Abstract
Myxococcus xanthus is widely used as a model system for studying gliding motility, multicellular development, and cellular differentiation. Moreover, M. xanthus is a rich source of novel secondary metabolites. The analysis of these processes has been hampered by the limited set of tools for inducible gene expression. Here we report the construction of a set of plasmid vectors to allow copper-inducible gene expression in M. xanthus. Analysis of the effect of copper on strain DK1622 revealed that copper concentrations of up to 500 μM during growth and 60 μM during development do not affect physiological processes such as cell viability, motility, or aggregation into fruiting bodies. Of the copper-responsive promoters in M. xanthus reported so far, the multicopper oxidase cuoA promoter was used to construct expression vectors, because no basal expression is observed in the absence of copper and induction linearly depends on the copper concentration in the culture medium. Four different plasmid vectors have been constructed, with different marker selection genes and sites of integration in the M. xanthus chromosome. The vectors have been tested and gene expression quantified using the lacZ gene. Moreover, we demonstrate the functional complementation of the motility defect caused by lack of PilB by the copper-induced expression of the pilB gene. These versatile vectors are likely to deepen our understanding of the biology of M. xanthus and may also have biotechnological applications.
INTRODUCTION
Myxococcus xanthus is a deltaproteobacterium, belonging to the myxobacteria, which exhibits a complex and unique life cycle. In the presence of nutrients, cells feed cooperatively, being able to prey on other microorganisms (16). In response to starvation, cells initiate a developmental program that culminates in the formation of multicellular fruiting bodies, within which cells differentiate into dormant myxospores (21). Due to this fascinating life cycle, M. xanthus is an excellent model organism to study cellular processes such as social behavior, gliding motility, cell-cell communication, signal transduction, regulation of gene expression, morphogenesis, and differentiation. Also, myxobacteria synthesize a large variety of secondary metabolites with diverse structures and a multitude of biological activities (20).
Although many tools for the genetic manipulation of M. xanthus are available, robust tools that allow the conditional expression of genes of interest have not been well developed so far. Two systems allow the constitutive overexpression of genes and are based on the use of the oar promoter (5, 12) or the pilA promoter (8, 22). Furthermore, three types of inducible vectors have been designed, although all of them have features that make them unsuitable for general use in a simple and controlled manner. One system relies on the light-inducible extracytoplasmic function σ factor CarQ (2, 11, 19). To express genes under the control of CarQ, cells have to be illuminated by blue light, while incubation in the dark represses transcription. The nature of the inducing signal requires a special incubator and complicates the reproducibility of the experiments. Moreover, light interferes with development. A second system is based on the PpilA-lacO-RBS promoter in combination with the LacI repressor and uses isopropyl β-d-1-thiogalactopyranoside (IPTG) as an inducer (9). Unfortunately, the expression level in the absence of IPTG is high, and the addition of the inducer raises the amount of protein produced only approximately 2-fold. The third system is based on the MerR-like repressor CarH (3). Maximum expression levels with this vector are obtained in the absence of vitamin B12 and with cells under constant illumination. This hampers the use of this expression system during development, because light blocks development.
The M. xanthus genome carries a large number of copper-inducible genes, which are differentially regulated (14, 15, 18). Among the different expression profiles observed for these genes, that exhibited by the multicopper oxidase cuoA offers the possibility of using its promoter to express genes in a highly controlled manner (18); i.e., no expression of the gene is detected in the absence of copper, and the level of expression increases linearly with the copper concentration in the culture medium.
In this report we describe the generation of a set of plasmid vectors based on the cuoA promoter that can be used to express genes in M. xanthus in a highly controlled manner by the addition of copper and with expression levels that range from undetectable in the absence of copper to high and constant levels that linearly depend on the copper concentration.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The M. xanthus and Escherichia coli strains used in this study are listed in Table 1. M. xanthus strains were grown at 30°C in liquid Casitone-Tris (CTT) medium with vigorous shaking as previously described (7). CTT agar plates containing 0.5% or 1.5% Bacto agar (Difco) were used to analyze social motility and adventurous motility, respectively. When necessary, media were supplemented with kanamycin (40 μg/ml), tetracycline (15 μg/ml), bromo-chloro-indolyl-galactopyranoside (40 μg/ml), or copper sulfate (at the concentrations indicated for each experiment). Specific growth conditions are described in Results. E. coli strains were grown at 37°C in Luria-Bertani medium, which was supplemented with ampicillin (50 μg/ml), kanamycin (25 μg/ml), tetracycline (25 μg/ml), or bromo-chloro-indolyl-galactopyranoside (40 μg/ml) when needed. The plasmids used in this study are listed in Table 2.
Table 1.
