Abstract
Halohydrin dehalogenase from Agrobacterium radiobacter AD1 (HheC) is a valuable tool in the preparation of R enantiomers of epoxides and β-substituted alcohols. In contrast, the halohydrin dehalogenase from Arthrobacter sp. AD2 (HheA) shows a low S enantioselectivity toward most aromatic substrates. Here, three amino acids (V136, L141, and N178) located in the two neighboring active-site loops of HheA were proposed to be the key residues for controlling enantioselectivity. They were subjected to saturation mutagenesis aimed at evolving an S-selective enzyme. This led to the selection of two outstanding mutants (the V136Y/L141G and N178A mutants). The double mutant displayed an inverted enantioselectivity (from S enantioselectivity [ES] = 1.7 to R enantioselectivity [ER] = 13) toward 2-chloro-1-phenylethanol without compromising enzyme activity. Strikingly, the N178A mutant showed a large enantioselectivity improvement (ES > 200) and a 5- to 6-fold-enhanced specific activity toward (S)-2-chloro-1-phenylethanol. Further analysis revealed that those mutations produced some interference for the binding of nonfavored enantiomers which could account for the observed enantioselectivities. Our work demonstrated that those three active-site residues are indeed crucial in modulating the enantioselectivity of HheA and that a semirational design strategy has great potential for rapid creation of novel industrial biocatalysts.
INTRODUCTION
Optically pure compounds are valuable precursors in pharmaceutical and fine chemical synthesis. In recent years, the biocatalytic conversion of nonnatural substrates has grown increasingly as an alternative to chemical asymmetric synthesis in the production of chiral compounds (8, 17). Bacteria represent a major source of those valuable biocatalysts, and up to now hundreds of bacteria and/or the corresponding enzymes have been successfully implemented in industry. Halohydrin dehalogenases, which occur in the biodegradation pathways of halogenated compounds, catalyze the dehalogenation of vicinal halohydrins and the reverse reaction of epoxide ring opening (3, 25). Recently these enzymes attracted great attention for their ability to produce optically pure epoxides and halohydrins (6, 10). Due to their high enantioselectivity and capability to accept a series of nonnatural nucleophiles (CN−, N3−, NO2−, OCN−, etc.), they can also be used in the preparation of chiral β-substituted alcohols (7, 11, 13).
Halohydrin dehalogenase from Agrobacterium radiobacter AD1 (HheC) has been studied extensively (15, 25). The enzyme displays a high R enantioselectivity toward various short-chain aliphatic and aromatic substrates, which is believed to be a result of nonproductive binding of the unfavored S enantiomer in a spatially limited substrate-binding pocket (4). Among all residues lining the pocket, the loop residue W139 represents a critical obstacle for the binding of the S enantiomer of bulky substrates. Indeed, the binding of the S enantiomer was largely enhanced when W139 was replaced by a phenylalanine (24). In addition, another two active-site loop residues, T134 and N176, that form a small bulge together with W139 were proved to be accountable for the high HheC enantioselectivity toward disubstituted epoxides in HheC, mainly due to the steric conflict between the small bulge and the S enantiomers of those substrates (14).
Recently, the X-ray structure of halohydrin dehalogenase from Arthrobacter sp. AD2 (HheA) was resolved (PDB code 1ZMO). The enzyme has 34% amino acid sequence similarity to and shares a similar quaternary and tertiary structure with HheC, but with a much more open substrate-binding pocket (2). This correlates well with the fact that HheA prefers C4-C5 halohydrins as substrates, whereas HheC prefers short-chain substrates (C2-C3). The low sequence similarity and different configurations of the active sites imply that the two enzymes are notably different. Recent studies revealed that HheA displays a high regioselectivity and a broad substrate range, as does HheC. However, it catalyzes dehalogenation and epoxide ring-opening reactions with a low to moderate S enantioselectivity (13). This makes HheA an attractive target for rational modifications, thus turning it in a highly S-selective enzyme. Such evolved mutants in addition to HheC would greatly facilitate the application of halohydrin dehalogenases in asymmetric synthesis of either product enantiomer.
Here, three identified “hot spots,” V136, L141, and N178, which lie in two neighboring active-site loops of HheA, were subjected to saturation mutagenesis analysis, resulting in the selection of two outstanding variants. The aromatic substrate 2-chloro-1-phenylethanol (2-CPE) was used as a model substrate for enantioselectivity screening since the wild-type HheA displayed a rather low S enantioselectivity [ES] toward 2-CPE (ES = 1.7). The kinetic resolution results showed that those three residues are critical for controlling the enantioselectivity of HheA. Moreover, steady-state kinetic studies and molecular docking analysis were performed to further understand the structural basis of the enantioselectivity of HheA.
