Background: C. difficile TcdA and TcdB glucosylate small GTPases.
Results: Structural and functional studies reveal comparable activities with Rho substrates, enhanced activities following autoprocessing, and TcdA-specific modification of Rap2A.
Conclusion: TcdA is a potent enzyme and modifies a broader array of GTPase substrates than TcdB.
Significance: These findings highlight the importance of autoprocessing for activity and reveal differences in target specificity between the toxins.
Keywords: Bacterial Toxins, Crystallography, Glycosyltransferases, Protein Structure, Rho GTPases, Clostridium difficile, Rap2A
Abstract
The principle virulence factors in Clostridium difficile pathogenesis are TcdA and TcdB, homologous glucosyltransferases capable of inactivating small GTPases within the host cell. We present crystal structures of the TcdA glucosyltransferase domain in the presence and absence of the co-substrate UDP-glucose. Although the enzymatic core is similar to that of TcdB, the proposed GTPase-binding surface differs significantly. We show that TcdA is comparable with TcdB in its modification of Rho family substrates and that, unlike TcdB, TcdA is also capable of modifying Rap family GTPases both in vitro and in cells. The glucosyltransferase activities of both toxins are reduced in the context of the holotoxin but can be restored with autoproteolytic activation and glucosyltransferase domain release. These studies highlight the importance of cellular activation in determining the array of substrates available to the toxins once delivered into the cell.
Introduction
Clostridium difficile is the leading cause of hospital-acquired diarrhea, and in recent years, the number of C. difficile-associated disease cases has increased dramatically. The primary virulence factors of C. difficile are the large, homologous exotoxins TcdA2 and TcdB (1, 2), members of the large clostridial toxin family, which also includes lethal toxin and hemorrhagic toxin from Clostridium sordellii and the α-toxin from Clostridium novyi. The enzymatic function of the toxins is carried out by a 63-kDa N-terminal glucosyltransferase domain (GTD) that acts on small GTPases involved in regulation of the cytoskeleton (3, 4). Delivery of the GTD into the cell is achieved by binding to the target cells (5, 6), endocytosis (7, 8), pH-induced pore formation (9–11), translocation of the GTD across the endosomal membrane, and release of the GTD by autoproteolysis (12, 13). Once delivered to the cytosol, the GTDs inactivate Rho and Ras family GTPases by glucosylation of a threonine residue in the switch I region (Thr-37 in RhoA) (14, 15). TcdA and TcdB have been reported to glucosylate RhoA, RhoB, RhoC, RhoG, Rac1-3, Cdc42, and TC10 using the co-substrate UDP-glucose (16, 17). Glucosylation of the GTPases prevents their interactions with multiple effectors and regulatory molecules and thereby prevents multiple signaling events in the host cell (18).
Although TcdA and TcdB have long been accepted as the primary virulence factors responsible for C. difficile-associated disease, there are conflicting data concerning the relative importance of each of the two toxins in causing disease (1, 2, 19–24). Given that the majority of pathogenic strains of C. difficile produce both TcdA and TcdB, it is unlikely that these two toxins have completely redundant functions. Although the roles of the toxins in human disease are unclear, a number of differences in activity have been noted for TcdA and TcdB in cells and in animal models. TcdA has been shown to be a potent enterotoxin in hamster and rabbit ileal loop assays, whereas TcdB is not (20, 21). Both toxins are reported to have potent enterotoxic effects on human tissues (19, 25). In cell culture, both TcdA and TcdB induce cell rounding, but TcdB is ∼1000 times more potent than TcdA in most cell lines (26, 27).
The molecular basis for the observed differences in TcdA and TcdB cytopathicity could include differences in the binding (26), pore-forming (9), autoproteolysis (28), and glucosyltransferase (26) activities. Multiple studies have demonstrated that TcdA and TcdB have different binding activities, suggesting that the toxins have distinct receptors (26, 29–31). Distinct binding targets almost certainly contribute to differences in potency toward various cells (26). However, in one of the few studies directly comparing the activities of TcdA and TcdB, Chaves-Olarte et al. (26) have reported that the difference in glucosyltransferase activity was the major determinant contributing to the difference in cytopathic potency between TcdA and TcdB. The authors showed that TcdB holotoxin (HT) is ∼100 times more active than TcdA HT at modifying substrate in vitro. In the absence of GTPase, TcdB also had a higher rate of UDP-glucose hydrolysis (26).
To understand the mechanistic basis for the differences in glucosyltransferase activities of TcdA and TcdB, we have determined structures of the TcdA GTD and compared them with those from other large clostridial toxins. Analysis of these structures along with biochemical comparisons of TcdA and TcdB glucosyltransferase activities are reported.
EXPERIMENTAL PROCEDURES
Plasmids
The nucleotide sequences encoding amino acids 1–2710 of TcdA (TcdA HT), 1–1832 of TcdA, 1–800 of TcdA, 1–542 of TcdA (TcdA GTD), or 1–543 of TcdB (TcdB GTD) were amplified from VPI 10463 and cloned into the Bacillus megaterium expression vector pC-His1622 (MoBiTec, BMEG20) using the restriction sites BsrGI and SphI. The gene encoding TcdB was cloned into BMEG20 as described previously (32). The GTPase and effector sequences were cloned into pGEX4T (RhoA (33) Rap2A, Ral guanine nucleotide dissociation stimulator (RalGDS)-receptor binding domain (RBD) (34)) or pGEX2T (Rac1 (33), Cdc42 (33)) using the sites BamHI and EcoRI.