Bacterial strains used in this study
| Species and strain | Genotype | Reference |
|---|---|---|
| E. coli JM109 | F′[traD36 proAB+lacIqlacZΔM15] recA1 supE44 endA1 hsdR17 gyrA96 relA1 thi Δ(lac-proAB) | 23 |
| M. xanthus | ||
| DK1622 | Wild type | 10 |
| JM10AZY | DK1622 PcuoA-lacZY(pNG10AZY) | This study |
| JMMAT3ZY | DK1622 PcuoA-lacZY(pMAT3ZY) | This study |
| DK10416 | DK1622 ΔpilB | 22 |
| SA4100 | DK1622 ΔpilB PcuoApilB(pMAT1) | This study |
Table 2.
Plasmids used in this study
| Name | Descriptiona | Plasmid from which derived | Reference or source |
|---|---|---|---|
| pUC19 | Apr | 23 | |
| pKY481 | lacZY Kanr | 1 | |
| pUC7(KmPstI−) | Kanr | Provided by S. Inouye | |
| pUC7Tc(B−S−) | Tetr | Provided by S. Inouye | |
| pNG1A | PcuoA Kanr | pUC19 | This study |
| pNG10A | PcuoA Tetr | pUC19 | This study |
| pSL105 | PpilA-pilB Kanr | 8 | |
| pSWU30 | Mx8 attP int Tetr | 22 | |
| pMAT3 | PcuoA Mx8 attP int Tetr | pSWU30 | This study |
| pMAT4 | PcuoA Mx8 attP int Kanr | pMAT1 | This study |
| pNG10AZY | PcuoAlacZY Tetr | pNG10A | This study |
| pMAT3ZY | PcuoAlacZY Mx8 attP int Tetr | pMAT3 | This study |
| pMAT1 | PcuoA-pilB Mx8 attP int Kanr | pSL105 | This study |
Kanr, Tetr, and Apr, resistance to kanamycin, tetracycline, and ampicillin, respectively.
Motility assays, developmental conditions, and spore quantification.
Motility assays were performed using cells from exponentially growing cultures. The cells were harvested and resuspended in CTT medium or TM buffer (10 mM Tris-HCl [pH 7.6], 1 mM MgSO4) to a calculated density of 5 × 109 cells/ml. Ten-microliter aliquots of cell suspensions were spotted on CTT medium supplemented with 1.5 or 0.5% agar and the indicated copper concentrations. After 24 h, colony morphology was observed visually in a Nikon SMZ800 stereomicroscope and the diameter of the spots measured. Clone fruiting (CF) medium (6) was used to induce development as previously reported (13). When required, copper was added at the concentrations indicated for each experiment. Specific developmental conditions are described in Results. For counting spores, 10 drops of 20 μl each were spotted onto CF plates with various copper concentrations. At different time points, the fruiting bodies of one plate were harvested, resuspended in 200 μl of TM buffer, and dispersed by sonication. Myxospores were counted in a Petroff-Hausser chamber.
Construction of the series of plasmid vectors.
Routine molecular biology techniques were used for the construction of the plasmid vectors (17). The plasmid vectors pNG10A and pNG1A, which harbor tetracycline and kanamycin resistance markers (Tetr and Kanr), respectively, were derived from pUC19. First, pUC19 was digested with SspI and DraI to eliminate a 0.94-kb DNA fragment containing the ampicillin resistance marker. The correct fragment was ligated to a 1.44-kb SalI-digested fragment (blunt ended with Klenow) containing the Tetr gene previously obtained from plasmid pUC7Tc(B−S−) (kindly provided by S. Inouye). The resulting plasmid was then digested with KpnI and BamHI and ligated to a 0.98-kb PCR-amplified DNA fragment containing the cuoA promoter. This cuoA promoter was previously amplified using M. xanthus DK1622 chromosomal DNA as a template. The final vector was designated pNG10A. To obtain pNG1A, the same procedure was followed, with the exception that a 1.35-kb Kanr marker was inserted instead of the Tetr gene, which was obtained from plasmid pUC7(KmPstI−) digested with SalI.
Vectors pMAT3 and pMAT4 are derived from pSWU30 (22) and pMAT1, respectively. pMAT1 was constructed by replacing the pilA promoter of pSL105 (8) with a cuoA promoter fragment including 825 bp upstream of the cuoA start codon. This fragment was amplified using M. xanthus DK1622 chromosomal DNA as a template. The PCR fragment was treated with EcoRI and XbaI and ligated into pSL105 restricted with the same enzymes. To construct pMAT4, the pilB gene from pMAT1 was removed by digestion with XbaI and HindIII and filling of the gap with a fragment of two partially complementary oligonucleotides. To obtain the plasmid pMAT3, the enzymes EcoRI and HindIII were used to cleave the cuoA promoter with the XbaI-BamHI-HindIII multilinker from pMAT4, and the obtained fragment was cloned into the EcoRI- and HindIII-restricted vector pSWU30 (22).
