membranes of Gram-negative bacteria and are essential for transport of macromolecular structures, such as proteins or DNA, across the outer membranes via diverse multiprotein systems (28–34). Secretin subunits exhibit multiple domains specifically acting in substrate recognition, interaction with other proteins, or regulation of transport, respectively, thereby forming characteristic cylindrical structures comprising a large N-terminal periplasmic vestibule and an extracellular chamber containing the secretin-specific periplasmic gate formed by the conserved C-terminal secretin domains (35). Recent crystallographic studies of domains of the type II protein secretion system secretin GspD from enterotoxigenic E. coli and the type III protein secretion system secretins EscC and InvG from enteropathogenic E. coli and Salmonella typhimurium provided insights into the domain structure, indicating general ring-building secondary structure motifs of the unconserved N-terminal parts among the secretins (36–40). It was shown that a so-called N0 subdomain with a βαββαββ fold and N1-like subdomains comprising βαββα folds each form ringlike structures, which are stacked and form the periplasmic vestibule.
Electron microscopy analysis of the secretin complex PilQ of T. thermophilus revealed an unusually long structure of ∼34 nm composed of a typical cuplike structure formed by the secretin domain containing the characteristic periplasmic gate and additionally six ringlike structures thought to be assembled by the N termini of the PilQ proteins (16). Analysis of the predicted secondary structure of the PilQ N terminus indicated putative N1-like ring-building motifs (βαββα folds) in the middle section, suggesting their assignment as potential subdomains N2–N5, forming the first four rings underneath the cuplike structure (Fig. 7).
In contrast, the N terminus of the PilQ protein (amino acid residue 1 to the start of subdomain N2) did not comprise any typical or clearly assignable N0 or N1-like ring-building subdomain folds. Our finding that residues 25–136 are essential for the first N-terminal ring structure leads to the conclusion that an ααβαββα motif forms the first N-terminal ring structure of PilQ (Fig. 7), which therefore was designated N0 subdomain.
Interestingly, a sequence comprising a βββαββ motif is located between the putative N2 motif and the designated N0 subdomain, suggesting an involvement of this domain in formation of the second ring structure and therefore being analogous to ring-building subdomain N1 (Fig. 7). Interestingly, βββαββ folds are known as so-called oligonucleotide/oligosaccharide binding motifs, which were shown to mediate binding of single-stranded DNA/RNA (41, 42). Oligonucleotide/oligosaccharide binding motifs do not share sequence similarities; substrate recognition is rather achieved by common topologies of the appropriate domains. Because the PilQ complex plays an essential role in DNA binding and translocation (16, 20), it is tempting to speculate that the N1 domain in PilQ is implicated in DNA binding.
Interestingly, different truncated PilQ derivatives that were processed to the identical PilQ proteins with the first 136 residues deleted were still able to assemble into a truncated PilQ complex missing subdomain N0 (PilQΔN0). This complex was shown to be functional in mediating transport of DNA. Thus, a specific protein quality control is supposed to lead to a controlled degradation of residues 65–136 and 126–136, whereas the remaining part is protected against degradation. Quality control of outer membrane proteins was described as a complex network of periplasmic proteins acting as chaperones and/or proteases and thereby preventing aggregation of unfolded proteins and facilitating correct folding, finally preventing the accumulation of unfolded proteins in the periplasm (43, 44). Because the two truncated PilQ derivatives were uniformly processed to identical PilQΔ136 proteins, a quality control resulting in defined degradation of N-terminal residues is suggested to take place. In E. coli, the protein DegP has been described to play a central role in quality control whereby it has a dual function as a protease and a chaperone, leading proteins to degradation or folding pathways (45). Genome inspection of the HB27 genome led to the identification of genes encoding potential trypsin-like serine proteases Do (TTC0956, TTC0417, and TTC1905), which exhibit 61–69% similarity to the DegP protein in E. coli. These Thermus proteins might function in a DegP-like manner in protein quality control and thereby enable the correct insertion/assembly of the post-translationally truncated PilQΔ136 in the outer membrane. Membrane insertion might then be facilitated by the Omp85 homologous protein (46) because PilQ complex assembly was also dependent on Omp85 in Neisseria meningitidis (47), but this has to be experimentally proven in T. thermophilus.
