Abstract
Listeria monocytogenes is responsible for the potentially life-threatening food-borne disease listeriosis. One epidemic-associated clonal group of L. monocytogenes, epidemic clone I (ECI), harbors a Sau3AI-like restriction-modification (RM) system also present in the same genomic region in certain strains of other lineages. In this study, we identified and characterized two other, novel type II RM systems, LmoJ2 and LmoJ3, at this same locus. LmoJ2 and LmoJ3 appeared to recognize GCWGC (W = A or T) and GCNGC, respectively. Both RM systems consisted of genes with GC content below the genome average and were in the same genomic region in strains of different serotypes and lineages, suggesting site-specific horizontal gene transfer. Genomic DNA from the LmoJ2 and LmoJ3 strains grown at various temperatures (4 to 42°C) was resistant to digestion with restriction enzymes recognizing GCWGC or GCNGC, indicating that the methyltransferases were expressed under these conditions. Phages propagated in an LmoJ2-harboring strain exhibited moderately increased infectivity for this strain at 4 and 8°C but not at higher temperatures, while phages propagated in an LmoJ3 strain had dramatically increased infectivity for this strain at all temperatures. Among the sequenced Listeria phages, lytic phages possessed significantly fewer recognition sites for these RM systems than lysogenic phages, suggesting that in lytic phages sequence content evolved toward reduced susceptibility to such RM systems. The ability of LmoJ2 and LmoJ3 to protect against phages may affect the efficiency of phages as biocontrol agents for L. monocytogenes strains harboring these RM systems.
INTRODUCTION
Listeria monocytogenes is a Gram-positive bacterium that can cause a severe food-borne disease (listeriosis) in humans and animals. Symptoms of listeriosis include septicemia, meningitis, encephalitis, stillbirths, and abortions. In the United States, listeriosis is estimated to result in ca. 1,600 cases and 250 deaths annually, with a fatality rate estimated at ca. 16% (32, 36). Control of L. monocytogenes has been challenging since this microorganism is ubiquitously present in the environment and exhibits a number of unique environmental adaptations, including the ability to grow at refrigeration temperatures, to form biofilms, and to resist disinfectants and phages (5, 12, 14, 19, 21, 36).
A few clusters of genetically related strains of L. monocytogenes have been responsible for multiple chronologically and geographically unrelated outbreaks (6, 13, 14). One of these clusters, epidemic clone I (ECI), has been extensively studied due to involvement in numerous outbreaks and sporadic cases in North America and Europe (6, 13). One such outbreak was in 1985 in California, involving Mexican-style cheese, and the genome sequence of the ECI strain F2365 implicated in that outbreak has been determined (27).
One of the characteristic attributes of ECI strains is that their genomic DNA exhibits resistance to the restriction endonuclease Sau3AI due to cytosine methylation at GATC sites (42). Genome sequence analysis of F2365 revealed a putative Sau3AI-like restriction-modification (RM) system consisting of three genes encoding a cytosine-5 methyltransferase, a restriction endonuclease, and a DNA-binding protein (see Fig. 1A) (27, 41). The involvement of this gene cassette in Sau3AI resistance was subsequently confirmed with deletion mutagenesis (40). In addition to ECI (which, along with most other serotype 4b strains, belongs to lineage I), this RM cassette was also harbored in the same genomic region by certain strains of other lineages, including strains of serotypes 1/2a (lineage II) and 4a (lineage III) (40).
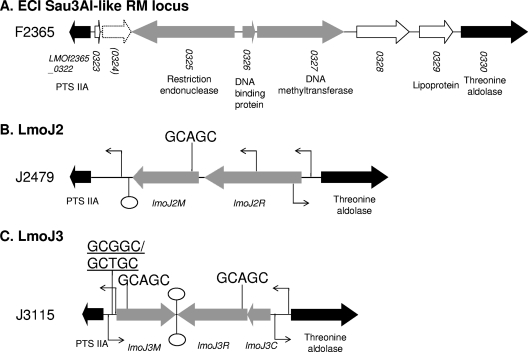
Fig 1.
Organization of the genomic region harboring LmoJ2 and LmoJ3. Gray arrows indicate genes comprising each RM system. Conserved flanking genes are shown as black arrows, and other genes unrelated to RM systems are indicated with white arrows. The stippled arrow indicates the pseudogene in strain F2365 (27). The location and direction of predicted promoters are marked with a bent arrow, and putative Rho-independent terminators are shown as lollipop symbols. Locations of recognition sites for LmoJ2 and LmoJ3 are indicated, and those that may be important in transcriptional control are underlined.
In RM systems, the restriction endonuclease cuts foreign, typically unmethylated DNA molecules, while the methyltransferase attaches methyl groups to the genomic DNA so that it is protected from restriction (35). Hence, RM systems can defend host cells against phage infection while also reducing horizontal gene transfer (34, 35). RM systems may therefore play important roles in the ecology and evolution of bacteria, including L. monocytogenes.
There is evidence for several additional RM systems in the genomic region that harbors the Sau3AI-like RM system in ECI and other L. monocytogenes strains. Some of these RM systems were identified in our laboratory via in silico analysis of sequenced Listeria genomes, and these include multiple type I RM systems and two type IV RM systems (McrB and Mrr), which will be described in detail in a separate presentation. In this report, we will focus on two novel type II RM systems (LmoJ2 and LmoJ3) that inhabit this location and that were identified in the process of investigating genomic content and diversity in this region. We describe strains possessing these RM systems and provide evidence for site-specific methylation of genomic DNA and for relative protection against phage infection in L. monocytogenes strains harboring these RM systems.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
L. monocytogenes isolates analyzed in this study are listed in Table 1. Bacteria were grown either in brain heart infusion (BHI) (Becton, Dickinson and Co., Sparks, MD) or on BHI plates containing 1.2% agar (Becton, Dickinson and Co.) until stationary phase (overnight at 25, 37, and 42°C, 7 days at 8°C, and approximately 27 days at 4°C).
Table 1.
