Background: Botryococcus braunii accumulates high levels of methylated triterpenes that contribute to the buoyancy of the algae and serve as highly valued chemical feedstocks.
Results: AdoMet-dependent methyltransferases catalyzing successive and regio-specific methylations of squalene or botryococcene are identified.
Conclusion: Methylation of squalene and botryococcene requires distinct methyltransferases.
Significance: Substrate selectivity and successive cycles of regio-specific catalysis by triterpene methyltransferases from B. braunii are reported.
Keywords: Algae, Biofuel, Metabolic Engineering, Natural Products, S-Adenosylmethionine (AdoMet), AdoMet-dependent Methyltransferase, Botryococcene, Squalene
Abstract
Botryococcus braunii race B is a colony-forming, green algae that accumulates triterpene oils in excess of 30% of its dry weight. The composition of the triterpene oils is dominated by dimethylated to tetramethylated forms of botryococcene and squalene. Although unusual mechanisms for the biosynthesis of botryococcene and squalene were recently described, the enzyme(s) responsible for decorating these triterpene scaffolds with methyl substituents were unknown. A transcriptome of B. braunii was screened computationally assuming that the triterpene methyltransferases (TMTs) might resemble the S-adenosyl methionine-dependent enzymes described for methylating the side chain of sterols. Six sterol methyltransferase-like genes were isolated and functionally characterized. Three of these genes when co-expressed in yeast with complementary squalene synthase or botryococcene synthase expression cassettes resulted in the accumulation of mono- and dimethylated forms of both triterpene scaffolds. Surprisingly, TMT-1 and TMT-2 exhibited preference for squalene as the methyl acceptor substrate, whereas TMT-3 showed a striking preference for botryococcene as its methyl acceptor substrate. These in vivo preferences were confirmed with in vitro assays utilizing microsomal preparations from yeast overexpressing the respective genes, which encode for membrane-associated enzymes. Structural examination of the in vivo yeast generated mono- and dimethylated products by NMR identified terminal carbons, C-3 and C-22/C-20, as the atomic acceptor sites for the methyl additions to squalene and botryococcene, respectively. These sites are identical to those previously reported for the triterpenes extracted from the algae. The availability of closely related triterpene methyltransferases exhibiting distinct substrate selectivity and successive catalytic activities provides important tools for investigating the molecular mechanisms responsible for the specificities exhibited by these unique enzymes.
Introduction
Botryococcus braunii is a colony-forming, freshwater green algae that has attracted considerable interest because it reportedly accumulates hydrocarbon oils from 30 to 86% (1) of its dry weight and because these oils are considered progenitors to oil and coal shale deposits (2–4). Although all B. braunii are morphologically similar, three distinct chemotypes of B. braunii have been reported depending on the type of hydrocarbons each accumulates (5). Race A accumulates fatty acid-derived alkadienes and alkatrienes (6); race L accumulates the tetraterpene lycopadiene (7); and race B amasses the linear triterpenes, botryococcene, squalene, and their methylated derivatives (8). Di- and tetramethylated botryococcenes are generally the most abundant oils accumulating in race B (9). However, lower amounts of tetramethylated squalene (10) and variable amounts of other structural derivatives of botryococcene ranging from C31 to C37 accumulate to various levels in different race B strains and in response to variable culture conditions (9, 11). The oils accumulate both in intracellular oil bodies and in association with an extracellular matrix (12), which in race B consists mainly of long-chain, cross-linked polyacetals formed in large part from acetalization of polymethylsqualene diols that account for ∼10% of the dry weight (13). Other polymethylsqualene derivatives have been described in race B, such as diepoxy-tetramethylsqualene (14), botryolins (15), and braunixanthins (16). The linear triterpenes, botryococcene, squalene, and their methylated derivatives, are hence common components of B. braunii race B and make up a large proportion of its total biomass.
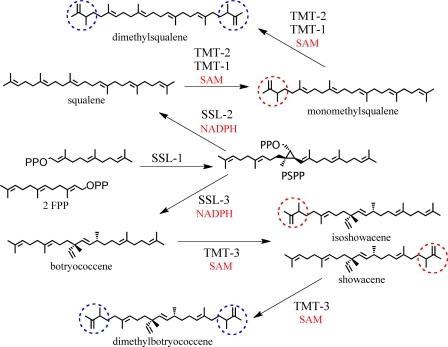
A unique mechanism for botryococcene biosynthesis was recently described by Niehaus et al. (17), in which two squalene synthase-like (SSL)3 enzymes perform the successive half-reactions that are normally catalyzed by a single enzyme in the case of squalene synthase. SSL-1 uses farnesyl diphosphate as a substrate to catalyze the production of pre-squalene diphosphate, which a second enzyme, SSL-3, converts to botryococcene in an NADPH-dependent manner. A third enzyme, SSL-2, catalyzes the biosynthesis of squalene from pre-squalene diphosphate produced by SSL-1 but cannot efficiently use farnesyl diphosphate as a substrate. Overall, it was suggested that the squalene and botryococcene produced by the SSL enzymes were channeled into the production of the liquid oils and the biosynthesis of squalene derivatives (17), such as the extracellular matrix, whereas the conventional B. braunii squalene synthase (18) appears to synthesize squalene destined for sterol biosynthesis.
It is not botryococcene and squalene, however, that accumulate to substantial levels in this algae, but the methylated forms of these triterpenes. For instance, although the liquid oil content of B. braunii race B is composed primarily of botryococcenes, generally less than 1% is in the nonmethylated C30 form, and the majority is dominated by dimethylated and tetramethylated forms, depending on the strain and culture conditions (9, 19, 20). Essentially all the squalene that accumulates is in methylated forms, accumulating in the oil fraction (less than 5% of the total oil (21)) or incorporated into a variety of other B. braunii natural products (13–16). Because B. braunii race B accumulates 30% or more of its dry weight as these triterpene components, we estimate that the methylated triterpenes can account for over 25% of the total algal biomass dry weight and contribute directly to the buoyancy that distinguishes these algal colonies. Unlike many green algae that are flagellated and phototaxic (22), the buoyancy characteristic of Botryococcus provides a means for it to float in its normal aqueous habitats and to intercept a greater amount of photosynthetic light. In addition to this purported physiological role, the methylated forms of botryococcene and squalene enhance their utility as feedstocks for petrochemical processing and chemical manufacturing (23). For instance, the increased branching evident in the methylated triterpenes improves their hydrocracking to chemical species of value for the synthesis of industrial polymers and other commodity-based chemicals (24) and yields high quality gasoline, kerosene, and diesel fuels upon distillation (25).
