Background: Deregulation of protein kinases MARK2 and PINK1 has been linked to neurodegenerative diseases.
Results: MARK2 was identified as an upstream regulator of PINK1.
Conclusion: The MARK2-PINK1 cascade provides new insights into the regulation of mitochondrial trafficking in neurons.
Significance: This protein kinase signaling axis might provide a link between neurodegenerative processes in Alzheimer and Parkinson diseases.
Keywords: Alzheimer Disease, Mitochondrial Transport, Neurodegeneration, Parkinson Disease, Protein Kinases
Abstract
The kinase MARK2/Par-1 plays key roles in several cell processes, including neurodegeneration such as Alzheimer disease by phosphorylating tau and detaching it from microtubules. In search of interaction partners of MARK2, we identified phosphatase and tensin homolog (PTEN)-induced kinase 1 (PINK1), which is important for the survival of neurons and whose mutations are linked to familial Parkinson disease (PD). MARK2 phosphorylated and activated the cleaved form of PINK1 (ΔN-PINK1; amino acids 156–581). Thr-313 was the primary phosphorylation site, a residue mutated to a non-phosphorylatable form (T313M) in a frequent variant of PD. Mutation of Thr-313 to Met or Glu in PINK1 showed toxic effects with abnormal mitochondrial distribution in neurons. MARK2 and PINK1 were found to colocalize with mitochondria and regulate their transport. ΔN-PINK1 promoted anterograde transport and increased the fraction of stationary mitochondria, whereas full-length PINK1 promoted retrograde transport. In both cases, MARK2 enhanced the effects. The results identify MARK2 as an upstream regulator of PINK1 and ΔN-PINK1 and provide insights into the regulation of mitochondrial trafficking in neurons and neurodegeneration in PD.
Introduction
Transport of vesicles and organelles is vital for cells, especially in neurons. Tau and other microtubule-associated proteins promote the assembly and stabilization of neuronal microtubule tracks and ensure microtubule-dependent transport. We showed previously that elevated tau leads to inhibition of traffic because of competition with motor proteins for microtubule binding. This inhibition can be relieved by MARK2,4 which phosphorylates microtubule-associated proteins and causes their detachment from microtubules (1). Consequently, MARK2 has a pivotal role in the regulation of microtubule dynamics and cellular transport.
Kinases of the MARK/Par-1 family perform diverse functions in neuronal polarity, transport, migration, and neurodegeneration (2). MARK/Par-1 comprises several domains, which offer multiple means of regulation. For example, the catalytic domain of MARK2 is activated by phosphorylation of Thr-208 by MARKK/TAO1 or LKB1 and inactivated by phosphorylation of Ser-212 by GSK3β (3–5). MARK2 activity can be inhibited by complex formation (e.g. with PAK5 (6)) or intramolecular interactions between domains (7, 8). Thus, in view of the importance of MARK for neuronal polarity and neurodegeneration, we set out to identify novel partners of MARK2 and herein describe the interaction of MARK2 with ΔN-PINK1 (aa 156–581)/full-length PINK1 (PINK1FL) and the effects of these kinases on mitochondrial transport and distribution.
PINK1 is a mitochondrially targeted serine/threonine kinase that promotes cell survival, particularly under conditions of oxidative/metabolic stress (9–11). PINK1 mutations are linked to PD and mostly result in loss of kinase activity. The molecular events responsible for neuronal death as well as physiological substrates or regulators of PINK1 are currently a matter of debate (12, 13). The mitochondrial localization of PINK1 (14, 15) and alterations in mitochondrial morphology, dynamics, and function caused by PINK1 deficiency (16–20) implicate a central role of PINK1 in normal mitochondrial function. But the location of PINK1 is still unclear. PINK1 has been reported to reside either in the inner mitochondrial membrane, the mitochondrial intermembrane space, the outer mitochondrial membrane (21, 22), or even the cytoplasm (15, 23). Other data support a model in which PINK1 spans the outer mitochondrial membrane with the N-terminal end inside the mitochondria and the C-terminal kinase domain facing the cytosol (24). This topology relies on a transmembrane domain located just after the mitochondrial targeting sequence (MTS) of PINK1 and allows substrate phosphorylation outside of the mitochondria. However, PINK1 protein is cleaved upon entry into the mitochondria to produce two N-terminally truncated protein fragments of 54 and 45 kDa (12) that localize more to the cytosolic than to the mitochondrial fraction (25). This suggests that cytosolic targets and signal transduction pathways may be modified by cleaved PINK1 to affect neuronal survival (23). The physiological function of these truncated PINK1 proteins is still unknown, although ΔN-PINK1 is quite stable, indicating that this is one of the predominant forms in the steady state (25). Here, we show that MARK2 interacted with and phosphorylated preferentially the 45-kDa fragment (termed ΔN-PINK1), thereby increasing its kinase activity. Both kinases colocalized with mitochondria in primary neurons, especially in the growth cone, and modified mitochondrial transport. The main site phosphorylated by MARK2 on ΔN-PINK1 was Thr-313, a frequent mutation site (T313M) in certain forms of familial PD (26). This suggests that failure of the MARK2-PINK1 signaling cascade contributes to the disease.
EXPERIMENTAL PROCEDURES
Plasmids
The full-length cDNA clone PINK1 wild-type (WT) pCMV-Sport6 was obtained from the RZPD (Ressourcenzentrum Primärdatenbank, Berlin, Germany). PINK1FL was amplified by PCR using oligonucleotides that introduce an NdeI restriction site at the start codon and an NheI restriction site behind the stop codon. The 45-kDa fragment ΔN-PINK1 was chosen to include residues 156–581 because this corresponds closely to the cellular fragment (whose exact cleavage site is not known (27)). The fragment was cloned into pEU-myc and examined by sequencing. PINK1 deletion and point mutants (e.g. ΔN-PINK1, aa 156–581) were generated by PCR using primers containing appropriate mutations and restriction sites. All PINK1-YFP constructs were generated by PCR using oligonucleotides that introduce a HindIII restriction site at the start codon and an NheI restriction site behind the stop codon of the PINK1 sequence. The fragment was cloned into the YFP-pShuttle fluorescence expression vector to generate adenovirus as described previously (28). PINK1FL-pVL-His was generated for recombinant expression in Sf9 cells using a modified pVL1393 vector (BD Pharmingen (3)) and the NdeI/NheI PINK1FL fragment. ΔN-PINK1-pGEX-4T-3 was generated via blunt-end cloning to express and purify recombinant protein. The ΔN-PINK1T313A-pGEX-4T-3, ΔN-PINK1T313E-pGEX-4T-3, ΔN-PINK1T313M-, ΔN-PINK1T313E-, and PINK1FL/T313M-pShuttle mutants were generated using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) and primers containing the appropriate sequence and point mutations. The cloning of MARK2-pGBKT7 constructs was reported previously (6). All sequences generated by PCR were verified by DNA sequencing.
Yeast Two-hybrid Analysis
The yeast two-hybrid screen and direct binding assays were performed according to the manufacturer's instructions for the MATCHMAKER GAL4 Two-Hybrid System (Clontech) and were described previously (6).
Protein Purification
Recombinant PINK1FL-pVL-His was expressed in Sf9 cells using the baculovirus system BaculoGold (BD Pharmingen). Recombinant GST-ΔN-PINK1, GST-ΔN-PINK1T313A, GST-ΔN-PINK1T313E, and His-MARK2 were expressed in Escherichia coli (BL21 DE3pLys; Merck) and purified with a nickel-nitrilotriacetic acid column (Qiagen, Hilden, Germany) for His-tagged proteins and with glutathione-Sepharose 4B beads (GE Healthcare) for GST-tagged proteins as described previously (3).
Immunoprecipitation and Immunoblot Analysis
Immunoprecipitation and immunoblot analysis were done by standard procedures as described previously (6).
Antibodies and Dyes
Antibodies, markers, immunofluorescence, and coupling methods are described in the supplemental Experimental Procedures.
Cell Fractionation
Preparations of mitochondrial fractions from N2A cells were kindly provided by A. M. Cuervo (Albert Einstein College, Bronx, NY) and are described elsewhere (29).
In Vitro Kinase Activity Assay
Purified PINK1 and MARK2 proteins were used at a concentration of 1 μm each. 150 μm histone H4 (Sigma-Aldrich) was used as a substrate for ΔN-PINK1. As substrate for MARK2, we used 150 μm TR1 peptide derived from the first repeat of tau protein containing the KXGS motif (30). The activity was assayed as described previously (3). Phosphorylated TR1 peptide was measured in a scintillation counter (Tricarb 1900 CA, Packard), and phosphorylated proteins were separated by SDS-PAGE and visualized by autoradiography using a bioimaging analyzer (BAS 3000, Raytest, Straubenhardt, Germany).
Preparation of in Vitro Phosphorylated PINK1 Peptides
The preparation of the in vitro phosphorylated PINK1 peptides is described in the supplemental Experimental Procedures.
Phosphopeptide Mapping
Two-dimensional phosphopeptide mapping was performed on thin-layer cellulose plates (Macherey-Nagel, Düren, Germany) according to Boyle et al. (31). Peptides were separated in the first dimension by electrophoresis at pH 1.9 in 88% formic acid, acetic acid, water (50:156:1794) and in the second dimension by thin-layer chromatography in n-butyl alcohol/pyridine/acetic acid/water (150:100:30:120, pH 3.5).
Mass Spectrometry
MALDI-TOF mass spectrometry was performed on a Voyager-DE STR time-of-flight instrument (Applied Biosystems, Darmstadt, Germany) equipped with a nitrogen laser operating at 337 nm. The peptides were analyzed in a positive ion delayed extraction reflector mode using α-cyano-4-hydroxycinnaminic acid (Sigma-Aldrich) (10 mg/ml in 0.05% TFA with 50% acetonitrile) as the UV light-absorbing matrix. Samples were dissolved in matrix solution and spotted onto the target plate, allowing them to air dry. All mass spectra were externally calibrated with standard peptides (New England Biolabs, Frankfurt, Germany) containing angiotensin, neurotensin, and various ACTH fragments. All spectra were analyzed using Data ExploreTM software (Applied Biosystems).
Cell Culture
Cell culture was performed with HEK293, CHO, PC12, and Sf9 cells following standard protocols at 37 °C with 5% CO2. Details are described in the supplemental Experimental Procedures.
