Background: Sulfolipid-1 (SL-1) is a Mycobacterium tuberculosis outer membrane lipid whose biosynthesis is not fully understood.
Results: Chp1 catalyzes two acyl transfer reactions to form SL-1. Sap modulates SL-1 levels and transmembrane transport.
Conclusion: The activities of Chp1 and Sap complete the SL-1 pathway.
Significance: Lipid biosynthesis and transport are coupled at the membrane interface by multiple proteins that may regulate substrate specificity and flux.
Keywords: Biosynthesis, Enzymes, Glycolipids, Metabolism, Microbiology, Mycobacterium tuberculosis
Abstract
Mycobacterium tuberculosis possesses unique cell-surface lipids that have been implicated in virulence. One of the most abundant is sulfolipid-1 (SL-1), a tetraacyl-sulfotrehalose glycolipid. Although the early steps in SL-1 biosynthesis are known, the machinery underlying the final acylation reactions is not understood. We provide genetic and biochemical evidence for the activities of two proteins, Chp1 and Sap (corresponding to gene loci rv3822 and rv3821), that complete this pathway. The membrane-associated acyltransferase Chp1 accepts a synthetic diacyl sulfolipid and transfers an acyl group regioselectively from one donor substrate molecule to a second acceptor molecule in two successive reactions to yield a tetraacylated product. Chp1 is fully active in vitro, but in M. tuberculosis, its function is potentiated by the previously identified sulfolipid transporter MmpL8. We also show that the integral membrane protein Sap and MmpL8 are both essential for sulfolipid transport. Finally, the lipase inhibitor tetrahydrolipstatin disrupts Chp1 activity in M. tuberculosis, suggesting an avenue for perturbing SL-1 biosynthesis in vivo. These data complete the SL-1 biosynthetic pathway and corroborate a model in which lipid biosynthesis and transmembrane transport are coupled at the membrane-cytosol interface through the activity of multiple proteins, possibly as a macromolecular complex.
Introduction
Mycobacterium tuberculosis, the causative agent of tuberculosis, is characterized by a complex cell wall that contributes to its pathogenesis and inherent resistance to therapeutics. The cell wall encompasses multiple layers exterior to the cytosolic membrane and comprises not only peptidoglycan but also arabinogalactan polysaccharide layers and an extremely hydrophobic bilayer known as the mycobacterial outer membrane. Abundant M. tuberculosis-specific surface lipids such as sulfatides, acyltrehaloses, and dimycocerosates are noncovalently assembled in the mycobacterial outer membrane and have been implicated in virulence and host immune responses (1–4). Sulfolipid-1 (SL-1),6 the most abundant sulfatide, is unique to pathogenic mycobacteria (see Fig. 1). The levels of this tetraacylated glycolipid have been positively correlated with strain virulence, but despite a half-century of research into this connection, the biological functions of SL-1 have remained elusive.
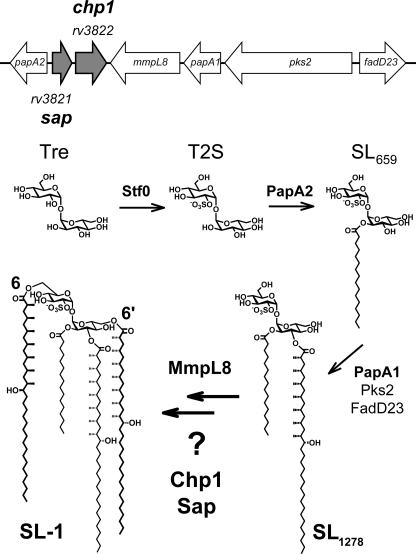
FIGURE 1.
Genes of unknown function in SL-1 locus may be involved in biosynthesis. Genes associated with SL-1 biosynthesis are clustered in the M. tuberculosis genome, but one putative operon contains two genes of unknown function, sap (rv3821) and chp1 (rv3822). After sulfation of trehalose (Tre) by Stf0, T2S is successively esterified by PapA2 and PapA1. MmpL8 participates in the final two esterifications by an unknown mechanism and is essential for sulfolipid transport; Chp1 and Sap may also be involved.
Various studies have implicated SL-1 in the inhibition of mitochondrial oxidative phosphorylation, alteration of phagosome-lysosome fusion, and stimulation as well as suppression of cytokine and reactive oxygen species production in host leukocytes (5–12). However, M. tuberculosis gene disruption strains lacking fully elaborated SL-1 do not appear to have consistent phenotypes or phenotypes distinguishable from wild-type M. tuberculosis in animal models of infection (13–17). In contrast, the diacyl sulfolipid SL1278, a biosynthetic precursor of SL-1, is a well documented active metabolite (see Fig. 1). SL1278 was found to bind to the MHC-like lipid receptor CD1b and to stimulate the cytokines IFN-γ and IL-2 in CD8+ T-cells from donors positive for the tuberculin skin test (18). Subsequent work using synthetic analogs of SL1278 showed that the ability of SL1278 to elicit a CD1-restricted T-cell response is dependent on the length of the fatty acid acyl chains, as well as the presence and number of methyl-branched substituents on the acyl chains (19).
Elucidating the biosynthetic pathway of SL-1 is a key aspect in understanding how M. tuberculosis regulates SL-1 and its precursors as a potential mechanism for host immune modulation. Many of the initial steps in SL-1 biosynthesis have been defined; in addition, SL-1 biosynthesis appears to be coupled to lipid transport across the cytosolic membrane (15–25). However, the machinery underlying the final biosynthetic steps is still not understood. The complete elucidation of SL-1 biosynthesis could provide additional avenues for targeted disruption of M. tuberculosis sulfolipids and a further means of dissecting their biological roles.
SL-1 comprises a trehalose-2-sulfate (T2S) core elaborated with four acyl groups: a straight-chain fatty acid (palmitate or stearate) and three multiply methyl-branched (hydroxy)phthioceranoic acids (see Fig. 1). The sulfotransferase Stf0 initiates SL-1 biosynthesis by sulfating the abundant disaccharide trehalose to form T2S. The acyltransferase PapA2 then catalyzes the esterification of T2S at the 2′-position to generate a monoacylated intermediate, SL659 (15). The polyketide synthase Pks2 synthesizes methyl-branched (hydroxy)phthioceranoyl chains using an activated fatty acid starter unit provided by the fatty acid AMP ligase FadD23 (also known as FAAL23) (20, 21). PapA1 transfers the product of Pks2 to the 3′-position of SL659, yielding diacylated SL1278 (15). Additional acylations at the 6- and 6′-positions of SL1278 are required to produce fully elaborated SL-1. These final steps are chemically similar to the reaction catalyzed by PapA1, but there is no in vitro evidence that PapA1 is capable of this activity.
Intriguingly, the lipid transporter MmpL8 has been implicated in SL-1 formation. MmpL8 belongs to the RND (resistance-nodulation-division) permease protein family and is hypothesized to transport SL-1 or SL1278 from the cytosolic leaflet to the periplasmic leaflet of the cytosolic membrane (18, 19). The M. tuberculosis ΔmmpL8 gene disruption mutant accumulates the diacyl precursor SL1278 in the cell membrane rather than the predicted SL-1, implying that MmpL8 is required for biosynthesis as well as transport (18, 19). However, no member of the RND permease family has been shown to have enzymatic activity, nor does MmpL8 contain any known conserved catalytic domains (22).