MATERIALS AND METHODS
1,3-Dicholoro-2-propanol (1,3-DCP), racemic 2-chloro-1-phenylethanol (rac-2-CPE) and its R enantiomer, and racemic styrene oxide and its R enantiomer were purchased from Alfa Aesar. (S)-2-Chloro-1-phenylethanol and (S)-styrene oxide were purchased from Sigma-Aldrich. l-Arabinose was from Lancaster. Escherichia coli strain MC1061 and rac-p-nitro-2-bromo-1-phenylethanol were kind gifts from Dick B. Janssen (Groningen University, The Netherlands). Recombinant strains were cultivated at 37°C in LB medium containing 100 μg/ml ampicillin. All the other materials were purchased from local businesses.
Construction of saturation mutagenesis libraries of HheA.
All saturation mutagenesis libraries were constructed using a Phusion high-fidelity PCR kit (New England BioLabs) according to the supplied manual. The V136X/L141X saturation mutagenesis library was constructed by overlapping PCR with the recombinant expression vector pBADHheA-WT, carrying the wild-type hheA gene (accession number AF397297) as a template. Partially overlapping primers, fwV136 (5′-GCAAGAAGCCGXXXGCCTACAAC-3′, where XXX is VHG, NDC, and TGG, respectively) and rvL141 (5′-GGCTTCTTGCCXXXTGAGGAGGTGAT-3′, where XXX is CDB, GHN, and CCA, respectively) were designed to randomize the two residues. Saturation mutagenesis libraries of V136Y/L141G/N178X and N178X were constructed by using pBADHheA-V136Y/L141G and pBADHheA-WT as templates, respectively. A pair of primers, fwN178 95′-TCGGTCCGNNSTTCTTCAATAACCC-3′) and rvN178 (5′-TGAAGAASNNCGGACCGATAGCGTA-3′) were designed for randomization at N178. The primers used in this study were supplied by Invitrogen (Shanghai, China). The 50-μl reaction system contained 1× high-fidelity (HF) buffer, 200 μM (each) deoxynucleoside triphosphate (dNTP), 1 mM Mg2+, 100 ng template, 2 μM (each) mixed primers, and 0.01 U/μl Phusion polymerase. The temperature program used was 3 min at 98°C followed by 30 cycles of 10 s at 98°C, 45 s at 50°C, and 2 min at 72°C, finishing with 10 min at 72°C. PCR amplification products were digested with DpnI to remove parental templates, after which the reaction mixtures were transformed into chemically competent E. coli MC1061.
Colorimetric assay for library screening.
The constructed small focused libraries were screened for halohydrin dehalogenase activity using the previously developed colorimetric assay in a 96-well plate format with some modifications (22). The expression was induced by adding 0.002% (wt/vol) l-arabinose when the culture reached an optical density at 600 nm (OD600) of 0.1. After incubation at 37°C for 15 h, cells were collected by centrifugation (3,000 × g, 10 min) at 4°C and washed with 2 mM HEPES buffer (pH 8.0) to eliminate pH shift caused by the remaining LB medium. Afterwards, cells were lysed with 0.1% Triton X-100 for 10 min. The colorimetric screening assay for activity was carried out in 96-well plates at 30°C by using 1,3-DCP (5 mM) as a model substrate, while 5 mM both (R)-2-CPE and (S)-2-CPE were used for enantioselectivity screening. One thousand colonies were screened for libraries containing two randomized sites, while 90 colonies were screened for libraries with a single randomized site.
Expression and purification of HheA variants.
The recombinant strain E. coli MC1061, carrying either pBADHheA-WT or pBADHheA-mutant genes, was incubated overnight at 37°C in 500 ml LB medium containing 100 μg/ml ampicillin and 0.05% (wt/vol) l-arabinose to reach an OD600 of 1.7 to 2.0. Cells were harvested by centrifugation (5,000 × g, 50 min) at 4°C and washed once with 10 mM Tris-SO4 buffer (pH 7.5). Both wild-type HheA and its mutants were purified by using a Q-Sepharose (20 ml; GE) column as described before (25). During purification, the halohydrin dehalogenase activity was determined using the chromogenic substrate rac-p-nitro-2-bromophenylethanol by monitoring the absorbance change at 310 nm in 50 mM Tris-SO4 buffer (pH 7.5) at 30°C on a Puxi-1900 double-beam UV-visible (UV/VIS) spectrophotometer (12). The concentrations of proteins were measured using Bradford's method.
Steady-state kinetic measurement.
Kinetic parameters of wild-type and mutant HheA toward both enantiomers of 2-CPE were determined in 50 mM Tris-SO4 buffer (pH = 8.0) at 30°C by monitoring halide liberation using purified enzymes (1). Less than 0.5% (vol/vol) dimethyl sulfoxide (DMSO) was used to facilitate the solubilization of the substrates. The reaction system is 5 ml, and the reaction was initiated by the addition of a certain amount of enzyme. Samples (0.25 ml each) were taken periodically from the reaction mixture and immediately added into 0.9 ml reagent I [30 mM NH4Fe(SO4)2 in 1 M HNO3] to stop the reaction, after which 0.1 ml reagent II [saturated solution of Hg(SCN)2 in absolute ethanol] was added. The absorbance of the mixture at 460 nm was measured on a Puxi Tu-1900 UV/VIS spectrometer. Initial rates were determined from the initial linear part of the reaction progress curves by varying the concentration of the substrate. Kinetic data were fitted with the Michaelis-Menten equation using the Sigmaplot software program (version 10.0) to obtain the steady-state kinetic parameters. All kcat values are defined as per monomer.