Immunostaining, Labeling, and Live Cell Light Microscopy
Cells were fixed in 3.7% paraformaldehyde for 15 min at room temperature. Cells were then washed twice with PBS to remove paraformaldehyde. Afterward, cells were permeabilized with 80% ice-cold methanol for 5 min at −20 °C and washed three times with PBS. Cells were incubated with primary antibody for 1 h at 37 °C after blocking with 10% goat serum for 30 min at 37 °C to avoid unspecific binding. Cells were washed three times before incubating with fluorescence-labeled secondary antibody for 1 h at 37 °C. Cells were then mounted with Permafluor (Beckman Coulter, Krefeld, Germany) and observed with a 63× oil immersion objective on an LSM510 Meta confocal microscope (Zeiss, Jena, Germany) using lasers, beam splitters, and filters according to the fluorophores. For time lapse imaging of mitochondria, cells were plated on glass bottom dishes. Cells were kept at 37 °C by air heating and supplied with 5% CO2. Images were taken every 2–15 s adapted to the best experimental settings for tracking of mitochondria using a 63× objective, beam path, and laser settings for Cy5 for mitochondrial observation and YFP and CFP for confirming transfection of retinal ganglion cells (RGCs).
Data Analysis
The Zeiss LSM510 Image Browser software was used to track individual particles. We interpreted all stained mitochondria that were transported toward the axonal growth cone as anterograde and those that moved to the cell body as retrograde. Kymographs were then generated and analyzed using ImageJ Kymograph. The observation time varied from 5 to 10 min. To subtract the contribution from random Brownian motions, a mitochondrion was considered as actively transported if its velocity was faster than 0.3 μm/s (32). The following parameters were measured: velocity v (μm/s), distance of transport (run length L; μm), direction (anterograde or retrograde), and pausing time. Mitochondria with velocities below 0.3 μm/s were defined to be pausing. If a mitochondrion stood idle for more than 80s it was classified as stationary. Each movement (anterograde, retrograde, pause, or stationary) was counted as one independent value. For each transfection, three independent movies were evaluated. The averages were analyzed for statistical significance by one- or two-way analysis of variance. N indicates the number of experiments, and n is the number of observed mitochondrial movements. p values of <0.05, <0.01, and <0.001 are indicated with one, two, or three asterisks, respectively.
RESULTS
Identification of PINK1 as MARK2-interacting Protein
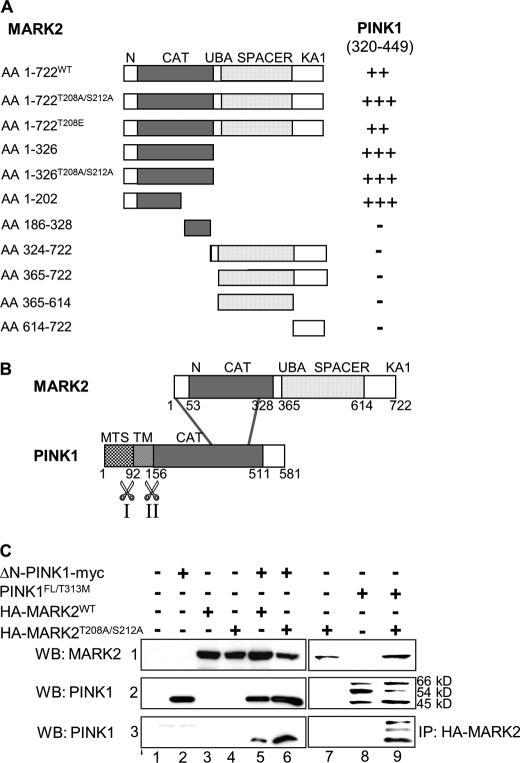
To identify new regulators or alternative substrates of MARK2, a human fetal brain cDNA library was screened using the MATCHMAKER yeast two-hybrid system. The bait was obtained by cloning the complete coding sequence of a kinase-dead MARK2 cDNA mutant into the two-hybrid vector. The MARK2T208A/S212A mutation was chosen because it mimics the unphosphorylated state of the kinase and keeps the substrate cleft in an open conformation (3, 8). Positive clones were further analyzed by DNA sequencing. One insert contained the DNA encoding part of the PINK1 kinase domain (aa 320–449). This suggested a possible interplay between these kinases in neurodegeneration because mutations in PINK1 are related to early onset Parkinsonism (33), and MARK2 is implicated in Alzheimer disease (2).
Fig. 1 illustrates the interactions between ΔN-PINK1 and MARK2. Different domains and mutants of MARK2 were checked in combination with the detected PINK1 fragment in direct two-hybrid interaction tests to map the binding domains of the kinases (Fig. 1A). Both proteins interact via their kinase domains (Fig. 1B). The C-terminal domain of MARK2 is not involved in binding the PINK1 fragment (aa 320–449) because this domain alone showed no interaction with the PINK1 construct. To confirm the results of the direct two-hybrid tests, a coimmunoprecipitation using HEK293 cells was carried out (Fig. 1C). We generated ΔN-PINK1 (residue 156–581) and different mutants to study its activity and interactions in the absence of possible influence by its adjacent domains (34). This construct corresponds closely to the most stable 45-kDa PINK1 cleavage product (25). We coexpressed wild-type or kinase-dead HA-tagged MARK2 in combination with ΔN-PINK1 or PINK1FL. When lysates of these cells were immunoprecipitated with anti-HA antibody, ΔN-PINK1 was detected in the HA-MARK2 immune complexes (Fig. 1C, row 3, lanes 5 and 6). The interaction with kinase-dead MARK2 was stronger than with MARK2WT, indicating that conformational changes of MARK2 in its activation loop may enhance the binding to ΔN-PINK1. An interaction of PINK1FL and MARK2 was not detectable by coimmunoprecipitation (data not shown). However, coexpression of kinase-dead HA-tagged MARK2 and a non-phosphorylatable form of PINK1FL (T313M mutant) led to binding of both kinases detectable by coimmunoprecipitation (Fig. 1C, row 3, lane 9), suggesting that the phosphorylation reaction modulates the strength of the interaction (for further details, see Figs. 3 and 4).
FIGURE 1.
Interaction between MARK2 and PINK1. A, mapping the interaction domains of MARK2 and PINK1 by direct yeast two-hybrid tests. Constructs are shown schematically, amino acids are in parentheses, and domains are distinguished by different shades. N, N-terminal header domain; CAT, catalytic domain; UBA, ubiquitin-associated domain; KA1, kinase-associated domain 1; TM, putative transmembrane domain. Results of the two-hybrid analysis are indicated by +++ (very strong interaction), ++ (strong), + (weak), and − (no interaction). Note that both kinases interact via their catalytic domains. B, interaction of the kinases MARK2 and ΔN-PINK1 via their catalytic domains. The putative cleavage site 1 (I) between aa 76 and 77 and site 2 (II) between aa 156 and 157 in PINK1 are indicated by scissors. Cleavage at aa 76/77 results in the 54-kDa fragment, and cleavage at aa 156/157 results in the 45-kDa ΔN-PINK1 fragment. C, HEK293 cells were transfected with plasmids encoding ΔN-PINK1-myc, PINK1FL/T313M, HA-tagged MARK2WT, or HA-MARK2T208A/S212A (inactive mutation) either singly (lanes 2-4, 7, and 8) or in combination (lane 5, 6, and 9). Empty pEU vector (lane 1) was used to make the total amount of transfected DNA equivalent. Cell lysates were immunoprecipitated with anti-HA antibody (row 3) and immunoblotted either with anti-HA antibody (row 1, WB: MARK) or anti-myc antibody (rows 2 and 3, WB: PINK1). ΔN-PINK1 immunoprecipitates more strongly with the inactive MARK2 mutant than with MARK2WT probably due to altered conformation after activation. In the case of PINK1FL, only a non-phosphorylatable form (T313M) binds sufficiently well to MARK2T208A/S212A. IP, immunoprecipitation; WB, Western blotting.
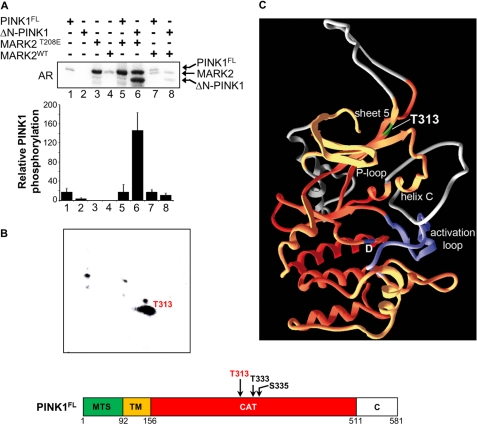
FIGURE 3.
MARK2 phosphorylates ΔN-PINK1 mainly on Thr-313. A, to examine the influence of MARK2 on PINK1 phosphorylation, both kinases (0.25 μg) were incubated in different combinations in the presence of [32P]ATP. The autoradiogram (AR) shows that active MARK2T208E strongly phosphorylates ΔN-PINK1 (lane 6). By contrast, MARK2WT does not phosphorylate ΔN-PINK1 (lanes 7 and 8), indicating that the phosphorylation of ΔN-PINK1 is dependent on MARK2 activity (lane 6; compare with lane 8). Because of the similar electrophoretic behavior of phosphorylated MARK2 and PINK1FL, the bands did not separate (lane 5). Value 5 of the bar graph was calculated by subtraction of the radioactive intensity of experiment 3 from that of experiment 5 and reveals only minor phosphorylation of PINK1FL. Error bars: S.E. from mean of 3 independent experiments. B, tryptic digest of the ΔN-PINK1 samples phosphorylated with recombinant active MARK2T208E was mapped by thin-layer electrophoresis (horizontal) followed by thin-layer chromatography (vertical). The main spot corresponds to peptides containing phosphorylated Thr-313, a residue known to be mutated to a non-phosphorylatable form in PD. The minor spots are in accordance with phosphorylated Thr-333 and Ser-335 of ΔN-PINK1 peptides. The identified phosphorylated residues are indicated in the schematic representation of PINK1FL. C, structural model of PINK1 by SwissModel using CaMK1 (Protein Data Bank code 1A06) as a template. The activation loop is shown in purple. Inserts that are not common in other kinases are gray; the catalytic aspartate is blue. Threonine 313, which is phosphorylated by MARK2, is located in β-strand 5 (green). Phosphorylation of this residue likely results in the interaction of strand 5 with helix C or its insert, thus stabilizing helix C. This helix is part of the scaffold that fixes the Mg-ATP underneath the P-loop and is a requirement for the activity of the kinase. CAT, catalytic domain; TM, transmembrane domain.