In addition to the genes described above, the SL-1 biosynthetic locus encompasses a putative operon with two ORFs, rv3821 and rv3822, both of which are annotated as conserved hypothetical proteins that we have named Sap and Chp1, respectively (Fig. 1). In this work, we demonstrate that the final steps in SL-1 biosynthesis and SL-1 transport require Sap and Chp1 in addition to MmpL8. Lipid analysis of M. tuberculosis gene disruption strains revealed that Sap, Chp1, and MmpL8 are all necessary for M. tuberculosis to produce wild-type levels of SL-1. Chp1 and MmpL8 are essential for SL-1 biosynthesis, whereas Sap and MmpL8 are required for sulfolipid transport. In vitro, Chp1 was specifically modified by an activity-based fluorophosphonate probe, identifying Chp1 as a serine hydrolase superfamily member. Chp1 can also use a fully synthetic diacyl sulfolipid analog as both an acyl donor and acceptor in two successive, regioselective reactions to form a tetraacylated sulfolipid. Using the combined activities of Chp1, PapA1, and PapA2, all four sulfolipid acylation reactions were reconstituted in a one-pot synthesis. Finally, in M. tuberculosis, the clinically approved lipase inhibitor tetrahydrolipstatin (THL) inhibited the activity of Chp1, but not that of PapA1 or PapA2. These data complete the SL-1 biosynthetic pathway and corroborate a model in which SL-1 biosynthesis and transmembrane transport are coupled through the activity of multiple proteins at the membrane-cytosol interface.
EXPERIMENTAL PROCEDURES
Reagents and Chemicals
T2S and the SL-1 model compound 6,6′-di-O-(2-methylarachidoyl)-3′-O-(2-methylstearoyl)-2′-O-palmitoyltrehalose-2-O-sulfate (SL-A) were synthesized as described (23, 24). The synthesis and characterization of 2-O-palmitoyl-3-O-stearoyl-α,α-d-trehalose-2′-O-sulfate (T2S-PS) and the construction of Chp1 heterologous expression vectors are detailed under supplemental “Experimental Procedures.”
Bacterial Strains and Growth Media
The M. tuberculosis Erdman strain (ATCC 35801) and Mycobacterium smegmatis mc2155 (ATCC 700084) were grown at 37 °C. The growth medium was 7H9 (liquid) or 7H11 (solid) with 0.5% glycerol and 0.05% Tween 80 plus 0.5% glucose or 10% albumin/dextrose/catalase for M. smegmatis and plus 10% oleate/albumin/dextrose/catalase for M. tuberculosis. For selective media, antibiotic concentrations were 100 μg/ml carbenicillin, 50 μg/ml kanamycin, or 100 μg/ml hygromycin for Escherichia coli and 20 μg/ml kanamycin or 50 μg/ml hygromycin for mycobacteria.
Sequence Homology Analysis and Structure Prediction
Amino acid sequences for Sap (Rv3821) and Chp1 (Rv3822) were obtained from TubercuList (25). Transmembrane helices were predicted by the TMHMM hidden Markov model (52). The Chp1 sequence was also submitted to the Phyre protein fold recognition server for protein fold and structure prediction (26).
Construction of Gene Disruption Mutants
The Δsap and Δchp1 mutant strains were created by homologous recombination using specialized phage transduction (27). These mutants replaced 429 bp of sap (amino acids (aa) 27–171) and 862 bp of chp1 (aa 44–331) with a hygromycin resistance cassette. Recombinant clones were confirmed by PCR (supplemental Fig. S7). Strains were complemented with integrating plasmids encoding the target gene with a native promoter (upstream 1 kb of the first gene in the putative operon).
Lipid Extraction and Mass Spectrometry Analysis
M. tuberculosis strains were grown for 3–5 days to late log phase. Cultures were diluted in Tween-free medium to A600 = 0.25–0.3 and grown for 2 days. Cells were harvested and extracted in 1 ml of hexane/50 ml of culture. The upper organic phase (“surface lipid” fraction) was removed and added to an equal volume of 1:1 chloroform/methanol. The remaining cell pellet and aqueous phase were extracted in 4 ml of 1:1 chloroform/methanol and incubated overnight at room temperature. Cell debris was pelleted by centrifugation, and the supernatant (“cell pellet” fraction) was decanted. All extractions were repeated in at least three independent experiments. The triacylated sulfolipid SL1868 was partially purified from wild-type H37Rv cells in an adapted protocol (see supplemental “Experimental Procedures”) (14).
High-resolution Fourier transform ion cyclotron resonance MS and MSn data were obtained on an Apex II FT-ICR mass spectrometer (Bruker Daltonics) as described previously (28) with the following modifications. Two sets of electrospray ionization source tuning parameters were used to acquire mass spectra. For the mass range m/z 300–1000, the capillary voltage was set to 4.5 kV, the capillary exit voltage was set to −300 V, the skimmer 1 voltage was set to −20 V, and the skimmer 2 voltage was set to −7 V. For the mass range m/z 1000–3000, the skimmer 2 voltage was lowered to approximately −1 to −3 V.
Additional MSn spectra were obtained on an LTQ mass spectrometer equipped with an electrospray ionization source (Thermo Finnigan) operating in the negative ion mode. Ions were introduced into the ion source via direct injection at a rate of 5–10 μl/min. Collision-induced dissociation was used for MSn experiments. The precursor ions were isolated with an isolation width of 1–3 Da, the ions were activated with a 26% normalized collision energy for 100 ms, and the qz value was maintained at 0.250.
Chp1 Subcellular Localization by Fractionation and Immunoblotting
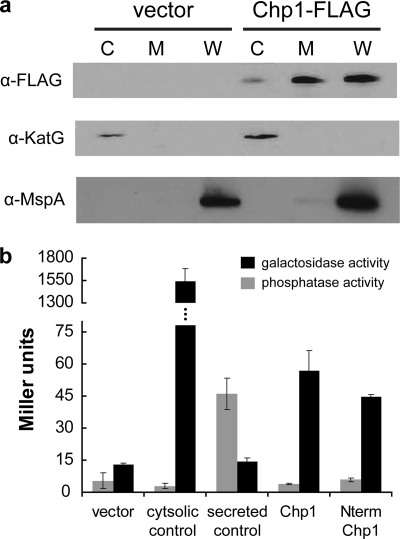
M. smegmatis expressing full-length Chp1 with a C-terminal 3×FLAG epitope tag was grown to late log phase (A600 = 1–1.5) and fractionated by sonication and differential centrifugation to generate cytosol-, membrane-, and cell wall-enriched fractions as described previously (29) and under supplemental “Experimental Procedures.” Protein concentrations were determined by the bicinchoninic acid protein assay, and 5 μg of protein from each fraction were separated by SDS-PAGE. For the anti-MspA blot, samples were extracted with 0.6% octyl thioglucoside as described (30), and 310 ng of each subcellular fraction were separated by SDS-PAGE. Blots were probed with anti-KatG (Colorado State University), anti-FLAG M2, anti-GroEL2 (Abcam ab20519), and anti-MspA (31) antibodies and visualized by chemiluminescence.
Enzymatic Activity of Chp1-Alkaline Phosphatase and Chp1-β-Galactosidase Fusions
Chp1-alkaline phosphatase (AP) and Chp1-β-gal fusion constructs were electroporated into M. smegmatis, and β-gal and AP activities were determined using the substrates 2-nitrophenyl-β-d-galactopyranoside and 4-nitrophenyl phosphate essentially as reported (35, 36). Activity is reported in Miller units (reaction A420/(culture A600 × Vs × min), where Vs = volume of the original culture used in the reaction). Control vectors encoding a secretion signal and secreted AP were a kind gift of Miriam Braunstein (University of North Carolina, Chapel Hill, NC).