Gene reporter studies.
To analyze and quantify the expression of genes under the control of PcuoA, E. coli lacZY genes (the permease is not encoded in the M. xanthus genome and has to be included in the plasmid for qualitative analyses) were amplified by PCR using plasmid pKY481 (1) as a template. The products were then digested with either BamHI-XbaI or BamHI-HindIII and ligated to pNG10A and pMAT3, respectively. The resulting plasmids (pNG10AZY and pMAT3ZY, respectively) were electroporated into M. xanthus DK1622, and several Tetr strains were checked by Southern blotting to confirm that the plasmids were integrated at the PcuoA site (in the case of pNG10AZY) or at the Mx8 attB site (in the case of pMAT3ZY).
Specific β-galactosidase activity in cell extracts obtained by sonication of the strains harboring lacZY genes was determined as previously described (18) and is expressed as nmol of o-nitrophenol produced per min per mg of protein. Measurements shown are the averages of data from triplicate experiments.
Western blotting.
Generally, cells from exponentially growing cultures were harvested, and total protein was separated by SDS-PAGE (proteins from 5 × 107 cells loaded per lane) and analyzed by Western blotting (17). PilB was detected using anti-PilB antibodies (8) and peroxidase-conjugated goat anti-rabbit secondary antibodies as recommended by the manufacturer (Roche). Blots were developed using SuperSignal West Pico chemiluminescence reagent (Pierce).
Nucleotide sequence accession numbers.
The complete nucleotide sequences of pNG1A, pNG10A, pMAT3, and pMAT4 have been deposited in the GenBank database under accession numbers JN940926, JN940923, JN940924, and JN940925, respectively.
RESULTS
Copper tolerance of M. xanthus DK1622.
Previous studies on the copper response in M. xanthus have been carried out using strain DZF1 because it does not agglutinate in liquid cultures. However, the wild-type strain most frequently used is DK1622, for which the complete genome sequence is available (4). For this reason, we studied the copper tolerance of DK1622 in order to establish the range of copper concentrations that do not significantly alter growth, motility, and development. To determine the effect of copper on growth, DK1622 cells were grown in liquid cultures containing CTT medium without copper and then diluted to an optical density at 600 nm (OD600) of 0.05 in fresh CTT liquid medium supplemented with the copper concentrations indicated in Fig. 1. Cultures were incubated for 24 h (only 4 to 5 generations) at 30°C before the OD600 was determined again. As shown in Fig. 1, cell yields of DK1622 with copper concentrations of up to 0.5 mM were similar to those observed in the absence of metal. Higher copper concentrations diminish cell yield, and no growth was observed with copper concentrations of between 0.9 and 1 mM. The copper tolerance of DK1622 during growth is similar to that reported for strain DZF1 (18).
Fig 1.
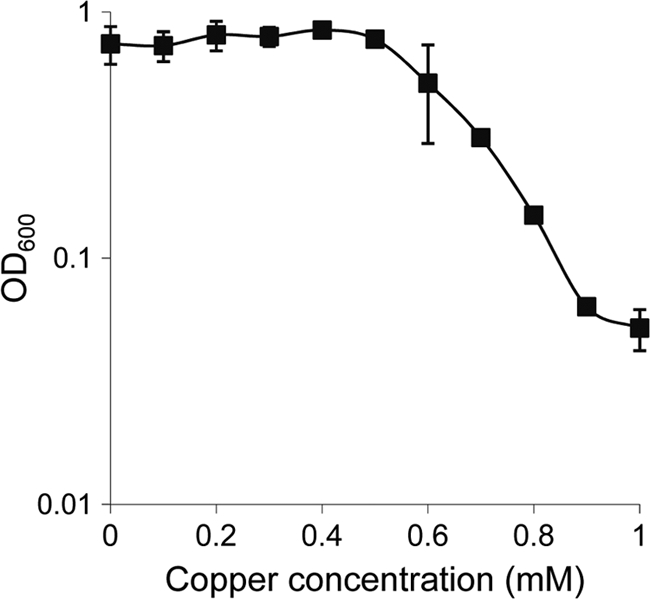
Sensitivity of M. xanthus DK1622 to copper during growth. Cells grown in the absence of copper were diluted to an OD600 of 0.05 in CTT medium with the indicated copper concentrations. Cell density was monitored after incubation at 30°C for 24 h. Experiments were performed in triplicate. Error bars indicate standard deviations.