Enzymatic kinetic resolution.
The kinetic resolution experiments were performed using crude extracts of both wild-type HheA and its variants in a sealed container at 30°C. All crude extracts were prepared by cell sonication followed by centrifugation (18,000 × g, 60 min), and their expression levels were estimated by using SDS-PAGE. The kinetic resolution of 5 mM rac-2-CPE in 18 ml Tris-SO4 buffer (200 mM, pH 8.0) was started by adding 2 ml of each crude extract (containing 5 mg protein in total). Wild-type HheA and the N178A mutant were also tested for the kinetic resolution of 5 mM racemic styrene oxide with 5 mM NaN3. The reaction was carried out in 200 mM Tris-SO4 buffer (pH 7.5) with 2 ml of crude extract (25 mg protein for wild-type HheA due to the low activity of the enzyme and 5 mg for the N178A mutant). All enzymatic reactions were validated by carrying out reactions under the same conditions in the absence of enzyme. Moreover, the effect of the E. coli expression background on the substrate stability of styrene oxide was also investigated by using the crude extract of E. coli carrying the empty pBAD vector. The enzymatic conversion was monitored by periodically taking samples (0.5 ml each) from the reaction mixture. The samples were extracted with 1 ml ethyl acetate containing mesitylene as an internal standard. Prior to GC analysis, the samples were dried with Na2SO4. The enantiomeric excess (e.e.) of the remaining substrates and products was determined by chiral GC under the following conditions: 100°C for 6 min, 10°C/min to 170°C, and 15 min at 170°C. All separations were carried out using a β-dex 225 chiral column (30 m × 0.25 mm × 0.25 μm; Supelco).
Molecular docking analysis.
The crystal structure of apo-HheA (PDB code 1ZMO) was used to generate the models of wild-type HheA and its variants after removal of all crystallographic bound waters. Hydrogen atoms were added to the protein, and residues were kept fixed in their crystallographic positions in all experiments. Variants were constructed by replacing the target amino acids (V136, L141, and N178) in silico and by energy minimization using the software program SWISS-MODEL. Ligand files were produced using the Prodrg Server. Gasteiger charges were added to the ligand. Proteins were prepared by adding the polar hydrogen atoms, followed by the addition of Kollman charges. The rigid dockings between proteins and ligands were performed using the AutoDock software program (version 4.02) using the Lamarckian genetic algorithm (16). The docking center was located at catalytic residue S134, and the docking box was settled to contain all desired active-site residues. The selection criteria for hits were as follows: the secondary hydroxyl moiety of Cα in the substrate was positioned within hydrogen bonding distance of the OH groups of the catalytic residues Y147 and S134, and the halide group of Cβ was placed in the direction of the water molecule that is present in the halide-binding site of HheA. The best docking conformation for each variant was discovered from the top 100 docking results with the productive conformation of HheA. Visualization and graphics were done using the PyMol software program (5).
RESULTS
Construction and screening of saturation mutagenesis libraries of HheA.
Inspection of the superimposed structures of HheA (PDB code 1ZMO) and HheC (PDB code 1PWZ) revealed the absence of C-terminal fragment in HheA, as well as profound structural differences around the active-site loop region that forms the substrate-binding pocket and the halide-binding site of the enzymes (data not shown). Strikingly, the residue at the position that is equivalent to the bulky W139 in HheC is leucine 141 in HheA, which could be responsible for the difference in enantioselectivity observed between HheA and HheC. Therefore, L141 and another two residues, V136 and N178, present in the equivalent positions of T134 and N176 in HheC, respectively, were chosen for saturation mutagenesis. For this, a focused library of V136X/L141X was first constructed according to a CASTing rule that two spatially close residues should be randomized simultaneously to allow for the exploration of synergistic effects between the two residues (18). After that, a combinatory saturation mutagenesis library of N178X was constructed by choosing the best variant of the V136/L141 library as a template.
The saturation mutagenesis libraries were screened using the previously developed high-throughput colorimetric activity assay on a 96-well plate format (22). The expression levels of each variant of the constructed libraries were estimated by measuring the corresponding OD600 values. 1,3-DCP was used as a model substrate for activity screening due to its low cost and relatively high stability. After 1,000 colonies were screened, 40 positive colonies were selected and subsequently subjected to enantioselectivity screening, resulting in 4 colonies which displayed activity toward (R)-2-CPE that was higher than that for the S enantiomer. The enantioselectivities of the resulting four variants were then pretested using the crude extract of the enzymes. All tested variants indeed exhibited an inverted R enantiopreference, with E values of 3 to 13. Sequencing results showed that the four variants are as follows: L141D (R enantioselectivity [ER] = 4.5), V136M/L141D (ER = 2.4), V136I/L141G (ER = 4.6), and V136Y/L141G (ER = 13.3). No replacement with a bulky amino acid was observed at position 141; instead, a replacement with glycine was found in two of the four selected mutants. The results might indicate either that the steric hindrance of the amino acid at position 141 is not the main enantiomer discriminator in HheA or that replacement with bulky amino acids has detrimental effects on enzyme activity. Therefore, such variants had already been eliminated from the first-round screening. The V136Y/L141G variant with the highest E-value was selected as a template for the construction of the N178X-V136Y/L141G saturation mutagenesis library.