Expression and Purification of Chp1 Catalytic Domain in E. coli
His-MBP-Chp1-cat, which is the putative catalytic domain of Chp1 fused to an N-terminal His6 tag and maltose-binding protein (32), was expressed in E. coli BL21(DE3). Following a 4-h induction with 1 mm isopropyl β-d-thiogalactopyranoside at 37 °C, cells were lysed in 50 mm sodium phosphate (pH 7.2) and 10% glycerol (buffer A). The clarified crude lysate was incubated in batch with 10 ml of amylose resin and washed with 100 ml of buffer A. Bound protein was eluted in buffer A plus 10 mm maltose, and fractions containing the His-MBP-Chp1-cat fusion protein were pooled, dialyzed, and incubated overnight at 4 °C with tobacco etch virus protease. Cleaved maltose-binding protein and other impurities were removed by incubation with amylose resin. The flow-through fraction containing purified Chp1-cat protein was concentrated and stored at −80 °C.
In Vitro Sulfolipid Biosynthetic Reactions
PapA2 and PapA1 were expressed and purified as described (15). For reconstitution of SL-1 analog biosynthesis, reactions contained 1 μm PapA2, 1 μm PapA1, 1 μm Chp1-cat, 50 μm palmitoyl-CoA, and 1 mm T2S in 100 μl of reaction buffer (100 mm sodium phosphate (pH 7.2), 1 mm DTT, and 10% glycerol). To test reactivity with T2S-PS, Chp1-cat was incubated at 1 μm with 0.1 mm T2S-PS in 100 μl of reaction buffer. Competition reactions also included either 0.1 mm palmitoyl-CoA or 0.1 mm SL-A. Reactions were incubated at room temperature for 12–16 h. Reactions were extracted with an equal volume of 1:1 chloroform/methanol. The organic phase was co-spotted with an equal volume of 10 mg/ml 2-(4′-hydroxybenzeneazo)benzoic acid suspended in 1:1 water/ethanol. Spots were analyzed by MALDI-TOF-MS in negative ion mode using a 20-kV accelerating voltage and 110-ns extraction delay with 500 shots/spectrum (QB3/Chemistry Mass Spectrometry Facility, University of California at Berkeley). For MSn analysis (performed as described above), 100 μl of each reaction in 100 mm ammonium bicarbonate (pH 7.2) were lyophilized and dissolved in methanol prior to analysis.
35S and 14C Metabolic Labeling and Lipid Analysis by TLC
M. tuberculosis strains were grown to late log phase. For 35S labeling, cells were resuspended at A600 ∼ 1 in 10 ml of PBS with 1% acetate and 100 μCi of [35S]sulfate. For 14C labeling, 5 μCi of [14C]propionic acid were added directly to 10 ml of culture at A600 ∼ 1. After overnight incubation, cell pellets were extracted sequentially in hexanes and 1:1 chloroform/methanol as described above. An equal volume of extracts resuspended in one-tenth or one-twentieth the original extraction volume was spotted on silica plates (HPTLC Silica Gel 60, EMD Chemicals) and developed in 60:12:1 chloroform/methanol/water, followed by phosphorimaging. For THL treatment experiments, THL in Me2SO was added to 10-ml cultures at 0, 10, 20, and 40 μg/ml for 6 h, followed by the addition of 5 μCi of [14C]propionic acid with further incubation, extraction, and analysis as described above. The lipid phthiocerol dimycocerosate was used as a loading control and resolved in 90:10 petroleum ether/hexanes.
RESULTS AND DISCUSSION
Bioinformatic Analysis of Sap and Chp1
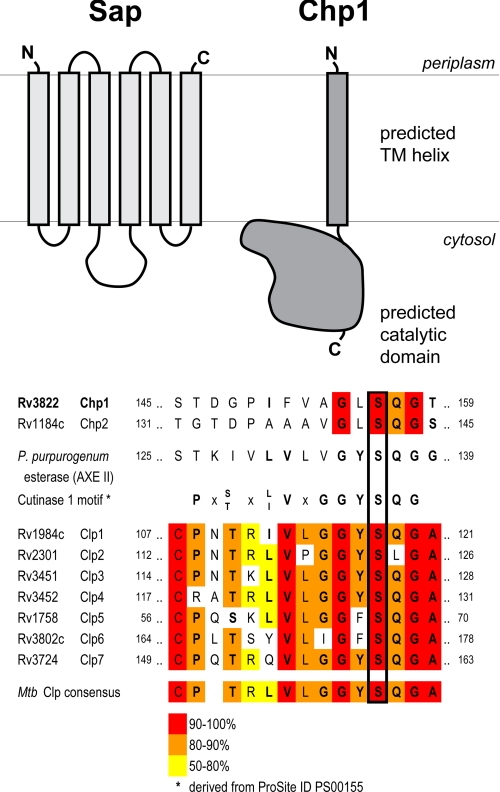
The rv3821 locus encodes a 237-amino acid integral membrane protein homologous to the M. smegmatis Gap protein, which is required for glycopeptidolipid transport to the cell surface (33). Because rv3821 may be analogously involved in SL-1 transport, we refer to the protein encoded by rv3821 as Sap (sulfolipid-1-addressing protein). Sap has six predicted transmembrane helices, with a hydrophilic domain between helices 3 and 4 (aa 93–134) that is highly variable among identified Gap-like proteins and has been hypothesized to be involved in substrate recognition (Fig. 2) (33). Sap shares 30% sequence identity with M. smegmatis Gap (MSMEG_0403); more distant homologs in M. tuberculosis include Rv1517, which is encoded by a locus linked to lipo-oligosaccharide biosynthesis in Mycobacterium marinum, and Rv3481c, whose gene is in the same operon as a putative triacylglycerol synthase. Sap also belongs to the LysE protein superfamily (Pfam ID PF01810), whose members have been implicated in small molecule transport in bacteria. For example, LysE from Corynebacterium glutamicum, which belongs to the same taxonomic suborder as M. tuberculosis, exports l-lysine and has two homologs in M. tuberculosis (Rv0488 and Rv1986) (34). Other LysE superfamily members are associated with antibiotic resistance and metal ion transport. These analyses support the hypothesis that Gap-like proteins are involved in the export of metabolites across the cell membrane and that Sap specifically may be involved in SL-1 transmembrane transport.
FIGURE 2.
Sap and Chp1 are membrane-associated proteins, and Chp1 is related to M. tuberculosis CLPs. Upper, a member of the Gap protein family, Sap is an integral membrane protein with six transmembrane helices and a hydrophilic domain (aa 93–134). Chp1 has a single predicted N-terminal transmembrane (TM) helix followed by a conserved C-terminal PE-PPE domain with an α/β-hydrolase fold. The domain structures and membrane orientations shown were predicted by the TMHMM algorithm. Lower, the Chp1 putative catalytic serine (black box) was identified by combined sequence/structure alignment with P. purpurogenum acetylxylan esterase II (AXE II). In both Chp1 and the related Chp2, the sequence surrounding this serine shares some similarity with the conserved cutinase motif (boldface residues). In contrast, the M. tuberculosis (Mtb) CLPs have an identifiable, albeit modified cutinase motif (38).
The rv3822 locus encodes a 404-aa protein with a predicted N-terminal transmembrane helix (aa 46–64). The C-terminal domain (aa 104–325) has predicted α/β-hydrolase secondary structure that is conserved among certain members of the M. tuberculosis PE and PPE protein families, but the native functions of these proteins are unknown (35, 36). The closest homolog of Rv3822 is Rv1184c, which shares the conserved C-terminal domain and is associated with the polyacyltrehalose biosynthetic locus. Rv3822 is more distantly related to M. tuberculosis proteins known as the cutinase-like proteins (CLPs) (Fig. 2), which have been shown to have phospholipase, esterase, and thioesterase activities on a variety of model substrates (37–39). One of the CLPs, Rv3802c, is associated with the mycolic acid biosynthetic gene locus and displays hydrolytic activity similar to that of other CLPs, but its possible role in mycolic acid biosynthesis has not been elucidated (37). Because of its similarity to CLPs, we refer to Rv3822 as a cutinase-like hydrolase protein (Chp1). From an in silico structural analysis, we identified a putative catalytic triad (Ser-156–Asp-232–His-255) in Chp1 based on the alignment of these residues with the known active site of a fungal acetylxylan esterase from Penicillium purpurogenum (36, 40). On the basis of these analyses, we hypothesized that Chp1 is a membrane-anchored acyltransferase involved in SL-1 biosynthesis.