Protein Expression and Purification
The plasmids encoding TcdA, TcdB, and toxin fragments were transformed into B. megaterium following the manufacturer's instructions (MoBiTec). Transformed B. megaterium were grown in LB containing 10 mg/liter tetracycline in LB. 30 ml of overnight culture was used to inoculate 1 liter of medium, and the cultures were placed at 37 °C and 230 rpm. When the cultures reached A600 = 0.3, expression was induced by the addition of 5 g of d-xylose. After 4.5 h, the cells were harvested by centrifugation and resuspended in 100 mm NaCl, 20 mm Tris, pH 8.0. Following French press lysis, the lysates were centrifuged at 48,000 × g for 20 min. Protein was purified from the supernatant by nickel affinity chromatography followed by gel filtration chromatography in 100 mm NaCl, 20 mm Tris, pH 8.
RalGDS-RBD and the GTP-binding proteins were expressed in Escherichia coli BL21 cells grown in LB containing 100 mg/liter ampicillin. 10 ml of overnight culture was used to inoculate 1 liter of medium, and the cultures were placed at 37 °C and 230 rpm. When the cultures reached A600 = 0.6, the temperature was changed to 21 °C, and expression was induced by the addition of 0.5 mm IPTG. After 16 h, the cells were harvested by centrifugation and resuspended in 100 mm NaCl, 20 mm Tris, pH 8.0. Following French press lysis, the lysates were centrifuged at 48,000 × g for 20 min. Protein was purified from the supernatant using glutathione-Sepharose 4B (GE Healthcare) followed by gel filtration chromatography. The GST tags were not removed.
Crystallization
TcdA GTD was concentrated to 16 mg/ml in 150 mm NaCl, 20 mm Tris, pH 8.0. TcdA GTD was crystallized by the sitting drop method at 21 °C with a 1:1 ratio of protein to mother liquor containing 0.2 m l-proline, 10% PEG 3350, and 0.1 m HEPES, pH 7.5. For co-crystallization of the GTD with substrate, 10 mm UDP-glucose and 2 mm MnCl2 were added to the protein, and hanging drops were prepared with mother liquor containing 20% PEG 6000 and 0.1 m Bicine, pH 8–9. Crystals were exchanged into the appropriate mother liquor containing either 15% glycerol (protein alone) or 20% ethylene glycol (protein plus UDP-glucose/Mn2+), mounted on cryo loops, and flash cooled in liquid nitrogen.
Structure Determination and Refinement
X-ray data were collected from single crystals on NE-CAT beamline 24 ID-C at the Advanced Photon Source (Argonne, IL) at 100 K and a wavelength of 1.0094 Å. Diffraction data were indexed, integrated, and scaled using HKL2000 (35). A starting model was obtained for the TcdA GTD without UDP-glucose by molecular replacement with the TcdB GTD (Protein Data Bank entry 2BVM) as a search model using Phenix. The model was iteratively built using Coot (36) and refined using Phenix (37) with 5 TLS groups per chain (supplemental Table S1). The structure with UDP-glucose bound was determined in the same way, except the apo structure was used as the search model for molecular replacement. In the final structures, 90.0 and 91.2% of the residues were in the most favored positions in the Ramachandran plot for the bound and apo structures, respectively (calculated by PROCHECK (38)). No residues were in disallowed regions. For the apo structure, the final model consists of residues 2–538, one manganese ion, and 233 water molecules. For the structure with UDP-glucose bound, the final model consists of residues 2–538, one manganese, one UDP-glucose, and 109 water molecules.
In Vitro Glucosyltransferase Assay
UDP-[14C]glucose (250 mCi/mmol) was obtained from PerkinElmer Life Sciences. GTD (0.1 nm) and GTP binding protein (2 μm) were mixed with 24 μm UDP-[14C]glucose in a buffer containing 50 mm HEPES, pH 7.5, 100 mm KCl, 1 mm MnCl2, 2 mm MgCl2, and 0.1 mg/ml BSA. The reactions were incubated at 37 °C for 5, 10, 15, 30, or 60 min. The reaction was stopped by the addition of loading buffer and heating, and the proteins were separated by SDS-PAGE. Glucosylation of GTPase was analyzed by phosphorimaging. For graphical representation, band densitometry was performed with ImageJ software, and the band intensities were normalized with Cdc42 modified by TcdA GTD set at 1.
Rap Activation Assay
HeLa cells were cultured in Dulbecco's modified Eagle medium supplemented with 10% FBS. Caco-2 cells were grown in minimum Eagle's medium with 10% FBS. Cells were grown to 80% confluence in 10-cm dishes and then treated with buffer, 10 nm TcdA, or 0.1 nm TcdB in serum-free medium. After 2 h, the cells were washed two times with PBS. The cells were lysed and resuspended in 0.4 ml of lysis buffer (50 mm Tris, pH 7.5, 500 mm NaCl, 10 mm MgCl2, 2% Igepal). The lysates were clarified by centrifugation and normalized for protein concentration. Activated Rap2A was pulled down with GST-RalGDS-RBD (50 μg) on glutathione-Sepharose 4B resin for 1 h at 4 °C. The resin was washed four times with 0.5 ml of wash buffer (25 mm Tris, pH 7.5, 40 mm NaCl, 30 mm MgCl2). 25 μl of loading buffer was added to the beads, and the sample was heated at 95 °C for 5 min. Bound Rap2A was detected by Western blot using a Rap2A antibody (610215, BD Transduction Laboratories).
RESULTS
Structure of TcdA GTD
TcdA GTD crystal structures were determined in the presence and absence of UDP-glucose at 2.6 Å (Protein Data Bank entry 3SRZ) and 2.2 Å (Protein Data Bank entry 3SS1) resolution, respectively (Fig. 1 and supplemental Table S1 and Figs. S1 and S2). As observed in GTD structures from other large clostridial toxins, the molecule is composed of a core GT-A fold surrounded by multiple helical projections (39). The N-terminal projection (Fig. 1A) is thought to act as a membrane localization domain (MLD), targeting the GTD to the site of membrane-bound GTPases (40, 41). The other projections at the top right and top left of the GTD “front” (Fig. 1A) are thought to contribute to GTPase substrate specificity (39).
