Abstract
This study aimed to investigate the possible influence of bacterial intra- and interspecies interactions on the ability of Listeria monocytogenes and Salmonella enterica to develop mixed-culture biofilms on an abiotic substratum, as well as on the subsequent resistance of sessile cells to chemical disinfection. Initially, three strains from each species were selected and left to attach and form biofilms on stainless steel (SS) coupons incubated at 15°C for 144 h, in periodically renewable tryptone soy broth (TSB), under either monoculture or mixed-culture (mono-/dual-species) conditions. Following biofilm formation, mixed-culture sessile communities were subjected to 6-min disinfection treatments with (i) benzalkonium chloride (50 ppm), (ii) sodium hypochlorite (10 ppm), (iii) peracetic acid (10 ppm), and (iv) a mixture of hydrogen peroxide (5 ppm) and peracetic acid (5 ppm). Results revealed that both species reached similar biofilm counts (ca. 105 CFU cm−2) and that, in general, interspecies interactions did not have any significant effect either on the biofilm-forming ability (as this was assessed by agar plating enumeration of the mechanically detached biofilm bacteria) or on the antimicrobial resistance of each individual species. Interestingly, pulsed-field gel electrophoresis (PFGE) analysis clearly showed that the three L. monocytogenes strains did not contribute at the same level either to the formation of mixed-culture sessile communities (mono-/dual species) or to their antimicrobial recalcitrance. Additionally, the simultaneous existence inside the biofilm structure of S. enterica cells seemed to influence the occurrence and resistance pattern of L. monocytogenes strains. In sum, this study highlights the impact of microbial interactions taking place inside a mixed-culture sessile community on both its population dynamics and disinfection resistance.
INTRODUCTION
Biofilms are commonly defined as communities of microorganisms attached to a surface or interface and producing an extracellular matrix in which these microorganisms are embedded (13, 66). Biofilm formation is a natural phenomenon which happens almost everywhere microorganisms and surfaces exist in close proximity (28). In the food industry, pathogenic biofilms have been of considerable interest in the context of food safety and have provoked the interest of many research groups (60, 78). Obviously, the attachment of pathogenic bacteria to food contact surfaces and the subsequent biofilm formation are undesirable since the detachment of cells from the biofilm structure can lead to the cross-contamination of food products and cause food-borne diseases (4). The risk becomes even more serious since the resistance of biofilm cells to antimicrobials is significantly increased compared with what is normally seen when the same cells are planktonic (21, 42).
Listeria monocytogenes is a ubiquitous Gram-positive facultative intracellular bacterial pathogen which provokes listeriosis, a severe disease with high hospitalization and mortality rates, with the consumption of contaminated food being the principle mode of its transmission to humans (18, 35). Salmonellae represent a group of Gram-negative bacteria that are recognized worldwide as major zoonotic pathogens for both humans and animals (6). Notably, many L. monocytogenes and Salmonella enterica strains are capable of adhering to various surfaces, both biotic (e.g., plant and animal tissues) and abiotic (e.g., stainless steel and plastic), and create biofilms (for recent reviews see references 19, 56, and 64). Attachment to surfaces is believed to be important for survival and persistence of these pathogens in food processing environments, with some strains being able to remain on equipment surfaces even for several years (7, 41, 48, 73).
Apparently, biofilms can be formed by a single bacterial species. However, in the majority of natural and man-made environments, monospecies biofilms are relatively rare. Conversely, microorganisms are associated with surfaces in complex multispecies communities (74, 75). Unfortunately, this complexity is not taken into consideration when microorganisms are grown in monocultures under laboratory conditions. The structural and functional dynamics of multispecies biofilms are largely due to the interactions between the different species (31, 47). Thus, spatial and metabolic interspecies interactions contribute to the organization of multispecies biofilms and the production of a dynamic local environment (22, 69). All of these interactions may often change the physiology of individual biofilm species, as well as the functions of the whole community (76). Indeed, cell-to-cell interactions have been demonstrated to play a key role in cell attachment and detachment from biofilms, as well as in the resistance of biofilm community members against antimicrobial treatments (1, 5, 25, 40, 55, 62, 70, 71).
The remaining microflora on inadequately cleaned and disinfected food processing surfaces is usually a complex community, contrary to the laboratory-studied pure-species biofilms (23, 43). Evidently, bacterial pathogens, such as L. monocytogenes and S. enterica, can be entrapped in multispecies sessile communities formed on such surfaces. In addition, attachment and biofilm formation by these two species have been shown to be influenced by either the natural in situ presence of other species or just their metabolic by-products (8, 10, 11, 24, 26, 27, 29, 32, 33, 37, 38, 39, 46, 58, 77). For instance, the presence of Staphylococcus xylosus and Pseudomonas fragi affected the numbers of L. monocytogenes biofilm cells on stainless steel (51), while compounds present in Hafnia alvei cell-free culture supernatant inhibited the early stage of S. enterica serovar Enteritidis biofilm formation on the same material (10).
A clearer understanding of the physiological behavior of multispecies biofilm communities formed by bacterial pathogens on abiotic surfaces in food processing environments could provide the information necessary to prevent their formation and therefore reduce the contamination of food products. Particularly challenging is the attempt to understand the complexity of interactions encountered within such communities and how this may influence the final community outcome and behavior (i.e., population, structure, composition, physiology, function, and resistance). Although L. monocytogenes and S. enterica are among the leading causes of food-borne illnesses worldwide, to the best of our knowledge, nothing is yet known about the formation of a dual-species biofilm composed of these two species.
To this aim, in this study, some selected L. monocytogenes and S. enterica strains (three strains per species) were left to form biofilms on stainless steel (SS) surfaces, under either monoculture or mixed-culture (mono-/dual-species) conditions. Mixed-culture biofilms were then subjected to disinfection by the application of some common chemical antimicrobial compounds (benzalkonium chloride, sodium hypochlorite, peracetic acid [PA], or a mixture of hydrogen peroxide [HP]-PA) (44). Additionally, the composition of mixed-culture sessile communities, whether on not exposed to disinfection, with regard to L. monocytogenes strain occurrence, was evaluated by using a promising pulsed-field gel electrophoresis (PFGE) approach.
MATERIALS AND METHODS
Bacterial strains, media, and preparation of inocula.
Bacterial strains used in this study are listed in Table 1. They consist of 11 L. monocytogenes and 8 S. enterica strains isolated from different origins. Before utilization, all the microorganisms were stored frozen (at −80°C) in bead vials (Protect; Technical Service Consultants, Ltd., Heywood, Lancashire, United Kingdom), and each one was then resuscitated by the addition of one bead to 100 ml of brain heart infusion (BHI) broth (Lab M International Diagnostics Group, Bury, Lancashire, United Kingdom) in a conical flask and incubation at 37°C for 24 h (precultures). Working cultures were prepared by the addition of a 100-μl aliquot of each preculture to 100 ml of BHI broth and incubation at 37°C for 16 h, at which time late exponential phase was attained for each strain. Cells from final working cultures were harvested by centrifugation (5,000 × g for 10 min at 4°C), washed twice with quarter-strength Ringer solution (Ringer's tablets; Merck, Darmstadt, Germany), and finally resuspended in quarter-strength Ringer solution (ca. 109 CFU ml−1), in order to be used as inoculum for the biofilm development assays. All strains had reached the stationary phase when harvested for the subsequent experiments (data not shown).
Table 1.