Next, the effect of copper on adventurous and social motility was examined. For this purpose, M. xanthus cells were grown in the absence of copper, concentrated, and spotted onto CTT agar plates containing either 0.5% agar (for social motility assays) or 1.5% agar (for adventurous motility assays) and different concentrations of copper. The results obtained revealed that copper concentrations of lower than 0.5 mM only slightly interfered with either type of motility. The diameters of the colonies in this range of copper concentrations were just around 10% smaller (for both A and S motility) than those observed in the absence of copper. Increasing copper concentrations caused a fairly linear regression in motility (Fig. 2). The effect of copper on motility was not studied before for strain DZF1.
Fig 2.
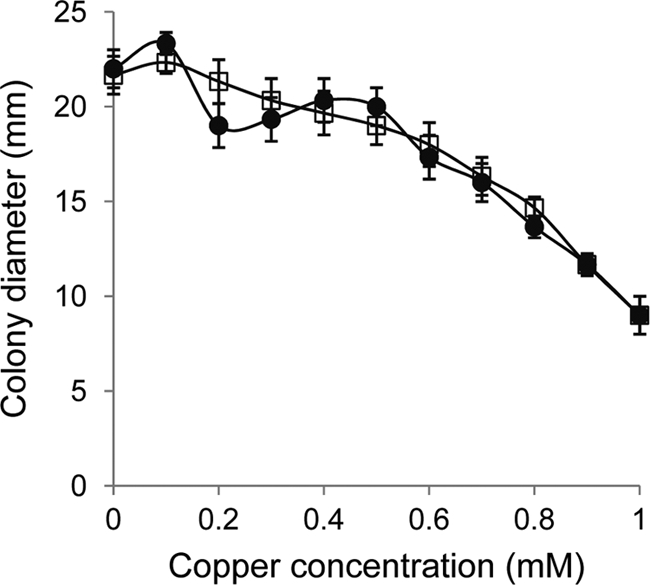
Effect of copper on motility. M. xanthus DK1622 cells were grown in the absence of copper and concentrated to an OD600 of 15. Ten microliters of these cells was spotted onto CTT agar plates containing 0.5% agar for S motility (circles) or 1.5% agar for A motility (squares) and the indicated copper concentrations. The diameter of the spots was determined at 96 h of incubation. Experiments were performed in triplicate. Error bars indicate standard deviations.
To study the effect of copper on development, liquid cultures of M. xanthus cells were concentrated and spotted onto CF agar plates (starvation medium) with different copper concentrations. The concentrations used were 10-fold lower than those used in rich medium because growing M. xanthus cells are more resistant to copper than developing cells (18). As shown in Fig. 3A, cells aggregated normally with copper concentrations of up to 60 μM. A delay in aggregation was observed with copper concentrations of between 60 and 120 μM. With higher concentrations, cells not only did not form fruiting bodies but also died. In contrast, and although aggregation seemed to be normal, the yield of myxospores diminished to 77% with a copper concentration of 20 μM and to 59% with 60 μM (Fig. 3B). According to these data, copper concentrations of up to 60 μM can be used when aggregation is studied. However, caution should be taken in sporulation analyses. M. xanthus DK1622 is slightly more resistant to copper than DZF1, which dies on CF agar plates with copper concentrations higher than 60 μM (18).