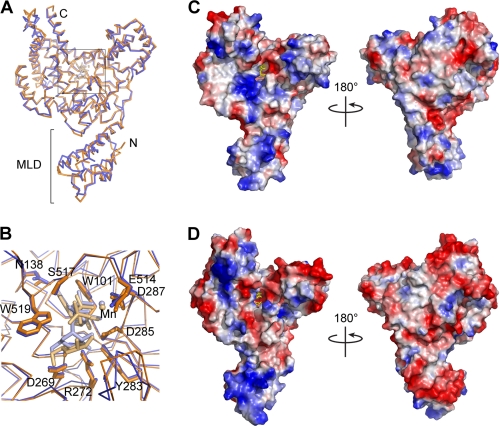
FIGURE 1.
Structure of the TcdA GTD. A–C, the UDP-glucose-bound TcdA GTD is shown with the core GT-A family fold in blue and the α-helical protrusions from the fold in green. The mobile 516–522 loop is in yellow, and the loop from the apo structure is superimposed on the structure in pink. Ser-517, Trp-519, and UDP-glucose are represented as sticks. B, a close-up view of the 516–522 loop; C, a “top” view (rotated 90° from the “front” view shown in A).
Comparison of the apo- and UDP-glucose-bound structures shows a significant difference in the position of the 516–522 loop (Fig. 1 and supplemental Fig. S2). This loop contains a conserved serine, Ser-517 in TcdA, which forms a hydrogen bond with the β-phosphate group in UDP-glucose, and a conserved tryptophan, Trp-519 in TcdA, which forms a hydrogen bond to the glycosidic oxygen (Fig. 1B and supplemental Fig. S3). In the apo structure, the loop is moved such that Trp-519 is located ∼10 Å away from its position in the UDP-glucose-bound structure. A similar conformational difference has been noted in a comparison of the apo structure of C. novyi α-toxin GTD and TcdB GTD bound to a hydrolyzed substrate (42). As described in the mammalian glucosyltransferases involved in carbohydrate synthesis (43), the loop acts as a “lid” covering the bound UDP-glucose when viewed from the “top” (Fig. 1C).
One difference between the TcdA and TcdB GTD structures is that the UDP-glucose is intact in the TcdA-GTD structure, whereas in the TcdB-GTD structure, it is hydrolyzed (Fig. 2). Although consistent with previously published studies showing that TcdB has a higher rate of UDP-glucose hydrolysis than TcdA (26, 44), the molecular explanation for this difference is not apparent from the structure. Because TcdA was previously reported to have a much lower glucosyltransferase activity than TcdB (26), we expected to see differences in the positions of the catalytic residues. However, other than the difference in the hydrolysis of the UDP-glucose, the enzymatic core is surprisingly similar between TcdA and TcdB. The residues and waters involved in UDP-binding and catalysis are highly conserved, and the binding of UDP-glucose is nearly identical (Fig. 2B).
FIGURE 2.
Comparison of the TcdA and TcdB GTD structures. The structures of the domains are similar in their overall architecture with a root mean square deviation of 1.6 Å for the α-carbon backbone. A, the aligned structures of the TcdA (blue) and TcdB (orange) GTDs are shown as backbone traces. B, a close-up view of the catalytic site with UDP-glucose coordinating residues shown as sticks. UDP-glucose (light blue), UDP (light orange), and glucose (light orange) are also shown as sticks. Manganese ions are shown as small spheres. Although the cores of the GTDs are conserved, the surfaces are highly divergent (C and D). The electrostatic surface potentials of the TcdA GTD (C) and the TcdB GTD (D) are shown with positively charged surfaces colored blue and negatively charged surfaces in red. The coordinated glyconucleosides are shown as yellow sticks.
Although the core structures of the TcdA and TcdB GTDs are conserved (Fig. 2A), the surface residues of these two enzymes are highly divergent. This is particularly notable on the “front” GTPase-binding surface adjacent to the UDP-glucose site (45). Amino acid changes in this region result in a significant change in the electrostatic properties of the surface and suggest that the TcdA and TcdB GTDs could have different substrate specificities within the cell (Fig. 2, C and D). In addition, there are significant differences in the electrostatic potential properties of the MLD. The TcdB MLD is markedly more charged than that of TcdA. The “front” surface (as shown in Fig. 2D, left) is dominated by a highly basic patch, whereas the opposite face is almost entirely acidic. TcdA has a smaller basic area on the front of the MLD and lacks the negatively charged patch on the back (Fig. 2C). These differences could further differentiate the activities of the TcdA and TcdB GTDs in cells.
Glucosyltransferase Activity of TcdA and TcdB GTDs
The striking differences between the TcdA and TcdB surfaces led us to perform a side-by-side comparison of TcdA and TcdB GTD activity against a panel of Rho and Ras family GTPase substrates. TcdA and TcdB GTD were incubated with purified GTPases in the presence of radiolabeled UDP-glucose. Transfer of glucose was detected by SDS-PAGE followed by phosphorimaging. We found that the TcdA and TcdB glucosyltransferase activities against Rho family substrates were comparable (Fig. 3). In time course experiments, TcdA was more efficient at modifying RhoA, whereas TcdB was faster at modifying Rac1. TcdA and TcdB modified Cdc42 to a similar extent with similar rates. Consistent with a previous report (26), we observed that TcdA could glucosylate Rap2A, whereas TcdB did not. Rap2A was not modified as efficiently as the Rho family substrates.
FIGURE 3.
Glucosyltransferase activity of the TcdA and TcdB GTDs. TcdA and TcdB GTDs (0.1 nm) were tested for their ability to glucosylate a panel of GTPases (2 μm) over the course of 1 h using UDP-[14C]glucose (24 μm) as a co-substrate. The band intensities for the representative experiments shown in the insets were quantified by densitometry. The means ± S.D. from three independent replicates are shown with TcdA in circles and TcdB in triangles. The data are scaled with the average value for Cdc42 modified by TcdA GTD set at 1.