Initial Characterization of Roles of Sap and Chp1 in Mycobacterial Sulfolipid Biosynthesis
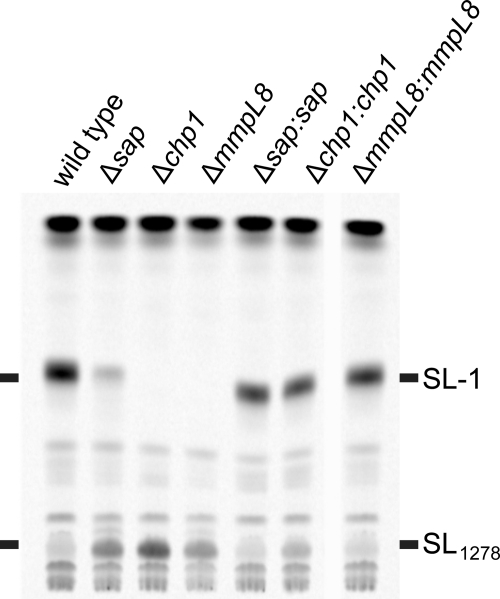
To test our hypothesis that Sap and Chp1 function in SL-1 biosynthesis, we compared the lipid profiles of M. tuberculosis Erdman wild-type cells and the gene disruption strains Δstf0, Δsap, Δchp1, and ΔmmpL8. Cells were grown in the presence of [14C]propionate, which preferentially labels methyl-branched lipids, and the cell pellet lipid extracts were analyzed by TLC. SL-1 was detected in wild-type cells, but not in ΔmmpL8 cells (Fig. 3), which accumulated SL1278 as observed previously (18, 19). The Δchp1 strain displayed a similar phenotype to ΔmmpL8. Intriguingly, Δsap not only accumulated SL1278 but also produced SL-1, albeit at reduced levels. The complementation of Δchp1 and Δsap with their corresponding genes restored the ability of these strains to synthesize SL-1. Thus, the reduced amount of SL-1 produced by the Δsap mutant is not due to a downstream effect of the tagged gene deletion on chp1 transcription. Metabolic labeling with [35S]sulfate to detect sulfated lipids yielded analogous results and further confirmed the identity of SL-1 and SL1278; neither metabolite was observed in Δstf0, which does not produce sulfated trehalose and therefore lacks sulfated glycolipids (see supplemental Fig. S1) (16).
FIGURE 3.
Chp1 is essential for SL-1 biosynthesis, and Sap modulates SL-1 levels. M. tuberculosis wild-type, Δsap, Δchp1, ΔmmpL8, and corresponding complemented strains were metabolically labeled with [14C]propionate. Cell pellet extracts were analyzed by TLC and phosphorimaging. SL-1 was observed at reduced levels in Δsap and not at all in Δchp1 and ΔmmpL8, and all three strains accumulated SL1278. The wild-type lipid profile was restored by complementation for all strains.
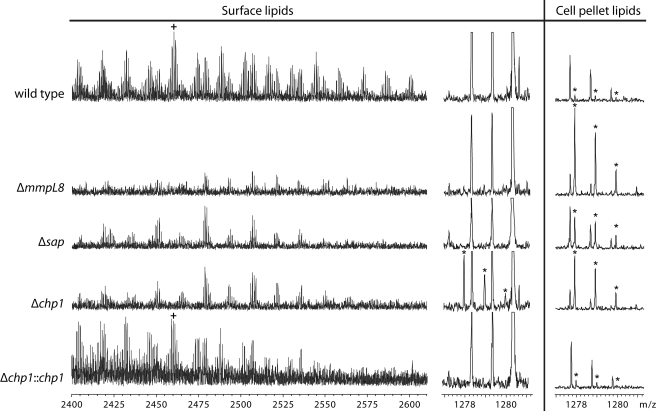
These results were corroborated by high-resolution mass spectrometry analysis of the surface lipid and cell pellet lipid fractions, which were obtained by extracting cells with hexanes and 1:1 chloroform/methanol, respectively (Fig. 4 and supplemental Fig. S2). Uniquely and unexpectedly, SL1278 was found in Δchp1 in the surface lipid fraction as well as in the cell pellet fraction. Both this SL1278 transport defect and SL-1 biosynthesis were restored by complementation with wild-type chp1, confirming that the presence of SL1278 in the surface lipid fraction is not due to a nonspecific loss of membrane integrity in Δchp1. The presence of SL1278 in the surface lipid fraction in Δchp1, but not in either Δsap or ΔmmpL8, suggests that both Sap and MmpL8 are necessary for sulfolipid transport.
FIGURE 4.
Δchp1 transports SL1278 to cell surface. Electrospray ionization Fourier transform ion cyclotron resonance MS analysis of lipid extracts from M. tuberculosis wild-type, Δsap, Δchp1, and ΔmmpL8 strains showed that the three knock-out strains lack SL-1 and accumulate SL1278. (To aid comparison, the m/z 2460 ion belonging to the SL-1 series is marked with a plus sign, and the m/z 1277.9, 1278.9, and 1279.9 ions belonging to the SL1278 series are marked with asterisks to distinguish them from other isobaric compounds in the lipid extracts). In addition, SL1278 was found in the surface lipid fraction only in Δchp1, and both SL-1 production and the SL1278 transport phenotype were restored by complementation with chp1.
Biochemical Characterization of Chp1 Function
To test our hypothesis that Chp1 is membrane-anchored, we determined its subcellular location by fractionating M. smegmatis cells expressing full-length Chp1. As predicted, Chp1 was found in the cytosolic membrane- and cell wall-enriched fractions (Fig. 5a). On the basis of the hypothesis that Chp1 is most likely associated with the cytosolic membrane, we determined the enzymatic activity of a set of Chp1 fusion constructs attached either to β-gal, which is folded and active only in the cytosol, or to AP, which is active only in the oxidizing environment of the periplasm (36, 45). Fusions of β-gal or AP to the C terminus of full-length Chp1 or the Chp1 putative transmembrane helix (aa 1–71) were expressed in M. smegmatis, and the cells were assayed for enzymatic activity. In all cases, activity above background was detected only when Chp1 was fused to β-gal (Fig. 5b). Similar results were obtained by qualitative examination of growth on agar containing a chromogenic substrate (supplemental Fig. S3). These data suggest that Chp1 is membrane-associated and oriented with the C-terminal catalytic domain in the cytosol (Fig. 2).
FIGURE 5.
Chp1 localizes to cell membrane with catalytic domain in cytosol. a, the immunoblot of subcellular fractions of M. smegmatis expressing Chp1 with a C-terminal 3×FLAG tag shows Chp1 enriched in the cell membrane and cell wall. KatG and MspA are markers for the cytosol- and cell wall-enriched fractions. C, cytosol; M, membrane; W, cell wall. b, in M. smegmatis strains expressing full-length Chp1 or the Chp1 N-terminal (Nterm) domain with C-terminal fusions to AP (gray bars) and β-gal (black bars), enzymatic activity was observed only with β-gal fusions. M. smegmatis strains transformed with empty vector or with AP or β-gal with or without an N-terminal secretion signal served as negative and positive controls. Turnover of colorimetric substrates is expressed in Miller units.