L. monocytogenes strains used in this study
| Strain | Yr of isolation | Origin | State, countrya | Serotypeb | Lineagec (MLGT) | Source or referenced |
|---|---|---|---|---|---|---|
| Strains with LmoJ2 | ||||||
| J2479 | 2003 | Clinical | MI, USA | 4b | III (Lm3.42) | CDC |
| 267 | 1999 | Clinical | NK | 4b | III (Lm3.49) | R. Kanenaka |
| NRRL B-33191 | 2001 | Animal | NK | 4b | III (Lm3.13) | 38 |
| SK2182 | 1954 | Clinical | Ontario, Canada | 4b | I (1.70_4b) | R. G. E. Murray |
| SK2107 | 1955 | Clinical | Nova Scotia, Canada | 4b | I (1.70_4b) | R. G. E. Murray |
| SK2108 | 1956 | Clinical | Nova Scotia, Canada | 4b | I (1.70_4b) | R. G. E. Murray |
| SK90 | 2004 | Processing plant | NC, USA | 1/2a | II (2.30_1/2a_T189) | 25 |
| SK1747 | 2005 | Processing plant | VA, USA | 1/2a | II (2.30_1/2a_T189) | 25 |
| LW-A75 | 2003 | Food | NK | 1/2a | II (2.20_1/2a_T492) | FDA |
| LW-A97 | 2004 | Food | NK | 1/2a | II (2.20_1/2a_T492) | FDA |
| 2008-392 | 2008 | Clinical | NC, USA | 4c | III (Lm3.46) | NCDHHS |
| NRRL B-33330 | 2002 | Food | NH, USA | ND | III (Lm3.17) | 38 |
| SK2166 | 1953 | Animal | Ontario, Canada | ND | III (Lm3.42) | R. G. E. Murray |
| NRRL B-33372 | 2004 | Animal | NK | ND | III (Lm3.26) | 38 |
| Strains with LmoJ3 | ||||||
| J3115 | 2004 | Clinical | VA, USA | 4b | I (1.73_4b) | CDC |
| 2007-584 | 2007 | Clinical | NC, USA | 4b | I (1.74_4b) | NCDHHS |
| 2010-0072B | 2010 | Clinical | NC, USA | 1/2a | II (2.92_1/2a) | NCDHHS |
NK, not known.
Serotype was determined either by serological assay or by the multiplex PCR described by Doumith et al. (10), according to which isolates listed as serotype 1/2a could be serotype 1/2a or 3a. “ND” indicates that no serotype designation could be assigned based on the multiplex PCR (typical for lineage III strains) (10).
Lineage was designated based on targeted MLGT (TMLGT) (39).
CDC, Centers for Disease Control and Prevention; FDA, U.S. Food and Drug Administration; NCDHHS, North Carolina Department of Health and Human Services.
PFGE and MLGT.
Pulsed-field gel electrophoresis (PFGE) was conducted with AscI (New England BioLabs, Ipswich, MA) and ApaI (Roche, Indianapolis, IN) as described previously (40), and BioNumerics (Applied Maths, Austin, TX) was employed for analysis of the PFGE profiles. The Dice coefficient was used for cluster analysis. Optimization was set at 1.5% for both enzymes, while the position tolerance was 1.5% for ApaI and 2.0% for AscI. For multilocus genotyping (MLGT), analyses of single nucleotide polymorphisms were conducted using a 49-probe version of the assay described before (11, 38). The reduced version of 49 probes was obtained by deleting 11 probes (ACC1, ACC6, AMI1, INLA2, INLA5, INLA9, INLA12, INLB2, INLB3, SIG1, and SIG3) that did not provide additional haplotype discrimination.
Extraction of bacterial genomic DNA, PCR, and DNA-DNA hybridizations.
Genomic DNA was extracted from broth or plate cultures using the DNeasy blood and tissue kit (Qiagen, Valencia, CA), following the directions provided by the manufacturer. Restriction endonucleases (HindIII, ApeKI, and Fnu4HI) were used as suggested by the vendor (New England BioLabs). Digested genomic DNAs were compared with uncut genomic DNA as described previously (40).
PCR was carried out with Ex Taq polymerase (TaKaRa, Madison, WI) in a thermocycler (Biometra, Goettingen, Germany). The PCR was initiated at 95°C for 5 min and was followed by 31 cycles (each 95°C 1 min, 52°C 1 min, and 72°C 1 min), with a final extension at 72°C for 10 min. Primers employed in this study were purchased from Eurofins (Huntsville, AL) and are listed in Table 2. DNA-DNA hybridizations were done as described previously (7). The LmoJ2 and LmoJ3 probes (J2479Res and J3115Met, respectively) were prepared by PCR using the primers J2479Res_F/J2479_3R and J3115_2F/J3115Met_R, respectively (Table 2). PCR amplicons were purified from the gel using the QIAquick gel extraction kit (Qiagen), denatured for 10 min in boiling water, and digoxigenin (DIG)-labeled (Roche).
Table 2.
DNA primers used in this study
| Primer | Sequence | Target |
|---|---|---|
| J2479Res_F | GACCTAGTAAAGCAGGTGCT | lmoJ2R |
| J2479_3R | AGGTACCCATTCGATAGTCG | lmoJ2R |
| J3115_2F | TATCAGGCTTTCCCTGTCAA | lmoJ3M |
| J3115Met_R | GGTACTACAACCGAATTCCC | lmoJ3M |
| H7858_0334R | GTTCCCGAATCATTTCCAC | LMOf2365_0322 |
| H7858_0338F | CTCGTGAATCTCCAAATGCG | LMOf2365_0330 |
DNA sequencing and analysis.
PCR products were obtained from L. monocytogenes strains J2479 and J3115 using primers annealing to the flanking genes (H7858_0334R and H7858_0338F) (Table 2). The PCR amplicons were gel purified with the QIAquick gel extraction kit (Qiagen) and sequenced by GeneWiz (South Plainfield, NJ). The sequencing results were manually assembled and annotated by utilizing the ORF Finder software tool (www.ncbi.nlm.nih.gov/projects/gorf/), BLASTp (2), and conserved domain searches (24) provided by the National Center for Biotechnology Information (NCBI). Recognition sites for the RM systems were inferred based on the specificity sequences of homologs in the REBASE database (http://rebase.neb.com/rebase/rebase.html) (30). Promoters were predicted by the Neural Network Promoter Prediction software program (Berkeley Drosophila Genome Project; http://www.fruitfly.org/seq_tools/promoter.html) (37). Putative Rho-independent terminators were identified by using the model of Lesnik et al. (22) and the RNAfold server (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi) to detect stem-loop structures. Ribosome binding sites (RBS) were identified on the basis of location and similarity to the consensus sequence AGGAGGTG. Conserved domains of cytosine-5 methyltransferases were identified based on domains of M.HhaI described by Kumar et al. (20).