Although the unique mechanisms for C30 botryococcene and squalene biosynthesis in B. braunii have been elucidated (17), the specific mechanism(s) by which these triterpenes are methylated was unknown. Small molecule methylation has been extensively characterized for many diverse compounds and typically consists of a methyltransferase (MT) that utilizes the universal methyl donor S-adenosyl methionine (AdoMet), and exhibits variable degrees of selectivity for a wide range of methyl acceptor molecules (26, 27). MTs are also distinguished as C-, O-, N-, S-, or halide methyltransferases, an indication of the methylation target within the acceptor substrate (26). Although MTs may only share limited overall amino acid sequence similarities, domains responsible for AdoMet binding appear to be broadly conserved (28), and highly conserved structural folds have served to associate MTs into five distinct structural classes (26). Most of the plant small molecule MTs fall into structural Class 1, consisting of β-sheets sandwiched between α-helices, and cluster phylogenetically on the basis of the chemical class of the methyl acceptor and the methylation target site (i.e. C-, O-, or N-methylation) (27, 29, 30). There are, of course, important exceptions to such clustering trends. An indole alkaloid MT, for instance, shows closer sequence similarity to tocopherol MTs than to other alkaloid-specific N-MTs (29). Clustering in this instance appears more related to the evolutionary origins of the MT and the propensity of duplicated MT genes to undergo neofunctionalization.
In the current work, we supposed that the methylation of botryococcene and squalene up to their tetramethylated forms could be catalyzed by a single, multifunctional MT or multiple MTs with each responsible for one particular methylation in the catalytic cascade of each triterpene. We also assumed that because squalene and botryococcene are chemically and biochemically related to sterols, the methylated forms of triterpenes in B. braunii possibly arose within the same evolutionary timeframe as sterols and the mechanisms for sterol biosynthesis. Consequently, we searched for homologs of sterol MTs (31, 32) within a transcriptome of B. braunii and provide here the first functional identification of several unique triterpene C-methyltransferases.
EXPERIMENTAL PROCEDURES
Cloning SMT-like Genes
The triterpene methyltransferase-3 (TMT-3) was identified through a random sequencing effort of expressed sequence tags using a B. braunii phage cDNA library as described previously (17). Briefly, phages were converted to their plasmid form using the mass excision protocol as described by the manufacturer (Stratagene), and ∼500 individual colonies were randomly selected for automated DNA sequencing using sequencing primers flanking the cDNA insertion sites. Manually assembled cDNA sequences were then screened against the National Center for Biotechnology Information (NCBI) tBlastn search function across all available databases, and TMT-3 was identified as exhibiting similarity to C-24-sterol methyltransferase (SMT) genes. All other SMT-like genes were identified in a B. braunii 454 transcriptomic dataset as described previously (17). This dataset was screened computationally using an NCBI BLAST search window with the Chlamydomonas reinhardtii SMT-1 protein sequence (EDP05221) and the Arabidopsis thaliana SMT-1 sequence (AAG28462) as the queries, which led to the identification of six full-length ORFs that were at least 42% identical and 59% similar to C. reinhardtii SMT. Full sequence data are available from GenBank (TMT-1, JN828962; TMT-2 JN828963; TMT-3, JN828964; SMT-1, JN828965; SMT-2, JN828966; SMT-3, JN828967).
Primers flanked by the BamHI and NotI or EcoRI and NotI restriction enzyme sites were designed to amplify each of the six SMT-like genes from B. braunii mRNA, the amplification products digested with the corresponding restriction enzymes, and then ligated into the standard yeast expression vectors YEp352-Ura or pESC-Leu (17). All constructs were verified by DNA sequencing.
Yeast Expression Studies
Yeast strains previously developed for high level accumulation of squalene and botryococcene were used for evaluating the putative triterpene methyltransferase genes (17, 33, 34). Strain TN-7 was created by insertional mutagenesis of the ERG1 (squalene epoxidase) gene in the Cali-7 yeast strain. CALI7-1 is a leu2, trp1, his3, ura3Δ, erg9Δ::HIS3, sue, dpp1 derivative from the parental strain ATCC 28383, which allows for sterol uptake under aerobic conditions and farnesyl diphosphate accumulation (33, 34). The TN-7 parental strain was transformed with expression vectors containing either the full-length Botryococcus squalene synthase (BSS) gene (18) or a fusion of the Botryococcus SSL-1 and SSL-3 genes (functional equivalent of botryococcene synthase) including a sequence encoding for the carboxyl-terminal membrane-targeting domain of the Botryococcus squalene synthase protein (SSL-1–3m) (17). The various methyltransferase expression vectors were introduced into these two yeast strains using the lithium acetate transformation protocol followed by selection for complementation of the uracil and leucine auxotrophic growth markers (34). Transformants were confirmed to possess the various expression vectors using colony PCR with primers selective for the methyltransferase genes. Individual colonies were subsequently grown in 30 ml of the appropriate Yeast Synthetic Drop-out medium (selection) containing 5 mg/liter ergosterol for the indicated time at 30 °C before analyzing the cultures for production of novel triterpene components. In brief, 1-ml aliquots of the culture were combined with 1 ml of acetone, mixed vigorously, and incubated at room temperature for 10 min. One ml of hexane was added and mixed vigorously for 60 s. The mixture was then centrifuged briefly at 500 × g to separate the phases, and an aliquot of the organic phase (1–3 μl) was analyzed by GC-MS with a Varian CP-3800 GC coupled to a Varian Saturn 2200 MS/MS (Varian Medical Systems) using a Supelco SLB-5ms fused silica capillary column (30 m × 0.25 mm × 0.25-μm film thickness, Supelco). The initial oven temperature was set at 220 °C for 1 min, ramped to 280 °C at 20 °C/min, and then ramped to 298 °C at 3 °C/min.
Purification of Mono- and Dimethylated Triterpenes
Yeast lines containing the respective triterpene synthase and TMT expression cassettes were grown in 1 liter of Yeast Synthetic Drop-out medium containing 5 mg/liter ergosterol at 28 °C for 8 days, after which hexane extracts were prepared. The crude extracts were then subject to HPLC separation on a Waters 2695 HPLC with a Waters 2996 photodiode array detector (Waters Corp.) and a Develosil 60-3, 250 × 20-mm column (Nomura Chemical), run in isocratic mode (100% n-hexane) at 8 ml/min. Under these conditions, C32 botryococcene, C31 botryococcene, C32 squalene, and C31 squalene eluted at ∼18, 22, 32, and 34 min, respectively. Repetitive chromatographic runs afforded further purification of the various compounds.
NMR of Methylated Triterpenes
1H and 13C NMR spectra were recorded on a JEOL Alpha selector 600 NMR spectrometer at 300 K. Chemical shifts were referenced relative to solvent peaks, namely dH 7.24 and dC 77.1 for CDCl3. Each product was identified as shown in Fig. 4 by reference to 13C chemical shifts for botryococcenes and methylsqualenes reported previously (10, 16, 20, 21, 35).
FIGURE 4.