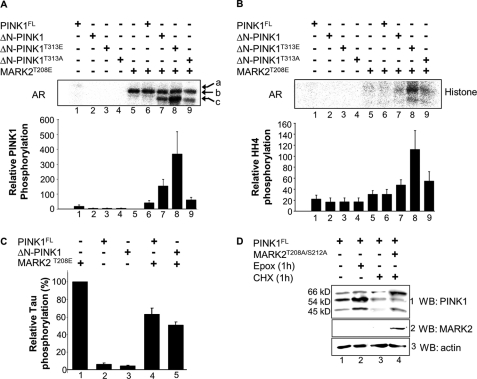
FIGURE 4.
MARK2 activates ΔN-PINK1 and stabilizes PINK1FL. A, to examine the response of the mutant ΔN-PINK1T313E and ΔN-PINK1T313A to active MARK2T208E, the kinases (0.25 μg) were incubated in different combinations in the presence of [32P]ATP. The autoradiogram (AR) shows that phosphorylation of ΔN-PINK1T313A is weak but still occurs, indicating that MARK2 also phosphorylates other sites of the kinase (lane 9). Phosphorylation of ΔN-PINK1T313E is strongly increased in comparison with the approach with non-mutated ΔN-PINK1 (lanes 7 and 8), indicating the importance of this residue for further phosphorylation events by MARK2. a, PINK1FL; b, MARK2; c, ΔN-PINK1. Error bars: S.E. from mean of 3 independent experiments. B, the effect of ΔN-PINK1 phosphorylation by MARK2T208E was measured via the phosphorylation of the in vitro substrate histone H4 by ΔN-PINK1 (28). Bacterially purified active GST-MARK2T208E phosphorylates histone H4 weakly (lane 5), but in combination with ΔN-PINK1, the histone phosphorylation is enhanced (lane 7). The ΔN-PINK1T313A mutant shows nearly the same MARK2-induced activation level (lane 7) as non-mutated ΔN-PINK1 (lane 9). However, activation of ΔN-PINK1 by active MARK2 is strongly enhanced by mutation of Thr-313 to glutamate (lane 8), indicating that this residue is important but not sufficient for kinase activation (compare lanes 3 and 8). Error bars: S.E. from mean of 3 independent experiments. C, to determine the impact of the PINK1-MARK2 interaction on Tau phosphorylation, both kinases (0.25 μg) were preincubated in different combinations in the presence of [32P]ATP, and the Tau peptide TR1 (30) was added as a substrate for MARK2. The kinase activity of MARK2T208E alone was normalized to 100% (lane 1). In combination with PINK1FL and ΔN-PINK1, the TR1 phosphorylation was reduced (lanes 4 and 5), probably because of inhibitory binding. Error bars: S.E. from mean of 3 independent experiments. D, CHO cells were transfected either with PINK1FL alone or in combination with MARK2T208A/S212A as indicated. 16 h after transfection, the cells were treated either with epoxomycin (Epox) to stop proteasomal degradation of proteins or with cycloheximide (CHX) to stop protein biosynthesis and harvested after 1 h. As a control, PINK1FL-transfected cells were treated with DMSO (lane 1). Cell lysates were immunoblotted either with anti-PINK1 antibody (row 1, WB: PINK1) or anti-MARK2 antibody (row 2, WB: MARK2). A β-actin immunoblot was used as a loading control (row 3). Both the proteasomal inhibitor (row 1, lane 2) and MARK2T208A/S212A (row 1, lane 4) increase the stability of PINK1FL and cleaved forms, whereby MARK2T208A/S212A particularly stabilizes PINKFL and the shortest cleaved 45-kDa PINK1 corresponding to ΔN-PINK1. WB, Western blot.
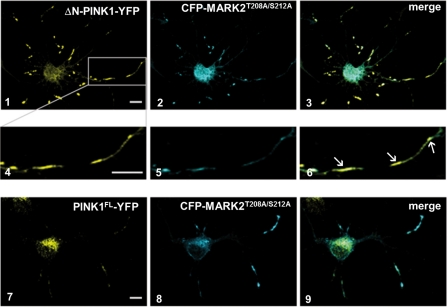
To examine the subcellular localization of MARK2 and ΔN-PINK1, primary rat cortical neurons were transfected with ΔN-PINK1-YFP and CFP-MARK2T208A/S212A by adenovirus and analyzed by immunofluorescence (Fig. 2). Both kinases have a vesicular appearance in axons and dendrites and a mainly cytoplasmic distribution in the cell soma. A strong colocalization was visible particularly in vesicle-like structures, confirming the two-hybrid and coimmunoprecipitation data (Figs. 2 and 4–6). Cortical neurons transfected with PINK1FL and MARK2T208A/S212A showed partial colocalization due to different expression patterns of PINK1FL and ΔN-PINK1 (Figs. 2 and 7–9). PINK1FL was more localized in the cell soma and to a lesser extent in axons and dendrites.
FIGURE 2.
MARK2 colocalizes to ΔN-PINK1 and PINK1FL. Subcellular localization of MARK2T208A/S212A and ΔN-PINK1 in primary rat cortex cells (E18) is shown. Neuronal cultures were transfected with MARK2T208A/S212A and ΔN-PINK1 adenoviruses, cultured for 24 h, and fixed. We chose the adenoviral expression system because the protein expression is weak, and the properties of the endogenous kinases are reflected by the exogenous kinases. The intracellular distribution of both proteins was visualized by fluorescence microscopy. A strong colocalization of ΔN-PINK1-YFP (yellow; panels 1 and 4) and CFP-MARK2T208A/S212A (inactive) (cyan; panels 2 and 5) in vesicle-like structures can be observed in the axon and dendrites of the neuron (merge; panels 3 and 6, arrows). PINK1FL-YFP (yellow; panels 7 and 9) is mainly expressed in the cell soma but also colocalizes with CFP-MARK2T208A/S212A (cyan; panels 8 and 9) in axons and dendrites. Scale bar, 10 μm.
FIGURE 5.
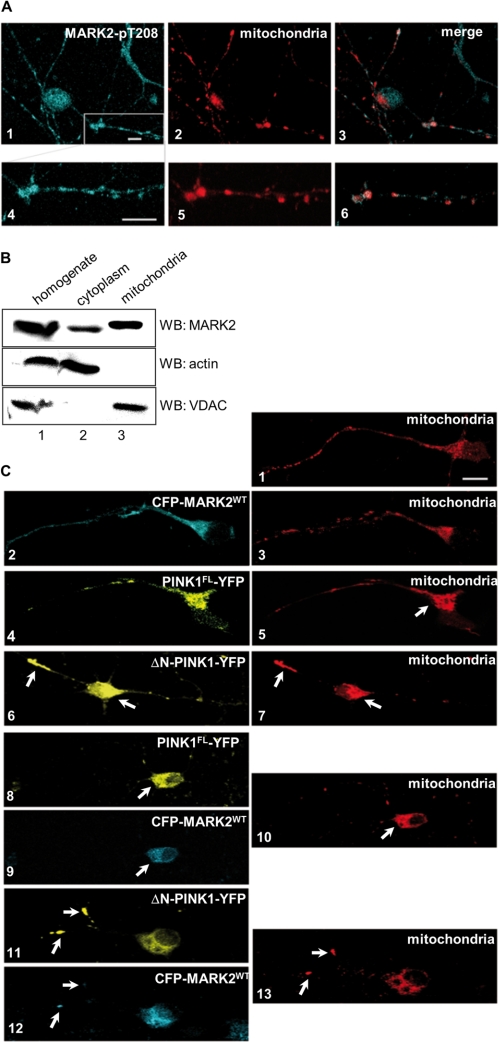
Subcellular localization of MARK in N2A cells and primary neurons. A, subcellular localization of endogenous active MARK2 and mitochondria was examined in primary rat cortex cells (E18). Neuronal cultures were stained for endogenous active MARK2 with the Thr(P)-208 antibody (cyan; panel 1) and mitochondria (red; panel 2). The kinase and the mitochondria are evenly distributed to the cell body, neurites, and growth cone and colocalize partially, especially in axons and dendrites (see magnification in panels 4–6). B, cytoplasmic and mitochondrial fractions or the whole cell homogenates of N2A cells were examined by the polyclonal MARK1–4 antibody SA4633 (raised against aa 1–377). The purity of the fractions was confirmed by Western blotting (WB) with anti-β-actin antibody (cytoplasmic marker) and the anti-voltage-dependent anion-selective channel (VDAC)/porin antibody (mitochondrial marker). MARK is found in the mitochondrial fraction (lane 3). C, subcellular distribution of mitochondria in primary rat cortex cells (E18). Untransfected and adenoviral CFP-MARK2WT-transfected neurons show normal and even distribution of mitochondria in neurites and soma (panels 1–3). Adenoviral transfection of the neurons with PINK1FL (yellow; panel 4) and staining for mitochondria show less mitochondria in the axon (red; panel 5). Expression of ΔN-PINK1-YFP (yellow; panel 6) and staining for mitochondria (red; panel 7) show less mitochondria in neurites and additionally mitochondrial accumulation (arrow), indicating transport inhibition. Cotransfection of PINK1FL (yellow; panel 8) and MARK2WT (cyan; panel 9) reveals a main localization of mitochondria in the cell body (red; panel 10, arrow), whereas cotransfection of ΔN-PINK1 (yellow; panel 11) and MARK2WT (cyan; panel 12) leads to local accumulations of mitochondria in axons and dendrites (red; panel 13, arrows). Scale bars, 10 μm.
FIGURE 6.