The sequences of various chromosomal fragments of F2365, as well as several Listeria phages, were retrieved from the NCBI database and examined for the frequencies of restriction sites for LmoJ2 and LmoJ3 (GCWGC [W = A or T] and GCNGC, respectively) (Table 3). The number of restriction sites for LmoJ2 and LmoJ3 was determined by enumerating the sites for ApeKI (recognition sequence, G↓CWGC) and Fnu4HI (recognition sequence, GC↓NGC), respectively, with the NEBcutter software tool (http://tools.neb.com/NEBcutter2/). The frequency of the restriction sites was calculated by dividing the number of the restriction sites by the length of the analyzed DNA, in kb.
Table 3.
Frequency of LmoJ2 and LmoJ3 recognition sites (GCWGC and GCNGC, respectively)
| DNAa | Family | Host serotype(s) | Phage type | Size (kb) | GC content (%) | No. of sites per kb |
GCNGC and GCWGC-rich gene | GC content (%) | No. of sites per kb |
Reference | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GCNGC | GCWGC | GCNGC per kb | GCWGC per kb | |||||||||
| F2365a | 130.0 | 38 | 2.35 | 1.28 | 27 | |||||||
| F2365b | 130.0 | 38 | 2.61 | 1.57 | 27 | |||||||
| F2365c | 130.0 | 37 | 2.32 | 1.51 | 27 | |||||||
| F2365d | 130.0 | 40 | 3.07 | 1.62 | 27 | |||||||
| F2365e | 130.0 | 39 | 2.52 | 1.52 | 27 | |||||||
| Avg | 2.57 | 1.50 | ||||||||||
| A500 | Siphoviridae | 4 | Temperate | 38.9 | 37 | 2.60 | 1.47 | LiPA500_gp016 (tmpb) | 40 | 8.20 | 3.73 | 9 |
| A118 | Siphoviridae | 1/2 | Temperate | 40.8 | 36 | 2.45 | 1.25 | A118p16 (tmp) | 39 | 7.43 | 4.46 | 3 |
| A006 | Siphoviridae | 1/2 | Temperate | 38.1 | 36 | 2.13 | 0.94 | LiPA006_gp15 (tmp) | 37 | 3.96 | 2.50 | 9 |
| B025 | Siphoviridae | 5, 6 | Temperate | 42.7 | 35 | 1.83 | 1.15 | LiPB025_gp14 (tmp) | 38 | 5.69 | 3.66 | 9 |
| PSA | Siphoviridae | 4 | Temperate | 37.6 | 35 | 1.30 | 0.80 | 2389gp14 (tmp) | 38 | 4.22 | 2.92 | 43 |
| B054 | Myoviridae | 5, 6 | Temperate | 48.2 | 36 | 1.97 | 1.58 | LiPB054_gp18 (tmp) | 39 | 2.54 | 2.12 | 9 |
| P100 | Myoviridae | 1/2, 4, 5 | Lytic | 131.4 | 36 | 0.26 | 0.22 | LBPV100_gp028 (tail lysin) | 42 | 3.84 | 3.29 | 3 |
| A511 | Myoviridae | 1/2, 4 | Lytic | 137.6 | 36 | 0.24 | 0.21 | LiPA511_gp028 (tail lysin) | 42 | 3.84 | 3.29 | 18 |
| P40 | Siphoviridae | 1/2, 4, 5, 6 | Lytic | 35.6 | 41 | 0.25 | 0.06 | P40_gp14 (tmp) | 45 | 2.74 | 1.09 | 9 |
| P35 | Siphoviridae | 1/2 | Lytic | 35.8 | 39 | 0.25 | 0.03 | LiPP35_gp14 (tmp) | 44 | 3.18 | 0.53 | 9 |
F2365a to -e correspond to randomly chosen DNA sequences between 70,000 and 200,000, 650,000 and 780,000, 1,230,000 and 1,360,000, 1,810,000 and 1,940,000, and 2,390, 000 and 2,520,000 nt, respectively, in the F2365 genome (NC_002973.6).
tmp, tape measure protein.
Phage infection assays.
For phage assays, we utilized Listeria phage 20422-1, isolated from the environment of a turkey processing facility (16). This phage was propagated in L. monocytogenes DP-L862 as described previously (16). The methylated phage derivatives 20422-1MJ2 and 20422-1MJ3 were obtained by propagating 20422-1 at least twice in strains J2479 and J3115, which harbor LmoJ2 and LmoJ3, respectively. Phage amplification and susceptibility assays were done as described previously (16). Efficiency of plaquing (EOP) was defined as the ratio of PFU per ml obtained from a specific strain to PFU/ml obtained with the reference strain, DP-L862. Assays were done in at least three independent trials and statistically analyzed with the SAS software program (SAS, Cary, NC) using the mixed analysis-of-variance (ANOVA) model.
Nucleotide sequence accession numbers.
The nucleotide sequences of LmoJ2 from L. monocytogenes J2749 and LmoJ3 from L. monocytogenes J3115 have been submitted to GenBank (accession no. JN235993 and JN235992, respectively).
RESULTS
Sequence analysis of LmoJ2.
The novel RM systems were localized between the PTS IIA and aldolase genes on the chromosome of L. monocytogenes. As discussed above, in strain F2365 (ECI), this region harbors a Sau3AI-like RM system (Fig. 1A). The two novel type II RM systems were designated LmoJ2 and LmoJ3 after the strains in which they were first identified (L. monocytogenes J2749 and J3115, respectively). Sequence analysis of the 4.5-kb PCR amplicon from L. monocytogenes strain J2479 revealed that LmoJ2 consists of two open reading frames (ORFs), lmoJ2M (1,144 bp) and lmoJ2R (1,659 bp) (Fig. 1B). The GC contents of lmoJ2M and lmoJ2R were 34% and 31%, respectively, noticeably lower than the genome average of 38% (27).
The deduced polypeptide encoded by lmoJ2M exhibited high similarity (59 to 75% identity) with DNA methyltransferases from various bacteria. Conserved domain searches suggested that this protein (designated M.LmoJ2P) was a cytosine-5 methyltransferase (pfam00145). Nine of the 10 conserved cytosine-5 methyltransferase motifs (20) were identified in M.LmoJ2P, with one (motif IX) missing (data not shown).