Enhanced Glucosyltransferase Activity following Autoprocessing
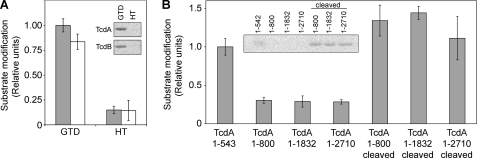
The highly conserved enzymatic core observed in the structure of the TcdA GTD provides no evidence that it is a deficient enzyme as compared with TcdB. Consistent with this view, our in vitro studies indicate that the GTDs of TcdA and TcdB modify similar amounts of Rho family GTPases. Previous studies on the glucosyltransferase activity of TcdA have used only TcdA HT. In cells, however, the GTD is released, and this isolated domain is thought to traffic to the membrane, where it acts on host GTPases (40, 46). We wondered whether using GTD versus HT might result in a difference in activity. Therefore, we tested the ability of GTD or HT to modify Cdc42 for both toxins (Fig. 4A). For both TcdA and TcdB, the HT modified less substrate than the free GTD. Therefore, in the context of the HT, the glucosyltransferase activity of TcdA and TcdB is somehow inhibited.
FIGURE 4.
Inhibition of glucosyltransferase activity in the holotoxin. Recombinant Cdc42 (2 μm) was incubated with UDP-[14C]glucose (24 μm) and the indicated toxin or toxin fragment (0.1 nm each) for 1 h. The proteins were resolved by SDS-PAGE, and the gels were analyzed by phosphorimaging. A, comparison of GTD and HT activity of TcdA (gray bars) and TcdB (white bars) (n = 4). Activity is inhibited in the holotoxins. B, TcdA GTD(1–542), TcdA(1–800), TcdA(1–1832), and TcdA HT(1–2710) were tested for their capacity to modify Cdc42 (n = 3). The activity of the 1–800, 1–1832, and 1–2710 proteins was reduced relative to GTD but was increased after induction of autoprocessing. Band intensities were quantified, and the data were scaled with the average value for Cdc42 modified by TcdA GTD set at 1. Error bars, S.D. Insets show representative experiments.
Structure of TcdA GTD in Context of Holotoxin
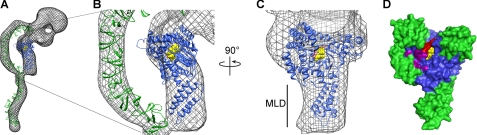
Because the HT has a lower activity than the GTD (Fig. 4), we sought to understand the structural determinants of the glucosyltransferase activity in the context of the holotoxin. Our laboratory has recently determined the structure of TcdA HT at 25 Å resolution using electron microscopy and random conical tilt (Fig. 5A) (32). The structure contains a large bilobed head with two extensions. We have shown that the larger of these extensions contains the RBD, and the smaller one contains the GTD (32). Fig. 5 shows the EM structure of TcdA with a model of the TcdA RBD (green) and the structure of the TcdA GTD (blue) placed into the density. The GTD fits into the map with the N-terminal MLD pointing away from the head and fitting into a narrower region of density. The other helical projections of the GTD fit into density “flaps” that project toward the RBD and provide confidence that the general orientation of the GTD is correct.
FIGURE 5.
Structure of the TcdA GTD alone and in the context of the TcdA holotoxin structure. A–C, three-dimensional reconstruction of TcdA (32) filtered to 25 Å is shown as a mesh surface with the crystal structure of the TcdA GTD (blue) and a model of the TcdA RBD (52) (green) placed into the density. UDP-glucose is shown as yellow spheres. In C, the model of the binding domain and the corresponding map density are removed. D, surface of the TcdA GTD shown in the same orientation as in C. The core GT-A fold is shown in blue with the additional α-helical regions in green (as in Fig. 1). The 516–522 loop is colored red, and UDP-glucose is represented as yellow spheres. Amino acids Lys-448, Gln-454, Glu-460, Arg-462, and Gly-471 are shown in purple. The corresponding residues in TcdB have been shown to be involved in substrate binding (45).
Oriented in this way (Fig. 5), the “front” surface involved in GTPase binding faces the RBD. Based on this arrangement, we predicted that the presence of the RBD sterically inhibits binding of GTPases in the context of the holotoxin. In addition, the model suggests that the “top” surface containing the mobile 516–522 loop will be occluded by the autoprocessing domain in the context of the holotoxin. This occlusion could affect the position of the 516–522 loop involved in UDP glucose binding, hydrolysis, and transfer.
To test these possibilities, we generated a TcdA fragment lacking the RBD (residues 1–1832) and a fragment corresponding to the glucosyltransferase and autoprocessing domains (residues 1–800) and tested these proteins for their capacity to modify Cdc42 (Fig. 4B). The glucosyltransferase reaction is inhibited in both the 1–800 and 1–1832 fragments and in the holotoxin (residues 1–2710). Releasing the GTD by initiating in vitro autoprocessing results in enhanced substrate modification for these proteins, comparable with that of the GTD alone (Fig. 4 and supplemental Fig. S4B).
Glucosylation of Rap2A in Cells
The increased activity of TcdA and TcdB following autoprocessing highlights the importance of the cellular context for cellular activity. Because our in vitro assay indicated that Rap proteins were modified at lower levels than Rho family proteins, we wanted to test whether TcdA-mediated Rap modification could occur in the context of cellular intoxication.
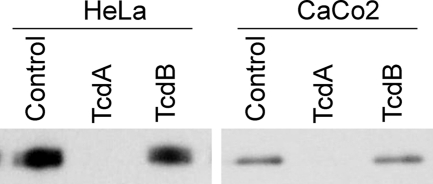
HeLa and Caco-2 cells were treated with 10 nm TcdA or 0.1 nm TcdB, doses that cause the cells to round but not die within 2 h. After 2 h, the cells were lysed, and activated Rap was pulled down using the effector RalGDS. Activated Rap2A was detected by Western blot in mock-treated and TcdB-treated cells (Fig. 6). However, no activated Rap2A was detected in TcdA-treated cells, suggesting that it had been inactivated by glucosylation.