Fig 3.
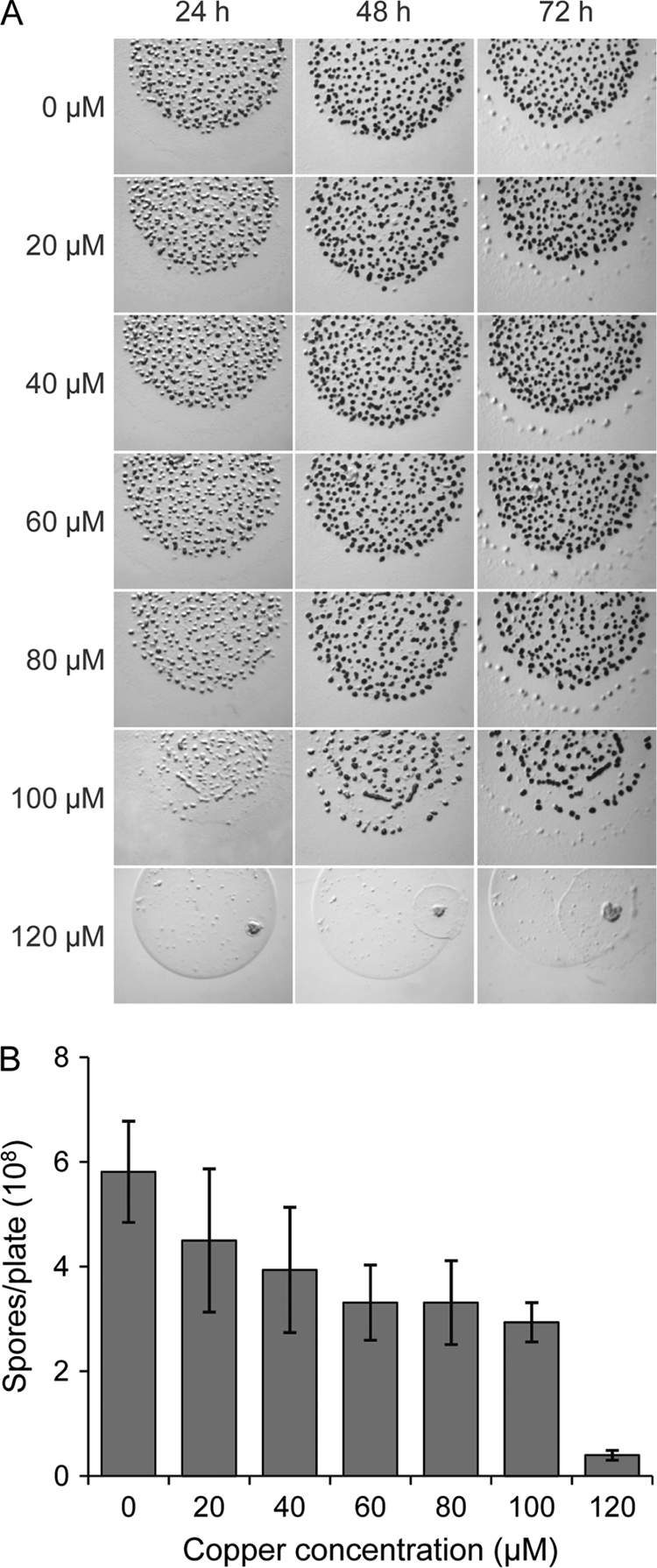
Effect of copper on development. (A) M. xanthus DK1622 cells were grown in the absence of copper and concentrated to an OD600 of 15. Ten microliters of cells was spotted onto CF agar plates containing the indicated copper concentrations. Aggregation was monitored by taking pictures under a dissecting microscope. (B) To quantify the number of spores, cells were concentrated as indicated in panel A. Ten drops of 20 μl each were then spotted onto CF agar plates containing the indicated copper concentrations. After incubation for 72 h, fruiting bodies were harvested from the plates and disrupted by sonication, and spores were quantified in a Petroff-Hausser chamber. Experiments were performed in triplicate. Error bars indicate standard deviations.
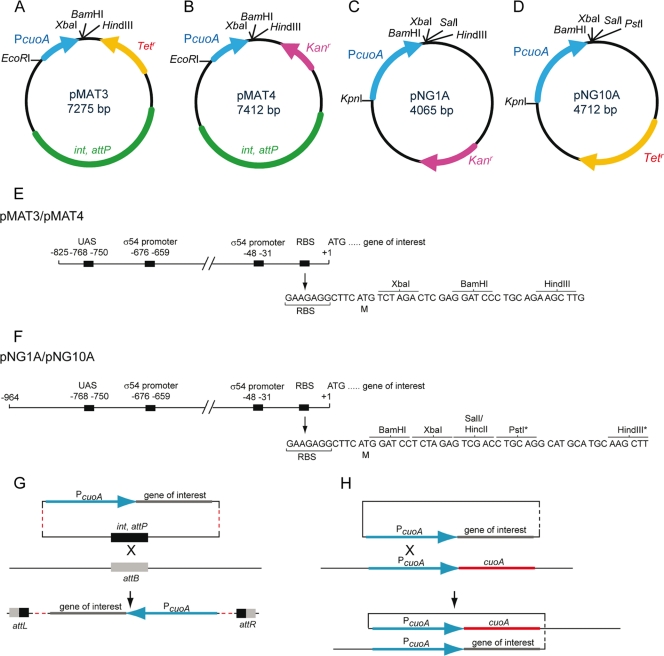
Construction of a set of plasmid vectors designed for the conditional expression of genes in M. xanthus by copper supplementation.
A set of plasmid vectors for the conditional expression of genes in M. xanthus was constructed. All the plasmids contain a DNA fragment with the PcuoA promoter followed by multiple restriction sites to allow the cloning of the gene of interest (Fig. 4). Two of these vectors, pNG10A and pMAT3, harbor a Tetr marker, while the other two, pNG1A and pMAT4, confer Kanr, to allow the simultaneous manipulation of M. xanthus cells with other vectors. The two vectors of the series pNG integrate at the PcuoA site by a single homologous recombination event, ensuring that all the cis elements required for the expression of the genes are present (Fig. 4C, D, F, and H). In contrast, the vectors of the pMAT series integrate at the chromosomal phage Mx8 attB site by specific recombination using the phage attachment site attP and the integrase, both of which are encoded by the two plasmids (Fig. 4A, B, E, and G). Having vectors that integrate at different positions in the M. xanthus genome increases the flexibility for strain constructions.