The deduced polypeptide encoded by the second ORF, lmoJ2R, was similar (45 to 59% identity) to proteins from various bacteria. Notably, the lmoJ2R homologs in these bacteria were located adjacent to lmoJ2M homologs, suggesting that these two genes were harbored on one cassette in these genomes. Conserved domain searches suggested that the product of lmoJ2R (designated R.LmoJ2P) belonged to a protein family represented by the AlwI restriction endonuclease (pfam09491).
The LmoJ2 recognition site was predicted to be GCWGC by BLAST searches using REBASE. Proteins with the highest homology to M.LmoJ2P in REBASE were M.Gsp412ORF3572P (YP_003254598.1) in Geobacillus sp. Y412MC61 plasmid pGYMC6101 and M.Bce98ORF752P (YP_001374089.1) in Bacillus cereus subsp. cytotoxis NVH 391-98. For R.LmoJ2P, the REBASE database homologs included Gsp412ORF3572P (YP_003254599.1) and Bce98ORF752P (YP_001374090.1), both of which were adjacent to their homologs to M.LmoJ2P. The predicted recognition site for these putative methyltransferases and restriction endonucleases was GCWGC; hence, an inference was drawn that LmoJ2 might also recognize GCWGC.
Sequence analysis revealed a putative promoter upstream of lmoJ2R along with a putative Rho-independent terminator downstream of lmoJ2M (Fig. 1B). Putative ribosome-binding sites were detected in front of lmoJ2M and lmoJ2R. Noticeably, the ribosome-binding site for lmoJ2R (AGGCTACT) deviated considerably from the consensus sequence.
RM recognition sites in the vicinity of the promoter region play an important role in transcription control of some RM systems by reducing transcription when the sites are methylated (26). The sole LmoJ2 recognition site (GCAGC) in LmoJ2 was located within the methyltransferase gene (Fig. 1B) and thus would not be expected to be involved in the regulation of LmoJ2.
Sequence analysis of LmoJ3.
LmoJ3 was identified from the analysis of the sequence of the 3.7-kb PCR product obtained from L. monocytogenes strain J3115. Three ORFs were identified, designated lmoJ3M (984 bp), lmoJ3R (1,170 bp), and lmoJ3C (384 bp) (Fig. 1C). The GC contents of these ORFs were 29% (lmoJ3M) and 32% (lmoJ3R and lmoJ3C). As described above for LmoJ2, this was noticeably below the genome average.
The deduced product of lmoJ3M shared similarity (58 to 65% identity) with DNA methyltransferases in other bacteria. Conserved domain searches of the deduced polypeptide (designated M.LmoJ3P) assigned it to the cytosine-5 methyltransferase protein family (pfam00145). Comparisons with M.HhaI revealed that M.LmoJ3P harbored all 10 conserved motifs of cytosine-5 methyltransferases (data not shown). The deduced product of lmoJ3R (designated R.LmoJ3P) belonged to a protein family represented by the restriction endonuclease NgoFVII (recognition sequence GCSGC, where S = C or G) (pfam09565) (33). The deduced product of the third ORF, lmoJ3C (designated C.LmoJ3P), harbored a helix-turn-helix motif (pfam01381), suggesting that it is a DNA-binding protein that may function as a control (C) protein in the regulation of the RM cassette (26).
BLAST analysis using REBASE identified several M.LmoJ3P homologs with specificity for GCNGC, including M.Bsp6I in Bacillus sp. plasmid pXH13 (CAA57293.1) and M.MagORF4250P in Mycoplasma agalactiae strain 5632 (YP_003515594.1). Several homologs of R.LmoJ3P with specificity for GCNGC were also identified. These results suggested that R.LmoJ3 might also recognize GCNGC.
A putative promoter was identified upstream of lmoJ3C and upstream of lmoJ3M, and bidirectional Rho-independent terminators were identified between lmoJ3M and lmoJ3R (Fig. 1C). These predictions suggest that two transcripts are generated from LmoJ3: one for lmoJ3C and lmoJ3R and a divergent transcript for lmoJ3M. Putative ribosome-binding sites were identified in front of each of the three ORFs of LmoJ3.
Four GCNGC sites were identified in the LmoJ3 region (Fig. 1C). Two of these sites (GCGGC and GCTGC) were close to each other between the putative lmoJ3M promoter and the start of lmoJ3M (Fig. 1C), raising the possibility that they might be involved in the regulation of this RM system.
LmoJ2 and LmoJ3 are harbored in the same genomic region by diverse strains of L. monocytogenes.
Hybridization of genomic DNA from 463 L. monocytogenes isolates in our strain collection with DNA probes derived from LmoJ2 and LmoJ3 identified several that putatively harbored these RM systems. LmoJ2 was detected in a total of 14 isolates (14/463; 3.0% of the total isolates) of diverse serotypes (including 4b, 1/2a, and 4c) (Table 1). Evidence for LmoJ3 was obtained for two additional isolates, of serotypes 4b and 1/2a (Table 1), suggesting a lower prevalence of LmoJ3-harboring isolates (3/463; 0.6% of the total isolates).
PCR with primers derived from flanking genes (LMOf2365_0322 and LMOf2365_0330) and genes internal to LmoJ2 and LmoJ3 indicated that LmoJ2 and LmoJ3 were flanked by the same genes in these isolates as in J2479 and J3115. Additionally, the size of the PCR products was conserved among the isolates harboring LmoJ2 or lmoJ3, suggesting a lack of detectable insertions or deletions in this region (Fig. 2 and data not shown).
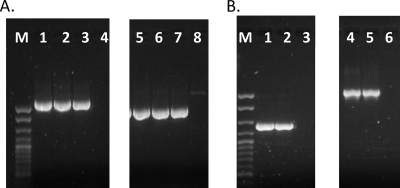
Fig 2.