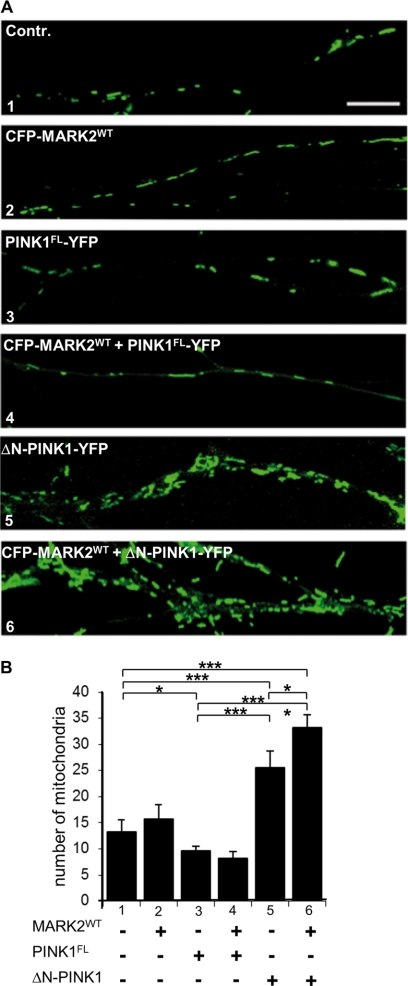
Density of mitochondria in RGC axons 20 h after transfection with PINK1FL, ΔN-PINK1, or MARK2WT by adenovirus. A, field of RGC axons either untransfected (Contr; panel 1) or adenoviral transfected with CFP-MARK2WT (panel 2), PINK1FL-YFP (panel 3), ΔN-PINK1-YFP (panel 5), or in combination with MARK2WT (panels 4 and 6) and stained for mitochondria. Untransfected and MARK2WT- or PINK1FL-transfected neurons show a normal and uniform distribution of mitochondria (panels 1–3). Mitochondria are frequent and tubular. By contrast, mitochondria in ΔN-PINK1-YFP-transfected neurons and even more in combination with MARK2WT show mitochondrial clustering/accumulation in swollen axons (panels 5 and 6). Scale bars, 10 μm. B, quantification of mitochondria measured in the proximal 50 μm of the axon. The number of mitochondria increases ∼2-fold in ΔN-PINK1-transfected cells (bar 5) compared with controls and MARK2WT-transfected cells (bars 1 and 2). This effect is further enhanced by coexpression of ΔN-PINK1 and MARK2WT (bar 6). PINK1FL-transfected and PINK1FL and MARK2WT-cotransfected cells show a lower density of axonal mitochondria (bars 3 and 4). Significant differences are indicated (p < 0.05; N ≥ 5; n ≥ 100; two-way analysis of variance). Error bars show S.E. p values of <0.05 and < 0.001 are indicated with one and three asterisks, respectively.
FIGURE 7.
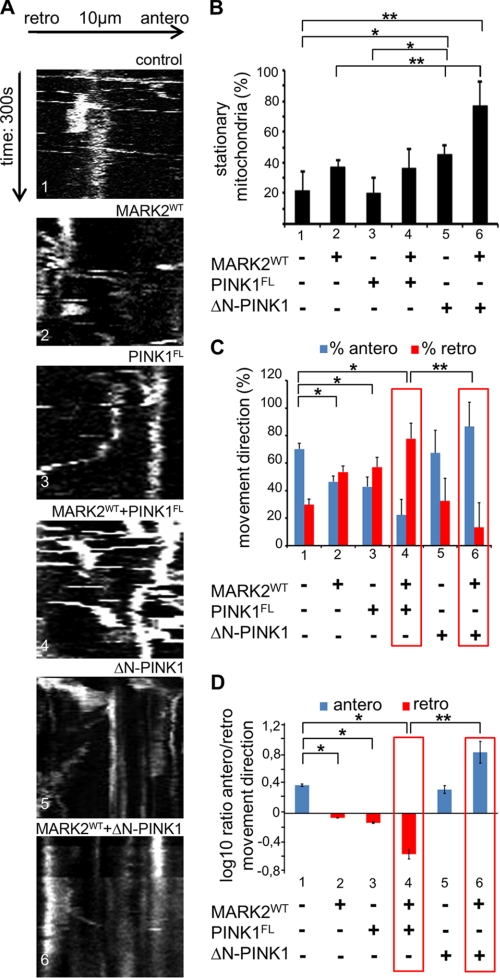
Analysis of mitochondrial movement in RGC axons 24 h after transfection with PINK1FL, ΔN-PINK1, or MARK2WT by adenovirus. A, kymographs showing that in untransfected cultures (panel 1) and MARK2WT (panel 2) and PINK1FL (panel 3) adenoviral transfected axons mitochondria can perform saltatory movements in both directions (horizontal arrow, 10 μm; vertical arrow, 300 s). MARK2WT and PINK1FL doubly transfected neurons show predominantly retrograde saltatory movements in comparison with control cells (panel 4). In axons transfected with ΔN-PINK1 alone (panel 5) or ΔN-PINK1 and MARK2WT (panel 6), mitochondria show almost no movement. B, quantification of stationary mitochondria in controls (bar 1) and adenoviral transfected RGCs (bars 2–6). In control cells, ∼20% of the mitochondria were stationary. Expression of ΔN-PINK1 or MARK2WT leads to a ∼2-fold increase of stationary mitochondria (bars 2 and 5). Coexpression of MARK2WT in combination with ΔN-PINK1 further enhances stationary mitochondria up to 75% (bar 6). Significant differences in B, C, and D are indicated (p < 0.05; N ≥ 5; n ≥ 100; two-way analysis of variance). Error bars show S.E. C, quantification of mitochondrial directionality in control cells and PINK1FL-, ΔN-PINK1-, and/or MARK2WT-transfected cells. In control cells, anterograde (antero) movements dominate (70%; blue bar 1). Expression of MARK2WT or PINK1FL reduces the amount of anterogradely moving mitochondria to ∼45% (blue bar 2) and increases retrograde (retro) transport so that both directions become comparable (red bars 2 and 3). This effect is further enhanced by MARK2WT because cotransfection of MARK2WT and PINK1FL changes the retrograde value to 80% (red bar 4). Cotransfection of MARK2WT and ΔN-PINK1 increases the anterograde mitochondrial movement (blue bar 6). Thus, MARK2WT amplifies the opposing effects of PINK1FL and ΔN-PINK1 (red boxed areas). D, ratios of anterograde versus retrograde mitochondrial movements shown in logarithmic scale. p values of <0.05 and <0.01 are indicated with one and two asterisks, respectively.
FIGURE 8.
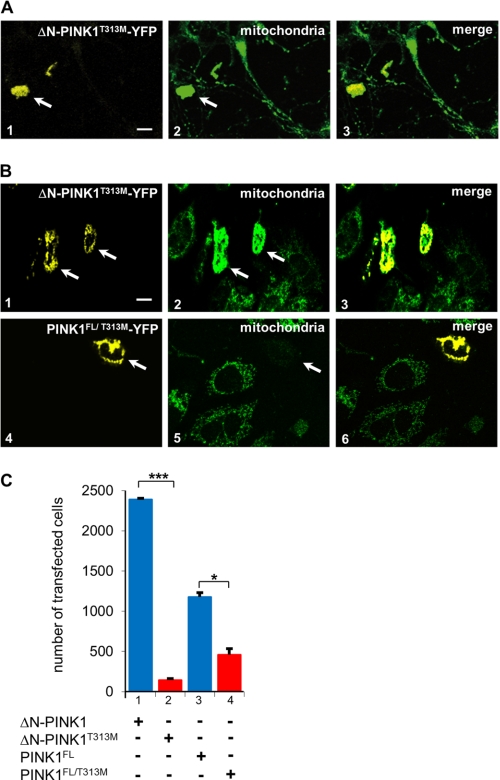
Effect of ΔN-PINK1T313M and PINK1FL/T313M on transfected cortex and CHO cells. A, subcellular distribution of mitochondria in primary rat cortex cells (E18) transfected by adenoviral ΔN-PINK1T313M. Expression of the PINK1 mutant is toxic for neurons. The cells show no neurites and a strong mitochondrial accumulation in the cell soma indicated by the arrow. B, subcellular distribution of ΔN-PINK1T313M, PINK1FL/T313M, and mitochondria in transfected CHO cells treated with Taxol. Expression of the PINK1 mutants is toxic for the cells. The cells are small, and in the case of transfection of ΔN-PINK1T313M, the mitochondria accumulate at the cell soma (B, panel 2, arrow). In the case of transfection of full-length mutant PINK1FL/T313M, the mitochondria are degraded (B, panel 5, arrow). Scale bars, 10 μm. C, quantification of the effects of ΔN-PINK1T313M and PINK1FL/T313M on the viability of transfected cells. The number of transfected cells per coverslip decreases significantly after PINK1FL/T313M expression (bar 4) compared with the cell number expressing PINK1FL (bar 3). This effect is much stronger in the case of ΔN-PINK1. Cells transfected with ΔN-PINK1T313M have survival rates of only 6% compared with cells expressing ΔN-PINK1 (bars 1 and 2). Significant differences are indicated (p < 0.05; N ≥ 5; n ≥ 100; two-way analysis of variance). Error bars show S.E. p values of <0.05 and < 0.001 are indicated with one and three asterisks, respectively.
FIGURE 9.
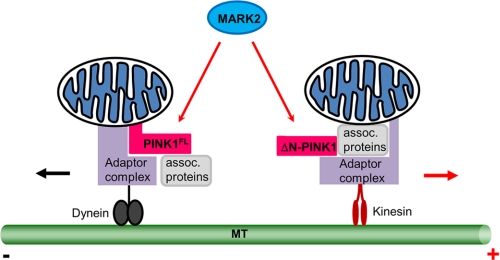
Model of interplay between MARK2 and PINK1FL/ΔN-PINK1 to regulate mitochondrial transport. The model shows microtubules (MT), motors, adaptor complexes, associated (assoc.) proteins, and mitochondria. The following observations are incorporated. Kinesin motors are linked to mitochondria by adaptor proteins like Miro and Milton (36) and regulate in association with ΔN-PINK1 the anterograde movement (red arrow), whereas Miro also has an effect on dynein-mediated retrograde movement (55) (black arrow). PINK1 is associated with the adaptor complex and exerts a regulatory role in mitochondrial transport (this work; Fig. 7). Active mitochondria tend to cause cleavage of PINK1FL to ΔN-PINK1 (42), which is phosphorylated at Thr-313 by MARK2 (this work; Fig. 3); both events promote anterograde movement by kinesin (Fig. 7, C, bars 6, and D, bar 6). In damaged mitochondria with low membrane potential, intact PINK1FL is not cleaved to ΔN-PINK1 and recruits parkin to mitochondria to promote mitophagy (45). Consistent with this, retrograde movement by dynein is promoted by PINK1FL and further increased by MARK2 (Fig. 7, C, bars 4, and D, bar 4).
MARK2 Phosphorylates ΔN-PINK1 at Thr-313 and Enhances Its Activity
Because the catalytic domains of the two kinases interact with one another, the question arose whether this influences their kinase activities. To examine this possibility, we performed in vitro phosphorylation assays to test the ability of MARK2 to phosphorylate PINK1FL or ΔN-PINK1 and vice versa. First, bacterially purified PINK1FL or ΔN-PINK1, alone or in combination with MARK2WT, and constitutively active MARK2T208E (3) were incubated with [32P]ATP.