Fig 4.
(A to D) A set of plasmid vectors for copper-inducible gene expression. Unique restriction sites for cloning genes under the control of the cuoA promoter are indicated. (E and F) Cloning sites and regulatory sequences in the cuoA promoter regions present in the pMAT and pNG vectors. The asterisks in the pNG set indicate that the HindIII and PstI sites are unique in pNG1A and pNG10A, respectively (see panels C and D). Two putative σ54 promoters, one putative upstream-activating sequence (UAS) where an enhancer-binding protein may bind, and one putative ribosome-binding site (RBS) identified upstream of cuoA are depicted. (G and H) Schematic illustrating chromosomal integration of the pMAT vectors (G) and the pNG vectors (H) by site-specific and homologous recombination, respectively. Note that the integrase catalyzing the site-specific integration of the pMAT vectors is encoded by the plasmids (indicated in green in A and B).
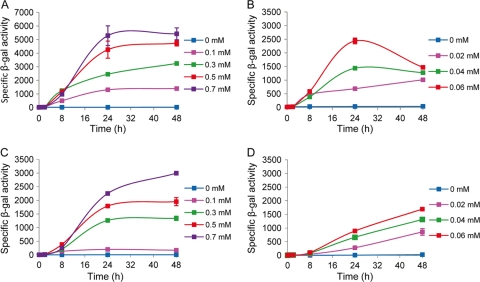
Analysis of promoter induction using the lacZ gene from E. coli as a reporter.
Plasmids pMAT3ZY and pNG10AZY contain the lacZY genes cloned downstream of the cuoA promoter in pMAT3 and pNG10A, respectively (Fig. 4A and D). Strains derived from M. xanthus DK1622, obtained after integration of these two plasmids at the proper sites, were grown in the absence and in the presence of different copper concentrations. Cell extracts obtained at different time points after copper induction were assayed for specific β-galactosidase activity. The two strains showed no β-galactosidase activity in the absence of copper (Fig. 5). The addition of copper caused the induction of lacZ expression, with a linear correlation between β-galactosidase activity and copper concentration for both vectors during growth (compare Fig. 5A and C). The profiles obtained were similar for the two vectors, with a plateau being reached at between 24 and 48 h after copper addition. However, 2-fold-higher levels of activity were obtained with the pMAT3 vector. During development, much lower copper concentrations were required to obtain levels of β-galactosidase activity similar to those observed during growth, as was already reported for strain DZF1 (18). Two differences could be observed in the expression of lacZ cloned in each type of vector (compare Fig. 5B and D) during development: (i) no plateau is obtained between 24 and 48 h in case of the pNG10A vector, and (ii) the highest levels of expression are again obtained with the pMAT3 vector at 24 h, although the values are very similar with both vectors at 48 h.
Fig 5.
Quantification of the expression of lacZ cloned under the control of the cuoA promoter. Cells harboring plasmid pMAT3ZY (A and B) or pNG10AZY (C and D) were either grown on CTT agar plates (A and C) or incubated on starvation medium CF (B and D) containing the copper concentrations indicated by the color code displayed in each panel. At each time point, cells were harvested and disrupted by sonication, and specific β-galactosidase activity was determined (expressed as nmol of o-nitrophenol produced per min per mg of protein). Experiments were performed in triplicate. Error bars indicate standard deviations.
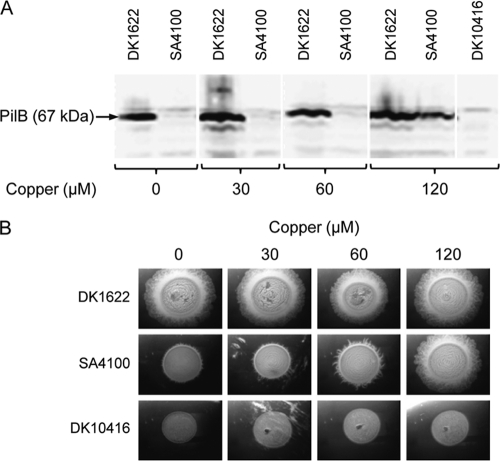
Expression of M. xanthus genes by copper induction.