The 19 bacterial strains used in this study
| Straina | Isolate origin(s) | Description or other strain identification(s) | Reference and/or sourceb |
|---|---|---|---|
| L. monocytogenes strains | |||
| FMCC_B-160 | Environment, food processing plant | Strain 19UD | 12; DSA |
| FMCC_B-125 | Human | Strain ScottA, serotype 4b | 59; ATO-DLO |
| FMCC_B-126 | Unknown | This study | |
| FMCC_B-130 | Food (dairy) | This study | |
| FMCC_B-165 | Environment, food processing plant | Strain 7UD | 12; DSA |
| FMCC_B-129 | Food (meat) | This study | |
| FMCC_B-164 | Environment, food processing plant | Strain 8UD | 12; DSA |
| FMCC_B-124 | Unknown | Strain NCTC10527, serotype 4b | NCTC |
| FMCC_B-169 | Environment, food processing plant | Strain 2UD | 12; DSA |
| FMCC_B-166 | Environment, food processing plant | Strain 6UD | 12; DSA |
| FMCC_B-127 | Food (chicken salad) | This study | |
| S. enterica strains | |||
| FMCC_B-17 | Unknown | This study | |
| FMCC_B-19 | Unknown | This study | |
| FMCC_B-42 | Food | Serovar Enteritidis | This study |
| FMCC_B-56 | Food (eggs) | Serovar Enteritidis, PT4, strain P167807 | Division of Enteric Pathogens, Central Public Health Laboratory, London, UK |
| FMCC_B-62 | Unknown | Serovar Typhimurium, strain DSM554 | DSMZ |
| FMCC_B-67 | Unknown | This study | |
| FMCC_B-137 | Human, salmonellosis outbreak | Serovar Typhimurium, strain DT193, multidrug resistant | 16 |
| FMCC_B-194 | Unknown | Serovar Typhimurium, strain JH3016 rpsM::gfp+ | 30 |
The six strains in boldface were the ones selected for biofilm formation on SS coupons and disinfection experiments. FMCC, Food Microbiology Culture Collection of the Laboratory of Microbiology and Biotechnology of Foods, Agricultural University of Athens, Greece.
DSA, collection of the Dipartimento di Scienze Degli Alimenti, Università Degli Studi di Udine, Italy; ATO-DLO, Agrotechnological Research Institute, Wageningen, Netherlands; NCTC, National Collection of Type Cultures; DSMZ, Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (German Collection of Microorganisms and Cell Cultures).
Biofilm formation on polystyrene microplates.
The ability of 19 bacterial strains to form biofilms on polystyrene (PS) microtiter plates under monoculture conditions was evaluated by using the method described by van Merode et al. (72), with some adaptations. Analytically, 200-μl of each bacterial suspension in quarter-strength Ringer solution, containing ca. 108 CFU ml−1, was transferred into a well of a sterile 96-well PS microtiter plate (655161; Greiner Bio-One/BioLine Scientific, Athens, Greece), and the plate was then incubated for 3 h at either 15 or 30°C, under static conditions, in order for the bacteria to attach on the parietes of the wells. Following the attachment step, the planktonic bacteria were removed (by using a multichannel pipette), and each well was then washed twice with quarter-strength Ringer solution to remove the loosely attached cells. Subsequently, 200-μl of growth medium (100% tryptone soy broth [TSB] or 10% TSB [dTSB]; Lab M) was added into each well, and the plate was then incubated for 48 h at either 15 or 30°C under static conditions to allow the attached bacteria to grow and create biofilms. Growth medium was renewed at 24 h of incubation. Before the addition of fresh growth medium, each well was washed with quarter-strength Ringer solution. At the end of incubation, planktonic bacteria were removed by violently turning over the plate, and each well was then washed twice with quarter-strength Ringer solution to remove the loosely attached cells. Subsequently, 200 μl of 1% (wt/vol) crystal violet solution (Sigma-Aldrich/Life Science Chemilab S.A., Athens, Greece) was added into each well, and the plate was then incubated for 30 min at room temperature (RT). After being washed three times with 200 μl of deionized water to remove excess stain, the crystal violet was solubilized in 200 μl of an ethanol-acetone mixture (80:20, vol/vol). Dye absorbance at 575 nm (A575) was measured using a microtiter plate reader (Sunrise; Tecan, Männedorf, Switzerland).
Each experiment (strain) included four replicate wells and was repeated twice using independent bacterial cultures. As a control, wells initially filled with sterile quarter-strength Ringer solution and subsequently submitted to exactly the same procedure were used. In order to calculate the net amount of biofilm formed by each strain, the absorbance value of the control was always subtracted.
Biofilm formation on SS coupons.
Stainless steel (SS) coupons (3 by 1 by 0.1 cm, type AISI-304; Halyvourgiki, Inc., Athens, Greece) were initially soaked in acetone (overnight) to remove any manufacturing process debris and grease. Coupons were then washed by soaking for 30 min at 50°C in a 2% (vol/vol) solution of the commercial detergent RBS 35 (Fluka/Life Science Chemilab, S.A.) with shaking, rinsed thoroughly with tap water followed by distilled water, and air dried. Finally, cleaned coupons were individually placed in empty glass test tubes (length, 10 cm; diameter, 1.5 cm) and autoclaved at 121°C for 15 min.
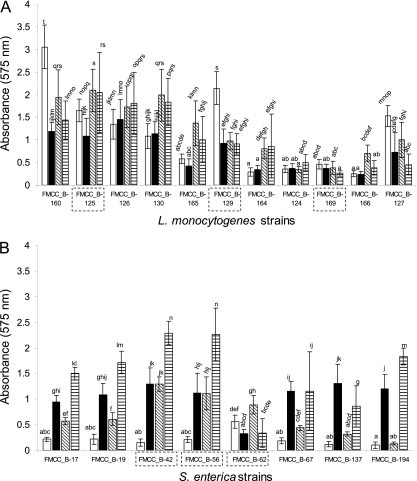
To produce biofilms on SS coupons, six strains were selected (three strains per species) and were left to produce biofilms under either monoculture or mixed-culture conditions. In the latter case, both mono- and dual-species conditions were included. Care was taken to select strains presenting different biofilm formation profiles (based on the results of biofilm formation on PS microplates) (Fig. 1) and also having different isolation origins (i.e., clinical, food, or environment) in an attempt to pursue variability. For biofilm formation, two subsequent steps were followed: sterile coupons were initially incubated in saline bacterial suspensions (ca. 108 CFU ml−1) (bacterial attachment step), and afterwards coupons carrying strongly attached bacteria were incubated in periodically renewable growth medium (biofilm formation step).
Fig 1.
Biofilm formation (A575) on PS microplates by the 11 L. monocytogenes strains (A) and the 8 S. enterica strains (B). Biofilm cells were indirectly quantified by crystal violet staining and absorbance measurements at 575 nm. Monoculture biofilms were left to be formed on PS microplates for 48 h under four different environmental conditions: at 15°C in TSB (□), at 15°C in dTSB (■), at 30°C in TSB (▧), and at 30°C in dTSB (▤). In all cases, growth medium was renewed at 24 h of incubation. The bars represent the mean values ± standard deviations (n = 8, two independent experiments, each performed four times). For each figure separately, mean values sharing at least one common lowercase letter shown above the bars are not significantly different at a P value of <0.05. Strains selected for biofilm formation on SS coupons are included in dashed rectangular boxes.