FIGURE 6.
Inactivation of Rap2A in cells treated with TcdA. HeLa or Caco2 cells were treated with buffer, 10 nm TcdA, or 0.1 nm TcdB for 2 h. Activated Rap2A was pulled down from the cell lysates using RalGDS-RBD and detected by Western blot.
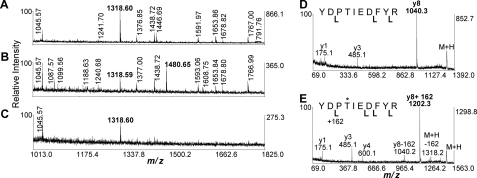
We have directly detected modification of Rap2A using a mass spectrometry approach (Fig. 7 and supplemental Fig. S5 and supplemental Methods). HeLa cells expressing FLAG-tagged Rap2A were either mock-treated or treated with 10 nm TcdA or TcdB. Rap2A was pulled down and analyzed by MALDI-TOF/TOF mass spectrometry. A peptide covering Rap2A amino acids 32–41 (m/z 1318.60 Da) was observed for Rap pulled down from mock-treated, TcdA-treated, and TcdB-treated cells. A related peptide (m/z 1480.65) was observed only for pull-downs from TcdA-treated cells, consistent with glucosylation (+162 shift) of Thr-35 (Fig. 7B). TOF/TOF fragmentation spectra of both species were consistent with the predicted sequence and mapped the site of glucosylation to Thr-35 (Fig. 7, D and E). Therefore, TcdA is not only capable of glucosylating Rap2A in vitro but can also glucosylate it in cells.
FIGURE 7.
Glucosylation of Rap2A in cells treated with TcdA. A–E, MALDI-TOF/TOF mass spectrometry indicating the glucosylation of the peptide YDPTIEDFYR. A–C, a portion of the MALDI-TOF peptide mass map (m/z 1013–1825) is shown to highlight the diagnostic singly charged peptide ions at m/z 1318.60 and 1480.65 (labeled in boldface type) that represent the peptide in the native and glucosylated state, respectively. The m/z 1480.65 is only found in the TcdA-treated samples (B). MALDI-TOF/TOF fragmentation spectra are shown for the m/z 1318.60 peptide (D) and for the glucosylated m/z 1480.65 form (E). Labeled y ions are denoted by cleavage brackets below the sequence. The y8 fragment ion that contains Thr-35 is diagnostic for the modification, which adds 162 Da. Ions are also observed that are consistent with neutral loss of glucose from the y8 and M + H ions.
DISCUSSION
We initiated this study with the goal of understanding differences in TcdA and TcdB glucosyltransferase activity and substrate specificity. The glucosylation of RhoA, Rac1, and Cdc42 by TcdB has been well characterized (14, 39, 45, 47); however, much less is known about TcdA and the interaction of TcdA with these substrates. One of the few studies directly comparing the enzymatic activities of TcdA and TcdB reports that TcdB is ∼100 times more active than TcdA at glucosylating RhoA, Rac1, and Cdc42 in vitro and has a higher rate of UDP-glucose hydrolysis in the absence of substrate (26). This study was done with the holotoxins and not with the isolated domains that are released into the cell.
Here we present crystal structures of the TcdA GTD with and without its co-substrate UDP-glucose at 2.6 and 2.2 Å, respectively. Because TcdA was reported to have a much lower glucosyltransferase activity than TcdB (26), we expected to see differences in the residues involved in UDP binding and catalysis, yet the structure reveals that these residues are highly conserved, not only in identity but also in their position within the GTD (Fig. 2B).
The conservation of the core of the GTDs led us to reinvestigate their activity toward a panel of GTPase substrates. Using experimental conditions similar to those of previous reports, we found that TcdA and TcdB modified similar amounts of Rho family substrates (Fig. 3). These results contradict the previous report that TcdB is 100 times more potent as a glucosyltransferase (26). We hypothesized that this difference might be due to the fact that previous studies used HT, whereas we used isolated GTD. Upon testing the ability of GTD and HT to modify Cdc42, we found that the glucosyltransferase activity is inhibited in the context of the HT (Fig. 4A). The activity of the HT can be restored through the initiation of autoprocessing (Fig. 4B), an inositol-hexakisphosphate-induced proteolysis event that releases the GTD from the rest of the toxin (12, 13). We now know that TcdB undergoes autoprocessing much more readily than TcdA (28), and, in our hands, TcdB is also more sensitive to degradation. A gel documenting the cleavage state of our reagents is included as supplemental Fig. S4A. It is possible that in the previous study, TcdB was more active than TcdA because the GTD was released by proteolysis, whereas TcdA remained locked in a less active conformation. Our findings suggest that the ∼1000-fold difference in cytopathic potency between TcdA and TcdB is not due to differences in the intrinsic glucosyltransferase activity but is more likely due to differences in binding and/or delivery of the GTD into the target cell.
The observation that GTD release results in an increase in glucosyltransferase activity led us to examine why the activity is restricted in the context of the holotoxin. Our first hypothesis, based on the placement of the TcdA GTD within the EM structure, was that GTPase binding would be sterically occluded by the location of the RBD. Our experiments with a TcdA truncation that lacks the RBD, however, suggest this is not the case; the glucosyltransferase activity of the TcdA(1–1832) protein is activated by autoproteolysis at levels similar to what we observed for the full-length toxin (Fig. 4B). Removal of the delivery domain through the generation of a TcdA(1–800) fragment also resulted in a protein with restricted activity until the GTD was released. These data suggest that the physical presence of the autoprotease domain (residues 543–800) is restricting the activity of the GTD. The autoprotease domain could affect the position of the 516–522 loop (Fig. 1, B and C), a possibility that will be investigated in future studies.