LmoJ2 and LmoJ3 are present in the same genomic locations in L. monocytogenes stranis of diverse serotypes. Confirmation of genomic location of LmoJ2 (A) was performed using PCR with primers H7858_0334R and J2479_3R (lanes 1 to 4) and with primers J2479Res_F and H7858_0338F (lanes 5 to 8). Lanes 1 and 5, SK2107; lanes 2 and 6, SK2108; lanes 3 and 7, J2479 (used as the positive control); and lanes 4 and 8, a negative control lacking LmoJ2 or LmoJ3. Confirmation of the genomic location of LmoJ3 (B) was done using PCR with primers H7858_0334R and J3115Met_R (lanes 1 to 3) and with primers J3115_2F and H7858_0338F (lanes 4 to 6). Lanes 1 and 4, 2010-0072B; lanes 2 and 5, J3115, used as the positive control; and lanes 3 and 6, a negative control lacking lmoJ2 or LmoJ3. M, exACTGene cloning DNA ladder (Fisher Scientific, Fair Lawn, NJ).
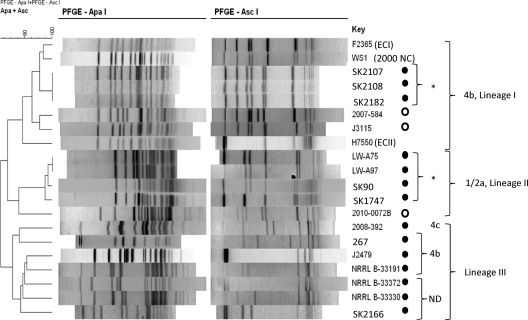
Relatedness of these isolates was examined via PFGE and MLGT. This analysis revealed two clonal groups harboring LmoJ2: one consisted of three serotype 4b isolates of lineage I with the same PFGE pattern and MLGT haplotype, while the other consisted of four food and environmental isolates of lineage II (serotype 1/2a or 3a) with identical or closely related PFGE profiles and MLGT haplotypes (Table 1 and Fig. 3). The serotype 4b clonal group was isolated from Canada in the mid-1950s, whereas the isolates belonging to lineage II were collected in the mid-2000s (Table 1). MLGT analysis revealed that these four lineage II isolates harbored premature stop codons in internalin A (inlA), suggesting virulence attenuation (MLGT types of these isolates were 2.20_1/2a_T492 and 2.30_1/2a_T189 [Table 1]). In addition to these two clonal groups, LmoJ2 was harbored by seven other isolates, which, intriguingly, were all of lineage III. These lineage III strains exhibited PFGE profiles markedly different from each other and from those of other isolates, even though two (J2479 and SK2166) had a common haplotype, Lm3.42 (Table 1 and Fig. 3).
Fig 3.
PFGE dendrogram of L. monocytogenes harboring LmoJ2 and LmoJ3. PFGE with ApaI and AscI and profile clustering were performed as described in Materials and Methods. Isolates harboring LmoJ2 and LmoJ3 are indicated with filled and open circles, respectively. Serotype 4b strains F2365 (ECI), H7550 (ECII), and WS1 (2000 North Carolina outbreak) were included as a reference. ND, serotype could not be determined using the multiplex PCR, as indicated in Table 1. “*” indicates closely related strains.
The two serotype 4b strains harboring LmoJ3 were of lineage I and were clearly distinguishable based on PFGE and MLGT (Table 1 and Fig. 3). Each of the LmoJ3 strains had a distinct MLGT haplotype, and all three haplotypes were novel. Thus, LmoJ2 and LmoJ3 were harbored by genetically heterogeneous isolates.
Genomic DNA from strains harboring LmoJ2 and LmoJ3 was methylated at GCWGC and GCNGC, respectively, when bacteria were grown planktonically or on agar at different temperatures, including 4°C.
To verify the recognition site for LmoJ2 and LmoJ3 and obtain evidence for expression of the DNA methyltransferases, genomic DNA was extracted from LmoJ2 or LmoJ3 isolates and digested with ApeKI (recognition sequence, G↓CWGC) and Fnu4HI (recognition sequence, GC↓NGC).
Genomic DNA from all LmoJ2 and LmoJ3 isolates grown at 37°C on agar was resistant to ApeKI and Fnu4HI, respectively (Fig. 4 and data not shown). Similar results were observed when isolates with LmoJ2 and LmoJ3 were grown at 37°C in liquid and at other temperatures (4, 8, 25, and 42°C) in liquid or on agar (data not shown). To confirm that LmoJ2 was specific for GCWGC and not GCNGC, DNA from LmoJ2 isolates was also digested with Fnu4HI and found to be susceptible (data not shown). The findings support the hypothesis that LmoJ2 and LmoJ3 recognize GCWGC and GCNGC, respectively; they also suggest that the methyltransferases were expressed at all tested temperatures and regardless of whether the bacteria were grown on agar or planktonically.
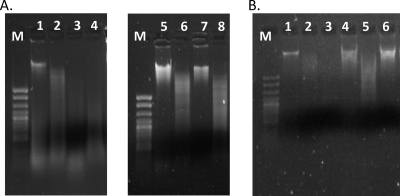
Fig 4.
LmoJ2 and LmoJ3 are associated with resistance to digestion by restriction enzymes recognizing GCWGC and GCNGC, respectively. Genomic DNA from F2365 (control), LW-A75 (LmoJ2), and J3115 (LmoJ3) was digested with ApeKI, Fnu4HI, and HindIII (quality control). DNA was extracted from cells grown on BHI agar at 37°C. (A) Lanes; 1, undigested genomic DNA of strain F2365 which lacks LmoJ2 or LmoJ3; 2 to 4, F2365 genomic DNA digested with HindIII, ApeKI, and Fnu4HI, respectively; 5, undigested genomic DNA from LmoJ2 isolate LW-A75; 6 to 8, genomic DNA of LW-A75 digested with HindIII, ApeKI, and Fnu4HI, respectively. (B) Lanes: 1, undigested F2365 genomic DNA; 2 and 3, F2365 genomic DNA digested with HindIII and Fnu4HI, respectively; 4, undigested genomic DNA from LmoJ3 isolate J3115; 5 and 6, genomic DNA of J3115 digested with HindIII and Fnu4HI, respectively. M, exACTGene cloning DNA ladder.
Evidence for expression of LmoJ2 and LmoJ3-associated restriction endonucleases.
Tentative evidence for expression of the putative restriction endonucleases was obtained by phage infection assays. It was hypothesized that if the restriction endonuclease is expressed, phage 20422-1 propagated in LmoJ2 or LmoJ3 strains would have higher EOP in these strains than when propagated in the reference strain DP-L862. The derivatives of 20422-1 propagated in J2479 and J3115 were designated 20422-1MJ2 and 20422-1MJ3, respectively.