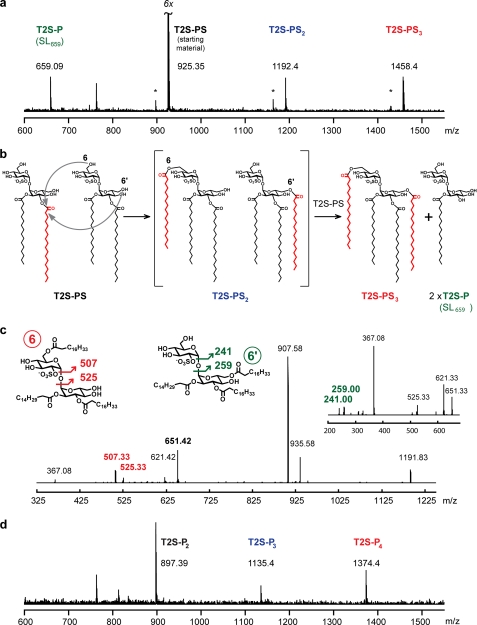
We next expressed and purified from E. coli the conserved C-terminal catalytic domain of Chp1 (Chp1-cat, aa 65–404) and sought to characterize its enzymatic activity in biochemical assays. We first tested for covalent labeling by a fluorescent fluorophosphonate activity-based probe (TAMRA-fluorophosphonate), which selectively modifies the catalytic residue of serine hydrolase superfamily members (41). Chp1-cat was effectively labeled with TAMRA-fluorophosphonate when properly folded, although not when denatured by heat, and mutation of the putative catalytic Ser-156 to Ala abrogated reactivity, confirming the assignment of Chp1 as a serine hydrolase (see supplemental “Experimental Procedures” and Fig. S4). To test Chp1-cat activity on sulfolipids, we synthesized the diacyl sulfolipid T2S-PS as an SL1278 analog. Surprisingly, when Chp1-cat was incubated with T2S-PS, products of lipid acyl transfer were detected in the reaction mixture by MALDI-MS, even in the absence of any acyl-CoA donor (Fig. 6a). The higher molecular weight components at m/z 1192 and 1458 were consistent with one and two additions of stearate to T2S-PS to form T2S-PS2 and T2S-PS3. These products indicate that Chp1 can use T2S-PS as both an acyl donor and acceptor. The observed products imply that Chp1 catalyzes the regiospecific transfer of a fatty acyl group (in this case, stearate) from the 3′-position of a donor molecule to the 6- or 6′-position of an acceptor molecule. By this mechanism, T2S-P (i.e. the monoacylated SL-1 precursor SL659) should be generated as a by-product, and indeed, this species is also observed in the mass spectrum. Palmitoyl-CoA did not successfully compete with T2S-PS as an acyl donor, as the products of palmitate addition to T2S-PS were not detected (supplemental Fig. S5A). Chp1-cat also did not appear to use SL-A, a tetraacylated analog of SL-1, as an acyl donor to T2S-PS but was capable of hydrolyzing SL-A (supplemental Fig. S5, B and C) (24).
FIGURE 6.
Chp1 uses diacyl-T2S as acyl donor and acceptor. a, MALDI-MS analysis revealed higher molecular weight reaction products when Chp1-cat was incubated with T2S-PS. The observed ions at m/z 659.1, 1192.4, and 1458.4 are consistent with the formation of the designated sulfolipid species. Ions labeled with asterisks indicate a minor T2S-P2 contaminant in the T2S-PS stock, as well as the corresponding T2S-P2S and T2S-PS2 product ions following stearate addition from T2S-PS. b, reaction scheme for Chp1 activity on T2S-PS. c, MS2 and subsequent MS3 (inset) analysis of the m/z 1192 ion showing fragmentation peaks consistent with stearate on either the glucose or acylsulfoglucose monomers (6- and 6′-positions of T2S, respectively). d, incubation of PapA2, PapA1, and Chp1 with T2S and palmitoyl-CoA yields ions at m/z 897.4, 1135.4, and 1374.4, consistent with the indicated acylation products.
Tandem MS (MSn) fragmentation of the T2S-PS3 ion at m/z 1192 revealed a mixture of regioisomers in which the additional stearoyl group was attached to either glucose monomer of T2S (Fig. 6b). SL1868, a triacylated T2S species (trehalose-2-sulfate-3′-palmitate-4′,6′-bisphthioceranoate) purified from wild-type H37Rv M. tuberculosis, was found by MSn analysis to comprise an analogous mixture of regioisomers (supplemental Fig. S6). On the basis of these data, we postulate that Chp1 catalyzes acyl transfer to either glucose monomer of the acceptor molecule and that this lack of regioselectivity with respect to the acceptor substrate is physiologically relevant.
Finally, we reconstituted the biosynthesis of SL-1 in a one-pot reaction by combining the acyltransferases PapA2 and PapA1 and Chp1-cat with palmitoyl-CoA as the acyl donor and T2S as the acceptor substrate. The product mixture included all of the products expected from a series of acyl transfer reactions, including the mono-, di-, tri-, and tetraacylated species (Fig. 6d). In the absence of Chp1, only the mono- and diacylated species were detected, as observed previously (15).
Chp1 Inhibition by THL in M. tuberculosis
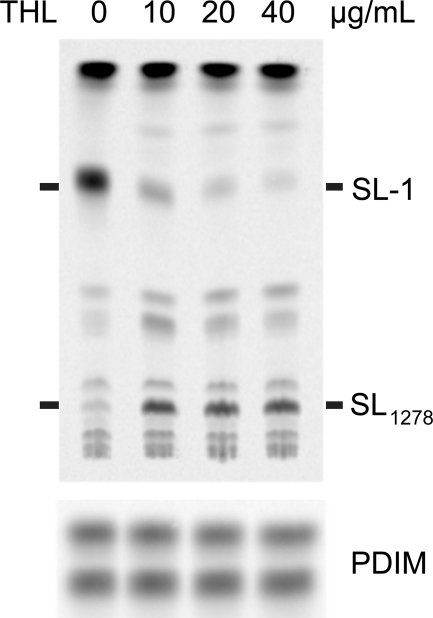
Because Chp1 is a serine hydrolase-type enzyme that recognizes a hydrophobic substrate, we hypothesized that it may be inhibited by THL, a clinically approved lipase inhibitor. More commonly known as Orlistat (Xenical, Roche Applied Science), THL is a lipophilic lactone active against gastric lipase and is approved for the treatment of obesity. THL has been shown to have bactericidal effects on some mycobacterial species. More specifically, THL inhibits general triacylglycerol lipase activity in Mycobacterium bovis bacillus Calmette-Guérin as well as the in vitro activity of the CLP family member Rv3802c and the extracellular lipase Rv0183 (37, 42–45). In this study, M. tuberculosis treated with THL and labeled with [14C]propionate showed a dose-dependent decrease in SL-1 production with a concomitant accumulation of SL1278 (Fig. 7). SL-1 synthesis was not completely suppressed even at THL concentrations close to the minimum inhibitory concentration of 50 μg/ml, consistent with the hypothesis that THL has multiple targets that contribute to its bactericidal activity. These results support our prediction that the lipophilic lactone THL preferentially inhibits Chp1 over the SL-1 acyltransferases PapA1 and PapA2 due to its higher affinity for Chp1 and/or its preferential partitioning into lipid membranes where Chp1 resides.
FIGURE 7.
THL treatment specifically inhibits conversion of SL1278 to SL-1. Upper panel, lipid extracts from M. tuberculosis treated with different concentrations of THL for 6 h followed by [14C]propionate labeling reveal the dose-dependent but incomplete inhibition of SL-1 formation and the accumulation of SL1278 by TLC and phosphorimaging. Lower panel, phthiocerol dimycocerosate (PDIM) loading control.