Enantioselectivity of the N178X-V136Y/L141G library was screened with both enantiomers of 2-CPE. One striking colony with much higher activity toward (S)-2-CPE than that of its R counterpart was obtained after screening 90 colonies. The sequencing result showed that this was due mainly to the introduction of one extra point mutation, N178A, into the V136Y/L141G variant, implying that the residue at position 178 might be crucial for controlling the S enantiopreference of HheA. To test this hypothesis, saturation mutagenesis was conducted at position 178 by using the wild-type hheA gene as a template. The enantioselectivity screening results showed that three colonies in the library presented a much higher activity toward the S enantiomer of 2-CPE than that of the V136Y/L141G/N178A variant. Sequencing results showed that all three variants were N178A, which further confirmed our hypothesis that N178A is the key mutation for evolving S-selective HheA.
Kinetic resolution studies.
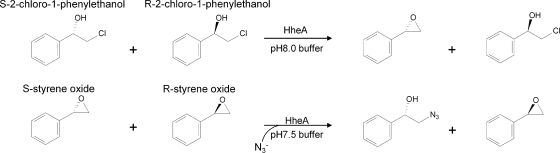
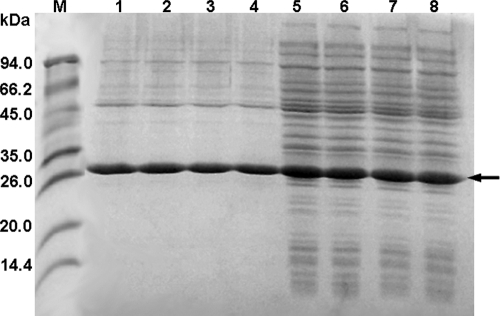
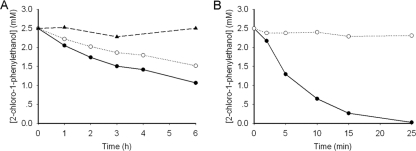
To further investigate the enantioselectivity patterns of those variants, studies of the kinetic resolution of rac-2-CPE and azide-mediated ring opening of styrene oxide were performed by using the crude extract of the enzymes (Fig. 1). Results are summarized in Table 1. The expression levels of those variants were comparable under our experimental conditions as judged by SDS-PAGE (Fig. 2). The kinetic resolution results clearly indicated that those three active-site residues are crucial for enantiomer recognition in HheA. Wild-type HheA showed a rather low S enantioselectivity in the kinetic resolution of rac-2-CPE (ES = 1.7); however, the V136Y/L141G variant exhibited a reversed enantioselectivity (ER = 13). By introducing the N178A mutation into the V136Y/L141G variant, enantiopreference in the V136Y/L141G/N178A triplet mutant was restored with an ES value of 23, which is 14-fold higher than that of the wild-type enzyme. Surprisingly, the N178A mutant exhibited complete enantioselectivity toward (S)-2-CPE (E > 200). Figure 3 shows the progress curves for the dehalogenation of rac-2-CPE catalyzed by both wild-type HheA (Fig. 3A) and the N178A mutant (Fig. 3B). Since no clear chemical conversion was observed within 6 h, the chemical background should have no major impact on the enantioselectivities of the wild-type enzyme and its variants in the dehalogenation of rac-2-CPE even when the reaction times were different. For the wild-type enzyme, the reaction reached about 33% and 50% of the total conversion after 3 h and 6 h, respectively, with an E value of 1.7. The e.e. values for both the substrate and the product were about 11% and 22%, respectively (Table 1). With the same amount of enzyme, the N178A variant catalyzed the conversion much faster than wild-type HheA, with the total conversion of 50% after 25 min, yielding an R substrate (98% e.e.) and an S product (97% e.e.) (Table 1).
Fig 1.
Kinetic resolution reactions catalyzed by HheA and its variants.
Table 1.
Kinetic resolution of dehalogenation of rac-2-CPE and the azide-mediated ring-opening reaction of rac-styrene oxide catalyzed by wild-type HheA and its variants
| Enzyme | Substrate | % conversion (h)a | e.e.(s) (%) | Config.b | e.e.(p) (%) | Config. | E valuec |
|---|---|---|---|---|---|---|---|
| HheA WT | rac-2-CPE | 33 (3) | 11 | R | 22 | S | 1.7 |
| V136Y/L141G variant | rac-2-CPE | 43 (2) | 57 | S | 77 | R | 13 |
| V136Y/L141G/N178A variant | rac-2-CPE | 40 (0.6) | 58 | R | 86 | S | 23 |
| N178A variant | rac-2-CPE | 50 (0.4) | 98 | R | 97 | S | >200 |
| HheA WT | rac-styrene oxide | 7 (0.4)d | 5 | R | 65 | S | 5 |
| N178A variant | rac-styrene oxide | 51 (0.4) | 96 | R | 93 | S | 108 |
The reaction time is indicated in parentheses. All reactions were carried out using the crude extract of the enzymes. The expression levels of HheA and its variants were estimated from SDS-PAGE.