To assess the functionality of copper-inducible gene expression, the pilB gene, which encodes an ATPase essential for social motility (8), was cloned under the control of PcuoA (plasmid pMAT1) and introduced into strain DK10416 (ΔpilB) to generate strain SA4100. As shown in Fig. 6A, PilB was not detectable in SA4100 in Western blot analysis using specific anti-PilB antibodies with copper concentrations of up to 60 μM. However, PilB accumulated to close to wild-type levels with 120 μM copper. When SA4100 cells were spotted onto plates with different copper concentrations, social motility was partially restored by 60 μM copper and was completely restored in the presence of 120 μM copper (Fig. 6B), clearly demonstrating the usefulness of these vectors for conditional complementation studies.
Fig 6.
Restoration of S motility by expression of the pilB gene under the control of the cuoA promoter. (A) Strains DK1622 (pilB+), SA4100 (ΔpilB PcuoA-pilB), and DK10416 (ΔpilB) were grown in CTT medium supplemented with the indicated copper concentrations for 24 h. Cells were harvested from exponentially growing cultures and analyzed by Western blot using anti-PilB antibodies. Proteins from an equal number of cells were loaded per lane. Copper concentrations higher than 120 μM were not tested. (B) Complementation of the ΔpilB S-motility defect by copper-induced gene expression of pilB+. Cells were grown as described for panel A, harvested, spotted onto CTT medium supplemented with 0.5% agar and the indicated copper concentrations, and imaged after 24 h. Shown in panels A and B are representative results from triplicate experiments.
DISCUSSION
A set of plasmids has been constructed to express genes in M. xanthus by copper supplementation, a genetic tool that was not so far well developed in this bacterium. The design of the vectors was based on previous work demonstrating that M. xanthus harbors several copper-dependent promoters that are expressed at high levels during both growth and development (14, 15, 18). Among the copper-inducible promoters, the cuoA promoter exhibits the properties required for the expression of other genes: (i) there is no detectable expression in the absence of copper (Fig. 5); (ii) expression linearly increases as a function of the copper concentration and reaches a plateau after 24 to 48 h, ensuring that the protein concentration in the cells will be maintained after 24 h of incubation in the presence of copper; and (iii) although the copper concentration that can be used during development is approximately 10-fold lower than that during growth, the copper dependency is similar during both stages of the life cycle. Other M. xanthus promoters that depend on copper either exhibit quite high expression levels in the absence of metal, do not reach a plateau at any time, or have diminished expression at 2 h after copper addition (5, 14, 15, 18).
Four different vectors have been designed, with two antibiotic resistance markers. Two encode resistance to kanamycin, while the other two encode resistance to tetracycline. One set of vectors integrates by homologous recombination at the native cuoA site, and two vectors have been designed to contain the Mx8 attP site for integration at the chromosomal attB site. The differences in gene expression observed when the same gene was cloned in vectors of the series pMAT or pNG could be explained by the location where they are integrated, i.e., the Mx8 attB site and the cuoA promoter, respectively. It is plausible that the control of gene expression achieved when the vector is integrated at the attB site will be different from that observed when the vector is integrated at the native site, because all the elements required for a tight regulation might be present only in the latter case. Since vectors integrated at the attB site yield higher levels of expression than those integrated at the native PcuoA site, these types of vectors can also be used to produce larger amounts of protein in the cells with lower copper concentrations.
According to all the data presented here, the vectors constructed in this study represent a clear improvement over all the other vectors that have previously been constructed to express genes in M. xanthus in a controlled manner. The fact that they are highly dependent on copper ensures no expression in the absence of metal, a clear advantage compared to vectors based on lacI (9). The vectors presented here exhibit several features that make them complementary to those based on vitamin B12. For instance, the amount of protein in the cells can be more easily controlled with copper than with B12 and light (3). As shown in Fig. 1, 2, and 3, many copper concentrations can be used to obtain high gene expression levels without interfering with growth, motility, or development. It is likely that it was due to these differences that the vector designed by García-Moreno et al. (3) was unsuitable for the construction of a conditional deletion mutation of the ftsZ gene. Experiments are in progress to test use of the vectors presented here to generate conditional mutations in genes such as ftsZ that have been unable to be deleted in M. xanthus using other expression systems.
ACKNOWLEDGMENTS
This work was supported by the Ministerio de Ciencia e Innovación (BFU2009-07565), program Consolider-Ingenio 2010 (CSD2009-00006), and the Max Planck Society. A.M.-M. and E.G.-B. are post- and predoctoral fellows from Plan Propio de la Universidad de Granada, respectively.
We thank S. Inouye for providing plasmids.