Conclusions
Based on the in vitro data presented here, the SL-1 biosynthetic pathway appears complete: Stf0 sulfates trehalose to generate T2S, which is then acylated once by PapA2, once by PapA1 using fatty acids from Pks2, and twice by Chp1. However, the lipid profiles of the ΔmmpL8 and Δsap mutants contradict this linear scheme. Following formation of the SL1278 precursor, SL-1 biosynthesis in M. tuberculosis is also dependent on the transport-associated membrane protein MmpL8 and Sap (Rv3821). Indeed, although ΔmmpL8, Δsap, and Δchp1 all accumulate SL1278, only in Δchp1 is SL1278 detected in the surface lipid fraction, implying that MmpL8 and Sap together mediate sulfolipid transport. However, whereas MmpL8 is essential for SL-1 formation, Sap appears to modulate flux through the pathway, similar to MmpS4-mediated modulation of glycopeptidolipid levels in M. smegmatis (46).
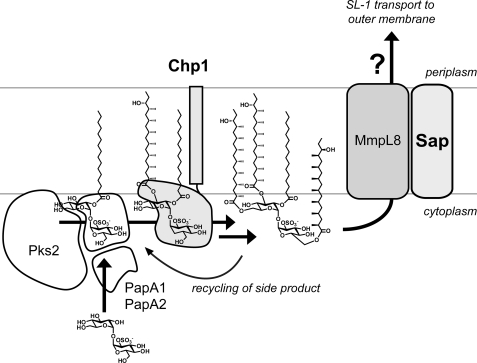
The accumulation of SL1278 in ΔmmpL8 and Δsap is consistent with two models for the coupling of biosynthesis and transport (Fig. 8) (13, 14). In the “sequential” model, MmpL8 transports SL1278, which is then processed to SL-1 in the periplasm. (We here assume that MmpL8 acts solely as a lipid flippase; the question of how SL-1 is transported from the periplasm to its ultimate location in the mycobacterial outer membrane will not be discussed further.) However, this model contradicts our Chp1 topology results and also requires retrograde transport of SL659 by-products for re-entry into the SL-1 pathway (Fig. 8).
FIGURE 8.
Proposed model for SL-1 biosynthesis and transmembrane transport. The data are most consistent with a model in which Chp1 completes SL-1 biosynthesis in the cytosolic leaflet, and MmpL8 and Sap transport SL-1 across the membrane. The membrane localization of Chp1 and the coupling of biosynthesis and transport via MmpL8 suggest that the SL-1 machinery may form a macromolecular complex to facilitate function. The mechanisms by which SL-1 is transported to the mycobacterial outer membrane are unknown. (Note that, in this figure, the hydroxyphthioceranoic groups on SL-1 are truncated.)
The data presented here and elsewhere more strongly support the “scaffolding” model, in which MmpL8 facilitates biosynthesis and then transports the final SL-1 product across the cytosolic membrane. MmpL8 could couple lipid biosynthesis and transport by acting as a scaffold that nucleates a macromolecular complex of cytosolic PapA1, PapA2, Pks2, and membrane-associated Chp1. The scaffold concept has a precedent in the M. tuberculosis phthiocerol dimycocerosate lipid biosynthetic pathway, in which the transporter MmpL7 and the biosynthetic enzyme PpsE were shown to interact in a yeast two-hybrid assay (47). In the SL-1 pathway, the close association of related enzymes could facilitate recycling of the Chp1 side product SL659 back into the biosynthetic pathway and thereby drive the Chp1 reaction forward. Importantly, a recent high-resolution structural analysis of lipids extracted from ΔmmpL8 revealed unexpected triacylated sulfolipids, suggesting that Chp1 is active in the absence of MmpL8, although at reduced levels (48).
Sap could be an additional component of the proposed scaffold. Although Sap is not absolutely essential for SL-1 biosynthesis, it appears to potentiate SL-1 levels and may confer specificity for sulfolipids over structurally similar glycolipids such as trehalose monomycolate and polyacyltrehalose. In this role, Sap may be functionally analogous to small integral membrane proteins that are substrate-specific components in bacterial vitamin transport (49). Close association between membrane-associated Chp1 and an MmpL8-Sap complex could aid transport of SL-1 away from Chp1. This action would prevent reverse hydrolysis, a Chp1 activity detected at low levels in vitro (supplemental Fig. S5C).
Chp1 catalysis of two successive acyl transfers is similar to the activity of the polyacyltrehalose enzyme PapA3, which esterifies trehalose with palmitic and mycolipenic acids (50). However, unlike the PapA enzymes, Chp1 does not use an activated thioester donor such as an acyl-CoA but rather catalyzes regioselective transesterification between two substrate molecules. This mechanism has precedent in M. tuberculosis with the antigen 85 complex (Ag85A, Ag85B, and Ag85C), a group of cell wall mycolyltransferases that synthesize trehalose dimycolate from two molecules of trehalose monomycolate (51). In comparison, the activity of Chp1 is more complex, with a combination of specificity and promiscuity that raises intriguing questions about how it achieves substrate recognition and chemical specificity. On the one hand, Chp1 is selective for diacyl over monoacyl sulfolipids and specific for the donor 3′-T2S position, yet it can accommodate two different triacyl regioisomers and catalyze ester formation at two chemically nonequivalent positions. As has been noted previously, the SL-1 and polyacyltrehalose biosynthetic loci are structurally similar (50). Indeed, the closest homolog of Chp1 is Chp2, encoded by rv1184c in the polyacyltrehalose locus (43% sequence identity), and preliminary results indicate that Chp2 is also essential for polyacyltrehalose biosynthesis.7
These data thus define a class of mechanistically similar glycolipid acyltransferases that comprise the Ag85 complex, Chp1, and possibly Chp2. Other members may include eight M. tuberculosis PE/PPE proteins that are the closest homologs of Chp1 and Chp2 (35). Although the PE/PPE protein family constitutes ∼10% of the M. tuberculosis genome, only Rv3097c (PE_PGRS63; LipY) has assigned enzymatic activity as a cell wall-associated triacylglycerol lipase (53). Whether our new insights into Chp1 acyltransferase activity will aid the functional assignment of these conserved proteins and whether they can be targeted as a group by inhibitory molecules such as THL await further investigation.
Supplementary Material
Acknowledgments
We thank Dr. Sloan Siegrist and Kimberly Sogi for helpful discussions and critical reading of the manuscript. The QB3/Chemistry Mass Spectrometry Facility at the University of California at Berkeley acknowledges National Institutes of Health Shared Instrumentation Grant 1S10RR017786-01. γ-Irradiated H37Rv whole cells and anti-KatG antibody were kindly provided as part of National Institutes of Health Contract HHSN266200400091C from NIAID (“Tuberculosis Vaccine Testing and Research Materials”), which was awarded to Colorado State University.
This work was supported, in whole or in part, by National Institutes of Health Grant AI51622 (to C. R. B.) and by Shared Instrumentation Grant 1S10RR017786-01 (to the QB3/Chemistry Mass Spectrometry Facility).

This article contains supplemental “Experimental Procedures” and Figs. S1–S23.
J. C. Seeliger, unpublished data.
- SL-1
- sulfolipid-1
- T2S
- trehalose 2-sulfate
- THL
- tetrahydrolipstatin
- SL-A
- 6,6′-di-O-(2-methylarachidoyl)-3′-O-(2-methylstearoyl)-2′-O-palmitoyltrehalose 2-O-sulfate
- T2S-PS
- 2-O-palmitoyl-3-O-stearoyl-α,α-d-trehalose
- aa
- amino acid(s)
- AP
- alkaline phosphatase
- CLP
- cutinase-like protein.