For the attachment step, 5 ml of each bacterial suspension in quarter-strength Ringer solution, containing ca. 108 CFU ml−1, was poured into each glass test tube containing a sterilized SS coupon and incubated at 15°C for 3 h under static conditions. Three different attachment conditions were studied: (i) monoculture using a bacterial suspension containing cells of only one strain; (ii) monospecies mixed culture using a bacterial suspension containing cells of three strains of the same species; and (ii) dual-species mixed culture using a bacterial suspension containing cells of all six strains of the two different species tested. In the last two cases, care was taken in order to ensure that the bacterial suspension contained approximately the same number of cells for each strain (ca. 108 CFU ml−1).
Following the attachment step, each coupon was carefully removed from the glass test tube using sterile forceps and was thereafter rinsed by immersing for 5 min in 5 ml of quarter-strength Ringer solution, with shaking, in order to remove the loosely attached cells. After the rinsing step, each coupon was individually introduced into a new sterile glass test tube containing 5 ml of TSB and subsequently incubated at 15°C for 6 days (144 h), under static conditions, to allow biofilm development on the coupon. Growth medium was renewed every 48 h. During medium renewal, loosely attached cells were removed by rinsing (as described above).
Each experiment included three replicate coupons and was repeated twice using independent bacterial cultures.
Disinfection of mixed-culture biofilms formed on SS coupons.
On the 6th day of incubation, each SS coupon carrying biofilm cells on it was carefully removed from the glass test tube using sterile forceps and was thereafter rinsed by pipetting two times with 10 ml of quarter-strength Ringer solution (each time) in order to remove the loosely attached cells. After being rinsed, the coupon was individually introduced into a new glass test tube containing 5 ml of disinfectant solution. Four different disinfectant solutions commonly used in the food industry were studied: (i) benzalkonium chloride (BC) at 50 ppm, (ii) sodium hypochlorite (NaClO) at 10 ppm, (iii) peracetic acid (PA) at 10 ppm, and (iv) a mixture of hydrogen peroxide and PA (HP-PA), each at 5 ppm. All chemical disinfectants were purchased from Sigma-Aldrich/Life Science Chemilab S.A., and their desired solutions were prepared in distilled water. Disinfection was carried out at 15°C for 6 min to imitate disinfection conditions encountered in real food processing areas.
Each experiment included three replicate coupons and was repeated twice using independent bacterial cultures.
Quantification of biofilm bacterial populations on SS coupons.
The enumeration of viable biofilm bacteria on SS coupons was performed after 6 days of incubation in the TSB and after the disinfection treatment (for 6 min), which was performed by using the bead vortexing method described by Giaouris and Nychas (20), with some minor adaptations. Briefly, each coupon was aseptically removed from the glass test tube and was then rinsed by pipetting twice with 10 ml of quarter-strength Ringer solution (each time). Between these two rinse steps, the coupon was immersed for 5 min in 5 ml of quarter-strength Ringer solution, with shaking, in order to better remove the loosely attached cells. After the second rinse, the coupon was transferred to a plastic centrifuge tube containing 6 ml of quarter-strength Ringer solution and 10 sterile glass beads (diameter, 3 mm) and left there for 10 min (at RT). In the case of enumeration of viable biofilm cells following disinfection, instead of quarter-strength Ringer solution, Dey-Engley neutralizing broth (BioChemica/Life Science Chemilab S.A.) was used. The plastic tube was then vortexed for 2 min at maximum speed to detach biofilm cells from the coupon. Detached cells were subsequently enumerated by agar plating after 10-fold serial dilutions. In the case of monospecies biofilm development, tryptone soy agar (TSA; Lab M) was used for the enumeration of bacteria. In the case of dual-species biofilm development, the cells of each species were enumerated using the following selective media: xylose lysine deoxycholate agar (XLD; Merck) for S. enterica and PALCAM Listeria Selective Agar (Merck) for L. monocytogenes. Plates were incubated at 37°C for 24 to 48 h.
Isolation of L. monocytogenes strains from mixed-culture biofilm communities formed on SS coupons.
In order to monitor the individual contribution of each L. monocytogenes strain in both the development and resistance of mixed-culture (mono- and dual species) biofilm communities, 20 colonies were randomly selected from the highest dilutions of PALCAM Listeria Selective agars (used to enumerate the viable bacteria recovered from SS coupons by the bead vortexing method). Out of a total number of 200 colonies picked, half were recovered from the monospecies biofilms (containing only L. monocytogenes strains), while the other half were recovered from the dual-species biofilms (containing both L. monocytogenes and S. enterica strains). In particular, from each of the two biofilm-forming conditions (mono- and dual species), 20 colonies were recovered from the control treatment (i.e., without disinfection), and 20 colonies were recovered from each of the four disinfection treatments (BC, NaClO, PA, and HP-PA). All isolated colonies were stored at −80°C in TSB containing 25% (vol/vol) glycerol (Serva GmbH, Heidelberg, Germany).
PFGE analysis.
Pulsed-field gel electrophoresis (PFGE) was performed according to Doulgeraki et al. (15). All reagents were purchased from Sigma-Aldrich/Life Science Chemilab S.A.), unless otherwise stated. Briefly, each L. monocytogenes isolate recovered from biofilm communities was subcultured twice in 1.5 ml of TSB (Lab M) at 37°C for 24 h and 16 h. Cells were harvested by centrifugation (5,000 x g for 10 min at 4°C), washed with 10 mM Tris-HCl (pH 7.6) containing 1 M NaCl, and resuspended in 100 μl of the same solution. Bacterial suspension was then heated at 37°C for 10 min, mixed with an equal volume of 2% (wt/vol) low-melting-point agarose (Bio-Rad Laboratories Inc., Athens, Greece) in 0.125 M EDTA (pH 7.6), pipetted into plug molds (Bio-Rad), and left there to solidify at 4°C. Bacterial cells in the solidified plugs were then lysed in situ in a solution containing 10 mg ml−1 lysozyme in 1 ml of EC buffer (6 mM Tris-HCl, 1 M NaCl, 100 mM EDTA, 1% [wt/vol] Sarkosyl, pH 7.6) for 16 h at 37°C. The lytic treatment was repeated with the same solution containing 2 U ml−1 mutanolysin. After treatment with 0.5 mg ml−1 proteinase K in 1 ml of 0.5 M EDTA containing 1% (wt/vol) Sarkosyl (pH 8) for 24 h at 55°C, the agarose plugs were washed twice for 1 h with 1 mM phenylmethylsulfonyl fluoride (PMSF) in 10 mM Tris-HCl containing 1 mM EDTA (pH 8.0) at 37°C and then stored at 4°C in 10 mM Tris-HCl containing 100 mM EDTA (pH 8.0) until further use.
The agarose plugs were cut with sterile coverslips, and slices (1 to 2 mm thick) of the plugs were washed three times at RT with 10 mM Tris-HCl containing 0.1 mM EDTA (pH 8.0) for 30 min with gentle agitation. Twenty units of the restriction enzyme ApaI (New England BioLabs/Bioline Scientific, Athens, Greece) was used for the restriction reaction, which was performed according to the manufacturer's recommendations. The digestion was stopped by adding 0.5 ml of 0.5 M EDTA (pH 8.0), and plugs were stored at 4°C until analysis.
Following digestion, slices were loaded into wells of a 1% PFGE-grade agarose gel (Bio-Rad), and the gel was run in 0.5× Tris-Borate buffer (45 mM Tris-HCl, 45 mM boric acid, 1 mM EDTA) using a CHEF (contour-clamped homogeneous electric field) DRII PFGE apparatus and cooling module (Bio-Rad) at 6 V/cm for 16 h, with a pulse time ramped from 1 to 10 s. Gels were then stained with ethidium bromide (0.5 μg ml−1) in water for 1 h and destained for 2 h before being photographed using a GelDoc system (Bio-Rad). Conversion, normalization, and further analysis were performed using the Pearson coefficient and unweighted pair group method using averages (UPGMA) cluster analysis with GelCompar software, version 4.0 (Applied Maths BVBA, Saint-Martens-Latem, Belgium; kindly provided by E. Tsakalidou, Dairy Laboratory, Agricultural University of Athens).