Moreover, this study provides novel insights into the role of pili in natural transformation of T. thermophilus. Former T. thermophilus mutant studies suggested an involvement of pilus structures in DNA uptake across the membranes because proteins essential for biogenesis of T4P, such as the inner membrane proteins PilC, PilD (prepilin peptidase), and PilF (AAA ATPase) or the pilin PilA4, are also essential for transformation (6, 15). Furthermore, pilD, pilF, or pilA4 mutants exhibited an increased DNA binding compared with the wild type, which suggested an involvement of these pilus proteins in DNA uptake across the outer membrane (20). Taken together, these results are consistent with results obtained from studies of the transformation systems in N. meningitidis or Neisseria gonorrhoeae, indicating a functional link between transformation and piliation (48–51), although the question whether the pilus structures are involved in DNA transport is still open. First hints that a functional pilus assembly apparatus but not pilus structures themselves are essential for transformation have been obtained by studies of the transformation systems in Neisseria species (52, 53). Our studies presented here revealed that the extremely low pilus expression of the mutant encoding the truncated PilQΔN0 complex allows for substantial natural transformation frequencies, which leads to the conclusion that the extended pili are not required for natural transformation.
The dimensions of the wild type PilQ complex together with the inner membrane association suggest that the six stacked ring structures span the periplasmic space, guiding the periplasmic pseudopilus through the outer membrane and thereby interacting with inner membrane proteins of the DNA translocator. Analysis of the membrane association of the truncated PilQΔN0 complex revealed an association with outer and inner membranes. The absence of pilus structures due to deletion of subdomain N0 indicated an essential function of this domain in interaction either with pilus or pseudopilus structures or with inner membrane proteins essential for pilus biogenesis/function such as PilC or PilMNOW. The deletion of the N0 subdomain might result in an incomplete opening of the secretin pore due to a defective conformational change in response to interactions with pilus structures. Because the mutants encoding the N0-truncated PilQ complex still exhibit high transformation frequencies and DNA uptake across the outer membrane is dependent on proteins essential for pilus biogenesis, such as PilA4 and PilF, it is tempting to speculate that the PilQΔN0 mutant might still assemble pilus structures within the periplasm. Periplasmic pilus assembly was also postulated for a pilQ/pilT mutant in N. meningitidis and N. gonorrhoeae (54, 55). Periplasmic pili in the huge periplasm of T. thermophilus might bind DNA as it has been shown for type IV pilus structures of Pseudomonas aeruginosa (56) and might guide the DNA to the inner membrane DNA translocator machinery in T. thermophilus.
Clues on the regulation of the translocation of DNA through a secretin pore might be derived from the postulated pseudopilus piston mechanism of type II protein secretion system secretins. Here it is suggested that the exotoxin sitting on the grown pseudopilus induces conformational changes in the secretin, which results in the opening of the periplasmic gate and the final release of the toxin (37, 57). If a pseudopilus triggers opening of the Thermus PilQ complex, thereby enabling DNA uptake, a direct interaction of the pseudopilus structures and the PilQ complex is a prerequisite. The deletion of subdomain N0 is suggested to cause either a permanent and partial opening of the secretin pore because DNA is still taken up, or the assembling pseudopilus induces a partial opening of the secretin pore, but this pore is defective in extruding pilus structures through the outer membrane. This partial opening of the secretin pore might also explain the reduced transformation frequencies of the mutants producing PilQΔN0 complexes from 10−3/10−2 to 10−4/10−3 compared with the wild type. Furthermore, the 10-fold reduced transformation frequency of the mutants producing the PilQΔN0 complex could also be due to the loss of an additional supply of external DNA bound by pilus structures and then delivered to the secretin pore.