PINK1FL or MARK2WT alone showed only slight autophosphorylation (Fig. 3A, lanes 1 and 4) that became strong in the case of MARK2T208E (Fig. 3A, lane 3). ΔN-PINK1 revealed no autophosphorylation (Fig. 3A, lane 2), but in combination with MARK2T208E, it was strongly phosphorylated (Fig. 3A, lane 6 and bar 6). The effect on PINK1FL was weak (Fig. 3A, bar 5), indicating a strong preference of MARK2 for the cleaved (cytoplasmic) ΔN-PINK1 or a conformational hindrance of PINK1FL in vitro. However, the results of the coimmunoprecipitation experiment with MARK2T208A/S212A and the non-phosphorylatable PINK1FL mutant indicate that MARK2 binds to the mutant but cannot complete the phosphorylation reaction in this situation.
To investigate the functional consequences of the phosphorylation of ΔN-PINK1 by MARK2, we first identified the phosphorylation sites by phosphopeptide mapping followed by mass spectrometry. Constitutively active MARK2T208E and ΔN-PINK1 were incubated with [32P]ATP. Phosphorylated ΔN-PINK1 was separated by SDS-PAGE followed by tryptic in-gel digestion and phosphopeptide enrichment. These peptides were mapped by thin-layer electrophoresis (horizontal) followed by ascending chromatography (vertical). The main spot shown in Fig. 3B corresponds to phosphorylated Thr-313 of ΔN-PINK1 peptides identified by MALDI-TOF MS analysis. The minor spots are in accordance with phosphorylated Thr-333 and Ser-335 of ΔN-PINK1 peptides. Because T313M is known as a frequent PD-associated mutation in the kinase domain of PINK1 (26), this residue may have an important function in the regulation of PINK1. The Thr-313, which is phosphorylated by MARK2, is located in β-strand 5 of PINK1 (Fig. 3C, marked in green). Phosphorylation of this residue could result in the interaction of this strand with helix C or its insert, thus stabilizing helix C. This helix is part of the scaffold that fixes the Mg-ATP underneath the P-loop and is a requirement for the activity of the kinase.
Because mutation of threonine to alanine prevents further phosphorylation/activation, whereas mutation to phosphorylation-mimicking glutamate produces active forms of many kinases (5), several PINK1 mutants were created replacing Thr-313 to confirm the specificity of this newly identified phosphorylation site. Bacterially purified ΔN-PINK1T313A was incubated alone or in combination with MARK2T208E in the presence of [32P]ATP. Indeed, this mutation caused a reduction (∼60%) of the phosphorylation of ΔN-PINK1 by active MARK2, but phosphorylation of ΔN-PINK1 still occurred, indicating that MARK2 also phosphorylates other sites of the kinase (Fig. 4A, compare lanes/bars 7 and 9). Although Thr-313 was mutated to glutamate in ΔN-PINK1, MARK2T208E increased further the phosphorylation by 67% in comparison with the approach with non-mutated ΔN-PINK1 (Fig. 4A, compare lanes/bars 7 and 8). This led to the assumption that Thr-313 is a “priming” phosphorylation site to prepare the ground for further phosphorylation of ΔN-PINK1.
To clarify whether this strong phosphorylation of ΔN-PINK1 by MARK2T208E influences the activity of ΔN-PINK1, phosphorylation assays using the artificial PINK1 substrate histone H4 were performed (34). The results indicate that MARK2T208E alone phosphorylates histone H4 only slightly (Fig. 4B, bar 5), but in combination with ΔN-PINK1, the substrate phosphorylation was enhanced (Fig. 4B, bar 7), indicating that MARK2 stimulates the kinase activity of ΔN-PINK1. ΔN-PINK1T313E was not constitutively active (Fig. 4B, bar 3), but MARK2T208E induced a 2.5-fold higher activation of the Glu mutant in comparison with the batch with non-mutated ΔN-PINK1 (Fig. 4B, bar 8). Accordingly, the ΔN-PINK1T313A mutant was not completely kinase-dead, but the activation could not be further enhanced by MARK2T208E in comparison with the non-mutated ΔN-PINK1 (Fig. 4B, bars 7 and 9). These results prove that phosphorylation of Thr-313 is important but not sufficient for full activation of ΔN-PINK1.
To exclude the possibility that MARK2 is activated by ΔN-PINK1 and thus causes the enhanced phosphorylation of histone, the kinase activity of MARK2 was measured by its phosphorylation of the tau peptide TR1 (derived from the first repeat sequence of tau protein (30)). Constitutively active MARK2T208E and PINK1FL or ΔN-PINK1 were mixed together with the TR1 peptide and incubated with [32P]ATP, and MARK2 activity was measured by the phosphorylation of TR1 using a scintillation counter. There was a ∼30–40% inhibition of MARK2 activity induced by PINK1FL or ΔN-PINK1 possibly due to binding of PINK1FL and ΔN-PINK1 (Fig. 4C, bars 4 and 5). A similar inhibitory mechanism was shown previously with MARK2 and PAK5 (6). PINK1FL also reduced MARK2 activity, although the MARK2-induced phosphorylation of PINK1FL was weak in vitro. To clarify whether PINK1FL is linked to MARK2 directly or indirectly, a protein stability assay was performed. CHO cells were individually transfected with PINK1FL alone or in combination with MARK2T208A/S212A (Fig. 4D). At 16 h after the transfection, the cells were treated for 1 h with 10 μm epoxomycin (proteasome inhibitor) or 50 μm cycloheximide (inhibitor of protein biosynthesis). Both the proteasomal inhibitor and MARK2T208A/S212A increased the stability of full-length and cleaved PINK1, arguing for a physiological relevance of this kinase-substrate interaction. MARK2T208A/S212A stabilized particularly PINK1FL and the shortest cleaved PINK1 corresponding to our ΔN-PINK1 (Fig. 4D, row 1, lane 4). Additionally, a phosphorylation assay was carried out. CHO cells were transfected with ΔN-PINK1 or PINK1FL alone or in combination with MARK2T208E. PINK1 was immunoprecipitated, and phosphorylation was detected by immunoblotting with anti-phosphoserine or anti-phosphothreonine antibody. As expected, ΔN-PINK1 was mainly phosphorylated at threonine sites in a MARK2-dependent manner (supplemental Fig. S1, lane 5), whereas PINK1FL coexpressed with MARK2 showed phosphorylation at serine- and threonine-specific sites (supplemental Fig. S1, lane 6). These results confirmed our assumption that even PINK1FL is modified by MARK2. In summary, we identified MARK2 as an upstream activating kinase of PINK1.
MARK2 Colocalizes with ΔN-PINK1 and Mitochondria in Primary Cortical Neurons
Endogenous PINK1 occurs both in the cytoplasm and on mitochondria despite the mitochondrial localization signal in PINK1 (23). But ΔN-PINK1 can also be found at mitochondria (35, 36). Nevertheless, the exact localization is still a matter of debate. MARK2 is a cytoplasmic kinase, but we had shown earlier that MARK2 has a dotted distribution with a diffuse background, suggesting that it is partially associated with vesicles (6, 37) (Fig. 2). To study the localization of both kinases and to determine in which cell compartment the interaction of both kinases occurs, we initially stained endogenous active MARK2 in primary cortical neurons using a phosphospecific MARK2 antibody (MARK Thr(P)-208) and costained for mitochondria (Fig. 5A, panels 1–6). Surprisingly, we found an extensive colocalization (up to 80%) of MARK2 not only with vesicles but also with mitochondria, particularly in axons and dendrites, emphasized at the higher magnification (Fig. 5A, panels 4–6). To confirm this association, we performed fractionation experiments (Fig. 5B) using whole cell homogenate (lane 1) and cytoplasmic (lane 2) and mitochondrial fractions (lane 3) of N2A cells. The mitochondrial fraction was verified for purity by detection of the mitochondrial transmembrane voltage-dependent anion-selective channel/porin protein and the absence of actin (cytoplasmic component). Fig. 5B clearly shows that MARK staining occurs in the cytoplasmic fraction and in the mitochondrial fraction (lanes 2 and 3), suggesting a possible role of MARK2 in mitochondrial function. To characterize the linkage of MARK2, PINK1FL, ΔN-PINK1, and mitochondria, primary cortical neurons were transfected with adenoviral YFP-MARK2, PINK1FL-YFP, or ΔN-PINK1-YFP and stained for mitochondria. As mentioned above, high expression of MARK2 leads to phosphorylation of tau, breakdown of the microtubule network, and cell death. The adenoviral expression system was chosen because the protein expression is weak and in these conditions hardly toxic. Fig. 5C illustrates the distribution of mitochondria in neurites and cell bodies. In untransfected primary cortical neurons, the mitochondria are evenly spread out in axons and concentrated at axon roots in the cell body (Fig. 5C, panel 1). Neurons transfected with CFP-MARK2 or PINK1FL-YFP by adenovirus show nearly the same mitochondrial distribution as untransfected cells (Fig. 5C, panels 2–5). Conspicuously, neurons transfected with PINK1FL show the highest mitochondrial density in the initial part of axons (Fig. 5C, panel 5, arrow). Expression of ΔN-PINK1 leads to a clear change of the distribution of mitochondria: they accumulate partially in neurites or are retained in the cell body (Fig. 5C, panels 6 and 7, arrows), indicating a transport disorder. Taken together, these results show that PINK1FL, ΔN-PINK1, and MARK2 colocalize in part with mitochondria and potentially influence mitochondrial transport.
Cotransfection of MARK2 and PINK1FL enhanced the concentration of mitochondria in the cell body (consistent with an inhibition of anterograde transport; see below). By contrast, mitochondria in neurons transfected with MARK2 and ΔN-PINK1 show local accumulations in axons (Fig. 5C, panels 11–13, arrows). The transfection rate was low, and transfected neurons tend to degenerate. It seems that overexpression of both kinases is toxic for the cell, and transport of mitochondria is impaired.