Statistical analysis.
All data were analyzed by analysis of variance (ANOVA) by the general linear model procedure of SPSS data analysis software for Windows (release 10.0.1; SPSS Inc., Chicago, IL). Least square means were separated by Fisher's least significant difference (LSD) test. All differences are reported at a significance level of an alpha of 0.05.
RESULTS
Monoculture biofilm formation on PS microplates under two temperatures and growth medium conditions.
The results of monoculture biofilm formation on PS microplates at two different temperatures (15, 30°C) and in two growth media (TSB, dTSB) are illustrated in Fig. 1A for the 11 L. monocytogenes strains and in Fig. 1B for the 8 S. enterica strains. It is apparent that, for both species, the influences of temperature and/or growth medium conditions on biofilm formation are strongly dependent on the strain studied. For instance, the L. monocytogenes FMCC_B-169 strain formed a low quantity of biofilm, independently of conditions (A577 measurements ranging from 0.26 to 0.46), while the FMCC_B-125 strain formed more biofilm at 30°C, compared to 15°C for both growth medium conditions (Fig. 1A). However, at 15°C, the latter strain formed more biofilm when cultivated in TSB than in dTSB. S. enterica strains formed, in general, more biofilm under low-nutrient conditions (dTSB) than under adequate-nutrient conditions (TSB) for both temperatures studied (Fig. 1B). However, the FMCC_B-62 strain seems to form more biofilm when cultivated under adequate nutrient conditions (TSB) than under low-nutrient conditions (dTSB) (Fig. 1B). In agreement with current results, a previous study of 122 Salmonella strains indicated that all had the ability to adhere to plastic microwell plates and that, generally, more biofilm was produced under low-nutrient conditions than under high-nutrient conditions (65). It is worth noting that at 15°C in TSB (conditions used for the study of biofilm formation on SS coupons), almost all the S. enterica strains form low quantities of biofilm on PS microplates (Fig. 1B).
Monoculture and mixed-culture (mono-/dual-species) biofilm formation on SS coupons.
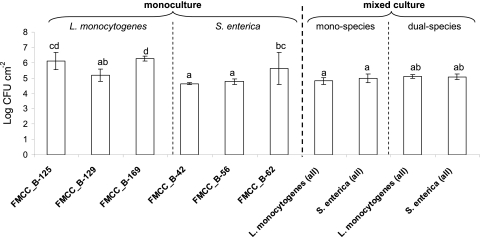
The results of biofilm formation on SS coupons by the three L. monocytogenes (FMCC_B-125, FMCC_B-129, and FMCC_B-169) and the three S. enterica (FMCC_B-42, FMCC_B-56, and FMCC_B-62) strains, cultured under either monoculture or mixed-culture conditions are presented in Fig. 2. In the latter case, biofilms were left to be formed either by the three strains belonging to the same species (monospecies) or by the six strains belonging to the two different species (dual-species; three strains per species). Regarding monoculture conditions, L. monocytogenes strains FMCC_B-125 and FMCC_B-169 reached a sessile population of 6.11 and 6.27 log CFU cm−2, respectively, while the biofilm population of strain FMCC_B-129 was approximately 10 times less (5.19 log CFU cm−2). S. enterica, strains FMCC_B-42 and FMCC_B-56 reached similar biofilm population levels (4.64 and 4.77 log CFU cm−2, respectively), while the biofilm counts of strain FMCC_B-62 were approximately 10 times more (5.63 log CFU cm−2). Regarding monospecies mixed-culture conditions, biofilm counts did not significantly differ between L. monocytogenes and S. enterica (4.82 and 4.98 log CFU cm−2, respectively). Likewise, in the case of dual-species mixed-culture conditions, both species presented similar biofilm populations (5.12 and 5.08 log CFU cm−2 for L. monocytogenes and S. enterica, respectively). Thus, the 51.7% of cells in the dual-species biofilm community belonged to L. monocytogenes strains, while the remaining 48.3% of cells belonged to S. enterica strains (see Fig. 4).
Fig 2.
Biofilm formation (log CFU cm−2) on SS coupons by L. monocytogenes (FMCC_B-125, FMCC_B-129, and FMCC_B-169) and S. enterica (FMCC_B-42, FMCC_B-56, and FMCC_B-62) strains. Biofilm cells were quantified by enumeration following detachment and agar plating. Biofilms were left to be formed on SS coupons incubated for 144 h in TSB at 15°C (with medium renewal every 48 h), under three different conditions: (i) monoculture (each strain individually), (ii) monospecies mixed culture (three strains of each one species together), and (ii) dual-species mixed culture (six strains of the two different species together). The bars represent the mean values ± standard deviations (n = 6, two independent experiments, each performed three times). Mean values sharing at least one common lowercase letter shown above the bars are not significantly different at a P value of <0.05.
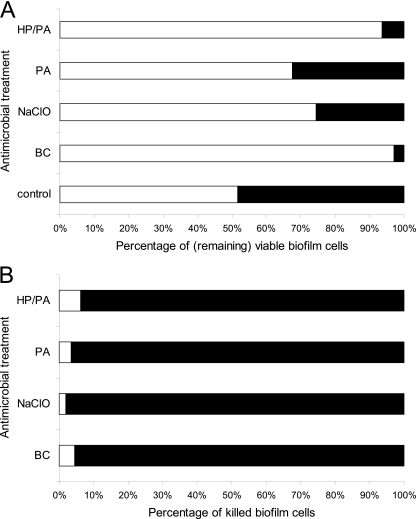
Fig 4.
Percentages of L. monocytogenes (open bars) and S. enterica (filled bars) biofilm cells in the dual-species sessile community remaining viable (A) and killed (B) following a 6-min exposure to the following disinfectants: BC at 50 ppm, NaClO at 10 ppm, PA at 10 ppm, and HP-PA at 5 ppm each. Control treatment is without disinfection. Dual-species biofilms were initially left to be formed on SS coupons incubated for 144 h in TSB at 15°C (with medium renewal every 48 h).
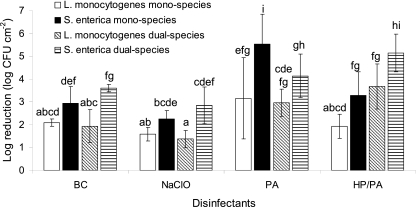
Chemical disinfection of mixed-culture biofilm communities formed on SS coupons.
The effect of each 6-min disinfection treatment (BC, NaClO, PA, and HP-PA) against L. monocytogenes and S. enterica biofilm cells, cultivated under either mono- or dual-species mixed-culture conditions on SS coupons, was expressed as biofilm population log reduction (difference in log CFU cm−2 values just before and after the treatment) (Fig. 3). In general, L. monocytogenes seems to be more resistant than S. enterica to the antimicrobial action of all tested disinfectants, independently of growth conditions (mono-/dual species). Results presented in Fig. 4 (i.e., percentages of remaining viable and killed biofilm cells) also support this. Thus, although both species reached, before disinfection, similar populations in the dual-species biofilm community (5.12 and 5.08 log CFU cm−2 or 51.7% and 48.3%, for L. monocytogenes and S. enterica, respectively), following disinfection, the dual-species community was composed mainly of L. monocytogenes (Fig. 4A). Thereby, the percentage of viable L. monocytogenes cells in the dual-species community ranged from 67.6% following disinfection with PA to 97.1% following disinfection with BC (Fig. 4A). Interestingly, all four disinfectants mainly killed S. enterica cells (Fig. 4B). Thus, the percentages of killed S. enterica biofilm cells out of the total number of killed cells ranged from 93.8% following disinfection with HP-PA to 98.1% following disinfection with NaClO.