In conclusion, our work has unraveled an ααβαββα fold as a ring-building motif in the DNA translocator secretin complex PilQ in T. thermophilus HB27. Functional analyses of the PilQ mutants indicate that the pilus structures on the cell surface are not essential for natural transformation. Whether pili or pseudopili are still assembled in the periplasmic space and whether they interact with the secretin triggering the opening of the secretin channel or whether the truncated secretin complex is constantly open, triggering DNA uptake, will be determined in future studies.
Supplementary Material
Acknowledgments
We are indebted to Deryck Mills and Friederike Joos (Max Planck Institute of Biophysics, Frankfurt, Germany) for the contribution to piliation analyses of HB27 cells by electron microscopy. José Berenguer (Universidad Autonomes de Madrid, Madrid, Spain) is gratefully acknowledged for providing antibodies against T. thermophilus HB27 S-layer protein that were used to analyze the purity of the membrane fractionation. We are also grateful to Bernd Ludwig (Goethe University, Frankfurt, Germany) for the kind gift of plasmid pDM12. We are also grateful to Volker Müller (Goethe University, Frankfurt, Germany) for the many stimulating discussions on secretin complex purification. We thank Werner Kühlbrandt and Hartmut Michel (Max Planck Institute of Biophysics, Frankfurt, Germany) for support. The Membrane protein Core Center at the Max Planck Institute of Biophysics was supported financially by the European Strategy Forum on Research Infrastructures Instruct program of the European Union.
The work was supported by Deutsche Forschungsgemeinschaft Grant AV9/6-1.

This article contains supplemental information, Figs. S1 and S2, and Tables S1 and S2.
- T4P
- type IV pili
- Bleo
- bleomycin
- SDM
- site-directed mutagenesis
- IM
- inner membrane
- AAA
- ATPases associated with diverse cellular activities.
REFERENCES
- 1. Jain R., Rivera M. C., Lake J. A. (1999) Horizontal gene transfer among genomes: the complexity hypothesis. Proc. Natl. Acad. Sci. U.S.A. 96, 3801–3806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lorenz M. G., Wackernagel W. (1994) Bacterial gene transfer by natural genetic transformation in the environment. Microbiol. Rev. 58, 563–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Koonin E. V., Makarova K. S., Aravind L. (2001) Horizontal gene transfer in prokaryotes: quantification and classification. Annu. Rev. Microbiol. 55, 709–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lawrence J. G. (1999) Gene transfer, speciation, and the evolution of bacterial genomes. Curr. Opin. Microbiol. 2, 519–523 [DOI] [PubMed] [Google Scholar]
- 5. Johnsborg O., Eldholm V., Håvarstein L. S. (2007) Natural genetic transformation: prevalence, mechanisms and function. Res. Microbiol. 158, 767–778 [DOI] [PubMed] [Google Scholar]
- 6. Averhoff B. (2009) Shuffling genes around in hot environments: the unique DNA transporter of Thermus thermophilus. FEMS Microbiol. Rev. 33, 611–626 [DOI] [PubMed] [Google Scholar]
- 7. Hidaka Y., Hasegawa M., Nakahara T., Hoshino T. (1994) The entire population of Thermus thermophilus cells is always competent at any growth phase. Biosci. Biotechnol. Biochem. 58, 1338–1339 [DOI] [PubMed] [Google Scholar]
- 8. Chen I., Dubnau D. (2004) DNA uptake during bacterial transformation. Nat. Rev. Microbiol. 2, 241–249 [DOI] [PubMed] [Google Scholar]
- 9. Hobbs M., Mattick J. S. (1993) Common components in the assembly of type 4 fimbriae, DNA transfer systems, filamentous phage and protein-secretion apparatus: a general system for the formation of surface-associated protein complexes. Mol. Microbiol. 10, 233–243 [DOI] [PubMed] [Google Scholar]