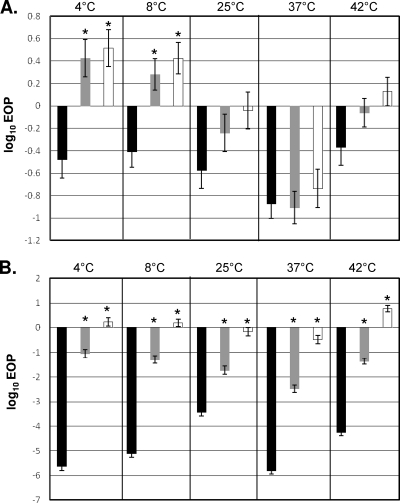
When J2479 grown at 4 or 8°C was infected with 20422-1MJ2 or 20422-1MJ3, EOP increased in comparison to that for infection with unmethylated phage (P < 0.05) (Fig. 5A). EOP changes with cells grown at 25, 37, or 42°C were not significant (Fig. 5A). A much more noticeable impact of methylation of the phage was observed with J3115. Infections of this strain with 20422-1 revealed that the EOP was 10−3 to 10−6, significantly lower than with the reference strain (P < 0.0001), at all tested growth temperatures. A pronounced (103- to 106-fold) increase in EOP was noted upon infection with 20422-1MJ3 (Fig. 5B). This effect was noted at all tested growth temperatures but was lowest when bacteria were grown at 25°C. A significant, albeit smaller, increase in EOP was also observed following infection of J3115 with 20422-1MJ2 (P < 0.0001) (Fig. 5B). The smaller increase in EOP with 20422-1MJ2 was expected, since only approximately 50% of the GCNGC sites would be methylated in such phage. Similar findings were obtained with an additional strain harboring LmoJ3 (2007-584) grown at 37°C (data not shown).
Fig 5.
Phage infection assay results for J2479 (A) or J3115 (B). Bars represent log10 EOP values following infection with Listeria phage 20422-1 propagated in DP-L862 (black), 20422-1MJ2 (gray), or 20422-1MJ3 (white). Error bars represent standard errors. Preparation of 20422-1MJ2 and 20422-1MJ3 and phage infection assays were done as described in Materials and Methods. “*” indicates a significant difference (P < 0.05) in EOP of the methylated phage in comparison to EOP of the unmethylated phage at the same temperature.
LmoJ2 and LmoJ3 restriction sites are significantly less frequent in the genomes of lytic versus temperate Listeria phages.
Analysis of sequenced Listeria phage genomes revealed that GCWGC and GCNGC sites were markedly less common in genomes of lytic phages than in temperate phages, regardless of phage family (Siphoviridae versus Myoviridae) or genome size. An extreme case of this trend was observed in the Siphoviridae lytic phages P35 and P40, which possessed only one and two GCWGC sites, respectively. In contrast, the frequency of LmoJ2 and LmoJ3 recognition sites in temperate phages was similar to that observed in chromosomal fragments of L. monocytogenes F2365 (Table 3).
Close scrutiny of the distribution of the GCWGC and GCNGC sites revealed that they were unevenly distributed in the phage genomes. In many phage genomes, such regions included a gene encoding a tape measure protein (Table 3). In Siphoviridae lytic phages, all GCWGC sites and the majority of GCNGC sites were located in this gene; in temperate phages, this gene also harbored considerably more GCWGC and GCNGC sites than the average for the phage genome. In Myoviridae lytic phages, most GCWGC and GCNGC sites were located in tail lysin genes (Table 3).
DISCUSSION
Here we have presented evidence for two novel Type II RM systems (LmoJ2 and LmoJ3) in the same locus of L. monocytogenes in which ECI strains harbor a Sau3AI-like RM system. Without exception, the presence of LmoJ2 and LmoJ3 was associated with resistance of the genomic DNA of the isolates to the restriction enzymes ApeKI and Fnu4HI, specific for GCWGC and GCNGC, respectively. LmoJ2 and LmoJ3 consisted of genes of unusually low GC content and were identified in genetically diverse strains which were of different serotypes and lineages. Even though the association of these RM systems with methylation of DNA at GCWGC and GCNGC sites is strong, further studies will be needed for unambiguous proof, especially since complete genome sequences of strains harboring these RM systems are currently not available. It will be of interest, for instance, to construct mutants lacking the RM gene cassettes or to express the methyltransferase in a heterologous host, as was recently done with two other RM systems of L. monocytogenes (17, 40).
Our findings imply that LmoJ2 and LmoJ3 may have been acquired through site-specific horizontal gene transfer, probably via multiple, independent events. Alternatively, these novel RM systems may have been acquired by an ancestral strain of L. monocytogenes but were maintained only in certain strains. Sequence analysis of the RM genes from strains of different lineages (e.g., strains of serotypes 4b and 1/2a, corresponding to lineages I and II, respectively) may help differentiate between these two scenarios, since ancestral origin would be reflected in sequence diversity levels comparable to those in other chromosomal loci.
Our bioinformatics data and methylation assays suggested that the recognition site of LmoJ2 (GCWGC) was included within those of LmoJ3 (GCNGC). In addition, for LmoJ3-harboring strains, phage DNA methylation by LmoJ2 increased infection efficiency compared with that of the unmethylated phage, albeit to a lesser degree than methylation by LmoJ3, providing further evidence for LmoJ2 recognition sites being a subset of those for LmoJ3. ApeKI and Fnu4HI are both sensitive to cytosine methylation at GpC sites (4), suggesting that LmoJ2 and LmoJ3 likely methylate cytosine at GpC sites within their recognition sequence.
The recognition sites of these RM systems attract attention due to three features. One is their relative GC richness, considering that average GC content for the genome of L. monocytogenes is 38% (27). Nonetheless, the L. monocytogenes chromosome harbored an average of at least one site per kb. As GC-rich as these sites are, their small size may result in this high frequency. Another noteworthy attribute is the inclusion of LmoJ2 restriction sequences within those for LmoJ3. These overlapping recognition sequences may have ecological implications for strains harboring these RM systems and occupying the same environmental niche. Although LmoJ2 might provide only moderate protection against phage and only under certain growth conditions (i.e., at low temperature), this RM system may convert its host to a biological reservoir for methylated phages that can counteract, albeit incompletely, restriction by LmoJ3, thus potentially offsetting any competitive edge conferred by LmoJ3. These interactions may promote coexistence of strains harboring these two RM systems. Last, cytosine methylation-susceptible restriction endonucleases recognizing GCWGC and GCNGC have been employed to identify cytosine methylation of CpG and GpC sites in human DNA within trinucleotide repeat tracks associated with neurodegenerative and neuromuscular diseases, such as Huntington's disease and fragile X syndrome (4, 29). The novel RM systems described here can expand the repertoire of tools to investigate biomedically relevant epigenetic changes.