Config., absolute configuration.
E values were calculated from the e.e. for the substrate [e.e.(s)] and that for the product [e.e.(p)] according to the formula E = ln[(1 − e.e.(s))/(1 + e.e.(s)/e.e.(p))]/ln[(1 + e.e.(s))/(1 + e.e.(s)/e.e.(p))].
The concentration of wild-type HheA used in this reaction is 5-fold higher than that of the enzymes used in all other reactions due to its low activity in the ring-opening reaction of styrene oxide with azide.
Fig 2.
SDS-PAGE analysis of the expression level and purity of wild-type HheA and its variants. The enzymes were expressed in E. coli MC1061 using the vector pBAD under the expression conditions described in Materials and Methods. Lane M, molecular size marker; lanes 1 to 4, purified enzymes of wild-type HheA and the V136Y/L141G, V136Y/L141G/N178A, and N178A variants, respectively; lanes 5 to 8, crude extracts of wild-type HheA and the V136Y/L141G, V136Y/L141G/N178A, and N178A variants, respectively. The arrow indicates the HheA band.
Fig 3.
Progress curves of the dehalogenation of 5 mM rac-2-CPE in 20 ml Tris-SO4 (200 mM, pH 8.0) at 30°C. Five-milligram crude extracts of each enzyme were used. (A) Reactions were carried out with wild-type HheA. Enantiomers of 2-CPE are shown as “●” for S and “○” for R. Chemical background was performed in the absence of enzyme and is indicated as “▲”. (B) The reaction was catalyzed by the N178A variant. Enantiomers of 2-CPE are shown as “●” for S and “○” for R.
Moreover, the kinetic resolution of the azide-mediated ring-opening reaction of styrene oxide was also performed with wild-type HheA and the N178A mutant. Due to the poor activity of the wild-type enzyme, a 5-fold-larger amount of protein relative to that of the N178A mutant was used. Independent of the amount of crude extract added, no effect of the E. coli expression background on substrate stability was observed (data not shown). Again, the N178A mutant exhibited a 22-fold-higher S enantioselectivity in the kinetic resolution of styrene oxide (ES = 108) than wild-type HheA (ES = 5), yielding the corresponding β-azidoalcohol with an e.e. of 93%. Thus, the mutant represents a valuable tool for the production of (S)-β-substituted alcohols. Importantly, the N178A mutation also increased the reaction rate in both dehalogenation and the ring-opening reaction, especially in the azide-mediated ring-opening reaction of styrene oxide, in which a conversion rate about 30-fold higher than that of wild-type HheA was obtained. The results demonstrated that the N178A mutation is crucial not only for enantioselectivity but also for the enzymatic activity.
Steady-state kinetic studies.
To gain insights into the improved and inverted enzyme enantioselectivity by the mutations in HheA, steady-state kinetic parameters for both the wild-type enzyme and the three HheA variants toward each enantiomer of 2-CPE were determined using purified enzymes (Table 2). The purities of those enzymes were about 80% as judged by SDS-PAGE (Fig. 2). The V136Y/L141G double mutant had a Km value for (R)-2-CPE that is about 3-fold lower than that of the wild-type enzyme and had about a 2-fold-higher Km value for its S enantiomer, whereas the double mutations did not show a significant effect on the kcat values. The results indicated that the inverted R enantioselectivity by the V136Y/L141G mutations in HheA was due mainly to the effects on the Km values toward both enantiomers of 2-CPE rather than the kcat values.
Table 2.
Steady-state kinetic parameters of purified wild-type HheA and its variants toward R and S enantiomers of 2-CPE
| Enzymea | Value for enantiomer |
Ecalb | |||||
|---|---|---|---|---|---|---|---|
| (R)-2-Chloro-1-phenylethanol |
(S)-2-Chloro-1-phenylethanol |
||||||
| Km (mM) | kcat (s−1) | kcat/Km (s−1 M−1) | Km (mM) | kcat (s−1) | kcat/Km (s−1 M−1) | ||
| HheA WT | 22.6 ± 3.0 | 0.72 ± 0.04 | 32 | 21.2 ± 1.5 | 0.94 ± 0.03 | 44 | 1.4 (S) |
| V136Y/L141G variant | 7.9 ± 0.4 | 0.70 ± 0.01 | 89 | 37.9 ± 6.0 | 0.67 ± 0.05 | 18 | 4.9 (R) |
| V136Y/L141G/N178A variant | >50 | >2.1 | 11 | 10.7 ± 0.5 | 2.87 ± 0.22 | 269 | 24.5 (S) |
| N178A variant | >50 | >1.0 | 4 | 4.8 ± 0.5 | 5.30 ± 0.14 | 1,104 | 276 (S) |
WT, wild type.