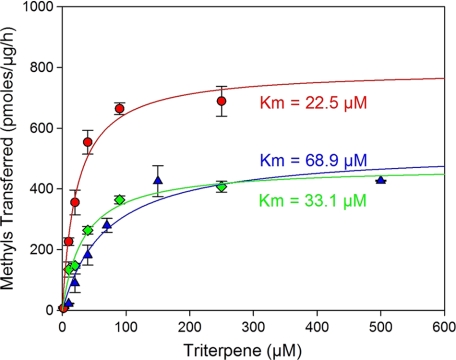
Michaelis-Menten enzyme kinetics for TMT-1 (green diamonds), TMT-2 (red circles), and TMT-3 (blue triangles). Enzyme assays contained 50 mm HEPES, pH 7.5, either 0.01% (TMT-3) or 0.1% (TMT-2 and TMT-3) 1,2-dicaproyl-sn-glycero-3-phosphocholine, the indicated concentration of acceptor substrate (botryococcene or squalene), 50 μm [3H]AdoMet (∼250 dpm/pmol), and 2-μl aliquots of microsomes (∼2 μg of total protein) in a 100-μl final reaction volume. Squalene and botryococcene were dissolved by sonication in the reaction buffer prior to the addition of [3H]AdoMet and microsomes. Microsomes were prepared from yeast overexpressing the gene for TMT-1, TMT-2 or TMT-3. Activities were determined as the amount of radioactivity incorporated in TLC-separated methylated triterpenes. Data are reported as pmol of methyl groups transferred to acceptor substrate per unit time and per μg of total microsomal protein. Data represent mean ± S.D., n = 2.
In Vitro Assays for Methyltransferase Activities
The various B. braunii SMT-like genes or empty vector control were expressed in TN-7 yeast and grown in 100 ml of selection medium for 3 days, after which microsomes were prepared according to the methods of Pompon et al. (36). Enzyme assays contained 50 mm HEPES, pH 7.5, either 0.01% (TMT-3) or 0.1% (all other samples) 1,2-dicaproyl-sn-glycero-3-phosphocholine, the indicated amounts of acceptor substrate (botryococcene, squalene, C32-botryococcene, C32-squalene, cycloartenol, zymosterol, or lanosterol), 50 μm [3H]AdoMet (∼150 dpm/pmol), 2 μl of microsomes (∼2–4 μg of total protein), in a 100-μl total volume. Assays were set up by first combining everything except [3H]AdoMet and microsomes and treating with a sonicating water bath (Branson 2510) for ∼1 min until the solution became cloudy due to micelle formation, after which [3H]AdoMet and microsomes were added and the reaction was incubated at 37 °C for 5 min. Reactions were stopped by adding an equal volume of 10% (w/v) KOH in methanol followed by extraction of hydrocarbon products with 400 μl of n-hexane. An aliquot of the organic phase was spotted on silica TLC plates and developed with n-hexane:methyl tert-butyl ether (25:1). Triterpenes were visualized with iodine vapor, and the corresponding zones were scraped and subjected to scintillation counting.
The experiments performed to determine the enzyme kinetics for TMT-1, -2, and -3 were slightly modified from the conditions described above. First, squalene and botryococcene were dissolved in isooctane, the appropriate amount was added to reaction vials, and the isooctane was evaporated before dissolving the triterpene substrate by sonication with the respective reaction buffer described above. Second, the reactions were incubated for 15 min rather than 5 min. The data were analyzed using SigmaPlot Enzyme Kinetics 1.3 software.
Methyltransferase mRNA Levels in Botryococcus
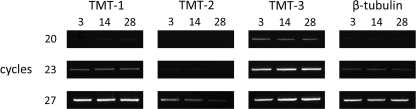
Cultures of B. braunii were grown under standard conditions for 3, 14, and 28 days after inoculation into fresh medium, and aliquots of algae were collected on nylon mesh, blotted dry, weighed, and stored at −80 °C until further analyzed (17). Total RNA was isolated using the RNeasy plant mini kit according to the manufacturer's directions (Qiagen). One μg of total RNA was then used for first-strand cDNA synthesis using SuperScript III (Invitrogen) according to the manufacturer's recommendation. PCR reactions were set up with equal amounts of the first-strand cDNA and primers (supplemental Table S1) specific for B. braunii TMT-1, TMT-2, TMT-3, and β-tubulin using Ex-taq (Takara) according to the manufacturer's recommendation. PCR reactions were performed for 20, 23, and 27 cycles, and equal aliquots were examined by agarose gel electrophoresis.
RESULTS
Identification of Triterpene Methyltransferase Candidate Genes
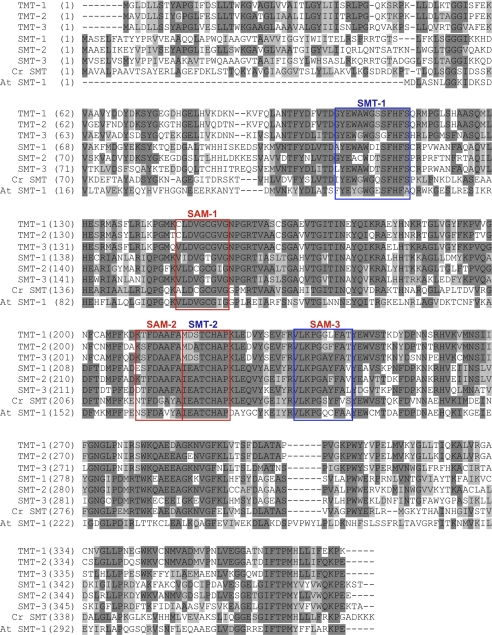
We predicted that a methyltransferase that could act on squalene or botryococcene might resemble a C-24 SMT because these enzymes act on the linear side chain of sterols. A B. braunii transcriptomic database (17) was screened computationally for cDNAs showing amino acid sequence similarities to the A. thaliana and C. reinhardtii SMT-1 enzymes. The BLAST search revealed six candidate genes that were greater than 42% identical and 59% similar to the C. reinhardtii SMT-1 (Fig. 1). For comparison, the A. thaliana genome contains three predicted SMT genes (37), and the C. reinhardtii genome contains only one SMT gene (38). These particular genes appear overrepresented in B. braunii when compared with other plants and algae, and this observation enhanced the prospects that these could be TMTs. Amino acid alignments revealed that all six candidate genes share three conserved AdoMet-binding sites as identified by Kagen and Clarke (28); however, the sterol-binding domain SMT-2, which is invariant in all known plant SMTs (31, 32), is absolutely conserved in three of the candidates (SMT-1, -2, and -3), but not so in the other three (TMT-1, -2, and -3) (Fig. 1). In contrast to other sterol methyltransferases (31, 32), the B. braunii MTs possess distinct amino-terminal hydrophobic regions within the first 50 amino acids indicative that these proteins might not behave as soluble proteins but rather might associate with membrane systems.
FIGURE 1.
Amino acid alignment of the six sterol C-24 methyltransferase-like genes from B. braunii race B along with those of C. reinhardtii (EDP05221) and A. thaliana (AAG28462). Conserved sterol-binding domains (SMT) and S-adenosyl methionine-binding domains (AdoMet) as identified by (27) are boxed and labeled in blue or red, respectively. Cr, C. reinhardtii; At, A. thaliana.