Fig 3.
Log reductions (log CFU cm−2) of L. monocytogenes and S. enterica biofilm cells following a 6-min exposure to disinfectants (BC, 50 ppm; NaClO, 10 ppm; PA, 10 ppm; HP-PA, 5 ppm each). Mixed culture biofilms were initially left to be formed on SS coupons incubated for 144 h in TSB at 15°C (with medium renewal every 48 h), either under monospecies or dual-species conditions (three strains per species). The bars represent the mean values ± standard deviations (n = 6, two independent experiments, each performed three times). Mean values sharing at least one common lower-case letter shown above the bars are not significantly different at a P value of < 0.05.
In general, interspecies interactions did not have any significant effect on the antimicrobial resistance of each individual species (Fig. 3). When a small effect was observed (between mono- and dual-species conditions), this was found to depend both on the species employed and the disinfectant applied (Fig. 3). For instance, the antimicrobial actions of either BC or NaClO were similar between mono- and dual-species conditions for both species tested. In contrast, in the case of disinfection with either PA or the mixture HP-PA, interspecies interactions were found to influence antimicrobial resistance. Thus, both species were found to be less resistant to HP-PA when cultured together than under monospecies conditions, whereas S. enterica seems to present higher resistance to PA action under dual-species conditions than under monospecies conditions.
Distribution of L. monocytogenes strains in the mixed culture biofilm communities.
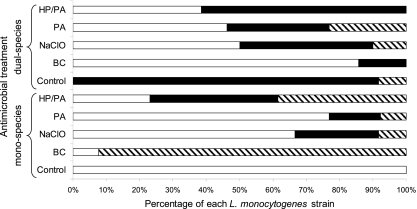
The individual contribution of each L. monocytogenes strain (FMCC_B-125, FMCC_B-129, and FMCC_B-169) in the composition of mono- and dual-species mixed-culture biofilm communities formed on SS coupons, whether or not exposed to disinfection, is illustrated in Fig. 5. Interestingly, PFGE analysis clearly revealed that the three L. monocytogenes strains did not contribute at the same levels to either the formation of the mixed-culture sessile communities or their antimicrobial resistance.
Fig 5.
Distribution of L. monocytogenes strains (FMCC_B-129, □; FMCC_B-169, ■; and FMCC_B-125, ▧) in mono- and dual-species mixed-culture biofilm communities remaining viable following a 6-min exposure to disinfectants (BC, 50 ppm; NaClO, 10 ppm; PA, 10 ppm; HP-PA, 5 ppm each). Control treatment is without disinfection. Mixed-culture biofilms were initially left to be formed on SS coupons incubated for 144 h in TSB at 15°C (with medium renewal every 48 h).
DISCUSSION
It is generally acknowledged that biofilms are the dominant lifestyle of bacteria in all environments, either natural or man-made. This sessile form of cell growth consists of slime matrix-embedded bacteria of either a single, but mostly of multiple, microbial species that form an interdependent structured community, capable of coordinated and collective behavior (49). Although microbial interactions among bacteria have been studied primarily in planktonic culture systems (68), these are more likely to occur in multispecies biofilms in which genetically distinct bacteria may become attached to one another via specific molecules (47, 57, 76). Bacterial interactions may be accomplished through extracellular compounds whose sole role is to influence gene expression, metabolic cooperativity and competition, physical contact, and the production of antimicrobial exoproducts (14). One or all of these may be occurring simultaneously and begin to influence a biofilm during the initial stages of formation, bacterial attachment, and surface colonization and continue to influence the structure and physiology of the biofilm as it develops (31).
However, until now, most of the mechanisms regarding biofilm formation have been revealed by means of studying monospecies biofilms. In this study, the simultaneous biofilm formation by six selected L. monocytogenes and S. enterica strains was investigated. Obtained results (Fig. 2) revealed that both species displayed similar biofilm counts (ca. 105 CFU cm−2), independently of culture conditions (i.e., mono-/dual species). Although numerous previous studies have already shown that both L. monocytogenes and S. enterica are able to form mixed-species biofilms with other species, the absence of any other published work on dual-species biofilms solely composed of these two species renders impossible any direct comparison. Leriche and Carpentier (38) studied the adhesion and subsequent development of L. monocytogenes on SS surfaces in both the absence and presence of a Staphylococcus sciuri biofilm and interestingly found that after 3 days of culture, Staphylococcus biofilms prevented the adherent L. monocytogenes population from increasing within the biofilm, leading to an average logarithmic CFU difference of 0.9 to 2.7 between the pure and mixed culture. Similarly, the single-species biofilm of L. monocytogenes showed higher cell numbers than the mixed-species biofilm with Pseudomonas fragi (51). In contrast, in another mixed-species biofilm of L. monocytogenes and Flavobacterium spp., the number of L. monocytogenes cells increased compared with a single-species biofilm, and, more importantly, L. monocytogenes was also recoverable for longer incubation periods (3).
A broad study of the effect of 29 bacterial strains isolated from the food processing environment on L. monocytogenes biofilm formation on SS showed that coculture with four strains resulted in increased biofilm formation, while the other strains showed no effect or decreased biofilm formation (8). Likewise, Zhao et al. (77) screened 413 microbial isolates from drains in food processing facilities for antilisterial properties and found two isolates (Enterococcus durans and Lactococcus lactis) that were highly inhibitory to L. monocytogenes when cocultivated in binary biofilms. Mixed-species biofilm formation experiments of L. monocytogenes EGD-e with seven different Staphylococcus aureus strains showed that, except for one S. aureus strain, the number of L. monocytogenes cells in the mixed-species biofilm was similar to the number of L. monocytogenes cells in the single-species biofilm (58). Chorianopoulos et al. (11) cocultured, on SS surfaces, five strains belonging to S. enterica, L. monocytogenes, Pseudomonas putida, Staphylococcus simulans, and Lactobacillus fermentum. Interestingly, in that study, mixed-culture biofilm was composed mainly of P. putida cells (97.8%), while S. enterica and L. monocytogenes represented together only the 2.2% of the population.
So far, limited studies have been conducted on the resistance of mixed-species biofilms to disinfectants (2, 11, 17, 50). However, most of the relevant studies performed did not include results of single-species biofilm resistance, making it impossible to judge whether there is any effect of interspecies interactions on the resistance of each individual species in the mixed community. In a recent study, Van der Veen and Abee (71) investigated the resistance of single- and mixed-species biofilms of L. monocytogenes and Lactobacillus plantarum, formed on PS microtiter plates, against benzalkonium chloride and peracetic acid. These colleagues showed that L. monocytogenes and L. plantarum grown in mixed-species biofilms were, under most conditions, more resistant to the disinfection treatments than single-species biofilms. In contrast, in current study, interspecies interactions did not seem to have any significant effect on the antimicrobial resistance of biofilm cells of each individual species (except for the case of HP-PA disinfection) (Fig. 3). Obviously, all of these contradictory results highlight, if anything, the impact of variability of experimental setup (i.e., disinfectant, species, abiotic substratum, and environmental growth conditions) on the results obtained. Consequently, special care should be taken in making conclusions based on the results of a single study since biofilms seem to be very diverse and unique, not just to the microorganism, but to the particular environment in which they are being formed.