The Ecal value was calculated using the equation Ecal = (kcat/Km)R/(kcat/Km)S (for the V136Y/L141G variant) or Ecal = (kcat/Km)S/(kcat/Km)R (for the others), where (kcat/Km)R and (kcat/Km)S are the values for the R and S enantiomers, respectively.
Furthermore, the V136Y/L141G/N178A triple mutant and the N178A single mutant had similar effect patterns for both kcat and Km values toward the two enantiomers of 2-CPE, indicating that the N178A mutation dominated the effects on those kinetic parameters. The results showed that the N178A mutation alone greatly decreased the Km value by more than 4-fold with (S)-2-CPE, but it increased the Km value by more than 2-fold with (R)-2-CPE, and most important, the mutation also greatly enhanced the kcat value by 5- to 6-fold in the dehalogenation of (S)-2-CPE, all of which made significant contributions to the high S enantioselectivity for the N178A mutant. The Km value of the N178A mutant toward (R)-2-CPE was too high to be determined due to the solubility of the substrate. The observed effect of the N178A mutation on the Km values of the two enantiomers was found to be in a contrast to what we observed with the V136Y/L141G double mutations, suggesting that those residues located in loop regions are an important area for modulating substrate binding and could be investigated further for evolving enantioselective HheA variants. The N178 residue, located in the halide-binding loop in HheA, has a dual effect on both substrate binding and activity. It is worth noting that previous studies have shown that halide release is rate limiting during the catalytic cycle of dehalogenation catalyzed by HheC (23), implying that HheA might share a kinetic mechanism similar to that of HheC. Moreover, the E values calculated (Ecal) on the basis of steady-state kinetic parameters are comparable to those obtained from kinetic resolution studies, indicating that the two experiments were conducted under similar conditions.
Molecule docking analysis.
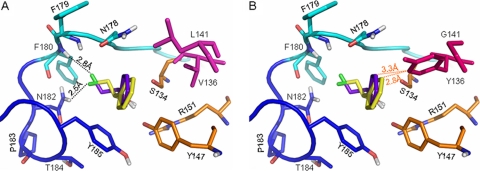
To gain insights into the nature of enantioselectivity of the enzymes, molecular docking of both enantiomers of 2-CPE to wild-type HheA and the two mutants (N178A and V136Y/L141G) was carried out using the program AutoDock (Fig. 4). In HheA, the enzyme uses a conserved catalytic triad (Ser134/Tyr147/Arg151) to catalyze the dehalogenation of halohydrins and the epoxide ring-opening reactions by forming a productive complex. Both enantiomers of 2-CPE could be well accommodated in the active site of wild-type HheA to form a productive complex (Fig. 4A). In the binding modes, the secondary hydroxyl moieties of the two enantiomers were positioned within the hydrogen bonding distance of the OH groups of the catalytic residues S134 and Y147 (2), and their phenyl groups pointed to the exterior of the active site with a slight difference in binding orientation. The most profound binding difference observed for the two enantiomers was orientation of the chloride atoms in the halide-binding site. In the R isomer it pointed to the 177Pro-Asn-Phe-Phe180 part, while in S it was orientated toward 181Asn-Asn-Pro-Thr-Tyr-185, resulting in local differences in the way that the enzyme stabilizes the bound chloride atom for the two enantiomers.
Fig 4.
Representation of the binding modes of the two enantiomers of 2-CPE in the active site of wild-type HheA (A) or the V136Y/L141G variant (B). The catalytic triad Ser134/Tyr147/Arg151 is shown in orange; V136 and L141 are in magenta, while Y136 and G141 are in hot pink; the 177Pro-Asn-Phe-Phe180 part of the halide-binding loop is in cyan, while the 181Asn-Asn-Pro-Thr-Tyr-185 part is in blue; the loop residues are shown in the corresponding cyan and blue, respectively. The docked substrates are shown in purple-blue for the R enantiomer and yellow for its S counterpart. Hydrogen bonds are indicated as black dotted lines. For clarity, van der Waals contacts are shown as orange dotted lines.