- 10. Averhoff B. (2004) DNA transport and natural transformation in mesophilic and thermophilic bacteria. J. Bioenerg. Biomembr. 36, 25–33 [DOI] [PubMed] [Google Scholar]
- 11. Sandkvist M. (2001) Biology of type II secretion. Mol. Microbiol. 40, 271–283 [DOI] [PubMed] [Google Scholar]
- 12. Koomey M. (1998) Competence for natural transformation in Neisseria gonorrhoeae: a model system for studies of horizontal gene transfer. APMIS Suppl. 84, 56–61 [DOI] [PubMed] [Google Scholar]
- 13. Rose I., Biukovi G., Aderhold P., Müller V., Grüber G., Averhoff B. (2011) Identification and characterization of a unique, zinc-containing transport ATPase essential for natural transformation in Thermus thermophilus HB27. Extremophiles 15, 191–202 [DOI] [PubMed] [Google Scholar]
- 14. Friedrich A., Rumszauer J., Henne A., Averhoff B. (2003) Pilin-like proteins in the extremely thermophilic bacterium Thermus thermophilus HB27: implication in competence for natural transformation and links to type IV pilus biogenesis. Appl. Environ. Microbiol. 69, 3695–3700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Friedrich A., Prust C., Hartsch T., Henne A., Averhoff B. (2002) Molecular analyses of the natural transformation machinery and identification of pilus structures in the extremely thermophilic bacterium Thermus thermophilus strain HB27. Appl. Environ. Microbiol. 68, 745–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Burkhardt J., Vonck J., Averhoff B. (2011) Structure and function of PilQ, a secretin of the DNA transporter from the thermophilic bacterium Thermus thermophilus HB27. J. Biol. Chem. 286, 9977–9984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hanahan D. (1983) Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166, 557–580 [DOI] [PubMed] [Google Scholar]
- 18. Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K. (eds) (1992) Current Protocols in Molecular Biology, Greene Publishing and Wiley-Interscience, New York [Google Scholar]
- 19. Koyama Y., Hoshino T., Tomizuka N., Furukawa K. (1986) Genetic transformation of the extreme thermophile Thermus thermophilus and of other Thermus ssp. J. Bacteriol. 166, 338–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schwarzenlander C., Haase W., Averhoff B. (2009) The role of single subunits of the DNA transport machinery of Thermus thermophilus HB27 in DNA binding and transport. Environ. Microbiol. 11, 801–808 [DOI] [PubMed] [Google Scholar]
- 21. Weber K., Osborn M. (1969) The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis. J. Biol. Chem. 244, 4406–4412 [PubMed] [Google Scholar]
- 22. Koyama Y., Hoshino T., Tomizuka N., Furukawa K. (1986) Genetic transformation of the extreme thermophile Thermus thermophilus and of other Thermus spp. J. Bacteriol. 166, 338–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. de Grado M., Castán P., Berenguer J. (1999) A high-transformation-efficiency cloning vector for Thermus thermophilus. Plasmid 42, 241–245 [DOI] [PubMed] [Google Scholar]
- 24. Ludtke S. J., Baldwin P. R., Chiu W. (1999) EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128, 82–97 [DOI] [PubMed] [Google Scholar]
- 25. van Heel M., Harauz G., Orlova E. V., Schmidt R., Schatz M. (1996) A new generation of the IMAGIC image processing system. J. Struct. Biol. 116, 17–24 [DOI] [PubMed] [Google Scholar]
- 26. Friedrich A., Hartsch T., Averhoff B. (2001) Natural transformation in mesophilic and thermophilic bacteria: identification and characterization of novel, closely related competence genes in Acinetobacter sp. strain BD413 and Thermus thermophilus HB27. Appl. Environ. Microbiol. 67, 3140–3148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rumszauer J., Schwarzenlander C., Averhoff B. (2006) Identification, subcellular localization and functional interactions of PilMNOWQ and PilA4 involved in transformation competency and pilus biogenesis in the thermophilic bacterium Thermus thermophilus HB27. FEBS J. 273, 3261–3272 [DOI] [PubMed] [Google Scholar]