In Vivo Functional Characterization of MT Activities
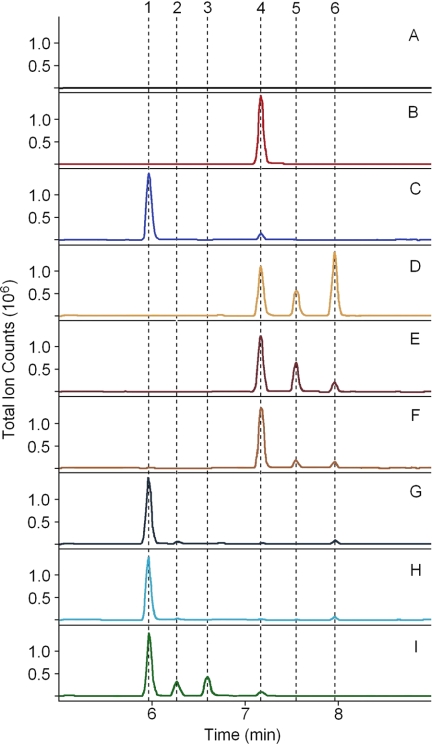
To screen the six candidates for TMT capabilities, we co-expressed the various SMT-like genes in TN-7 yeast engineered with either BSS or a construct in which SSL-1 and SSL-3 are fused with a (GSGG)3 amino acid linker that also contains the 73 C-terminal amino acids of BSS fused to its C terminus (SSL-1–3m). TN-7 yeast engineered with BSS or SSL-1–3m can accumulate squalene or botryococcene, respectively, to levels above 100 mg/liter (supplemental Figs. S1 and 2, B and C). When SMT-1, -2, or -3 were co-expressed with either BSS or SSL-1–3m, no distinct products could be detected in organic extracts by GC-MS analysis (data not shown); however, co-expression of TMT-1, -2, or -3 all resulted in the accumulation of several unique products (Fig. 2, D–I). Mass spectral analysis of the unique peaks showed parent ions of 424 and 438 atomic mass units (supplemental Fig. S2), suggesting mono- and dimethylated triterpenes, respectively.
FIGURE 2.
Functional characterization of B. braunii race B TMT genes. Yeast expressing various combinations of triterpene synthase and TMTs were grown in a shake flask for 5 days, and organic extracts were analyzed by GC-MS (chromatograms shown). TMT genes were co-expressed with BSS (squalene synthase) (TMT-1 (D), TMT-2 (E), and TMT-3 (F)) or with SSL-1–3m (botryococcene synthase) (TMT-1 (G), TMT-2 (H), and TMT-3 (I)). Yeast expressing only BSS (B) or SSL-1–3m (C) or only harboring empty expression vectors (A) serve as background controls. The chromatograms are annotated for the elution behavior of botryococcene (1), C31-botryococcene (2), C32-botryococcene (3), squalene (4), C31-squalene (5), and C32-squalene (6).
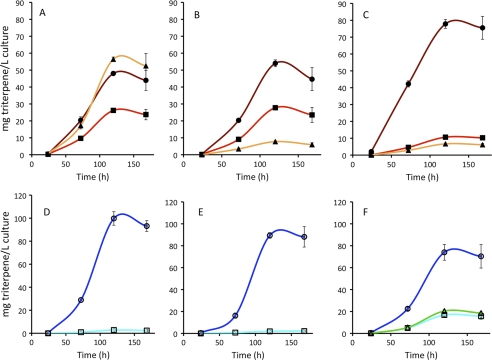
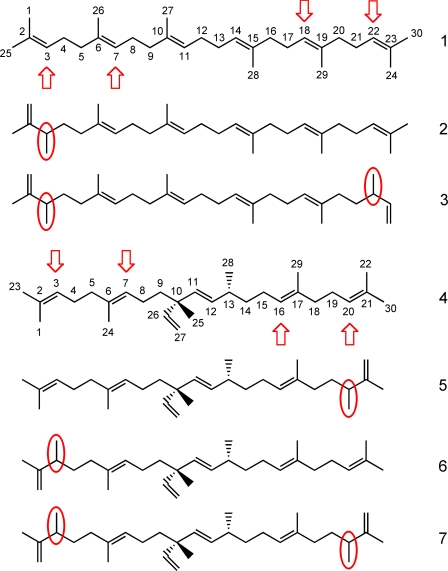
When TMT-1 was co-expressed with BSS, 63% of the total squalenes accumulated as methyl derivatives with 43% accumulating as dimethyl squalene (Figs. 2D and 3A). However, when coexpressed with SSL-1–3m, only 3% of the total botryococcenes accumulated as monomethyl botryococcene, and no dimethyl botryococcene was detected (Figs. 2G and 3D). Similarly, when TMT-2 was co-expressed with BSS, 40% of squalenes accumulated as methyl derivatives with 31% accumulating as monomethyl squalene (Figs. 2E and 3B). Only 2% of total botryococcenes accumulated as monomethyl botryococcene when co-expressed with SSL-1–3m (Fig. 2H and 3E). When TMT-3 was co-expressed with BSS, ∼18% of the total accumulating squalene was converted to its methyl derivatives, with 11% of that as monomethyl squalene (Figs. 2F and 3C). When TMT-3 was co-expressed with SSL-1–3m, 33% of the accumulating botryococcene was methylated with greater than half of that in the dimethyl botryococcene form (Figs. 2I and 3F).
FIGURE 3.
Accumulation of triterpenes in yeast engineered with various triterpene synthases and TMTs. A–C, yeast were engineered with BSS and TMT-1 (A), TMT-2 (B), or TMT-3 (C) on separate plasmids, and accumulation of squalene (closed circles), C31 squalene (closed squares), and C32 squalene (closed triangles) was measured. D–F, yeast were engineered with the botryococcene synthase (SSL-1–3m) construct and TMT-1 (D), TMT-2 (E), or TMT-3 (F) on separate plasmids, and accumulation of botryococcene (open circles), C31 botryococcene (open squares), and C32 botryococcene (open triangles) was measured. Yeast were grown in shake flasks at 30 °C for the indicated times, and organic extracts were analyzed by GC-MS. Data represent mean ± S.E. of three replicates.
Although the conversion of botryococcene and squalene to their mono- and dimethyl derivatives was readily detected, no further methylated products (tri- and tetramethylated) accumulated. We considered the possibility that multiple methyltransferases might act successively and cooperatively in the formation of C34 triterpenes, with one methyltransferase catalyzing the C30 to C32 conversion and another using C32 as a substrate to form a C34 triterpene. To test this possibility, yeast expressing either BSS or SSL-1–3m with TMT-1, TMT-2, or TMT-3 as well as each of the remaining five other SMT-like B. braunii genes were evaluated for their triterpene content. No unique products other than the C31 and C32 triterpenes observed in the yeast lines expressing only TMT-1, -2, or -3 (Fig. 2) were detected by GC-MS analysis (data not shown).
In Vitro Biochemical Confirmation
To verify the in vivo results with in vitro determinations, the six SMT-like genes were expressed in yeast, and microsomal preparations were used as the source of the enzymes in assays containing [3H]AdoMet and either botryococcene or squalene as substrates. TMT-1 and TMT-2 readily catalyzed the transfer of a methyl group from AdoMet to squalene but showed less than 1/100 of those levels of activity with botryococcene as the acceptor (Table 1). In contrast, TMT-3 favored botryococcene as the methyl acceptor and exhibited only very modest activity with squalene. None of the other three SMT-like genes showed any measurable methyltransferase activity with botryococcene or squalene as substrates. None of the six enzymes was able to methylate C32 botryococcene or C32 squalene, possible intermediates to the tetramethylated forms (see below). Equally surprising, none of the six B. braunii SMT-like genes methylated cycloartenol, zymosterol, or lanosterol (data not shown), which suggested that we have yet to find the proper substrate(s) for these MTs, that we have not been able to provide these hydrophobic substrates in a form available for catalytic turnover, or that these MTs were not catalytically competent under these in vitro conditions.