Although the TcdA and TcdB GTDs modified comparable amounts of RhoA, Rac1, and Cdc42 in our assays, TcdA was able to glucosylate the additional substrate Rap2A. To test whether this family of substrates was modified within cells treated with holotoxin, we analyzed the levels of activated Rap2A in cells treated with TcdA or TcdB. Activated Rap2A, which is present in mock-treated cells, was undetectable in HeLa or Caco-2 cells that had been treated with 10 nm TcdA for 2 h. The glucosylation of the switch I Thr of FLAG-tagged Rap2A could be detected in TcdA-treated HeLa cells by mass spectrometry. These data suggest that TcdA is capable of modifying a Ras family GTPase not only in vitro but also in cells.
Whereas TcdB and C. novyi α-toxin (Tcnα) target only Rho family substrates, TcdA and C. sordellii lethal toxin (TcsL) can glucosylate at least some members of both the Rho and Ras families. When the structures are compared, TcdA and TcsL have some striking similarities on the putative substrate-binding surface, most notably a large positively charged pocket adjacent to the UDP-glucose binding pocket (supplemental Fig. S6). In TcdB and Tcnα, however, an acidic patch replaces this basic pocket. It is important to note that these characteristics are not merely due to relatedness of the proteins. In fact, the GTDs of TcdB and TcsL are more closely related (75% identity) than the GTDs of TcdA and TcsL (47%). We hypothesize that this basic pocket may partially account for the ability of TcdA to modify Rap family substrates. In preliminary studies, mutation of either Lys-448 or Gly-471, residues that make up the pocket, to glutamate (the corresponding residues in TcdB) decreased the activity toward all of the substrates tested (data not shown). Efforts to delineate the combination of residues required for substrate specificity are on-going.
The ability of TcdA to modify Ras family substrates may have important implications for its role in pathogenesis. Rap proteins are known to be important in the regulation of cell-cell junctions (49). Thus, inactivation of Rap may be important for disrupting the integrity of the intestinal epithelium. Other Ras family members are involved in complex pathways regulating cell proliferation and survival (49). It is interesting to note that all TcdB sequences from the pathogenic TcdA−TcdB+ strains characterized thus far have mutations within the GTD that allow it to modify Ras family substrates, including Rap (50, 51). It is tempting to speculate that modification of Ras substrates by TcdA is a key process in C. difficile-associated disease and that the mutated TcdB in TcdA−TcdB+ strains allows it fulfill the role of TcdA in its absence. A more complete understanding of the glucosyltransferase activities of these two toxins and downstream effects of glucosylation of specific substrates will be essential in understanding the molecular mechanisms important in C. difficile-associated disease.
Supplementary Material
Acknowledgments
We gratefully acknowledge Jiong Shi, Sireesha Kollipara, and Chris Aiken (Vanderbilt) for assistance with generating a stable cell line and Eric Skaar (Vanderbilt) for providing laboratory space for experiments involving radioactivity. Plasmids for expression of GST-tagged human RhoA, Rac1, and Cdc42 were obtained from Seema Mattoo and Jack Dixon (University of California, San Diego). The Rap2A construct was provided by Alfred Wittinghofer (Max-Planck-Institut für Molekulare Physiologie, Dortmund, Germany).
This work was supported, in whole or in part, by National Institutes of Health Grant P30DK058404 (Vanderbilt Digestive Disease Research Center) and National Institutes of Health, NIAID, Grant AI095755-01. This work was also supported by an Investigator in Pathogenesis of Infectious Disease award from the Burroughs Wellcome Foundation (to D. B. L.). Use of the Advanced Photon Source at Argonne National Laboratory was supported by the United States Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract DE-AC02-06CH11357.

This article contains supplemental Table S1, Figs. S1–S6, Methods, and References.
The atomic coordinates and structure factors (codes 3SRZ and 3SS1) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- TcdA
- C. difficile toxin A
- TcdB
- C. difficile toxin B
- GTD
- glucosyltransferase domain
- HT
- holotoxin
- MLD
- membrane localization domain
- RalGDS
- Ral guanine nucleotide dissociation stimulator
- RBD
- receptor binding domain
- Tcnα
- C. novyi α-toxin
- TcsL
- C. sordellii lethal toxin
- Bicine
- N,N-bis(2-hydroxyethyl)glycine.