- 28. Koster M., Bitter W., de Cock H., Allaoui A., Cornelis G. R., Tommassen J. (1997) The outer membrane component, YscC, of the Yop secretion machinery of Yersinia enterocolitica forms a ring-shaped multimeric complex. Mol. Microbiol. 26, 789–797 [DOI] [PubMed] [Google Scholar]
- 29. Koster M., Bitter W., Tommassen J. (2000) Protein secretion mechanisms in Gram-negative bacteria. Int. J. Med. Microbiol. 290, 325–331 [DOI] [PubMed] [Google Scholar]
- 30. Johnson T. L., Abendroth J., Hol W. G., Sandkvist M. (2006) Type II secretion: from structure to function. FEMS Microbiol. Lett. 255, 175–186 [DOI] [PubMed] [Google Scholar]
- 31. Han X., Kennan R. M., Parker D., Davies J. K., Rood J. I. (2007) Type IV fimbrial biogenesis is required for protease secretion and natural transformation in Dichelobacter nodosus. J. Bacteriol. 189, 5022–5033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hager A. J., Bolton D. L., Pelletier M. R., Brittnacher M. J., Gallagher L. A., Kaul R., Skerrett S. J., Miller S. I., Guina T. (2006) Type IV pili-mediated secretion modulates Francisella virulence. Mol. Microbiol. 62, 227–237 [DOI] [PubMed] [Google Scholar]
- 33. Cornelis G. R. (2010) The type III secretion injectisome, a complex nanomachine for intracellular 'toxin' delivery. Biol. Chem. 391, 745–751 [DOI] [PubMed] [Google Scholar]
- 34. Thanassi D. G. (2002) Ushers and secretins: channels for the secretion of folded proteins across the bacterial outer membrane. J. Mol. Microbiol. Biotechnol. 4, 11–20 [PubMed] [Google Scholar]
- 35. Korotkov K. V., Gonen T., Hol W. G. (2011) Secretins: dynamic channels for protein transport across membranes. Trends Biochem. Sci. 36, 433–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Korotkov K. V., Pardon E., Steyaert J., Hol W. G. (2009) Crystal structure of the N-terminal domain of the secretin GspD from ETEC determined with the assistance of a nanobody. Structure 17, 255–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Reichow S. L., Korotkov K. V., Hol W. G., Gonen T. (2010) Structure of the cholera toxin secretion channel in its closed state. Nat. Struct. Mol. Biol. 17, 1226–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Spreter T., Yip C. K., Sanowar S., André I., Kimbrough T. G., Vuckovic M., Pfuetzner R. A., Deng W., Yu A. C., Finlay B. B., Baker D., Miller S. I., Strynadka N. C. (2009) A conserved structural motif mediates formation of the periplasmic rings in the type III secretion system. Nat. Struct. Mol. Biol. 16, 468–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tarry M., Jääskeläinen M., Paino A., Tuominen H., Ihalin R., Högbom M. (2011) The extra-membranous domains of the competence protein HofQ show DNA binding, flexibility and a shared fold with type I KH domains. J. Mol. Biol. 409, 642–653 [DOI] [PubMed] [Google Scholar]
- 40. Schraidt O., Lefebre M. D., Brunner M. J., Schmied W. H., Schmidt A., Radics J., Mechtler K., Galán J. E., Marlovits T. C. (2010) Topology and organization of the Salmonella typhimurium type III secretion needle complex components. PLoS Pathog. 6, e1000824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Arcus V. (2002) OB-fold domains: a snapshot of the evolution of sequence, structure and function. Curr. Opin. Struct. Biol. 12, 794–801 [DOI] [PubMed] [Google Scholar]
- 42. Theobald D. L., Mitton-Fry R. M., Wuttke D. S. (2003) Nucleic acid recognition by OB-fold proteins. Annu. Rev. Biophys. Biomol. Struct. 32, 115–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tommassen J. (2010) Assembly of outer-membrane proteins in bacteria and mitochondria. Microbiology 156, 2587–2596 [DOI] [PubMed] [Google Scholar]