TABLE 1.
Substrate preference of the various B. braunii SMT-like enzymes
The various B. braunii SMT-like genes or empty vector control were constitutively expressed in yeast for 3 days, after which microsomes were prepared according to the methods of Pompon et al. (36(). Enzyme assays contained 50 mm HEPES, pH 7.5, either 0.01% (TMT-3) or 0.1% (all other samples) 1,2-dicaproyl-sn-glycero-3-phosphocholine, 2 mm acceptor substrate (botryococcene, squalene, C32 botryococcene, C32 squalene), 50 μm [3H]AdoMet (∼150 dpm/pmol), 2-μl aliquots of microsomes (∼4 μg of total protein) in 100-μl final reaction volume. Assays were set up by first combining everything except [3H]AdoMet and microsomes and sonicating the mixture until the solution became cloudy due to micelle formation. The [3H]AdoMet and microsomes were added, and the reaction was incubated at 37 °C for 5 min. Reactions were stopped by adding an equal volume of 10% (w/v) KOH in methanol followed by extraction of hydrocarbon products with 400 μl of n-hexane. Aliquots of the organic phase were separated by TLC, and radioactivity incorporated into the triterpene fractions was determined by scintillation counting. Data are reported as pmol of methyl groups transferred to acceptor substrate per unit time and per μg of microsomal protein. Data represent mean ± S.E. of three replicates.
| Enzyme activity (pmol/h/μg) |
|||||||
|---|---|---|---|---|---|---|---|
| Empty | TMT-1 | TMT-2 | TMT-3 | SMT-1 | SMT-2 | SMT-3 | |
| Substrate | |||||||
| Squalene | 0 | 513.7 ± 8.6 | 862.2 ± 59.9 | 35.4 ± 3.0 | 0 | 0 | 0 |
| Botryococcene | 0 | 3.3 ± 1.3 | 4.5 ± 1.3 | 434.9 ± 31.8 | 0 | 0 | 0 |
| C32 squalene | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| C32 botryococcene | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Steady-state enzyme kinetics for the three TMTs were also performed using microsomes prepared from yeast overexpressing the respective genes (Fig. 4). TMT-1 and TMT-2 activities exhibited typical saturation kinetics with squalene as substrate, as did TMT-3 with botryococcene. The apparent Km values for the corresponding triterpene substrates varied from 22 to 69 μm with maximum velocities ranging from 400 to 750 pmol of methyls transferred per μg of microsomal protein per h. Although the TMTs were epitope-tagged with carboxyl-terminal hexahistidine residues, we were unsuccessful in detecting and quantifying the respective protein levels and hence are unable to report the catalytic turnover rate (kcat) for each of these enzymes. Microsomes from yeast harboring the expression vector without any inserted methyltransferase gene served as the negative control in all these experiments without any triterpene methyltransferase detected under these conditions.
Chemical Identification of Reaction Products
To determine the specific methylation sites on squalene and botryococcene, the mono- and dimethylated squalenes and botryococcenes produced in vivo by the engineered yeast were purified and subjected to 1H and 13C NMR analyses (see supplemental Table S2 for complete 13C NMR assignments). The monomethylated squalenes produced by yeast expressing TMT-1, -2, or -3 were all identical based on their NMR signals and methylated at the C-3 position of squalene (Fig. 5, compound 2). Similarly, all the dimethylated squalenes produced by all three yeast lines gave identical NMR signals indicative of methylation at the C-3 and C-22 positions (Fig. 5, compound 3). In contrast, the monomethylated botryococcene produced by yeast expressing TMT-3 occurred at two positions, either the C-20 position yielding showacene (Fig. 5, compound 5) or the C-3 position yielding isoshowacene (Fig. 5, compound 6). Based on the relative intensity of the NMR signals for the methyl substituent at C-20 in showacene (Fig. 5, compound 5) versus that for C-3 in isoshowacene (Fig. 5, compound 6), showacene accounts for more of the total monomethylated products than isoshowacene. Dimethylated botryococcene produced by TMT-3 had methyl groups at the C-3 and C-20 positions (Fig. 5, compound 7). The very small amounts of methylated botryococcenes produced in yeast expressing TMT-1 or TMT-2 were not sufficient for NMR analysis; however, the GC-MS patterns of monomethylated botryococcene produced by TMT-1, -2, and -3 were all identical (supplemental Fig. S2). These findings suggest that all the TMTs methylate botryococcene at same positions.
FIGURE 5.
Structures of the various triterpenes accumulating in yeast expressing squalene synthase or botryococcene synthase in combination with TMT-1, -2, or -3. Yeast expressing the squalene synthase (BSS) gene accumulates squalene (1), C31 monomethylated squalene (2), and C32 dimethylated squalene (3) when co-expressed with the TMT-1, TMT-2, or TMT-3 genes. Yeast expressing the botryococcene synthase expression cassette (SSL-1–3m) accumulates botryococcene (4), a mixture of C31 monomethylated isomers, showacene (5) and isoshowacene (6), and C32 dimethylated botryococcene (7) when co-expressed with TMT-3. Squalene and botryococcene have their carbons labeled, and the common sites of methylation are indicated with red arrows. The mono- and dimethylation sites within the triterpenes that accumulate in the respective yeast lines are highlighted with red circles. Methylation sites were assigned according to NMR signatures of the isolated compounds (supplemental Table S2) with reference to those previously reported (16, 20, 31, 34, 35).
TMT Expression in B. braunii
To determine whether expression of any of the TMT genes was regulated in B. braunii, the steady-state levels of the TMT mRNAs were determined over a culture cycle of 28 days. Three days after inoculation of new cultures, the fresh weight of the cultures was 568 mg/liter, increasing to 1,195 mg/liter by day 14 and 1,684 mg/liter by day 28. The oil content of these cultures increased from 98 mg/liter on day 3 to 172 mg/liter by day 14 and 239 mg/liter at day 28. Very little difference in the chemical profile of these oils was noted over this time course with less than 1% as C30 botryococcene, but the majority of the oil (>70%) was composed of di- to tetramethylated botryococcene. Consistent with the methylated triterpenes being synthesized constitutively throughout the culture cycle, TMT-1 and TMT-3 mRNA levels were relatively constant when normalized for total RNA (Fig. 6). In contrast, TMT-2 mRNA levels were significantly higher at day 3 and declined thereafter (Fig. 6).
FIGURE 6.