Phage infection assays suggested that the LmoJ3 restriction endonuclease was expressed at all tested temperatures, including 4 and 8°C. The LmoJ2 restriction enzyme also appeared to be expressed at low temperatures. Activity of the restriction endonucleases at low temperatures and the accompanying impact on protection against phage infection may suggest a fitness advantage associated with the presence of the RM systems. Given that refrigeration is widely utilized in food processing and distribution, this advantage may promote dissemination of LmoJ2- and LmoJ3-bearing strains in the food chain. However, the phage infection assays employed in the current study reflect a number of attributes in the phage infection process. Further studies (e.g., employing quantitative reverse transcriptase PCR or proteomics methods) are needed to characterize expression of the restriction endonuclease and investigate the possible impact of temperature. Further studies are also needed to assess fitness of the LmoJ2- and LmoJ3-harboring strains under different conditions.
Analysis of the frequency of LmoJ2 and LmoJ3 recognition sites within the genomes of temperate and lytic Listeria phages revealed a significantly lower frequency in genomes of lytic phages than in genomes of temperate phages, similarly to trends in restriction enzyme prevalence found in other studies (31). This trend is worth contemplating given that the majority of Listeria phages are temperate. The expected high susceptibility of temperate phages to the LmoJ2 and LmoJ3 restriction enzymes may reduce horizontal gene transfer frequently mediated by temperate phages (23, 28).
All phage genomes analyzed in this study had a high density of GCWGC and GCNGC sites in certain genes, such as the tape measure protein gene in temperate phages and lytic phages of the Siphoviridae and the tail lysin gene in lytic phages of the Myoviridae. Tape measure proteins determine the length of the bacteriophage tail, and there is a linear relationship between the size of this protein and tail length (1, 8). In the lactococcal bacteriophage Tuc2009, the tail lysin Tal2009 is located at the end of the tail and has been speculated to contribute to degradation of the bacterial cell wall, thus facilitating the injection of the phage DNA (15). It is intriguing that the tape measure protein and tail lysin are both associated with the phage tail; however, it still remains unknown why these restriction sites were densely packed in genes encoding these proteins.
In conclusion, we have demonstrated that two novel type II RM systems (LmoJ2 and LmoJ3) are harbored by diverse strains of L. monocytogenes at the same genomic location that harbors the Sau3AI-like RM system in ECI strains, suggesting a site-specific mechanism for acquisition of these RM systems in L. monocytogenes. LmoJ2 and LmoJ3 recognize the closely related DNA sequences GCWGC and GCNGC, and both RM systems appeared to be active at low temperatures. Further studies of LmoJ2, LmoJ3, and other RM systems in this locus will enhance our understanding of the ecological and evolutionary roles of genetic elements that bestow DNA methylation and phage resistance to L. monocytogenes and will provide information needed to assess the effectiveness of Listeria phages as a biocontrol agent.
ACKNOWLEDGMENTS
This work was partially supported by a grant from the American Meat Institute Foundation, USDA grant 2006-35201-17377, and the U.S. Department of Agriculture's Agricultural Research Service.
We thank B. Swaminathan, L. Graves, R. G. E. Murray, W. Lin, R. Kanenaka, and L. Wolf for strains used in this study. We are thankful to Jason Osborne for his assistance in the statistical analysis of the data and to all other members of our laboratory for their support and encouragement.
Footnotes
Published ahead of print 10 February 2012
REFERENCES
- 1. Abuladze NK, Gingery M, Tsai J, Eiserling FA. 1994. Tail length determination in bacteriophage T4. Virology 199:301–310 [DOI] [PubMed] [Google Scholar]
- 2. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 3. Carlton RM, Noordman WH, Biswas B, de Meester ED, Loessner MJ. 2005. Bacteriophage P100 for control of Listeria monocytogenes in foods: genome sequence, bioinformatic analyses, oral toxicity study, and application. Regul. Toxicol. Pharmacol. 43:301–312 [DOI] [PubMed] [Google Scholar]
- 4. Castel AL, Nakamori M, Thornton CA, Pearson CE. 2011. Identification of restriction endonucleases sensitive to 5-cytosin methylation at non-CpG sites, including expanded (CAG)n/(CTG)n repeats. Epigenetics 6:1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chan YC, Wiedmann M. 2009. Physiology and genetics of Listeria monocytogenes survival and growth at cold temperatures. Crit. Rev. Food Sci. Nutr. 49:237–253 [DOI] [PubMed] [Google Scholar]
- 6. Cheng Y, Siletzky RM, Kathariou S. 2008. Genomic divisions/ lineages, epidemic clones, and population structure, p 337–358 In Liu D. (ed), Handbook of Listeria monocytogenes, 1st ed CRC Press, Boca Raton, FL [Google Scholar]
- 7. Cheng Y, Kim JW, Lee S, Siletzky RM, Kathariou S. 2010. DNA probes for unambiguous identification of Listeria monocytogenes epidemic clone II strains. Appl. Environ. Microbiol. 76:3061–3068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cornelis GR, Agrain C, Sorg I. 2006. Length control of extended protein structures in bacteria and bacteriophages. Curr. Opin. Microbiol. 9:201–206 [DOI] [PubMed] [Google Scholar]
- 9. Dorscht J, et al. 2009. Comparative genome analysis of Listeria bacteriophages reveals extensive mosaicism, programmed translational frameshifting, and a novel prophage insertion site. J. Bacteriol. 191:7206–7215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Doumith M, Buchrieser C, Glaser P, Jacquet C, Martin P. 2004. Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J. Clin. Microbiol. 42:3819–3822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ducey TF, et al. 2007. A single-nucleotide-polymorphism-based multilocus genotyping assay for subtyping lineage I isolates of Listeria monocytogenes. Appl. Environ. Microbiol. 73:133–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gandhi M, Chikindas ML. 2007. Listeria: a foodborne pathogen that knows how to survive. Int. J. Food Microbiol. 113:1–15 [DOI] [PubMed] [Google Scholar]