- 44. Merdanovic M., Clausen T., Kaiser M., Huber R., Ehrmann M. (2011) Protein quality control in the bacterial periplasm. Annu. Rev. Microbiol. 65, 149–168 [DOI] [PubMed] [Google Scholar]
- 45. Meltzer M., Hasenbein S., Mamant N., Merdanovic M., Poepsel S., Hauske P., Kaiser M., Huber R., Krojer T., Clausen T., Ehrmann M. (2009) Structure, function and regulation of the conserved serine proteases DegP and DegS of Escherichia coli. Res. Microbiol. 160, 660–666 [DOI] [PubMed] [Google Scholar]
- 46. Nesper J., Brosig A., Ringler P., Patel G. J., Müller S. A., Kleinschmidt J. H., Boos W., Diederichs K., Welte W. (2008) Omp85(Tt) from Thermus thermophilus HB27: an ancestral type of the Omp85 protein family. J. Bacteriol. 190, 4568–4575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Voulhoux R., Bos M. P., Geurtsen J., Mols M., Tommassen J. (2003) Role of a highly conserved bacterial protein in outer membrane protein assembly. Science 299, 262–265 [DOI] [PubMed] [Google Scholar]
- 48. Wolfgang M., Lauer P., Park H. S., Brossay L., Hébert J., Koomey M. (1998) PilT mutations lead to simultaneous defects in competence for natural transformation and twitching motility in piliated Neisseria gonorrhoeae. Mol. Microbiol. 29, 321–330 [DOI] [PubMed] [Google Scholar]
- 49. Drake S. L., Koomey M. (1995) The product of the pilQ gene is essential for the biogenesis of type IV pili in Neisseria gonorrhoeae. Mol. Microbiol. 18, 975–986 [DOI] [PubMed] [Google Scholar]
- 50. Tønjum T., Freitag N. E., Namork E., Koomey M. (1995) Identification and characterization of pilG, a highly conserved pilus-assembly gene in pathogenic Neisseria. Mol. Microbiol. 16, 451–464 [DOI] [PubMed] [Google Scholar]
- 51. Aas F. E., Wolfgang M., Frye S., Dunham S., Løvold C., Koomey M. (2002) Competence for natural transformation in Neisseria gonorrhoeae: components of DNA binding and uptake linked to type IV pilus expression. Mol. Microbiol. 46, 749–760 [DOI] [PubMed] [Google Scholar]
- 52. Helm R. A., Barnhart M. M., Seifert H. S. (2007) pilQ Missense mutations have diverse effects on PilQ multimer formation, piliation, and pilus function in Neisseria gonorrhoeae. J. Bacteriol. 189, 3198–3207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Long C. D., Tobiason D. M., Lazio M. P., Kline K. A., Seifert H. S. (2003) Low-level pilin expression allows for substantial DNA transformation competence in Neisseria gonorrhoeae. Infect. Immun. 71, 6279–6291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Carbonnelle E., Helaine S., Nassif X., Pelicic V. (2006) A systematic genetic analysis in Neisseria meningitidis defines the Pil proteins required for assembly, functionality, stabilization and export of type IV pili. Mol. Microbiol. 61, 1510–1522 [DOI] [PubMed] [Google Scholar]
- 55. Wolfgang M., van Putten J. P., Hayes S. F., Dorward D., Koomey M. (2000) Components and dynamics of fiber formation define a ubiquitous biogenesis pathway for bacterial pili. EMBO J. 19, 6408–6418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. van Schaik E. J., Giltner C. L., Audette G. F., Keizer D. W., Bautista D. L., Slupsky C. M., Sykes B. D., Irvin R. T. (2005) DNA binding: a novel function of Pseudomonas aeruginosa type IV pili. J. Bacteriol. 187, 1455–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Douzi B., Ball G., Cambillau C., Tegoni M., Voulhoux R. (2011) Deciphering the Xcp Pseudomonas aeruginosa type II secretion machinery through multiple interactions with substrates. J. Biol. Chem. 286, 40792–40801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mattick J. S. (2002) Type IV pili and twitching motility. Annu. Rev. Microbiol. 56, 289–314 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.