Relative expression levels of TMT-1, TMT-2, and TMT-3 mRNAs during a culture cycle of B. braunii. Cultures of B. braunii were grown under standard conditions for 3, 14, and 28 days. Total RNA was prepared from each sample and converted to first-strand cDNA, and equal aliquots were then used for 20, 23, or 27 cycles of PCR amplification with primers specific for B. braunii TMT-1, TMT-2, TMT-3, and β-tubulin. Relative abundance of the amplicons was examined by gel electrophoresis.
DISCUSSION
The large accumulation of triterpene oils by B. braunii race B has provided the impetus for considerable interest in elucidating the biosynthesis of these seemingly simple molecules. The oil is composed largely of linear, branched-chain triterpenes resembling squalene, yet the triterpene scaffold, botryococcene, is synthesized by the successive action of two enzymes rather than a single enzyme like that typical for squalene biosynthesis (17). Although small amounts of the C30 botryococcene and squalene triterpenes do accumulate, methylated forms of these molecules predominate and accumulate to upwards of 30% of the total algal dry weight. Hence, these algae must possess a robust mechanism(s) for converting the triterpene scaffolds to their methylated forms, which also lends these molecules to a variety industrial applications (23).
In the current effort, we identified three triterpene methyltransferases contributing to the methylation status of botryococcene and squalene, and we were surprised by several of the biochemical properties uncovered during this investigation. Although we identified three triterpene MTs genes exhibiting sequence similarity to sterol methyltransferases (Fig. 1), two of these encoded enzymes showed specificity for squalene methylation. The third TMT appears to have specificity for botryococcene methylation. The specificity for squalene or botryococcene was unexpected because these molecules have similar physical features. Nonetheless, the B. braunii TMTs were found to discriminate between the two methyl acceptors, as depicted in Fig. 7, and this must arise from the ability of the respective enzymes to recognize differences in the internal linkages within squalene and botryococcene. TMT-3 must be able to recognize the internal ethyl, methyl substituents at C-10 of botryococcene, whereas TMT-1 and -2 must prefer the straight-chain linkage across C-11–14 of squalene.
FIGURE 7.
Proposed methylated triterpene biosynthetic pathways in B. braunii. SSL-1 converts two farnesyl diphosphate molecules (FPP) to pre-squalene diphosphate (PSPP), which is converted in an NADPH-dependent manner to either squalene or botryococcene by SSL-2 or SSL-3, respectively. TMT-1 and TMT-2 can transfer a methyl group from AdoMet (SAM) to squalene to form mono- or dimethylated squalene, whereas TMT-3 acts on botryococcene to form mono- or dimethylated botryococcene.
The substrate specificity of the TMTs was also unexpected when one considers the symmetry and asymmetry of squalene, botryococcene, and the monomethylated intermediates and the successive nature of these catalytic events. Most small molecule MTs catalyze monomethylation reactions, with some notable exceptions such as the trimethylation of phosphoethanolamine in the biosynthesis of the choline head group in phospholipid biosynthesis (39). However, the successive methylation of the sterol side chain at C-24 requires distinct, successively acting enzymes, sterol methyltransferase 1 and 2 (31, 40). The successive nature of the B. braunii TMTs appears to represent yet another twist in the activities of this diverse family of enzymes. For TMT-1 and -2, the symmetry of squalene affords equal probability of methylation at either end of the molecule, but these enzymes also introduce a second methylation at the equivalent position on the opposite side of the molecule. Although TMT-1 appears to perform this second methylation with great facility, this is not the case for TMT-2. The accumulation of dimethylated squalene exceeds that for monomethylated squalene greater than 2-fold in yeast expressing the TMT-1 gene (Fig. 3A), but dimethylated squalene only accumulates to ∼20% of that for monomethylated squalene in yeast expressing TMT-2 (Fig. 3B). TMT-3 functionally resembles TMT-1 with regard to the ease with which it introduces the second methylation into the botryococcene backbone. That is, the accumulation of dimethyl botryococcene slightly exceeded that of monomethyl botryococcene (Fig. 3F). Based on NMR analysis of the monomethylated botryococcene produced in yeast, showacene accumulated to higher levels than isoshowacene. It is unclear whether this arises from a preference for methylating botryococcene at C-20 rather than the C-3 position with both monomethylated botryococcenes serving as equal substrates in the second methylation reaction or whether both the C-3 and the C-20 positions of botryococcene are methylated with equal efficiency but isoshowacene (methylated at C-3) is the preferred substrate for the second methylation reaction, or a combination of both possibilities. Regardless, a ratio of showacene to isoshowacene of ∼1.7 to 1.0 is seen in monomethylated botryococcenes isolated from B. braunii (35), suggesting that yeast expressing TMT-3 and SSL-1–3m recapitulate the same biochemical bias as observed in B. braunii.
Although AdoMet-dependent methyltransferases are a very broad class of enzymes, the atomic acceptor sites for the majority these enzymes are either oxygen or nitrogen atoms and are common modifications in primary as well as specialized metabolites (26). C-Methylation is not unique but seems to have evolved for a more narrow class of compounds. For instance, the side chains of sterols in plants and fungi (41) and the aromatic rings of tocopherol (42), ubiquinones/menaquinones (43), and uroporphyrinogens (44) in prokaryotes and eukaryotes are all C-methylated. However, C-methylation of specialized metabolites is much less common than O- or N-methylation reactions. The polyketides oxytetracycline and mithramycin from Streptomyces rimosus and Streptomyces argillaceus, for instance, are C-methylated (45, 46), as is geranyl diphosphate by Streptomyces lasaliensis, which produces the soil-like odor methyl isobutyl alcohol from this methylated precursor (47). The triterpene methyltransferases described here represent a third example of C-specific methyltransferases that contribute to the biosynthesis of specialized compounds serving a unique physiological role, but in a eukaryote rather than a prokaryote system. Moreover, based on the close phylogenetic relatedness of TMT-1, -2, and -3 to sterol methyltransferases (supplemental Fig. S3), TMT-1, -2, and -3 for specialized triterpene metabolism may have arisen from the duplication and neofunctionalization of genes associated with sterol side-chain methylation by mechanisms as suggested by Gang (48) and Han et al. (49) for other O-MTs associated with specialized metabolism in plants. Given the large collection of highly conserved Class 1 MT crystal structures (26) and the utility of these structures for molecular modeling and mapping residues important for catalysis in the wider family of MT enzymes (50, 51), a similar strategy might be applied to better define those residues or domains of the Botryococcus MTs specifying substrate selectivity (sterol side chain versus linear triterpene) and target site selection (regio-positioning along the linear triterpene chain) for methylation.
Our inability to determine catalytic activities for SMT-1, -2, and -3 raises obvious questions about these putative enzymes. Are they actual MTs, and have we exercised sufficient caution to produce properly folded protein and provide substrate in a proper context? Without any positive activity results to report for these particular SMT-like proteins, we are simply unable to adequately address such concerns. However, technical details concerning these proteins and the assays are worth considering in this regard. First, all the candidate genes were expressed in yeast engineered to facilitate both in vivo assessments of enzyme activity (methylated triterpene accumulation) as well as providing cell-free preparations for highly sensitive, in vitro radiolabel assays. Unlike many MTs, which are functionally soluble enzymes, all the MTs evaluated here harbor significant hydrophobic regions that might serve to associate these enzymes with membranes. In fact, the three B. braunii TMTs investigated here were associated with microsomal preparations from yeast, and we were not able to generate soluble enzyme activities when the respective genes were expressed in bacteria (data not shown). Second, the in vitro assays with hydrophobic substrates such as squalene and botryococcene required the use of detergents and sonication to solubilize the substrates. Such assay conditions are clearly artificial and raise additional conceptual questions about the physiological relevance of the assays. We readily acknowledge such concerns and hence have relied on the in vivo characterization of the genes expressed in yeast as the first line of evidence for their functional identification.