Footnotes
Published ahead of print 27 January 2012
REFERENCES
- 1. Cho K, Zusman DR. 1999. AsgD, a new two-component regulator required for A-signalling and nutrient sensing during early development of Myxococcus xanthus. Mol. Microbiol. 34:268–281 [DOI] [PubMed] [Google Scholar]
- 2. Crawford EW, Jr, Shimkets LJ. 2000. The stringent response in Myxococcus xanthus is regulated by SocE and the CsgA C-signaling protein. Genes Dev. 14:483–492 [PMC free article] [PubMed] [Google Scholar]
- 3. García-Moreno D, et al. 2009. A vitamin B12-based system for conditional expression reveals dksA to be an essential gene in Myxococcus xanthus. J. Bacteriol. 191:3108–3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Goldman BS, et al. 2006. Evolution of sensory complexity recorded in a myxobacterial genome. Proc. Natl. Acad. Sci. U. S. A. 103:15200–15205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gómez-Santos N, Pérez J, Sánchez-Sutil MC, Moraleda-Muñoz A, Muñoz-Dorado J. 2011. CorE from Myxococcus xanthus is a copper-dependent RNA polymerase sigma factor. PLoS Genet. 7:e1002106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hagen DC, Betscher AP, Kaiser D. 1978. Synergism between morphogenetic mutants of Myxococcus xanthus. Dev. Biol. 64:284–296 [DOI] [PubMed] [Google Scholar]
- 7. Hodgkin J, Kaiser D. 1979. Genetics of gliding motility in Myxococcus xanthus (Myxobacterales): genes controlling movement of single cells. Mol. Gen. Genet. 171:167–176 [Google Scholar]
- 8. Jakovljevic V, Leonardy S, Hoppert M, Søgaard-Andersen L. 2008. PilB and PilT are ATPases acting antagonistically in type IV pili function in Myxococcus xanthus. J. Bacteriol. 190:2411–2421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jelsbak L, Kaiser D. 2005. Regulating pilin expression reveals a threshold for S motility in Myxococcus xanthus. J. Bacteriol. 187:2105–2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kaiser D. 1979. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc. Natl. Acad. Sci. U. S. A. 76:5952–5956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Letouvet-Pawlak B, Monnier C, Barray S, Hodgson DA, Guespin-Michel JF. 1990. Comparison of β-galactosidase production by two inducible promoters in Myxococcus xanthus. Res. Microbiol. 141:425–435 [DOI] [PubMed] [Google Scholar]
- 12. Martínez-Cañamero M, Muñoz-Dorado J, Farez-Vidal E, Inouye M, Inouye S. 1993. Oar, a 115-kilodalton membrane protein required for development of Myxococcus xanthus. J. Bacteriol. 175:4756–4763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Moraleda-Muñoz A, Carrero-Lérida J, Extremera AL, Arias JM, Muñoz-Dorado J. 2001. Glycerol 3-phosphate inhibits swarming and aggregation of Myxococcus xanthus. J. Bacteriol. 183:6135–6139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Moraleda-Muñoz A, Pérez J, Extremera AL, Muñoz-Dorado J. 2010. Expression and physiological role of three Myxococcus xanthus copper-dependent P1B-type ATPases during bacterial growth and development. Appl. Environ. Microbiol. 76:6077–6084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Moraleda-Muñoz A, Pérez J, Extremera AL, Muñoz-Dorado J. 2010. Differential regulation of six heavy metal efflux systems in the response of Myxococcus xanthus to copper. Appl. Environ. Microbiol. 76:6069–6076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pérez J, et al. 2011. Myxococcus xanthus induces actinorhodin overproduction and aerial mycelium formation by Streptomyces coelicolor. Microb. Biotechnol. 4:175–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY [Google Scholar]
- 18. Sánchez-Sutil MC, et al. 2007. Differential expression of the three multicopper oxidases from Myxococcus xanthus. J. Bacteriol. 189:4887–4898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Singer M, Kaiser D. 1995. Ectopic production of guanosine penta- and tetraphosphate can initiate early developmental gene expression in Myxococcus xanthus. Genes Dev. 9:1633–1644 [DOI] [PubMed] [Google Scholar]
- 20. Wenzel SC, Müller R. 2009. The biosynthetic potential of myxobacteria and their impact in drug discovery. Curr. Opin. Drug Discov. Dev. 12:220–230 [PubMed] [Google Scholar]
- 21. Whitworth DE. (ed). 2008. Myxobacteria: multicellularity and differentiation. ASM Press, Washington, DC [Google Scholar]
- 22. Wu SS, Kaiser D. 1997. The Myxococcus xanthus pilT locus is required for social gliding motility although pili are still produced. Mol. Microbiol. 23:109–121 [DOI] [PubMed] [Google Scholar]
- 23. Yanisch-Perron C, Vieira J, Messing J. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103–119 [DOI] [PubMed] [Google Scholar]