In current study, L. monocytogenes was found to present higher resistance than S. enterica to the antimicrobial action of all tested chemical disinfectants, independently of growth conditions (mono-/dual species) (Fig. 3 and 4). This was something quite expected, given the well-known greater capability of L. monocytogenes to adapt to survive and grow under a wider range of environmental conditions than S. enterica (18). In order to discriminate between the three different L. monocytogenes strains with regard to their ability to contribute to the formation and maintenance of the mixed culture biofilm communities (mono-/dual species) as well as to their antimicrobial recalcitrance, a promising PFGE approach, already used by our team (15, 34), was followed. In this study, PFGE analysis revealed interesting information regarding strain occurrence following the different treatments (control, BC, NaClO, PA, and HP-PA) (Fig. 5). It is thus evident that although the initial inoculum used for biofilm formation contained the same number of cells for each strain (ca. 108 CFU ml−1), mature mixed-culture biofilms did not contain equal amounts of cells for each strain.
The question remains why some L. monocytogenes strains were found to dominate in each mixed-culture biofilm community. Obviously, results based on monoculture biofilm formation (Fig. 2) cannot provide any sufficient explanation. The higher initial attachment ability, the higher specific growth rate, and/or the better entrapment ability in the developing biofilm structure (and as thus released detachment) of some strains than other strains also present are quite probably accounting for this. A future study able to address some or all of these intriguing questions would therefore be of great interest. Additionally, PFGE results revealed that the three L. monocytogenes strains enclosed in the mixed-culture biofilm communities do not present the same resistance to the disinfectants tested, while the simultaneous existence of S. enterica cells seems to influence the occurrence and resistance pattern of L. monocytogenes strains (Fig. 5).
Undoubtedly, microbial interactions among the different strains and species have a significant influence on the growth potential, survival, and more generally on the individual behavior of each strain (31). One of the most common interactions in multispecies biofilms is competition (24, 53, 61, 67). Microorganisms compete for nutrients, oxygen, and available space to colonize and subsequently try to inhibit the growth of other species/strains in biofilms. Toxic substances are secreted by many microbial species to kill or inhibit the growth of other species. In addition to competitive interactions, cooperative interactions also widely exist in multispecies biofilms and are essential for the overall biofilm fitness (5, 9, 37, 54). A mutualism type of synergy in biofilm formation between S. enterica serovar Enteritidis and Klebsiella pneumoniae has also been reported (33). Additionally, bacterial cells are able to communicate by releasing and sensing small diffusible signal molecules (autoinducers [AI]) in a process commonly known as quorum sensing (QS) (45, 63). As biofilms typically contain high concentrations of cells, AI activity and QS regulation of gene expression are considered essential components of biofilm physiology (36, 52).
In summary, microbial intra- and interspecies interactions encountered inside mono- and dual-species biofilms of L. monocytogenes and S. enterica were found to have a profound effect on both the population dynamics and the resistance pattern of each L. monocytogenes strain present. However, interspecies interactions did not significantly influence either the biofilm-forming ability or the antimicrobial resistance of each individual species. Following disinfection of dual-species sessile community with four common chemical antimicrobial compounds, the vast majority of cells killed belonged to S. enterica, and as a consequence the remaining viable community was mainly composed of L. monocytogenes cells.
However, in real food processing environments, the presence of many other microbial species clearly adds additional complexity to the behavior of multispecies biofilms since all incorporated microorganisms are able to compete, cooperate, and communicate with each other. Undoubtedly, further research is needed using a larger composite of strains and species to simulate better the conditions encountered in real, complex food processing ecosystems. The improved understanding of the physiology of multispecies biofilms formed by food-related bacteria could probably facilitate the development of methods for controlling bacterial biofilms in food areas.
ACKNOWLEDGMENTS
This work was partly financially supported by the European Union project ProSafeBeef (Food-CT-2006-36241) within the 6th Framework Programme.
We are grateful to Agapi Doulgeraki for her valuable assistance in PFGE analysis.
Footnotes
Published ahead of print 3 February 2012
REFERENCES
- 1. Annous BA, Fratamico PM, Smith JL. 2009. Scientific status summary. J. Food Sci. 74:24–37 [DOI] [PubMed] [Google Scholar]
- 2. Bremer PJ, Monk I, Butler R. 2002. Inactivation of Listeria monocytogenes/Flavobacterium spp. biofilms using chlorine: impact of substrate, pH, time and concentration. Lett. Appl. Microbiol. 35:321–325 [DOI] [PubMed] [Google Scholar]
- 3. Bremer PJ, Monk I, Osborne CM. 2001. Survival of Listeria monocytogenes attached to stainless steel surfaces in the presence or absence of Flavobacterium spp. J. Food Prot. 64:1369–1376 [DOI] [PubMed] [Google Scholar]
- 4. Brooks JD, Flint SH. 2008. Biofilms in the food industry: problems and potential solutions. Int. J. Food Sci. Technol. 43:2163–2176 [Google Scholar]
- 5. Burmølle M, et al. 2006. Enhanced biofilm formation and increased resistance to antimicrobial agents and bacterial invasion are caused by synergistic interactions in multispecies biofilms. Appl. Environ. Microbiol. 72:3916–3923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Callaway TR, Edrington TS, Anderson RC, Byrd JA, Nisbet DJ. 2008. Gastrointestinal microbial ecology and the safety of our food supply as related to Salmonella. J. Anim. Sci. 86:163–172 [DOI] [PubMed] [Google Scholar]
- 7. Carpentier B, Cerf O. 2011. Review—persistence of Listeria monocytogenes in food industry equipment and premises. Int. J. Food Microbiol. 145:1–8 [DOI] [PubMed] [Google Scholar]
- 8. Carpentier B, Chassaing D. 2004. Interactions in biofilms between Listeria monocytogenes and resident microorganisms from food industry premises. Int. J. Food Microbiol. 97:111–122 [DOI] [PubMed] [Google Scholar]
- 9. Castonguay M-H, et al. 2006. Biofilm formation by Escherichia coli is stimulated by synergistic interactions and co-adhesion mechanisms with adherence-proficient bacteria. Res. Microbiol. 157:471–478 [DOI] [PubMed] [Google Scholar]