It is also worth noting that we have not identified all the mechanisms responsible for in vivo methylation of botryococcene and squalene in B. braunii race B. We have identified several genes encoding for methyltransferases capable of introducing terminal methyl substituents at C-3 and C-22/C-20 of squalene and botryococcene and hence yielding dimethylated triterpenes. However, botryococcene and squalene accumulate in B. braunii largely in their di- and tetramethylated forms with ratios dependent on growth conditions (9, 10, 19). Hence, additional MTs or other mechanisms for the complete methylation pattern of these triterpenes remain to be discovered.
Supplementary Material
Acknowledgments
We thank members of the Chappell laboratory for critical comments during the course of this work.
This work was supported by grants from the National Science Foundation (CBET-0828817) and Sapphire Energy (to J. C.) and Grant-in-Aid for Scientific Research (B) (21380130) from the Japanese Society for the Promotion of Science (to S. O.).

This article contains supplemental Tables S1 and S2 and Figs. S1–S3.
- SSL
- squalene synthase-like
- MT
- methyltransferase
- TMT
- triterpene methyltransferase
- SMT
- sterol methyltransferase
- BSS
- Botryococcus squalene synthase
- AdoMet
- S-adenosyl methionine.
REFERENCES
- 1. Brown A. C., Knights B. A., Conway E. (1969) Hydrocarbon content and its relationship to physiological state in the green alga Botryococcus braunii. Phytochemistry 8, 543–547 [Google Scholar]
- 2. Derenne S., Largeau C., Hetényi M., BruknerWein A., Connan J., Lugardon B. (1997) Chemical structure of the organic matter in a Pliocene maar-type shale: implicated Botryococcus race strains and formation pathways. Geochim. Cosmochim. Acta 61, 1879–1889 [Google Scholar]
- 3. Glikson M., Lindsay K., Saxby J. (1989) Botryococcus, a planktonic green alga, the source of petroleum through the ages: transmission electron microscopical studies of oil shales and petroleum source rocks. Org. Geochem. 14, 595–608 [Google Scholar]
- 4. Mastalerz M., Hower J. C. (1996) Elemental composition and molecular structure of Botryococcus alginate in Westphalian cannel coals from Kentucky. Org. Geochem. 24, 301–308 [Google Scholar]
- 5. Metzger P., Largeau C. (2005) Botryococcus braunii: a rich source for hydrocarbons and related ether lipids. Appl. Microbiol. Biotechnol. 66, 486–496 [DOI] [PubMed] [Google Scholar]
- 6. Gelpi E., Oró J., Schneider H. J., Bennett E. O. (1968) Olefins of high molecular weight in two microscopic algae. Science 161, 700–702 [DOI] [PubMed] [Google Scholar]
- 7. Metzger P., Allard B., Casadevall E., Berkaloff C., Couté A. (1990) Structure and chemistry of a new chemical race of Botryococcus braunii (Chlorophyceae) that produces lycopadiene, a tetraterpenoid hydrocarbon. J. Phycol. 26, 258–266 [Google Scholar]
- 8. Okada S., Murakami M., Yamaguchi K. (1995) Hydrocarbon composition of newly isolated strains of the green microalga Botryococcus braunii. J. Appl. Phycol. 7, 555–559 [Google Scholar]
- 9. Metzger P., Casadevall E., Coute A. (1988) Botryococcene distribution in strains of the green alga Botryococcus braunii. Phytochemistry 27, 1383–1388 [Google Scholar]
- 10. Huang Z., Poulter C. D. (1989) Tetramethylsqualene, a triterpene from Botryococcus braunii var. showa. Phytochemistry 28, 1467–1470 [Google Scholar]
- 11. Metzger P., Berkaloff C., Casadevall E., Coute A. (1985) Alkadiene- and botryococcene-producing races of wild strains of Botryococcus braunii. Phytochemistry 24, 2305–2312 [Google Scholar]
- 12. Weiss T. L., Chun H. J., Okada S., Vitha S., Holzenburg A., Laane J., Devarenne T. P. (2010) Raman spectroscopy analysis of botryococcene hydrocarbons from the green microalga Botryococcus braunii. J. Biol. Chem. 285, 32458–32466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Metzger P., Rager M. N., Largeau C. (2007) Polyacetals based on polymethylsqualene diols, precursors of algaenan in Botryococcus braunii race B. Org. Geochem. 38, 566–581 [Google Scholar]
- 14. Metzger P. (1999) Two terpenoid diepoxides from the green microalga Botryococcus braunii: their biomimetic conversion to tetrahydrofurans and tetrahydropyrans. Tetrahedron 55, 167–176 [Google Scholar]
- 15. Metzger P., Rager M. N., Largeau C. (2002) Botryolins A and B, two tetramethylsqualene triethers from the green microalga Botryococcus braunii. Phytochemistry 59, 839–843 [DOI] [PubMed] [Google Scholar]
- 16. Okada S., Tonegawa I., Matsuda H., Murakami M., Yamaguchi K. (1997) Braunixanthins 1 and 2, new carotenoids from the green microalga Botryococcus braunii. Tetrahedron 53, 11307–11316 [Google Scholar]
- 17. Niehaus T. D., Okada S., Devarenne T. P., Watt D. S., Sviripa V., Chappell J. (2011) Identification of unique mechanisms for triterpene biosynthesis in Botryococcus braunii. Proc. Natl. Acad. Sci. U.S.A. 108, 12260–12265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Okada S., Devarenne T. P., Chappell J. (2000) Molecular characterization of squalene synthase from the green microalga Botryococcus braunii, race B. Arch. Biochem. Biophys. 373, 307–317 [DOI] [PubMed] [Google Scholar]
- 19. Metzger P., Casadevall E. (1983) Structure of three new botryococcenes synthesized by a laboratory-cultured strain of Botryococcus braunii. Tetrahedron Lett. 24, 4013–4016 [Google Scholar]
- 20. Metzger P., Casadevall E., Pouet M. J., Pouet Y. (1985) Structures of some botryococcenes: branched hydrocarbons from the b-race of the green alga Botryococcus braunii. Phytochemistry 24, 2995–3002 [Google Scholar]
- 21. Achitouv E., Metzger P., Rager M. N., Largeau C. (2004) C31-C34 methylated squalenes from a Bolivian strain of Botryococcus braunii. Phytochemistry 65, 3159–3165 [DOI] [PubMed] [Google Scholar]
- 22. Ueki N., Matsunaga S., Inouye I., Hallmann A. (2010) How 5000 independent rowers coordinate their strokes in order to row into the sunlight: phototaxis in the multicellular green alga Volvox. BMC Biol. 8, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Banerjee A., Sharma R., Chisti Y., Banerjee U. C. (2002) Botryococcus braunii: a renewable source of hydrocarbons and other chemicals. Crit. Rev. Biotechnol. 22, 245–279 [DOI] [PubMed] [Google Scholar]