Movement of Mitochondria Is Perturbed in MARK2/ΔN-PINK1-transfected Retinal Ganglion Cells
A proper mitochondrial localization is essential for neuronal function because of the high demand of energy, especially in active growth cones (38). Considering that MARK2 is involved in the regulation of microtubule-dependent transport (1) and that PINK1 is important for the anterograde transport of mitochondria along microtubules (36), we next investigated mitochondrial traffic in axons. Chicken RGCs were transfected with adenovirus encoding PINK1FL or ΔN-PINK1 and MARK2WT either alone or in combination. The advantage of these cells is that the polarities of the axons are well defined so that transport directions are readily analyzed. 20 h after transfection, the neurons were stained with marker dye reporting on total mitochondria. Control neurons display a uniform distribution of mitochondria in axons similar to neurons transfected with MARK2WT (Fig. 6A, panels 1 and 2). Likewise, PINK1FL is mainly colocalized with uniformly distributed mitochondria. By contrast, mitochondria in neurons transfected with ΔN-PINK1 are mostly accumulated in axonal swellings characteristic of degeneration (Fig. 6A, panel 5), indicating an inhibition of axonal transport. This effect was enhanced by cotransfection of ΔN-PINK1 in combination with MARK2WT (Fig. 6A, panel 6). Quantification of mitochondrial density revealed that the number of axonal mitochondria increases ∼2-fold in ΔN-PINK1-transfected cells (Fig. 6B, bar 5) in comparison with controls (Fig. 6B, bar 1) or MARK2WT-transfected cells (Fig. 6B, bar 2) evaluated in the proximal 50 μm of the axons. The mitochondrial density in axons of PINK1FL-transfected cells dropped slightly (Fig. 6B, bar 3), and this effect was further enhanced by MARK2WT (Fig. 6B, bar 4). Cotransfection of ΔN-PINK1 with MARK2WT led to a much higher mitochondrial density compared with the effect of ΔN-PINK1 alone (Fig. 6B, bars 5 and 6), supporting the hypothesis of a MARK2WT/ΔN-PINK1-induced mitochondrial transport disorder.
Indeed, the change in distribution of mitochondria in axons corresponds to a change in mobility. We analyzed mitochondrial movement by live cell confocal microscopy. Untransfected cultures display the typical discontinuous saltatory movements of mitochondria with pauses often followed by a reversal of direction (Fig. 7, A, panel 1, and C, bar 1). In the case of MARK2WT-, PINK1FL,- or ΔN-PINK1-transfected cells, the saltatory movements remain mostly intact (Fig. 7A, panels 2, 3, and 5). By contrast, in MARK2WT- and ΔN-PINK1-cotransfected cells, the movement of mitochondria was strongly inhibited. At 20 h after transfection, the mitochondria become mostly stationary (Fig. 7A, panel 6), whereas cotransfection of MARK2WT and PINK1FL had no influence on mitochondrial motion (Fig. 7A, panel 4).
For time periods of 10–20 min, we recorded the parameters of speed, run length, direction, pausing time, and their dependence on PINK1FL, ΔN-PINK1, and MARK2WT. Quantification showed that in control cells 20% of the stained particles are stationary (Fig. 7B, bar 1). The fraction of immobile mitochondria did not change significantly in PINK1FL-transfected cells (Fig. 7B, bar 3) and increased to ∼35% in cells transfected with MARK2WT alone or PINK1FL in combination with MARK2WT (Fig. 7B, bars 2 and 4). A further increase of stationary mitochondria to ∼45% can be observed in ΔN-PINK1-transfected cells (Fig. 7B, bar 5). This effect is boosted to ∼76% in ΔN-PINK1 plus MARK2WT-transfected cells (Fig. 7B, bar 6). Thus, ΔN-PINK1 forces mitochondria to become stationary in cooperation with MARK2.
ΔN-PINK1 Promotes Anterograde and PINK1FL Promotes Retrograde Mitochondrial Transport, Both Depending on MARK2
The next question was whether the moving fraction of mitochondria displays changes in their transport direction dependent on PINK1FL, ΔN-PINK1, and MARK2 transfection. This was tested in adenoviral transfected retinal ganglion cell axons. Overall in control cultures, most of the mitochondria moved anterogradely (Fig. 7C, bar 1; ∼70%). By contrast, in MARK2WT- or in PINK1FL-transfected cells the antero- and retrograde mitochondrial transport rates were nearly balanced with a slight excess of retrograde events (Fig. 7C, bars 2 and 3). Cotransfection of MARK2WT and PINK1FL strongly enhanced retrograde events to nearly 80% (Fig. 7C, bar 4, red box). However, in contrast to PINK1FL, ΔN-PINK1 reversed these effects and returned the ratio to the wild-type prevalence of anterograde movements, especially in the presence of MARK2WT (Fig. 7C, compare bars 1, 5, and 6). In summary, our data suggest that PINK1FL, ΔN-PINK1, and MARK2WT regulate the mitochondrial transport. PINK1FL pushes the balance toward retrograde movements, whereas ΔN-PINK1 promotes anterograde movements; in both cases, MARK2WT enhances these effects (illustrated by the anterograde/retrograde ratios in Fig. 7D, bars 4 and 6, red boxes). The enhancement of anterograde movements by ΔN-PINK1 is consistent with the observed mitochondrial accumulations and axonal swellings (see Fig. 6A, panels 5 and 6).
Mutation of MARK2 Target Site Thr-313 in ΔN-PINK1 and PINK1FL Is Toxic in Primary Cortical and CHO Cells
PINK1Thr-313 is frequently mutated in PD, and it is the main phosphorylation site of the upstream kinase MARK2. To clarify the importance of the site, we created different PINK1Thr-313 mutants and observed their subcellular localization as well as their effects on mitochondria in neuronal and CHO cells. Expression of PINK1T313E or PINK1T313M was toxic for HEK293 and CHO cells. It was therefore not possible to harvest a sufficient amount of mutated PINK1 adenovirus because most of the transfected cells died. Expression of ΔN-PINK1T313M in primary cortical neurons led to abnormal mitochondrial accumulations in the cell soma and retraction of axons and dendrites (Fig. 8A, panels 1–3, arrows). Similar effects were observed even when the cellular microtubules were stabilized by Taxol (Fig. 8B, panels 1–3, arrow). Expression of PINK1FL/T313M in Taxol-treated CHO cells led to the degradation of mitochondria (Fig. 8B, panels 4–6, arrow). This mutant is not inactive and can still be phosphorylated by MARK2 (see Fig. 4A), explaining its toxicity. The quantification of viable transfected cells confirms these results. The survival rate drops to 40% in the case of PINK1FL/T313M and to 6% in the case of ΔN-PINK1T313M expression compared with the expression of the non-mutated form (see Fig. 8C). The T313E mutant was toxic to cells both for full-length and truncated PINK1 because this mutant showed a higher activity in combination with its activating kinase MARK2 (see Fig. 4, A and B).
DISCUSSION
This report describes the interaction of two protein kinases, MARK2 and PINK1, both of which are implicated in neurodegenerative diseases (Alzheimer disease and PD). One of the main known functions of MARK2 is to phosphorylate tau and detach it from microtubules. This is an early event in Alzheimer disease, suggesting that overactivation of MARK2 might play a role in the breakdown of microtubules and the interruption of axonal transport (39). Conversely, in neurons with tau elevated by transfection, MARK2 can rescue transport inhibition by phosphorylating and detaching tau from microtubules (1). To analyze the MARK2 signaling pathways in greater detail, we set out to identify new regulatory partners of MARK2. One conspicuous hit was PINK1, a mitochondrially targeted serine/threonine kinase with neuroprotective features (9, 10). Loss-of-function mutations in the PINK1 gene result in recessive heritable forms of parkinsonism (40). Recent studies have shown that PINK1 plays a crucial role in the regulation of mitochondrial dynamics, transport, and function (12, 41). Upon entry into mitochondria, full-length PINK1 protein can be cleaved by voltage-sensitive proteolysis (42). The most stable cleavage product is the 45-kDa fragment, corresponding roughly to ΔN-PINK1 (25). Cytoplasmic actions of ΔN-PINK1 are critical for cell survival because expression of ΔN-PINK1 protects neurons against the dopaminergic neurotoxin 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridin (23). However, the mechanism of this ΔN-PINK1 function is unknown. A major limitation in understanding the different functions of this protein kinase with dual localization is the lack of reliable PINK1 antibodies and low levels of endogenous expression (43). Therefore, much of the existing data has been based on the expression of tagged, exogenous PINK1.
We found an interaction of MARK2 especially with ΔN-PINK1 that is based on their catalytic domains (Fig. 1B). Consistent with this, primary cortical neurons show colocalization of overexpressed ΔN-PINK1 and MARK2 in dotted structures in axons and dendrites identified as mitochondria (Figs. 2 and 5A). PINK1FL is predominantly localized in the mitochondrial outer membrane with the kinase domain facing the cytoplasm (24), allowing binding and potentially further modification of PINKFL by MARK2.
Nevertheless, the questions arose why several cleavage fragments of PINK1 are found in different cell compartments and how they become active. Recent studies suggest that PINK1FL is active in the PINK1/Parkin pathway. Parkin is a cytoplasmic E3 ubiquitin ligase and can be phosphorylated by PINK1FL (44). Both proteins cooperate to control mitochondrial clearance, known as mitophagy (for reviews, see Refs. 38 and 45 and see Ref. 46). However, neither the exact molecular mechanism how PINK1 recruits Parkin nor the possible role of Parkin phosphorylation are known in detail.
The interaction of MARK2 with ΔN-PINK1 and PINK1FL has distinct consequences for their kinase activities in vitro. MARK2 phosphorylates and activates preferentially ΔN-PINK1 (Figs. 3A and 4, A and B). In the case of PINK1FL, its interaction with MARK2 during the phosphorylation reaction was too transient to be detectable by coimmunoprecipitation, but it could be observed with the non-phosphorylatable mutant PINK1FL/T313M, which locks the two partners in an inactive complex (an analogous situation was observed previously for the interaction between MARK2 and PAK5 (6)). In addition, MARK2 induced the phosphorylation of PINK1FL in transfected CHO cells (supplemental Fig. S1). Phosphopeptide mapping followed by mass spectrometry analysis revealed that MARK2-dependent phosphorylation of ΔN-PINK1 occurs mainly at residue Thr-313, which lies in the kinase domain and possibly leads to a stabilization of the scaffold that fixes the Mg-ATP, a requirement for the activity of the kinase (Fig. 3C). The mutation T313M in PINK1 is associated with familial PD (26); although mutation of this site to alanine decreased the phosphorylation of ΔN-PINK1 by MARK2, it was not completely abolished (Fig. 4A, lane/bar 9), and activation could still occur (Fig. 4B, lane/bar 9), probably via conformational changes and phosphorylation of additional sites (Fig. 3B). However, mutation of Thr-313 to glutamate further enhanced the phosphorylation and activation of ΔN-PINK1 by MARK2 (Fig. 4, A, lane/bar 8, and B, lane/bar 8). Thr-313 could be a priming phosphorylation site, changing the conformation of the kinase and preparing it for further modifications. The importance of this PINK1 phosphorylation site is further emphasized by the fact that expression of PINK1 (T313E) showed severe toxicity for cells (both CHO and neuronal cells). Mutation of the phosphorylation site to Met led to abnormal mitochondrial accumulations in the cell soma (ΔN-PINK1T313M; Fig. 8B, panel 2, arrow) or degradation of mitochondria (PINK1FL/T313M; Fig. 8B, panel 5, arrow). The data argue that MARK2 is an upstream regulator of ΔN-PINK1 and suggest that failure of this cascade contributes to the development of PD. The mutation could also have effects on known PINK1 substrates like Omi/HtrA2 or TRAP-1 (22, 30), but it is questionable whether these proteins are phosphorylated by ΔN-PINK1 under physiological conditions because both are mitochondrial and not cytoplasmic.