In the binding mode as shown in Fig. 4A, the bound chloride atom of (R)-2-CPE was mainly stabilized by forming a halogen bond with the backbone NH group of F180, while that of (S)-2-CPE formed a halogen bond with the side chain amide group of N182. Inspecting the halide-binding site of HheA revealed that the side chain configuration of N182 was stabilized by a dense hydrogen bonding network formed with the carbonyl oxygen atom of F180, the backbone NH groups of T184 and Y185. Moreover, those interactions were further maintained by the sharp turn of the loop at position P183. Although N178 did not show any interactions with the bound chloride atom in either of the enantiomers, its side chain Oδ1 atom hydrogen bonded with the backbone NH group of F179. The N178A mutation would eliminate such a contact. Moreover, the N178A mutation was also involved in the change from a polar residue to a hydrophobic one, which could affect the surrounding microenvironment. Thus, it is more likely that the N178A mutation affected the backbone orientation of F179, as well as that of its neighboring F180, resulting in a weak binding of the chloride atom of the R enantiomer. The effect on the side chain conformation of N182 is much less significant. This could well explain the considerably increased S enantioselectivity of the N178A variant. On the other hand, we could speculate that the increased activity of the N178A variant is more likely due to the effect of the N178A mutation on the overall conformation of the halide-binding site, since the halide release step was proved to be rate limiting in the dehalogenation of halohydrins in HheC. The possibility that HheA and HheC follow a different kinetic mechanism could not be excluded either.
In the V136Y/L141G variant (Fig. 4B), the L141G mutation might provide a good accommodation to the V136Y mutation to prevent significant structure disturbance caused by the two mutations. Further inspection of the possible interactions between the two amino acids and the two enantiomers revealed that the distances between the substrate phenyl group and the side chain of Y136 were 2.8 Å and 3.3 Å for (S)-2-CPE and (R)-2-CPE, respectively. The difference in distance was mainly due to the difference in binding orientations of the two substrate phenyl groups in the active site. The reported van der Waals radii for both the Phe and Tyr rings is about 1.7 Å (21), and thus the distance of 2.8 Å is too short for van der Waals contacts between the two groups and repulsion will be produced instead, whereas 3.3 Å is the corresponding van der Waals distance, producing a favorable recognition of the R enantiomer in the V136Y/L141G double mutant. This is in agreement with the observed Km values and could explain the reversed enantiopreference of the mutant. Taken together, the substrate docking studied provided structural details for understanding the observed effects on the enantioselectivity of HheA by those mutations.
Conclusions.
In this work, we demonstrated that the three active-site residues (V136, L141, and N178) play crucial roles in controlling the enantioselectivity of HheA from Arthrobacter sp. AD2. The residues at positions 136 and 141 were identified as key residues to sterically control the binding conformation of the substrate phenyl group, and mutants with inverted R enantiopreference were obtained by the mutations at the two positions. Strikingly, a highly S-selective enzyme in the kinetic resolution of the dehalogenation of 2-CPE was successfully evolved by a single mutation of N178A in the halide-binding loop of the enzyme. The well-studied halohydrin dehalogenase from Agrobacterium radiobacter AD1 (HheC) displays a high R enantioselectivity (11), and no studies concerning the inverted enantioselectivity of the enzyme have been reported. To the best of our knowledge, up to now this is the first reported totally S-selective halohydrin dehalogenase. Enantioselectivity modification has been considered a major challenge in protein engineering, and normally series of mutations are needed in many cases (9, 19, 20, 26). It is quite amazing that such a pronounced effect on enantioselectivity was even accompanied by a 5- to 6-fold improvement in activity, which was produced by a single mutation. Overall, a semirational design approach was proved to be an efficient means of enzyme evolution.
ACKNOWLEDGMENTS
This research was supported by the National Natural Science Foundation of China (no. 20872014).
We are grateful for the discussion with our colleague Peng Zhou on molecular docking results.
Footnotes
Published ahead of print 20 February 2012
REFERENCES
- 1. Bergmann JG, Sanik J. 1957. Determination of trace amounts of chlorine in naphtha. Anal. Chem. 29:241–243 [Google Scholar]