- 24. Morrison R. T., Boyd R. N. (1973). in Organic Chemistry, 3rd Ed., pp. 109–110, Allyn and Bacon, Boston, MA [Google Scholar]
- 25. Hillen L. W., Pollard G., Wake L. V., White N. (1982) Hydrocracking of the oils of Botryococcus braunii to transport fuels. Biotechnol. Bioeng. 24, 193–205 [DOI] [PubMed] [Google Scholar]
- 26. Schubert H. L., Blumenthal R. M., Cheng X. (2003) Many paths to methyltransfer: a chronicle of convergence. Trends Biochem. Sci. 28, 329–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Noel J. P., Dixon R. A., Pichersky E., Zubieta C., Ferrer J. L. (2003) Structural, functional, and evolutionary basis for methylation of plant small molecules. Rec. Adv. Phytochem. 37, 37–58 [Google Scholar]
- 28. Kagan R., Clarke S. (1995) Widespread occurrence of three sequence motifs in diverse S-adenosylmethionine-dependent methyltransferases suggests a common structure for these enzymes. Arch. Biochem. Biophys. 316, 657–657 [DOI] [PubMed] [Google Scholar]
- 29. Liscombe D. K., Usera A. R., O'Connor S. E. (2010) Homolog of tocopherol C methyltransferases catalyzes N-methylation in anticancer alkaloid biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 107, 18793–18798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zubieta C., Ross J. R., Koscheski P., Yang Y., Pichersky E., Noel J. P. (2003) Structural basis for substrate recognition in the salicylic acid carboxyl methyltransferase family. Plant Cell 15, 1704–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Diener A. C., Li H., Zhou W., Whoriskey W. J., Nes W. D., Fink G. R. (2000) Sterol methyltransferase 1 controls the level of cholesterol in plants. Plant Cell 12, 853–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Grebenok R. J., Galbraith D. W., Penna D. D. (1997) Characterization of Zea mays endosperm C-24 sterol methyltransferase: one of two types of sterol methyltransferase in higher plants. Plant Mol. Biol. 34, 891–896 [DOI] [PubMed] [Google Scholar]
- 33. Song L. (2003) Detection of farnesyl diphosphate accumulation in yeast ERG9 mutants. Anal. Biochem. 317, 180–185 [DOI] [PubMed] [Google Scholar]
- 34. Takahashi S., Yeo Y., Greenhagen B. T., McMullin T., Song L., Maurina-Brunker J., Rosson R., Noel J. P., Chappell J. (2007) Metabolic engineering of sesquiterpene metabolism in yeast. Biotechnol. Bioeng. 97, 170–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Huang Z., Poulter C. D. (1989) Isoshowacene, a C31 hydrocarbon from Botryococcus braunii var. showa Phytochemistry 28, 3043–3046 [Google Scholar]
- 36. Pompon D., Louerat B., Bronine A., Urban P. (1996) Yeast expression of animal and plant P450s in optimized redox environments. Methods Enzymol 272, 51–64 [DOI] [PubMed] [Google Scholar]
- 37. Carland F., Fujioka S., Nelson T. (2010) The sterol methyltransferases SMT1, SMT2, and SMT3 influence Arabidopsis development through nonbrassinosteroid products. Plant Physiol. 153, 741–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Merchant S. S., Prochnik S. E., Vallon O., Harris E. H., Karpowicz S. J., Witman G. B. (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318, 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ridgway N. D., Vance D. E. (1987) Purification of phosphatidylethanolamine N-methyltransferase from rat liver. J. Biol. Chem. 262, 17231–17239 [PubMed] [Google Scholar]
- 40. Malhotra H. C., Nes W. R. (1971) The mechanism of introduction of alkyl groups at C-24 of sterols. IV. Inhibition by triparanol. J. Biol. Chem. 246, 4934–4937 [PubMed] [Google Scholar]
- 41. Nes W. D. (2000) Sterol methyl transferase: enzymology and inhibition. Biochim. Biophys. Acta 1529, 63–88 [DOI] [PubMed] [Google Scholar]
- 42. Shintani D., DellaPenna D. (1998) Elevating the vitamin E content of plants through metabolic engineering. Science 282, 2098–2100 [DOI] [PubMed] [Google Scholar]
- 43. Barkovich R. J., Shtanko A., Shepherd J. A., Lee P. T., Myles D. C., Tzagoloff A., Clarke C. F. (1997) Characterization of the COQ5 gene from Saccharomyces cerevisiae: evidence for a C-methyltransferase in ubiquinone biosynthesis. J. Biol. Chem. 272, 9182–9188 [DOI] [PubMed] [Google Scholar]
- 44. Leustek T., Smith M., Murillo M., Singh D. P., Smith A. G., Woodcock S. C., Awan S. J., Warren M. J. (1997) Siroheme biosynthesis in higher plants: analysis of an S-adenosyl-l-methionine-dependent uroporphyrinogen III methyltransferase from Arabidopsis thaliana. J. Biol. Chem. 272, 2744–2752 [DOI] [PubMed] [Google Scholar]
- 45. Rodríguez D., Quirós L. M., Salas J. A. (2004) MtmMII-mediated C-methylation during biosynthesis of the antitumor drug mithramycin is essential for biological activity and DNA-drug interaction. J. Biol. Chem. 279, 8149–8158 [DOI] [PubMed] [Google Scholar]
- 46. Zhang W., Watanabe K., Wang C. C., Tang Y. (2007) Investigation of early tailoring reactions in the oxytetracycline biosynthetic pathway. J. Biol. Chem. 282, 25717–25725 [DOI] [PubMed] [Google Scholar]
- 47. Komatsu M., Tsuda M., Omura S., Oikawa H., Ikeda H. (2008) Identification and functional analysis of genes controlling biosynthesis of 2-methylisoborneol. Proc. Natl. Acad. Sci. U.S.A. 105, 7422–7427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gang D. R. (2005) Evolution of flavors and scents. Annu. Rev. Plant Biol. 56, 301–325 [DOI] [PubMed] [Google Scholar]
- 49. Han Y., Gasic K., Korban S. S. (2007) Multiple-copy cluster-type organization and evolution of genes encoding O-methyltransferases in the apple. Genetics 176, 2625–2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gang D. R., Lavid N., Zubieta C., Chen F., Beuerle T., Lewinsohn E., Noel J. P., Pichersky E. (2002) Characterization of phenylpropene O-methyltransferases from sweet basil: facile change of substrate specificity and convergent evolution within a plant O-methyltransferase family. Plant Cell 14, 505–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yang Y., Yuan J. S., Ross J., Noel J. P., Pichersky E., Chen F. (2006) An Arabidopsis thaliana methyltransferase capable of methylating farnesoic acid. Arch. Biochem. Biophys. 448, 123–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.