Within neurons, endogenous PINK1 and MARK2 partly colocalize on mitochondria, especially in axons and dendrites (Fig. 5, A and C, and supplemental Fig. S2). Fractionation studies revealed that endogenous MARK is found in the mitochondrial fraction of N2a cells (Fig. 5B). From these findings, we hypothesize that MARK2 may be recruited to mitochondria via ΔN-PINK1 and/or PINK1FL. This would alter the mitochondrial distribution because MARK2 in combination with PINK1 changes mitochondrial transport parameters (Figs. 5–7). New aspects on the relationship between PINK1 and mitochondrial transport regulation have been published recently (47). PINK1 phosphorylates the GTPase Miro, a component of a kinesin-adaptor complex, and thus induces its Parkin-dependent degradation. The resulting decrease in mitochondrial movement may represent a quality control mechanism of defective mitochondria (47). In our hands, particularly ΔN-PINK1 induced an arrest of mitochondrial motility, which was even further enhanced by MARK2 coexpression (Fig. 7B). In addition, ΔN-PINK1 increased anterograde movement and eventually led to enhanced mitochondrial density in axons. MARK2 reinforced this effect, possibly by phosphorylation and activation of ΔN-PINK1. Consistent with this, a high membrane potential promotes the anterograde transport of mitochondria (48) and also promotes the proteolysis of full-length PINK1 into ΔN-PINK1 (42). Conversely, retrograde transport is favored for mitochondria with low membrane potential destined for mitophagy (49); in this case, the cleavage of PINK1FL is inhibited in agreement with the transport characteristics induced by PINK1FL + MARK2 (Fig. 7). Mounting evidence indicates that Parkin is recruited via PINK1FL to defective mitochondria, inducing their degradation (for a review, see Ref. 50). Thus, Parkin-decorated mitochondria assemble as large clusters primarily in the lysosome-rich perinuclear area (42, 51). This effect seems to be influenced by MARK2 (Fig. 7, C and D). MARK2 could phosphorylate PINK1FL and consequently enhance the binding and possibly the phosphorylation of Parkin and Miro by PINK1FL. This is also suggested by the results of a protein stability assay. MARK2T208/S212A enhanced particularly the stability of PINK1FL (Fig. 4D). Conversely, healthy mitochondria promote the cleavage of PINK1FL, leading to high levels of ΔN-PINK1, which is not able to translocate Parkin to impaired mitochondria (51). This suppresses mitophagy and could explain why mitochondria accumulate in the cell soma of ΔN-PINK1T313M-transfected cells (Fig. 8).
Mitochondria are transported along microtubules by motor proteins kinesin (forward) and dynein (reverse). The kinesin-adaptor complex attached to the outer membrane comprises the GTPase Miro, kinesin heavy chain, and the adaptor protein Milton (52). PINK1FL is also attached to this complex, and even ΔN-PINK1 can be targeted to it despite the lack of the mitochondrial targeting sequence (24, 36). These observations argue that ΔN-PINK1 can participate in the control of anterograde mitochondrial trafficking and influence kinesin indirectly via Milton, consistent with the enhanced anterograde transport in cells overexpressing ΔN-PINK1 (Fig. 7, C and D). This assumption is supported by the observation that PINK1ΔMTS (equivalent to our ΔN-PINK1) has much stronger effects on anterograde transport compared with the retrograde transport of mitochondria (47).
In summary, our results suggest a model with PINK1 as a molecular switch between anterograde and retrograde mitochondrial transport (Fig. 9). A related mechanism has been shown for huntingtin, which controls the direction of vesicular transport in neurons (53). We propose that transport direction of neuronal mitochondria is regulated by PINK1 cleavage dependent on the mitochondrial membrane potential and PINK1 binding/phosphorylation by MARK2. Alterations in mitochondrial homeostasis have been implicated as an important source of many neurodegenerative diseases. Suppression of PINK1 kinase activity and/or down-regulation of PINK1 transcription contributes to PD pathogenesis, but enhanced PINK1 kinase activity also induces neuronal cell death. A similar relationship was demonstrated recently for the leucine-rich repeat kinase 2 (LRRK2), which is also involved in PD (54). Together, our findings confirm the importance of a tight regulation of PINK1 and demonstrate that PINK1FL and ΔN-PINK1 have distinct functions regarding mitochondrial transport, depending on MARK2.
Supplementary Material
Acknowledgments
We are grateful to Jacek Biernat for adenoviral constructs of MARK2 and PINK1; Edda Thies for introduction to primary cell culture and mitochondrial transport experiments; Alexander Marx for help with data analysis; Cindy Johne for some PINK1 constructs; Xiaoyu Li for help with confocal microscopy; Yipeng Wang, Ulrike Krüger, Konstanze Winklhofer (University of Munich), and Eckhard Mandelkow for discussions during this work; and Sabrina Hübschmann for excellent technical assistance. We thank P. Seubert (Elan Pharmaceuticals, South San Francisco, CA) for providing the 12E8 antibody and A. M. Cuervo (Albert Einstein College, Bronx, NY) for providing preparations of subcellular fractions from N2A cells.
This work was supported in part by the Deutsche Forschungsgemeinschaft, Bundesministerium für Bildung und Forschung (Kompetenznetz Degenerativer Demenzen), and European Union (FP7-MEMOSAD project).

This article contains supplemental Experimental Procedures and Figs. S1 and S2.
- MARK
- microtubule-associated protein/microtubule affinity-regulating kinase (isoforms 1–4)
- aa
- amino acid(s)
- CFP
- cyan fluorescent protein
- FL
- full-length
- MTS
- mitochondrial targeting sequence
- PD
- Parkinson disease
- PINK1
- PTEN-induced kinase 1
- PTEN
- phosphatase and tensin homolog
- RGC
- retinal ganglion cell.
REFERENCES
- 1. Thies E., Mandelkow E. M. (2007) Missorting of tau in neurons causes degeneration of synapses that can be rescued by the kinase MARK2/Par-1. J. Neurosci. 27, 2896–2907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Matenia D., Mandelkow E. M. (2009) The tau of MARK: a polarized view of the cytoskeleton. Trends Biochem. Sci. 34, 332–342 [DOI] [PubMed] [Google Scholar]
- 3. Timm T., Li X. Y., Biernat J., Jiao J., Mandelkow E., Vandekerckhove J., Mandelkow E. M. (2003) MARKK, a Ste20-like kinase, activates the polarity-inducing kinase MARK/PAR-1. EMBO J. 22, 5090–5101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Timm T., Balusamy K., Li X., Biernat J., Mandelkow E., Mandelkow E. M. (2008) Glycogen synthase kinase (GSK) 3β directly phosphorylates Serine 212 in the regulatory loop and inhibits microtubule affinity-regulating kinase (MARK) 2. J. Biol. Chem. 283, 18873–18882 [DOI] [PubMed] [Google Scholar]
- 5. Lizcano J. M., Göransson O., Toth R., Deak M., Morrice N. A., Boudeau J., Hawley S. A., Udd L., Mäkelä T. P., Hardie D. G., Alessi D. R. (2004) LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J. 23, 833–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Matenia D., Griesshaber B., Li X. Y., Thiessen A., Johne C., Jiao J., Mandelkow E., Mandelkow E. M. (2005) PAK5 kinase is an inhibitor of MARK/Par-1, which leads to stable microtubules and dynamic actin. Mol. Biol. Cell 16, 4410–4422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Elbert M., Rossi G., Brennwald P. (2005) The yeast par-1 homologs kin1 and kin2 show genetic and physical interactions with components of the exocytic machinery. Mol. Biol. Cell 16, 532–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Panneerselvam S., Marx A., Mandelkow E. M., Mandelkow E. (2006) Structure of the catalytic and ubiquitin-associated domains of the protein kinase MARK/Par-1. Structure 14, 173–183 [DOI] [PubMed] [Google Scholar]
- 9. Deng H., Jankovic J., Guo Y., Xie W., Le W. (2005) Small interfering RNA targeting the PINK1 induces apoptosis in dopaminergic cells SH-SY5Y. Biochem. Biophys. Res. Commun. 337, 1133–1138 [DOI] [PubMed] [Google Scholar]
- 10. Valente E. M., Abou-Sleiman P. M., Caputo V., Muqit M. M., Harvey K., Gispert S., Ali Z., Del Turco D., Bentivoglio A. R., Healy D. G., Albanese A., Nussbaum R., González-Maldonado R., Deller T., Salvi S., Cortelli P., Gilks W. P., Latchman D. S., Harvey R. J., Dallapiccola B., Auburger G., Wood N. W. (2004) Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science 304, 1158–1160 [DOI] [PubMed] [Google Scholar]
- 11. Wood-Kaczmar A., Gandhi S., Yao Z., Abramov A. Y., Abramov A. S., Miljan E. A., Keen G., Stanyer L., Hargreaves I., Klupsch K., Deas E., Downward J., Mansfield L., Jat P., Taylor J., Heales S., Duchen M. R., Latchman D., Tabrizi S. J., Wood N. W. (2008) PINK1 is necessary for long term survival and mitochondrial function in human dopaminergic neurons. PLoS One 3, e2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Deas E., Plun-Favreau H., Wood N. W. (2009) PINK1 function in health and disease. EMBO Mol. Med. 1, 152–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pogson J. H., Ivatt R. M., Whitworth A. J. (2011) Molecular mechanisms of PINK1-related neurodegeneration. Curr. Neurol. Neurosci. Rep. 11, 283–290 [DOI] [PubMed] [Google Scholar]
- 14. Beilina A., Van Der Brug M., Ahmad R., Kesavapany S., Miller D. W., Petsko G. A., Cookson M. R. (2005) Mutations in PTEN-induced putative kinase 1 associated with recessive parkinsonism have differential effects on protein stability. Proc. Natl. Acad. Sci. U.S.A. 102, 5703–5708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Weihofen A., Ostaszewski B., Minami Y., Selkoe D. J. (2008) Pink1 Parkinson mutations, the Cdc37/Hsp90 chaperones and Parkin all influence the maturation or subcellular distribution of Pink1. Hum. Mol. Genet. 17, 602–616 [DOI] [PubMed] [Google Scholar]