REFERENCES
- 1. Lyras D., O'Connor J. R., Howarth P. M., Sambol S. P., Carter G. P., Phumoonna T., Poon R., Adams V., Vedantam G., Johnson S., Gerding D. N., Rood J. I. (2009) Toxin B is essential for virulence of Clostridium difficile. Nature 458, 1176–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kuehne S. A., Cartman S. T., Heap J. T., Kelly M. L., Cockayne A., Minton N. P. (2010) The role of toxin A and toxin B in Clostridium difficile infection. Nature 467, 711–713 [DOI] [PubMed] [Google Scholar]
- 3. Hofmann F., Busch C., Prepens U., Just I., Aktories K. (1997) Localization of the glucosyltransferase activity of Clostridium difficile toxin B to the N-terminal part of the holotoxin. J. Biol. Chem. 272, 11074–11078 [DOI] [PubMed] [Google Scholar]
- 4. Rupnik M., Pabst S., Rupnik M., von Eichel-Streiber C., Urlaub H., Söling H. D. (2005) Characterization of the cleavage site and function of resulting cleavage fragments after limited proteolysis of Clostridium difficile toxin B (TcdB) by host cells. Microbiology 151, 199–208 [DOI] [PubMed] [Google Scholar]
- 5. von Eichel-Streiber C., Sauerborn M. (1990) Clostridium difficile toxin A carries a C-terminal repetitive structure homologous to the carbohydrate binding region of streptococcal glycosyltransferases. Gene 96, 107–113 [DOI] [PubMed] [Google Scholar]
- 6. Dove C. H., Wang S. Z., Price S. B., Phelps C. J., Lyerly D. M., Wilkins T. D., Johnson J. L. (1990) Molecular characterization of the Clostridium difficile toxin A gene. Infect. Immun. 58, 480–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Florin I., Thelestam M. (1986) Lysosomal involvement in cellular intoxication with Clostridium difficile toxin B. Microb. Pathog. 1, 373–385 [DOI] [PubMed] [Google Scholar]
- 8. Papatheodorou P., Zamboglou C., Genisyuerek S., Guttenberg G., Aktories K. (2010) Clostridial glucosylating toxins enter cells via clathrin-mediated endocytosis. PLoS One 5, e10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Giesemann T., Jank T., Gerhard R., Maier E., Just I., Benz R., Aktories K. (2006) Cholesterol-dependent pore formation of Clostridium difficile toxin A. J. Biol. Chem. 281, 10808–10815 [DOI] [PubMed] [Google Scholar]
- 10. Barth H., Pfeifer G., Hofmann F., Maier E., Benz R., Aktories K. (2001) Low pH-induced formation of ion channels by clostridium difficile toxin B in target cells. J. Biol. Chem. 276, 10670–10676 [DOI] [PubMed] [Google Scholar]
- 11. Qa'Dan M., Spyres L. M., Ballard J. D. (2000) pH-induced conformational changes in Clostridium difficile toxin B. Infect. Immun. 68, 2470–2474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Egerer M., Giesemann T., Jank T., Satchell K. J., Aktories K. (2007) Autocatalytic cleavage of Clostridium difficile toxins A and B depends on cysteine protease activity. J. Biol. Chem. 282, 25314–25321 [DOI] [PubMed] [Google Scholar]
- 13. Reineke J., Tenzer S., Rupnik M., Koschinski A., Hasselmayer O., Schrattenholz A., Schild H., von Eichel-Streiber C. (2007) Autocatalytic cleavage of Clostridium difficile toxin B. Nature 446, 415–419 [DOI] [PubMed] [Google Scholar]
- 14. Just I., Selzer J., Wilm M., von Eichel-Streiber C., Mann M., Aktories K. (1995) Glucosylation of Rho proteins by Clostridium difficile toxin B. Nature 375, 500–503 [DOI] [PubMed] [Google Scholar]
- 15. Just I., Wilm M., Selzer J., Rex G., von Eichel-Streiber C., Mann M., Aktories K. (1995) The enterotoxin from Clostridium difficile (ToxA) monoglucosylates the Rho proteins. J. Biol. Chem. 270, 13932–13936 [DOI] [PubMed] [Google Scholar]
- 16. Jank T., Giesemann T., Aktories K. (2007) Rho-glucosylating Clostridium difficile toxins A and B. New insights into structure and function. Glycobiology 17, 15R–22R [DOI] [PubMed] [Google Scholar]
- 17. Genth H., Dreger S. C., Huelsenbeck J., Just I. (2008) Clostridium difficile toxins. More than mere inhibitors of Rho proteins. Int. J. Biochem. Cell Biol. 40, 592–597 [DOI] [PubMed] [Google Scholar]
- 18. Schirmer J., Aktories K. (2004) Large clostridial cytotoxins. Cellular biology of Rho/Ras-glucosylating toxins. Biochim. Biophys. Acta 1673, 66–74 [DOI] [PubMed] [Google Scholar]
- 19. Savidge T. C., Pan W. H., Newman P., O'brien M., Anton P. M., Pothoulakis C. (2003) Clostridium difficile toxin B is an inflammatory enterotoxin in human intestine. Gastroenterology 125, 413–420 [DOI] [PubMed] [Google Scholar]
- 20. Mitchell T. J., Ketley J. M., Haslam S. C., Stephen J., Burdon D. W., Candy D. C., Daniel R. (1986) Effect of toxin A and B of Clostridium difficile on rabbit ileum and colon. Gut 27, 78–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lyerly D. M., Saum K. E., MacDonald D. K., Wilkins T. D. (1985) Effects of Clostridium difficile toxins given intragastrically to animals. Infect. Immun. 47, 349–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lyerly D. M., Barroso L. A., Wilkins T. D., Depitre C., Corthier G. (1992) Characterization of a toxin A-negative, toxin B-positive strain of Clostridium difficile. Infect. Immun. 60, 4633–4639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Giannasca P. J., Warny M. (2004) Active and passive immunization against Clostridium difficile diarrhea and colitis. Vaccine 22, 848–856 [DOI] [PubMed] [Google Scholar]
- 24. Babcock G. J., Broering T. J., Hernandez H. J., Mandell R. B., Donahue K., Boatright N., Stack A. M., Lowy I., Graziano R., Molrine D., Ambrosino D. M., Thomas W. D., Jr. (2006) Human monoclonal antibodies directed against toxins A and B prevent Clostridium difficile-induced mortality in hamsters. Infect. Immun. 74, 6339–6347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Riegler M., Sedivy R., Pothoulakis C., Hamilton G., Zacherl J., Bischof G., Cosentini E., Feil W., Schiessel R., LaMont J. T. (1995) Clostridium difficile toxin B is more potent than toxin A in damaging human colonic epithelium in vitro. J. Clin. Invest. 95, 2004–2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chaves-Olarte E., Weidmann M., Eichel-Streiber C., Thelestam M. (1997) Toxins A and B from Clostridium difficile differ with respect to enzymatic potencies, cellular substrate specificities, and surface binding to cultured cells. J. Clin. Invest. 100, 1734–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sullivan N. M., Pellett S., Wilkins T. D. (1982) Purification and characterization of toxins A and B of Clostridium difficile. Infect. Immun. 35, 1032–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kreimeyer I., Euler F., Marckscheffel A., Tatge H., Pich A., Olling A., Schwarz J., Just I., Gerhard R. (2011) Autoproteolytic cleavage mediates cytotoxicity of Clostridium difficile toxin A. Naunyn Schmiedebergs Arch. Pharmacol. 383, 253–262 [DOI] [PubMed] [Google Scholar]