- 10. Chorianopoulos NG, Giaouris ED, Kourkoutas Y, Nychas G-JE. 2010. Inhibition of the early stage of Salmonella enterica serovar Enteritidis biofilm development on stainless steel by cell-free supernatant of a Hafnia alvei culture. Appl. Environ. Microbiol. 76:2018–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chorianopoulos NG, Giaouris ED, Skandamis PN, Haroutounian SA, Nychas G-JE. 2008. Disinfectant test against monoculture and mixed-culture biofilms composed of technological, spoilage and pathogenic bacteria: bactericidal effect of essential oil and hydrosol of Satureja thymbra and comparison with standard acid-base sanitizers. J. Appl. Microbiol. 104:1586–1596 [DOI] [PubMed] [Google Scholar]
- 12. Cocolin L, et al. 2005. Analysis of PCR-based methods for characterization of Listeria monocytogenes strains isolated from different sources. Int. J. Food Microbiol. 103:167–178 [DOI] [PubMed] [Google Scholar]
- 13. Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711–745 [DOI] [PubMed] [Google Scholar]
- 14. Davey ME, O'Toole GA. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64:847–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Doulgeraki AI, Paramithiotis S, Kagkli DM, Nychas G-JE. 2010. Lactic acid bacteria population dynamics during minced beef storage under aerobic or modified atmosphere packaging conditions. Food Microbiol. 27:1028–1034 [DOI] [PubMed] [Google Scholar]
- 16. Dourou D, Ammor MS, Skandamis PN, Nychas G-J. 2011. Growth of Salmonella Enteritidis and Salmonella Typhimurium in the presence of quorum sensing signalling compounds produced by spoilage and pathogenic bacteria. Food Microbiol. 28:1011–1018 [DOI] [PubMed] [Google Scholar]
- 17. Fatemi P, Frank JF. 1999. Inactivation of Listeria monocytogenes/Pseudomonas biofilms by peracid sanitizers. J. Food Prot. 62:761–765 [DOI] [PubMed] [Google Scholar]
- 18. Gandhi M, Chikindas ML. 2007. Listeria: a foodborne pathogen that knows how to survive. Int. J. Food Microbiol. 113:1–15 [DOI] [PubMed] [Google Scholar]
- 19. Giaouris E, Chorianopoulos N, Skandamis P, Nychas G-J. 2011. Attachment and biofilm formation by Salmonella in food processing environments, p 157–180 In Mahmoud BSM. (ed), Salmonella: a dangerous foodborne pathogen. Intech Open Access Publisher, Rijeka, Croatia [Google Scholar]
- 20. Giaouris ED, Nychas G-JE. 2006. The adherence of Salmonella Enteritidis PT4 to stainless steel: the importance of the air-liquid interface and nutrient availability. Food Microbiol. 23:747–752 [DOI] [PubMed] [Google Scholar]
- 21. Gilbert P, Allison DG, McBain AJ. 2002. Biofilms in vitro and in vivo: do singular mechanisms imply cross-resistance? J. Appl. Microbiol. 92(Suppl):98–110 [PubMed] [Google Scholar]
- 22. Goller CC, Romeo T. 2008. Environmental influences on biofilm development. Curr. Top. Microbiol. Immunol. 322:37–66 [DOI] [PubMed] [Google Scholar]
- 23. Gounadaki AS, Skandamis PN, Drosinos EH, Nychas G-JE. 2008. Microbial ecology of food contact surfaces and products of small-scale facilities producing traditional sausages. Food Microbiol. 25:313–323 [DOI] [PubMed] [Google Scholar]
- 24. Guillier L, Stahl V, Hezard B, Notz E, Briandet R. 2008. Modelling the competitive growth between Listeria monocytogenes and biofilm microflora of smear cheese wooden shelves. Int. J. Food Microbiol. 128:51–57 [DOI] [PubMed] [Google Scholar]
- 25. Habimana O, Heir E, Langsrud S, Asli AW, Møretrø T. 2010. Enhanced surface colonization by Escherichia coli O157:H7 in biofilms formed by an Acinetobacter calcoaceticus isolate from meat-processing environments. Appl. Environ. Microbiol. 76:4557–4559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Habimana O, Meyrand M, Meylheuc T, Kulakauskas S, Briandet R. 2009. Genetic features of resident biofilms determine attachment of Listeria monocytogenes. Appl. Environ. Microbiol. 75:7814–7821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Habimana O, et al. 2010. Micro ecosystems from feed industry surfaces: a survival and biofilm study of Salmonella versus host resident flora strains. BMC Vet. Res. 6:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hall-Stoodley L, Costerton JW, Stoodley P. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2:95–9108 [DOI] [PubMed] [Google Scholar]
- 29. Hassan AN, Birt DM, Frank JF. 2004. Behavior of Listeria monocytogenes in a Pseudomonas putida biofilm on a condensate-forming surface. J. Food Prot. 67:322–327 [DOI] [PubMed] [Google Scholar]
- 30. Hautefort I, Proença MJ, Hinton JC. 2003. Single-copy green fluorescent protein gene fusions allow accurate measurement of Salmonella gene expression in vitro and during infection of mammalian cells. Appl. Environ. Microbiol. 69:7480–7491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. James GA, Beaudette L, Costerton JW. 1995. Interspecies bacterial interactions in biofilms. J. Ind. Microbiol. 15:257–262 [Google Scholar]
- 32. Jeong DK, Frank JF. 1994. Growth of Listeria monocytogenes at 10°C in biofilms with microorganisms isolated from meat and dairy processing environments. J. Food Prot. 57:576–586 [DOI] [PubMed] [Google Scholar]
- 33. Jones K, Bradshaw SB. 1997. Synergism in biofilm formation between Salmonella enteritidis and a nitrogen-fixing strain of Klebsiella pneumoniae. J. Appl. Microbiol. 82:663–668 [DOI] [PubMed] [Google Scholar]
- 34. Kagkli D-M, Iliopoulos V, Stergiou V, Lazaridou A, Nychas G-J. 2009. Differential Listeria monocytogenes strain survival and growth in Katiki, a traditional Greek soft cheese, at different storage temperatures. Appl. Environ. Microbiol. 75:3621–3626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kathariou S. 2002. Listeria monocytogenes virulence and pathogenicity, a food safety perspective. J. Food Prot. 65:1811–1829 [DOI] [PubMed] [Google Scholar]
- 36. Kjelleberg S, Molin S. 2002. Is there a role for quorum sensing signals in bacterial biofilms? Curr. Opin. Microbiol. 5:254–258 [DOI] [PubMed] [Google Scholar]
- 37. Leriche V, Briandet R, Carpentier B. 2003. Ecology of mixed biofilms subjected daily to a chlorinated alkaline solution: spatial distribution of bacterial species suggests a protective effect of one species to another. Environ. Microbiol. 5:64–71 [DOI] [PubMed] [Google Scholar]
- 38. Leriche V, Carpentier B. 2000. Limitation of adhesion and growth of Listeria monocytogenes on stainless steel surfaces by Staphylococcus sciuri biofilms. J. Appl. Microbiol. 88:594–605 [DOI] [PubMed] [Google Scholar]
- 39. Leriche V, Chassaing D, Carpentier B. 1999. Behaviour of L. monocytogenes in an artificially made biofilm of a nisin-producing strain of Lactococcus lactis. Int. J. Food Microbiol. 51:169–182 [DOI] [PubMed] [Google Scholar]
- 40. Lindsay D, Brozel VS, Mostert JF, von Holy A. 2002. Differential efficacy of a chlorine dioxide-containing sanitizer against single species and binary biofilms of a dairy-associated Bacillus cereus and a Pseudomonas fluorescens isolate. J. Appl. Microbiol. 92:352–361 [DOI] [PubMed] [Google Scholar]
- 41. Lundén JM, Miettinen MK, Autio TJ, Korkeala HJ. 2000. Persistent Listeria monocytogenes strains show enhanced adherence to food contact surface after short contact times. J. Food Prot. 63:1204–1207 [DOI] [PubMed] [Google Scholar]
- 42. Mah T-FC, O'Toole GA. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9:34–39 [DOI] [PubMed] [Google Scholar]