- 16. Clark I. E., Dodson M. W., Jiang C., Cao J. H., Huh J. R., Seol J. H., Yoo S. J., Hay B. A., Guo M. (2006) Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature 441, 1162–1166 [DOI] [PubMed] [Google Scholar]
- 17. Exner N., Treske B., Paquet D., Holmström K., Schiesling C., Gispert S., Carballo-Carbajal I., Berg D., Hoepken H. H., Gasser T., Krüger R., Winklhofer K. F., Vogel F., Reichert A. S., Auburger G., Kahle P. J., Schmid B., Haass C. (2007) Loss-of-function of human PINK1 results in mitochondrial pathology and can be rescued by parkin. J. Neurosci. 27, 12413–12418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gautier C. A., Kitada T., Shen J. (2008) Loss of PINK1 causes mitochondrial functional defects and increased sensitivity to oxidative stress. Proc. Natl. Acad. Sci. U.S.A. 105, 11364–11369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Park J., Lee S. B., Lee S., Kim Y., Song S., Kim S., Bae E., Kim J., Shong M., Kim J. M., Chung J. (2006) Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature 441, 1157–1161 [DOI] [PubMed] [Google Scholar]
- 20. Poole A. C., Thomas R. E., Andrews L. A., McBride H. M., Whitworth A. J., Pallanck L. J. (2008) The PINK1/Parkin pathway regulates mitochondrial morphology. Proc. Natl. Acad. Sci. U.S.A. 105, 1638–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gandhi S., Muqit M. M., Stanyer L., Healy D. G., Abou-Sleiman P. M., Hargreaves I., Heales S., Ganguly M., Parsons L., Lees A. J., Latchman D. S., Holton J. L., Wood N. W., Revesz T. (2006) PINK1 protein in normal human brain and Parkinson's disease. Brain 129, 1720–1731 [DOI] [PubMed] [Google Scholar]
- 22. Pridgeon J. W., Olzmann J. A., Chin L. S., Li L. (2007) PINK1 protects against oxidative stress by phosphorylating mitochondrial chaperone TRAP1. PLoS Biol. 5, e172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Haque M. E., Thomas K. J., D'Souza C., Callaghan S., Kitada T., Slack R. S., Fraser P., Cookson M. R., Tandon A., Park D. S. (2008) Cytoplasmic Pink1 activity protects neurons from dopaminergic neurotoxin MPTP. Proc. Natl. Acad. Sci. U.S.A. 105, 1716–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhou C., Huang Y., Shao Y., May J., Prou D., Perier C., Dauer W., Schon E. A., Przedborski S. (2008) The kinase domain of mitochondrial PINK1 faces the cytoplasm. Proc. Natl. Acad. Sci. U.S.A. 105, 12022–12027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lin W., Kang U. J. (2008) Characterization of PINK1 processing, stability, and subcellular localization. J. Neurochem. 106, 464–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mills R. D., Sim C. H., Mok S. S., Mulhern T. D., Culvenor J. G., Cheng H. C. (2008) Biochemical aspects of the neuroprotective mechanism of PTEN-induced kinase-1 (PINK1). J. Neurochem. 105, 18–33 [DOI] [PubMed] [Google Scholar]
- 27. Silvestri L., Caputo V., Bellacchio E., Atorino L., Dallapiccola B., Valente E. M., Casari G. (2005) Mitochondrial import and enzymatic activity of PINK1 mutants associated to recessive parkinsonism. Hum. Mol. Genet. 14, 3477–3492 [DOI] [PubMed] [Google Scholar]
- 28. Stamer K., Vogel R., Thies E., Mandelkow E., Mandelkow E. M. (2002) Tau blocks traffic of organelles, neurofilaments, and APP vesicles in neurons and enhances oxidative stress. J. Cell Biol. 156, 1051–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yu W. H., Cuervo A. M., Kumar A., Peterhoff C. M., Schmidt S. D., Lee J. H., Mohan P. S., Mercken M., Farmery M. R., Tjernberg L. O., Jiang Y., Duff K., Uchiyama Y., Näslund J., Mathews P. M., Cataldo A. M., Nixon R. A. (2005) Macroautophagy—a novel β-amyloid peptide-generating pathway activated in Alzheimer's disease. J. Cell Biol. 171, 87–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Drewes G., Ebneth A., Preuss U., Mandelkow E. M., Mandelkow E. (1997) MARK, a novel family of protein kinases that phosphorylate microtubule-associated proteins and trigger microtubule disruption. Cell 89, 297–308 [DOI] [PubMed] [Google Scholar]
- 31. Boyle W. J., van der Geer P., Hunter T. (1991) Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol. 201, 110–149 [DOI] [PubMed] [Google Scholar]
- 32. Trinczek B., Ebneth A., Mandelkow E. M., Mandelkow E. (1999) Tau regulates the attachment/detachment but not the speed of motors in microtubule-dependent transport of single vesicles and organelles. J. Cell Sci. 112, 2355–2367 [DOI] [PubMed] [Google Scholar]
- 33. Klein C., Schlossmacher M. G. (2007) Parkinson disease, 10 years after its genetic revolution: multiple clues to a complex disorder. Neurology 69, 2093–2104 [DOI] [PubMed] [Google Scholar]
- 34. Sim C. H., Lio D. S., Mok S. S., Masters C. L., Hill A. F., Culvenor J. G., Cheng H. C. (2006) C-terminal truncation and Parkinson's disease-associated mutations down-regulate the protein serine/threonine kinase activity of PTEN-induced kinase-1. Hum. Mol. Genet. 15, 3251–3262 [DOI] [PubMed] [Google Scholar]
- 35. Plun-Favreau H., Klupsch K., Moisoi N., Gandhi S., Kjaer S., Frith D., Harvey K., Deas E., Harvey R. J., McDonald N., Wood N. W., Martins L. M., Downward J. (2007) The mitochondrial protease HtrA2 is regulated by Parkinson's disease-associated kinase PINK1. Nat. Cell Biol. 9, 1243–1252 [DOI] [PubMed] [Google Scholar]
- 36. Weihofen A., Thomas K. J., Ostaszewski B. L., Cookson M. R., Selkoe D. J. (2009) Pink1 forms a multiprotein complex with Miro and Milton, linking Pink1 function to mitochondrial trafficking. Biochemistry 48, 2045–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schmitt-Ulms G., Matenia D., Drewes G., Mandelkow E. M. (2009) Interactions of MAP/microtubule affinity regulating kinases with the adaptor complex AP-2 of clathrin-coated vesicles. Cell. Motil. Cytoskeleton 66, 661–672 [DOI] [PubMed] [Google Scholar]
- 38. Detmer S. A., Chan D. C. (2007) Functions and dysfunctions of mitochondrial dynamics. Nat. Rev. Mol. Cell Biol. 8, 870–879 [DOI] [PubMed] [Google Scholar]
- 39. Augustinack J. C., Schneider A., Mandelkow E. M., Hyman B. T. (2002) Specific tau phosphorylation sites correlate with severity of neuronal cytopathology in Alzheimer's disease. Acta Neuropathol. 103, 26–35 [DOI] [PubMed] [Google Scholar]
- 40. Cookson M. R., Bandmann O. (2010) Parkinson's disease: insights from pathways. Hum. Mol. Genet. 19, R21–R27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Winklhofer K. F., Haass C. (2010) Mitochondrial dysfunction in Parkinson's disease. Biochim. Biophys. Acta 1802, 29–44 [DOI] [PubMed] [Google Scholar]
- 42. Narendra D. P., Jin S. M., Tanaka A., Suen D. F., Gautier C. A., Shen J., Cookson M. R., Youle R. J. (2010) PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 8, e1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lin W., Kang U. J. (2010) Structural determinants of PINK1 topology and dual subcellular distribution. BMC Cell Biol. 11, 90–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kim Y., Park J., Kim S., Song S., Kwon S. K., Lee S. H., Kitada T., Kim J. M., Chung J. (2008) PINK1 controls mitochondrial localization of Parkin through direct phosphorylation. Biochem. Biophys. Res. Commun. 377, 975–980 [DOI] [PubMed] [Google Scholar]
- 45. Narendra D. P., Youle R. J. (2011) Targeting mitochondrial dysfunction: role for PINK1 and Parkin in mitochondrial quality control. Antioxid. Redox Signal. 14, 1929–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Deas E., Wood N. W., Plun-Favreau H. (2011) Mitophagy and Parkinson's disease: the PINK1-parkin link. Biochim. Biophys. Acta 1813, 623–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang X., Winter D., Ashrafi G., Schlehe J., Wong Y. L., Selkoe D., Rice S., Steen J., LaVoie M. J., Schwarz T. L. (2011) PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell 147, 893–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Miller K. E., Sheetz M. P. (2004) Axonal mitochondrial transport and potential are correlated. J. Cell Sci. 117, 2791–2804 [DOI] [PubMed] [Google Scholar]
- 49. Jin S. M., Lazarou M., Wang C., Kane L. A., Narendra D. P., Youle R. J. (2010) Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J. Cell Biol. 191, 933–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vives-Bauza C., Przedborski S. (2011) Mitophagy: the latest problem for Parkinson's disease. Trends Mol. Med. 17, 158–165 [DOI] [PubMed] [Google Scholar]
- 51. Geisler S., Holmström K. M., Skujat D., Fiesel F. C., Rothfuss O. C., Kahle P. J., Springer W. (2010) PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 12, 119–131 [DOI] [PubMed] [Google Scholar]
- 52. Goldstein A. Y., Wang X., Schwarz T. L. (2008) Axonal transport and the delivery of pre-synaptic components. Curr. Opin. Neurobiol. 18, 495–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Colin E., Zala D., Liot G., Rangone H., Borrell-Pagès M., Li X. J., Saudou F., Humbert S. (2008) Huntingtin phosphorylation acts as a molecular switch for anterograde/retrograde transport in neurons. EMBO J. 27, 2124–2134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sen S., Webber P. J., West A. B. (2009) Dependence of leucine-rich repeat kinase 2 (LRRK2) kinase activity on dimerization. J. Biol. Chem. 284, 36346–36356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Russo G. J., Louie K., Wellington A., Macleod G. T., Hu F., Panchumarthi S., Zinsmaier K. E. (2009) Drosophila Miro is required for both anterograde and retrograde axonal mitochondrial transport. J. Neurosci. 29, 5443–5455 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.