- 2. de Jong RM, Kalk KH, Tang L, Janssen DB, Dijkstra BW. 2006. The X-ray structure of the haloalcohol dehalogenase HheA from Arthrobacter sp. strain AD2: insight into enantioselectivity and halide binding in the haloalcohol dehalogenase family. J. Bacteriol. 188:4051–4056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. de Jong RM, et al. 2003. Structure and mechanism of a bacterial haloalcohol dehalogenase: a new variation of the short-chain dehydrogenase/reductase fold without an NAD(P)H binding site. EMBO J. 22:4933–4944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. de Jong RM, et al. 2005. Structural basis for the enantioselectivity of an epoxide ring opening reaction catalyzed by halo alcohol dehalogenase HheC. J. Am. Chem. Soc. 127:13338–13343 [DOI] [PubMed] [Google Scholar]
- 5. DeLano WL. 2002. The PyMOL molecular graphics system. DeLano Scientific, San Carlos, CA [Google Scholar]
- 6. Haak RM, et al. 2007. Synthesis of enantiopure chloroalcohols by enzymatic kinetic resolution. Org. Biomol. Chem. 5:318–323 [DOI] [PubMed] [Google Scholar]
- 7. Hasnaoui G, et al. 2005. Nitrite-mediated hydrolysis of epoxides catalyzed by halohydrin dehalogenase from Agrobacterium radiobacter AD1: a new tool for the kinetic resolution of epoxides. Tetrahedron Asymmetry 16:1685–1692 [Google Scholar]
- 8. Janssen DB, Majerić Elenkov M, Hasnaoui G, Hauer B, Lutje Spelberg JH. 2006. Enantioselective formation and ring-opening of epoxides catalysed by halohydrin dehalogenases. Biochem. Soc. Trans. 34:291–295 [DOI] [PubMed] [Google Scholar]
- 9. Liebeton K, et al. 2000. Directed evolution of an enantioselective lipase. Chem. Biol. 7:709–718 [DOI] [PubMed] [Google Scholar]
- 10. Lutje Spelberg JH, Tang L, van Gelder M, Kellogg RM, Janssen DB. 2002. Exploration of the biocatalytic potential of a halohydrin dehalogenase using chromogenic substrates. Tetrahedron Asymmetry 13:1083–1089 [Google Scholar]
- 11. Lutje Spelberg JH, van Hylckama Vlieg JET, Bosma T, Kellogg RM, Janssen DB. 1999. A tandem enzyme reaction to produce optically active halohydrins, epoxides and diols. Tetrahedron Asymmetry 10:2863–2870 [Google Scholar]
- 12. Lutje Spelberg JH, van Hylckama Vlieg JET, Tang L, Janssen DB, Kellogg RM. 2001. Highly enantioselective and regioselective biocatalytic azidolysis of aromatic epoxides. Org. Lett. 3:41–43 [DOI] [PubMed] [Google Scholar]
- 13. Majerić Elenkov M, Hauer B, Janssen DB. 2006. Enantioselective ring opening of epoxides with cyanide catalysed by halohydrin dehalogenases: a new approach to non-racemic β-hydroxy nitriles. Adv. Synth. Catal. 348:579–585 [Google Scholar]
- 14. Majerić Elenkov M, Hoeffken HW, Tang L, Hauer B, Janssen DB. 2007. Enzyme-catalyzed nucleophilic ring opening of epoxides for the preparation of enantiopure tertiary alcohols. Adv. Synth. Catal. 349:2279–2285 [Google Scholar]
- 15. Majerić Elenkov M, Tang L, Hauer B, Janssen DB. 2006. Sequential kinetic resolution catalyzed by halohydrin dehalogenase. Org. Lett. 8:4227–4229 [DOI] [PubMed] [Google Scholar]
- 16. Morris GM, et al. 1998. Automated docking using a Lamarckian genetic algorithm and empirical binding free energy function. J. Comput. Chem. 19:1639–1662 [Google Scholar]
- 17. Pollard DJ, Woodley JM. 2007. Biocatalysis for pharmaceutical intermediates: the future is now. Trends Biotechnol. 25:66–73 [DOI] [PubMed] [Google Scholar]
- 18. Reetz MT, Bocola M, Carballeira JD, Zha D, Vogel A. 2005. Expanding the range of substrate acceptance of enzymes: combinatorial active-site saturation test. Angew. Chem. Int. Ed. 117:4264–4268 [DOI] [PubMed] [Google Scholar]
- 19. Reetz MT, et al. 2009. Directed evolution of an enantioselective epoxide hydrolase: uncovering the source of enantioselectivity at each evolutionary stage. J. Am. Chem. Soc. 131:7334–7343 [DOI] [PubMed] [Google Scholar]
- 20. Reetz MT, Wilensek S, Zha D, Jaeger KE. 2001. Directed evolution of an enantioselective enzyme through combinatorial multiple-cassette mutagenesis. Angew. Chem. Int. Edit. 40:3589–3591 [DOI] [PubMed] [Google Scholar]
- 21. Richards FM. 1974. The interpretation of protein structures: total volume, group volume distributions and packing density. J. Mol. Biol. 82:1–14 [DOI] [PubMed] [Google Scholar]
- 22. Tang L, Li Y, Wang X. 2010. A high-throughput colorimetric assay for screening halohydrin dehalogenase saturation mutagenesis libraries. J. Biotechnol. 147:164–168 [DOI] [PubMed] [Google Scholar]
- 23. Tang L, Lutje Spelberg JH, Fraaije MW, Janssen DB. 2003. Kinetic mechanism and enantioselectivity of halohydrin dehalogenase from Agrobacterium radiobacter. Biochemistry 42:5378–5386 [DOI] [PubMed] [Google Scholar]
- 24. Tang L, van Merode AEJ, Lutje Spelberg JH, Fraaije MW, Janssen DB. 2003. Steady-state kinetics and tryptophan fluorescence properties of halohydrin dehalogenase from Agrobacterium radiobacter. Roles of W139 and W249 in the active site and halide-induced conformational change. Biochemistry 42:14057–14065 [DOI] [PubMed] [Google Scholar]
- 25. van Hylckama Vlieg JET, et al. 2001. Halohydrin dehalogenases are structurally and mechanistically related to short-chain dehydrogenases/reductases. J. Bacteriol. 183:5058–5066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zha D, Wilensek S, Hermes M, Jaeger KE, Reetz MT. 2001. Complete reversal of enantioselectivity of an enzyme-catalyzed reaction by directed evolution. Chem. Commun. 2001:2664–2665 [Google Scholar]