- 43. Martin B, Garriga M, Aymerich T. 2011. Prevalence of Salmonella spp. and Listeria monocytogenes at small-scale Spanish factories producing traditional fermented sausages. J. Food Prot. 74:812–815 [DOI] [PubMed] [Google Scholar]
- 44. McDonnell G, Russell AD. 1999. Antiseptics and disinfectants: activity, action, and resistance. Clin. Microbiol. Rev. 12:147–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Miller MB, Bassler BL. 2001. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55:165–199 [DOI] [PubMed] [Google Scholar]
- 46. Minei CC, Gomes BC, Ratti RP, D'Angelis CE, De Martinis EC. 2008. Influence of peroxyacetic acid and nisin and coculture with Enterococcus faecium on Listeria monocytogenes biofilm formation. J. Food Prot. 71:634–638 [DOI] [PubMed] [Google Scholar]
- 47. Moons P, Michiels CW, Aertsen A. 2009. Bacterial interactions in biofilms. Crit. Rev. Microbiol. 35:157–168 [DOI] [PubMed] [Google Scholar]
- 48. Møretrø T, Langsrud S. 2004. Listeria monocytogenes: biofilm formation and persistence in food-processing environments. Biofilms 1:107–121 [Google Scholar]
- 49. Nadell CD, Xavier JB, Foster KR. 2009. The sociobiology of biofilms. FEMS Microbiol. Rev. 33:206–224 [DOI] [PubMed] [Google Scholar]
- 50. Norwood DE, Gilmour A. 2000. The growth and resistance to sodium hypochlorite of Listeria monocytogenes in a steady-state multispecies biofilm. J. Appl. Microbiol. 88:512–520 [DOI] [PubMed] [Google Scholar]
- 51. Norwood DE, Gilmour A. 2001. The differential adherence capabilities of two Listeria monocytogenes strains in monoculture and multispecies biofilms as a function of temperature. Lett. Appl. Microbiol. 33:320–324 [DOI] [PubMed] [Google Scholar]
- 52. Parsek MR, Greenberg EP. 2005. Sociomicrobiology: the connections between quorum sensing and biofilms. Trends Microbiol. 13:27–33 [DOI] [PubMed] [Google Scholar]
- 53. Rao D, Webb JS, Kjelleberg S. 2005. Competitive interactions in mixed-species biofilms containing the marine bacterium Pseudoalteromonas tunicata. Appl. Environ. Microbiol. 71:1729–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Reisner A, Holler BM, Molin S, Zechner EL. 2006. Synergistic effects in mixed Escherichia coli biofilms: conjugative plasmid transfer drives biofilm expansion. J. Bacteriol. 188:3582–3588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Remis JP, Costerton JW, Auer M. 2010. Biofilms: structures that may facilitate cell-cell interactions. ISME J. 4:1085–1087 [DOI] [PubMed] [Google Scholar]
- 56. Renier S, Hebraud M, Desvaux M. 2011. Molecular biology of surface colonization by Listeria monocytogenes: an additional facet of an opportunistic Gram-positive foodborne pathogen. Environ. Microbiol. 13:835–850 [DOI] [PubMed] [Google Scholar]
- 57. Rickard AH, Gilbert P, High NJ, Kolenbrander PE, Handley PS. 2003. Bacterial coaggregation: an integral process in the development of multi-species biofilms. Trends Microbiol. 11:94–100 [DOI] [PubMed] [Google Scholar]
- 58. Rieu A, Lemaitre J-P, Guzzo J, Piveteau P. 2008. Interactions in dual species biofilms between Listeria monocytogenes EGD-e and several strains of Staphylococcus aureus. Int. J. Food Microbiol. 126:76–82 [DOI] [PubMed] [Google Scholar]
- 59. Schlech WF, III, et al. 1983. Epidemic listeriosis—evidence for transmission by food. N. Engl. J. Med. 308:203–206 [DOI] [PubMed] [Google Scholar]
- 60. Shi X, Zhu X. 2009. Biofilm formation and food safety in food industries. Trends Food Sci. Technol. 20:407–413 [Google Scholar]
- 61. Simões M, Simões LC, Pereira MO, Vieira MJ. 2008. Antagonism between Bacillus cereus and Pseudomonas fluorescens in planktonic systems and in biofilms. Biofouling 24:339–349 [DOI] [PubMed] [Google Scholar]
- 62. Simões M, Simões LC, Vieira MJ. 2008. Physiology and behavior of Pseudomonas fluorescens single and dual strain biofilms under diverse hydrodynamics stresses. Int. J. Food Microbiol. 128:309–316 [DOI] [PubMed] [Google Scholar]
- 63. Smith JL, Fratamico PM, Novak JS. 2004. Quorum sensing: a primer for food microbiologists. J. Food Prot. 67:1053–1070 [DOI] [PubMed] [Google Scholar]
- 64. Steenackers H, Hermans K, Vanderleyden J, De Keersmaecker SCJ. 22 January 2011, posting date Salmonella biofilms: an overview on occurrence, structure, regulation and eradication. Food Res. Int. doi:10.1016/j.foodres.2011.01.038 [Google Scholar]
- 65. Stepanović S, Cirković I, Ranin L, Svabić-Vlahović M. 2004. Biofilm formation by Salmonella spp. and Listeria monocytogenes on plastic surface. Lett. Appl. Microbiol. 38:428–432 [DOI] [PubMed] [Google Scholar]
- 66. Stoodley P, Sauer K, Davies DG, Costerton JW. 2002. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56:187–209 [DOI] [PubMed] [Google Scholar]
- 67. Tait K, Sutherland IW. 2002. Antagonistic interactions amongst bacteriocin-producing enteric bacteria in dual species biofilms. J. Appl. Microbiol. 93:345–352 [DOI] [PubMed] [Google Scholar]
- 68. Taylor-Robinson JD, Child M, Pickup R, Strike P, Edwards C. 2003. Cell-cell interactions influence resistance and survival of Salmonella serotype Typhimurium to environmental stress. J. Appl. Microbiol. 94:95–9102 [DOI] [PubMed] [Google Scholar]
- 69. Tolker-Nielsen T, Molin S. 2000. Spatial organization of microbial biofilm communities. Microb. Ecol. 40:75–84 [DOI] [PubMed] [Google Scholar]
- 70. Uhlich GA, Rogers DP, Mosier DA. 2010. Escherichia coli serotype O157:H7 retention on solid surfaces and peroxide resistance is enhanced by dual-strain biofilm formation. Foodborne Pathog. Dis. 7:935–943 [DOI] [PubMed] [Google Scholar]
- 71. van der Veen S, Abee T. 2011. Mixed species biofilms of Listeria monocytogenes and Lactobacillus plantarum show enhanced resistance to benzalkonium chloride and peracetic acid. Int. J. Food. Microbiol. 144:421–431 [DOI] [PubMed] [Google Scholar]
- 72. van Merode AEJ, van der Mei HC, Busscher HJ, Krom BP. 2006. Influence of culture heterogeneity in cell surface charge on adhesion and biofilm formation by Enterococcus faecalis. J. Bacteriol. 188:2421–2426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Vestby LK, Moretro T, Ballance S, Langsrud S, Nesse LL. 2009. Survival potential of wild type cellulose deficient Salmonella from the feed industry. BMC Vet. Res. 5:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wimpenny J. 2009. Microbial metropolis. Adv. Microb. Physiol. 56:29–84 [DOI] [PubMed] [Google Scholar]
- 75. Wimpenny J, Manz W, Szewzyk U. 2000. Heterogeneity in biofilms. FEMS Microbiol. Rev. 24:661–671 [DOI] [PubMed] [Google Scholar]
- 76. Yang L, et al. 2011. Current understanding of multi-species biofilms. Int. J. Oral Sci. 3:74–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Zhao T, Doyle MP, Zhao P. 2004. Control of Listeria monocytogenes in a biofilm by competitive-exclusion microorganisms. Appl. Environ. Microbiol. 70:3996–4003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Zottola EA, Sasahara KC. 1994. Microbial biofilms in the food processing industry—should they be a concern? Int. J. Food Microbiol. 23:125–148 [DOI] [PubMed] [Google Scholar]