- 13. Kathariou S. 2003. Foodborne outbreaks of listeriosis and epidemic-associated lineages of Listeria monocytogenes, p 243–256 In Torrence ME, Isaacson RE. (ed), Microbial food safety in animal agriculture, 1st ed Iowa State Press, Ames, IA [Google Scholar]
- 14. Kathariou S. 2002. Listeria monocytogenes virulence and pathogenicity, a food safety perspective. J. Food Prot. 65:1811–1829 [DOI] [PubMed] [Google Scholar]
- 15. Kenny JG, McGrath S, Fitzgerald GF, van Sinderen D. 2004. Bacteriophage Tuc2009 encodes a tail-associated cell wall-degrading activity. J. Bacteriol. 186:3480–3491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim JW, Siletzky RM, Kathariou S. 2008. Host ranges of Listeria-specific bacteriophages from the turkey processing plant environment in the United States. Appl. Environ. Microbiol. 74:6623–6630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim JW, et al. 13 January 2012. A novel restriction-modification system is responsible for temperature-dependent phage resistance in Listeria monocytogenes ECII. Appl. Environ. Microbiol. doi:10.1128/AEM.07086-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Klumpp J, et al. 2008. The terminally redundant, nonpermuted genome of Listeria bacteriophage A511: a model for the SPO1-like myoviruses of gram-positive bacteria. J. Bacteriol. 190:5753–5765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kornacki JL, Gurtler JB. 2007. Incidence and control of Listeria in food processing facilities, p 681–766 In Ryser ET, Marth EH. (ed), Listeria, listeriosis and food safety, 3rd ed CRC Press, Boca Raton, FL [Google Scholar]
- 20. Kumar S, et al. 1994. The DNA (cytosine-5) methyltransferases. Nucleic Acids Res. 22:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lemon KP, Higgins DE, Kolter R. 2007. Flagellar motility is critical for Listeria monocytogenes biofilm formation. J. Bacteriol. 189:4418–4424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lesnik EA, et al. 2001. Prediction of rho-independent transcriptional terminators in Escherichia coli. Nucleic Acids Res. 29:3583–3594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Loessner MJ, Rees CED. 2005. Listeria phages: basics and applications, p 362–379 In Waldor KM, Friedman DI, Adhya SL. (ed), Phages: their role in bacterial pathogenesis and biotechnology. ASM Press, Washington, DC [Google Scholar]
- 24. Marchler-Bauer A, et al. 2011. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 39:D225–D229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mullapudi S, Siletzky RM, Kathariou S. 2008. Heavy-metal and benzalkonium chloride resistance of Listeria monocytogenes isolates from the environment of turkey-processing plants. Appl. Environ. Microbiol. 74:1464–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nagornykh MO, et al. 2008. Regulation of gene expression in type II restriction-modification system. Genetika 44:606–615 [DOI] [PubMed] [Google Scholar]
- 27. Nelson KE, et al. 2004. Whole genome comparisons of serotype 4b and 1/2a strains of the food-borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Res. 32:2386–2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ochman H, Lawrence JG, Groisman EA. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299–304 [DOI] [PubMed] [Google Scholar]
- 29. Ramsahoye BH, Burnett AK, Taylor C. 1997. Restriction endonuclease isoschizomers ItaI, BsoFI and Fsp4HI are characterised by differences in their sensitivities to CpG methylation. Nucleic Acids Res. 25:3196–3198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Roberts RJ, Vincze T, Posfai J, Macelis D. 2010. REBASE—a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 38:D234–D236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rocha EP, Danchin A, Viari A. 2001. Evolutionary role of restriction/modification systems as revealed by comparative genome analysis. Genome Res. 11:946–958 [DOI] [PubMed] [Google Scholar]
- 32. Scallan E, et al. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg. Infect. Dis. 17:7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stein DC, Gunn JS, Radlinska M, Piekarowicz A. 1995. Restriction and modification systems of Neisseria gonorrhoeae. Gene 157:19–22 [DOI] [PubMed] [Google Scholar]
- 34. Thomas CM, Nielsen KM. 2005. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 3:711–721 [DOI] [PubMed] [Google Scholar]
- 35. Tock MR, Dryden DT. 2005. The biology of restriction and anti-restriction. Curr. Opin. Microbiol. 8:466–472 [DOI] [PubMed] [Google Scholar]
- 36. Wagner M, MacLauchlin J. 2008. Biology, p 3–26 In Liu D. (ed), Handbook of Listeria monocytogenes, 1st ed CRC Press, Boca Raton, FL [Google Scholar]
- 37. Waibel AH, Hanazawa T, Hinton GE, Shikano K, Lang KJ. 1989. Phoneme recognition using time-delay neural networks. IEEE Trans. Acoust. 37:328–339 [Google Scholar]
- 38. Ward TJ, Ducey TF, Usgaard T, Dunn KA, Bielawski JP. 2008. Multilocus genotyping assays for single nucleotide polymorphism-based subtyping of Listeria monocytogenes isolates. Appl. Environ. Microbiol. 74:7629–7642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ward TJ, Usgaard T, Evans P. 2010. A targeted multilocus genotyping assay for lineage, serogroup, and epidemic clone typing of Listeria monocytogenes. Appl. Environ. Microbiol. 76:6680–6684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yildirim S, et al. 2010. Conservation of genomic localization and sequence content of Sau3AI-like restriction-modification gene cassettes among Listeria monocytogenes epidemic clone I and selected strains of serotype 1/2a. Appl. Environ. Microbiol. 76:5577–5584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yildirim S, et al. 2004. Epidemic clone I-specific genetic markers in strains of Listeria monocytogenes serotype 4b from foods. Appl. Environ. Microbiol. 70:4158–4164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zheng W, Kathariou S. 1997. Host-mediated modification of Sau3AI restriction in Listeria monocytogenes: prevalence in epidemic-associated strains. Appl. Environ. Microbiol. 63:3085–3089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zimmer M, Sattelberger E, Inman RB, Calendar R, Loessner MJ. 2003. Genome and proteome of Listeria monocytogenes phage PSA: an unusual case for programmed + 1 translational frameshifting in structural protein synthesis. Mol. Microbiol. 50:303–317 [DOI] [PubMed] [Google Scholar]