REFERENCES
- 1. Middlebrook G., Coleman C. M., Schaefer W. B. (1959) Sulfolipid from virulent tubercle bacilli. Proc. Natl. Acad. Sci. U.S.A. 45, 1801–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gangadharam P. R., Cohn M. L., Middlebrook G. (1963) Infectivity, pathogenicity, and sulfolipid fraction of some Indian and British strains of tubercle bacilli. Tubercle 44, 452–455 [DOI] [PubMed] [Google Scholar]
- 3. Glickman M. S., Cox J. S., Jacobs W. R. (2000) A novel mycolic acid cyclopropane synthetase is required for cording, persistence, and virulence of Mycobacterium tuberculosis. Mol. Cell 5, 717–727 [DOI] [PubMed] [Google Scholar]
- 4. Cox J. S., Chen B., McNeil M., Jacobs W. R. (1999) Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature 402, 79–83 [DOI] [PubMed] [Google Scholar]
- 5. Goren M. B., D'Arcy Hart P., Young M. R., Armstrong J. A. (1976) Prevention of phagosome-lysosome fusion in cultured macrophages by sulfatides of Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 73, 2510–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pabst M. J., Gross J. M., Brozna J. P., Goren M. B. (1988) Inhibition of macrophage priming by sulfatide from Mycobacterium tuberculosis. J. Immunol. 140, 634–640 [PubMed] [Google Scholar]
- 7. Zhang L., English D., Andersen B. R. (1991) Activation of human neutrophils by Mycobacterium tuberculosis-derived sulfolipid-1. J. Immunol. 146, 2730–2736 [PubMed] [Google Scholar]
- 8. Kato M., Goren M. B. (1974) Synergistic action of cord factor and mycobacterial sulfatides on mitochondria. Infect. Immun. 10, 733–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Goren M. B., Vatter A. E., Fiscus J. (1987) Polyanionic agents as inhibitors of phagosome-lysosome fusion in cultured macrophages: evolution of an alternative interpretation. J. Leukocyte Biol. 41, 111–121 [DOI] [PubMed] [Google Scholar]
- 10. Zhang L., Goren M. B., Holzer T. J., Andersen B. R. (1988) Effect of Mycobacterium tuberculosis-derived sulfolipid-1 on human phagocytic cells. Infect. Immun. 56, 2876–2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brozna J. P., Horan M., Rademacher J. M., Pabst K. M., Pabst M. J. (1991) Monocyte responses to sulfatide from Mycobacterium tuberculosis: inhibition of priming for enhanced release of superoxide, associated with increased secretion of interleukin-1 and tumor necrosis factor α, and altered protein phosphorylation. Infect. Immun. 59, 2542–2548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brodin P., Poquet Y., Levillain F., Peguillet I., Larrouy-Maumus G., Gilleron M., Ewann F., Christophe T., Fenistein D., Jang J., Jang M. S., Park S. J., Rauzier J., Carralot J. P., Shrimpton R., Genovesio A., Gonzalo-Asensio J. A., Puzo G., Martin C., Brosch R., Stewart G. R., Gicquel B., Neyrolles O. (2010) High-content phenotypic cell-based visual screen identifies Mycobacterium tuberculosis acyltrehalose-containing glycolipids involved in phagosome remodeling. PLoS Pathog. 6, e1001100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Converse S. E., Mougous J. D., Leavell M. D., Leary J. A., Bertozzi C. R., Cox J. S. (2003) MmpL8 is required for sulfolipid-1 biosynthesis and Mycobacterium tuberculosis virulence. Proc. Natl. Acad. Sci. U.S.A. 100, 6121–6126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Domenech P., Reed M. B., Dowd C. S., Manca C., Kaplan G., Barry C. E., 3rd (2004) The role of MmpL8 in sulfatide biogenesis and virulence of Mycobacterium tuberculosis. J. Biol. Chem. 279, 21257–21265 [DOI] [PubMed] [Google Scholar]
- 15. Kumar P., Schelle M. W., Jain M., Lin F. L., Petzold C. J., Leavell M. D., Leary J. A., Cox J. S., Bertozzi C. R. (2007) PapA1 and PapA2 are acyltransferases essential for the biosynthesis of the Mycobacterium tuberculosis virulence factor sulfolipid-1. Proc. Natl. Acad. Sci. U.S.A. 104, 11221–11226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mougous J. D., Petzold C. J., Senaratne R. H., Lee D. H., Akey D. L., Lin F. L., Munchel S. E., Pratt M. R., Riley L. W., Leary J. A., Berger J. M., Bertozzi C. R. (2004) Identification, function, and structure of the mycobacterial sulfotransferase that initiates sulfolipid-1 biosynthesis. Nat. Struct. Mol. Biol. 11, 721–729 [DOI] [PubMed] [Google Scholar]
- 17. Rousseau C., Turner O. C., Rush E., Bordat Y., Sirakova T. D., Kolattukudy P. E., Ritter S., Orme I. M., Gicquel B., Jackson M. (2003) Sulfolipid deficiency does not affect the virulence of Mycobacterium tuberculosis H37Rv in mice and guinea pigs. Infect. Immun. 71, 4684–4690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gilleron M., Stenger S., Mazorra Z., Wittke F., Mariotti S., Böhmer G., Prandi J., Mori L., Puzo G., De Libero G. (2004) Diacylated sulfoglycolipids are novel mycobacterial antigens stimulating CD1-restricted T cells during infection with Mycobacterium tuberculosis. J. Exp. Med. 199, 649–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guiard J., Collmann A., Garcia-Alles L. F., Mourey L., Brando T., Mori L., Gilleron M., Prandi J., De Libero G., Puzo G. (2009) Fatty acyl structures of Mycobacterium tuberculosis sulfoglycolipid govern T cell response. J. Immunol. 182, 7030–7037 [DOI] [PubMed] [Google Scholar]
- 20. Sirakova T. D., Thirumala A. K., Dubey V. S., Sprecher H., Kolattukudy P. E. (2001) The Mycobacterium tuberculosis pks2 gene encodes the synthase for the hepta- and octamethyl-branched fatty acids required for sulfolipid synthesis. J. Biol. Chem. 276, 16833–16839 [DOI] [PubMed] [Google Scholar]
- 21. Gokhale R. S., Saxena P., Chopra T., Mohanty D. (2007) Versatile polyketide enzymatic machinery for the biosynthesis of complex mycobacterial lipids. Nat. Prod. Rep. 24, 267–277 [DOI] [PubMed] [Google Scholar]
- 22. Tseng T. T., Gratwick K. S., Kollman J., Park D., Nies D. H., Goffeau A., Saier M. H. (1999) The RND permease superfamily: an ancient, ubiquitous, and diverse family that includes human disease and development proteins. J. Mol. Microbiol. Biotechnol. 1, 107–125 [PubMed] [Google Scholar]
- 23. Langston S., Bernet B., Vasella A. (1994) Temporary protection and activation in the regioselective synthesis of saccharide sulfates. Helv. Chim. Acta 77, 2341–2353 [Google Scholar]
- 24. Leigh C. D., Bertozzi C. R. (2008) Synthetic studies toward Mycobacterium tuberculosis sulfolipid-I. J. Org. Chem. 73, 1008–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cole S. T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S. V., Eiglmeier K., Gas S., Barry C. E., 3rd, Tekaia F., Badcock K., Basham D., Brown D., Chillingworth T., Connor R., Davies R., Devlin K., Feltwell T., Gentles S., Hamlin N., Holroyd S., Hornsby T., Jagels K., Krogh A., McLean J., Moule S., Murphy L., Oliver K., Osborne J., Quail M. A., Rajandream M. A., Rogers J., Rutter S., Seeger K., Skelton J., Squares R., Squares S., Sulston J. E., Taylor K., Whitehead S., Barrell B. G. (1998) Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544 [DOI] [PubMed] [Google Scholar]