- 29. Krivan H. C., Clark G. F., Smith D. F., Wilkins T. D. (1986) Cell surface binding site for Clostridium difficile enterotoxin. Evidence for a glycoconjugate containing the sequence Gal α 1–3Gal β 1–4GlcNAc. Infect. Immun. 53, 573–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dingle T., Wee S., Mulvey G. L., Greco A., Kitova E. N., Sun J., Lin S., Klassen J. S., Palcic M. M., Ng K. K., Armstrong G. D. (2008) Functional properties of the carboxyl-terminal host cell-binding domains of the two toxins, TcdA and TcdB, expressed by Clostridium difficile. Glycobiology 18, 698–706 [DOI] [PubMed] [Google Scholar]
- 31. Frisch C., Gerhard R., Aktories K., Hofmann F., Just I. (2003) The complete receptor-binding domain of Clostridium difficile toxin A is required for endocytosis. Biochem. Biophys. Res. Commun. 300, 706–711 [DOI] [PubMed] [Google Scholar]
- 32. Pruitt R. N., Chambers M. G., Ng K. K., Ohi M. D., Lacy D. B. (2010) Structural organization of the functional domains of Clostridium difficile toxins A and B. Proc. Natl. Acad. Sci. U.S.A. 107, 13467–13472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Worby C. A., Mattoo S., Kruger R. P., Corbeil L. B., Koller A., Mendez J. C., Zekarias B., Lazar C., Dixon J. E. (2009) The fic domain. Regulation of cell signaling by adenylylation. Mol. Cell. 34, 93–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. de Rooij J., Zwartkruis F. J., Verheijen M. H., Cool R. H., Nijman S. M., Wittinghofer A., Bos J. L. (1998) Epac is a Rap1 guanine-nucleotide-exchange factor directly activated by cyclic AMP. Nature 396, 474–477 [DOI] [PubMed] [Google Scholar]
- 35. Otwinowski Z, Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 36. Emsley P., Cowtan K. (2004) Coot. Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 37. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) PHENIX. Building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 38. Morris A. L., MacArthur M. W., Hutchinson E. G., Thornton J. M. (1992) Stereochemical quality of protein structure coordinates. Proteins 12, 345–364 [DOI] [PubMed] [Google Scholar]
- 39. Reinert D. J., Jank T., Aktories K., Schulz G. E. (2005) Structural basis for the function of Clostridium difficile toxin B. J. Mol. Biol. 351, 973–981 [DOI] [PubMed] [Google Scholar]
- 40. Geissler B., Tungekar R., Satchell K. J. (2010) Identification of a conserved membrane localization domain within numerous large bacterial protein toxins. Proc. Natl. Acad. Sci. U.S.A. 107, 5581–5586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mesmin B., Robbe K., Geny B., Luton F., Brandolin G., Popoff M. R., Antonny B. (2004) A phosphatidylserine-binding site in the cytosolic fragment of Clostridium sordellii lethal toxin facilitates glucosylation of membrane-bound Rac and is required for cytotoxicity. J. Biol. Chem. 279, 49876–49882 [DOI] [PubMed] [Google Scholar]
- 42. Ziegler M. O., Jank T., Aktories K., Schulz G. E. (2008) Conformational changes and reaction of clostridial glycosylating toxins. J. Mol. Biol. 377, 1346–1356 [DOI] [PubMed] [Google Scholar]
- 43. Qasba P. K., Ramakrishnan B., Boeggeman E. (2005) Substrate-induced conformational changes in glycosyltransferases. Trends Biochem. Sci. 30, 53–62 [DOI] [PubMed] [Google Scholar]
- 44. Ciesla W. P., Jr., Bobak D. A. (1998) Clostridium difficile toxins A and B are cation-dependent UDP-glucose hydrolases with differing catalytic activities. J. Biol. Chem. 273, 16021–16026 [DOI] [PubMed] [Google Scholar]
- 45. Jank T., Giesemann T., Aktories K. (2007) Clostridium difficile glucosyltransferase toxin B-essential amino acids for substrate binding. J. Biol. Chem. 282, 35222–35231 [DOI] [PubMed] [Google Scholar]
- 46. Pfeifer G., Schirmer J., Leemhuis J., Busch C., Meyer D. K., Aktories K., Barth H. (2003) Cellular uptake of Clostridium difficile toxin B. Translocation of the N-terminal catalytic domain into the cytosol of eukaryotic cells. J. Biol. Chem. 278, 44535–44541 [DOI] [PubMed] [Google Scholar]
- 47. Spyres L. M., Daniel J., Hensley A., Qa'Dan M., Ortiz-Leduc W., Ballard J. D. (2003) Mutational analysis of the enzymatic domain of Clostridium difficile toxin B reveals novel inhibitors of the wild-type toxin. Infect. Immun. 71, 3294–3301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deleted in proof
- 49. Raaijmakers J. H., Bos J. L. (2009) Specificity in Ras and Rap signaling. J. Biol. Chem. 284, 10995–10999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Huelsenbeck J., Dreger S., Gerhard R., Barth H., Just I., Genth H. (2007) Difference in the cytotoxic effects of toxin B from Clostridium difficile strain VPI 10463 and toxin B from variant Clostridium difficile strain 1470. Infect. Immun. 75, 801–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chaves-Olarte E., Löw P., Freer E., Norlin T., Weidmann M., von Eichel-Streiber C., Thelestam M. (1999) A novel cytotoxin from Clostridium difficile serogroup F is a functional hybrid between two other large clostridial cytotoxins. J. Biol. Chem. 274, 11046–11052 [DOI] [PubMed] [Google Scholar]
- 52. Ho J. G., Greco A., Rupnik M., Ng K. K. (2005) Crystal structure of receptor-binding C-terminal repeats from Clostridium difficile toxin A. Proc. Natl. Acad. Sci. U.S.A. 102, 18373–18378 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.