- 26. Kelley L. A., Sternberg M. J. (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 27. Larsen M., Biermann K. E., Tandberg S., Hsu T., Jacobs W. R., Jr. (2007) Genetic manipulation of Mycobacterium tuberculosis. Curr. Protoc. Microbiol. 6, Unit 10A.2.1–10A.2.21 [DOI] [PubMed] [Google Scholar]
- 28. Holsclaw C. M., Sogi K. M., Gilmore S. A., Schelle M. W., Leavell M. D., Bertozzi C. R., Leary J. A. (2008) Structural characterization of a novel sulfated menaquinone produced by stf3 from Mycobacterium tuberculosis. ACS Chem. Biol. 3, 619–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rezwan M., Lanéelle M. A., Sander P., Daffé M. (2007) Breaking down the wall: fractionation of mycobacteria. J. Microbiol. Methods 68, 32–39 [DOI] [PubMed] [Google Scholar]
- 30. Heinz C., Niederweis M. (2000) Selective extraction and purification of a mycobacterial outer membrane protein. Anal. Biochem. 285, 113–120 [DOI] [PubMed] [Google Scholar]
- 31. Niederweis M., Ehrt S., Heinz C., Klöcker U., Karosi S., Swiderek K. M., Riley L. W., Benz R. (1999) Cloning of the mspA gene encoding a porin from Mycobacterium smegmatis. Mol. Microbiol. 33, 933–945 [DOI] [PubMed] [Google Scholar]
- 32. Busso D., Delagoutte-Busso B., Moras D. (2005) Construction of a set Gateway-based destination vectors for high-throughput cloning and expression screening in Escherichia coli. Anal. Biochem. 343, 313–321 [DOI] [PubMed] [Google Scholar]
- 33. Sondén B., Kocíncová D., Deshayes C., Euphrasie D., Rhayat L., Laval F., Frehel C., Daffé M., Etienne G., Reyrat J. M. (2005) Gap, a mycobacterial specific integral membrane protein, is required for glycolipid transport to the cell surface. Mol. Microbiol. 58, 426–440 [DOI] [PubMed] [Google Scholar]
- 34. Vrljic M., Garg J., Bellmann A., Wachi S., Freudl R., Malecki M. J., Sahm H., Kozina V. J., Eggeling L., Saier M. H., Jr. (1999) The LysE superfamily: topology of the lysine exporter LysE of Corynebacterium glutamicum, a paradyme for a novel superfamily of transmembrane solute translocators. J. Mol. Microbiol. Biotechnol. 1, 327–336 [PubMed] [Google Scholar]
- 35. Adindla S., Guruprasad L. (2003) Sequence analysis corresponding to the PPE and PE proteins in Mycobacterium tuberculosis and other genomes. J. Biosci. 28, 169–179 [DOI] [PubMed] [Google Scholar]
- 36. Sultana R., Tanneeru K., Guruprasad L. (2011) The PE-PPE domain in mycobacterium reveals a serine α/β hydrolase fold and function: an in-silico analysis. PLoS ONE 6, e16745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Parker S. K., Barkley R. M., Rino J. G., Vasil M. L. (2009) Mycobacterium tuberculosis Rv3802c encodes a phospholipase/thioesterase and is inhibited by the antimycobacterial agent tetrahydrolipstatin. PLoS ONE 4, e4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. West N. P., Chow F. M.., Randall E. J., Wu J., Chen J., Ribeiro J. M., Britton W. J. (2009) FASEB J. 23, 1694–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schué M., Maurin D., Dhouib R., Bakala N'Goma J. C., Delorme V., Lambeau G., Carrière F., Canaan S. (2010) Two cutinase-like proteins secreted by Mycobacterium tuberculosis show very different lipolytic activities reflecting their physiological function. FASEB J. 24, 1893–1903 [DOI] [PubMed] [Google Scholar]
- 40. Ghosh D., Sawicki M., Lala P., Erman M., Pangborn W., Eyzaguirre J., Gutierrez R., Jornvall H., Thiel D. J. (2001) Multiple conformations of catalytic serine and histidine in acetylxylan esterase at 0.90 Å. J. Biol. Chem. 276, 11159–11166 [DOI] [PubMed] [Google Scholar]
- 41. Patricelli M. P., Giang D. K., Stamp L. M., Burbaum J. J. (2001) Direct visualization of serine hydrolase activities in complex proteomes using fluorescent active site-directed probes. Proteomics 1, 1067–1071 [DOI] [PubMed] [Google Scholar]
- 42. Kremer L., de Chastellier C., Dobson G., Gibson K. J., Bifani P., Balor S., Gorvel J. P., Locht C., Minnikin D. E., Besra G. S. (2005) Identification and structural characterization of an unusual mycobacterial monomeromycolyl-diacylglycerol. Mol. Microbiol. 57, 1113–1126 [DOI] [PubMed] [Google Scholar]
- 43. Crellin P. K., Vivian J. P., Scoble J., Chow F. M., West N. P., Brammananth R., Proellocks N. I., Shahine A., Le Nours J., Wilce M. C., Britton W. J., Coppel R. L., Rossjohn J., Beddoe T. (2010) Tetrahydrolipstatin inhibition, functional analyses, and three-dimensional structure of a lipase essential for mycobacterial viability. J. Biol. Chem. 285, 30050–30060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Côtes K., Dhouib R., Douchet I., Chahinian H., de Caro A., Carrière F., Canaan S. (2007) Characterization of an exported monoglyceride lipase from Mycobacterium tuberculosis possibly involved in the metabolism of host cell membrane lipids. Biochem. J. 408, 417–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Low K. L., Rao P. S., Shui G., Bendt A. K., Pethe K., Dick T., Wenk M. R. (2009) Triacylglycerol utilization is required for regrowth of in vitro hypoxic nonreplicating Mycobacterium bovis bacillus Calmette-Guérin. J. Bacteriol. 191, 5037–5043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Deshayes C., Bach H., Euphrasie D., Attarian R., Coureuil M., Sougakoff W., Laval F., Av-Gay Y., Daffé M., Etienne G., Reyrat J. M. (2010) MmpS4 promotes glycopeptidolipid biosynthesis and export in Mycobacterium smegmatis. Mol. Microbiol. 78, 989–1003 [DOI] [PubMed] [Google Scholar]
- 47. Jain M., Cox J. S. (2005) Interaction between polyketide synthase and transporter suggests coupled synthesis and export of virulence lipid in M. tuberculosis. PLoS Pathog. 1, e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Layre E., De Paepe D., Larrouy-Maumus G., Vaubourgeix J., Mundayoor S., Lindner B., Puzo G., Gilleron M. (2011) Deciphering sulfoglycolipids of Mycobacterium tuberculosis. J. Lipid Res. 52, 1098–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rodionov D. A., Hebbeln P., Eudes A., ter Beek J., Rodionova I. A., Erkens G. B., Slotboom D. J., Gelfand M. S., Osterman A. L., Hanson A. D., Eitinger T. (2009) A novel class of modular transporters for vitamins in prokaryotes. J. Bacteriol. 191, 42–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hatzios S. K., Schelle M. W., Holsclaw C. M., Behrens C. R., Botyanszki Z., Lin F. L., Carlson B. L., Kumar P., Leary J. A., Bertozzi C. R. (2009) PapA3 is an acyltransferase required for polyacyltrehalose biosynthesis in Mycobacterium tuberculosis. J. Biol. Chem. 284, 12745–12751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Belisle J. T., Vissa V. D., Sievert T., Takayama K., Brennan P. J., Besra G. S. (1997) Role of the major antigen of Mycobacterium tuberculosis in cell wall biogenesis. Science 276, 1420–1422 [DOI] [PubMed] [Google Scholar]
- 52. Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. L. (2001) Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 305, 567–580 [DOI] [PubMed] [Google Scholar]
- 53. Deb C., Daniel J., Sirakova T. D., Abomoelak B., Dubey V. S., Kolattukudy P. E. (2006) A novel lipase belonging to the hormone-sensitive lipase family induced under starvation to utilize stored triacylglycerol in Mycobacterium tuberculosis. J. Biol. Chem. 281, 3866–3875 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.