Abstract
Previous studies have shown that BrpA plays a major role in acid and oxidative stress tolerance and biofilm formation by Streptococcus mutans. Mutant strains lacking BrpA also display increased autolysis and decreased viability, suggesting a role for BrpA in cell envelope integrity. In this study, we examined the impact of BrpA deficiency on cell envelope stresses induced by envelope-active antimicrobials. Compared to the wild-type strain UA159, the BrpA-deficient mutant (TW14D) was significantly more susceptible to antimicrobial agents, especially lipid II inhibitors. Several genes involved in peptidoglycan synthesis were identified by DNA microarray analysis as downregulated in TW14D. Luciferase reporter gene fusion assays also revealed that expression of brpA is regulated in response to environmental conditions and stresses induced by exposure to subinhibitory concentrations of cell envelope antimicrobials. In a Galleria mellonella (wax worm) model, BrpA deficiency was shown to diminish the virulence of S. mutans OMZ175, which, unlike S. mutans UA159, efficiently kills the worms. Collectively, these results suggest that BrpA plays a role in the regulation of cell envelope integrity and that deficiency of BrpA adversely affects the fitness and diminishes the virulence of OMZ175, a highly invasive strain of S. mutans.
INTRODUCTION
The oral cavity is a dynamic environment in which frequent and often rapid fluctuations in pH and the concentrations of antimicrobial agents and other stressors occur. Dental care products, such as toothpastes and mouth rinses, contain a variety of antibacterial compounds, including hydrogen peroxide, sodium lauryl sulfate, and chlorhexidine. Many bacteria in the highly complex oral flora can produce hydrogen peroxide and antibacterial peptides, better known as bacteriocins, allowing the producers to ensure their presence in the community by killing competing organisms (24). To survive in the relatively hostile environment of oral biofilms, bacteria must be able to sense, respond to, and cope with these insults. The cell envelope plays a vital role during these processes, as it protects the cell from the environment, maintains cell shape, acts as a molecular sieve, and provides a platform for components of the cell involved in sensing and transmission of environmental signals. Ensuring envelope integrity is therefore crucial for bacterial cells to survive.
Streptococcus mutans, a primary causative agent of human dental caries, lives almost exclusively in biofilms on the tooth surface. This bacterium is known for its ability to survive and adapt to environmental insults, including mounting protective responses in reaction to various stimuli (10, 28). Multiple pathways are utilized by S. mutans to modulate its capacity to cope with stresses, but certain two-component signal transduction systems (TCS), including CiaHR, VicRK, and LiaSR, play integral roles in survival and adaptation to low pH, reactive oxygen species (ROS), and cell envelope stress induced by antimicrobial agents (5, 6, 11, 38, 40). For example, mutants lacking LiaSR in S. mutans displayed increased susceptibility to lipid II cycle-interfering antibiotics and chemicals that perturb cell membrane integrity (40). In addition, BrpA (for biofilm regulatory protein A) is involved in acid and oxidative stress tolerance responses and biofilm development by S. mutans (42, 43). Relative to the parent strain, S. mutans strains lacking BrpA had a limited ability to grow and accumulate on a surface and displayed enhanced sensitivity to low pH and hydrogen peroxide.
A predicted surface-associated protein, BrpA contains a region homologous to the LytR-CpsA-Psr (LCP) domain of the LCP family of proteins. The LCP family of proteins is widely distributed among Gram-positive bacteria, and its members are generally annotated as cell wall-associated transcriptional regulators (17). Originally, the LytR protein of Bacillus subtilis was identified as an autogenous transcriptional attenuator that also regulated the promoter of the divergently transcribed lytABC operon, which encodes a lipoprotein (LytA), an N-acetylmuramoyl-l-alanine amidase (autolysin, LytC), and a modifier protein of LytC (LytB) (26). The LytR paralogue CpsA of Streptococcus agalactiae was subsequently shown to function as a transcriptional activator of the capsule operon (14, 16). Recently, LytR in Streptococcus pneumoniae was reported to be essential for normal septum formation (20), with the mutant displaying variability in size and shape. The lytR mutants were also found to form multiple asymmetrical septa. Similar functions were also observed with MsrR, a Psr-like protein in Staphylococcus aureus (36). A mutant lacking MsrR was reported to have a 4-fold decrease in MIC against oxacillin and a 2-fold reduction in MIC against teicoplanin compared to those of the parental strain.
Previously, we showed that BrpA deficiency in S. mutans causes major defects in biofilm formation and acid and oxidative stress responses (42, 43). Relative to the parent strain, the deficient mutant also had an increased rate of autolysis and a decreased viability, suggesting compromise in cell envelope biogenesis/homeostasis. In this study, we used reporter gene fusion and antibacterial susceptibility assays to further characterize S. mutans strains lacking BrpA. Results showed that S. mutans strains lacking BrpA were more susceptible to cell envelope-targeting antimicrobials and that cell envelope and environmental stresses enhanced the expression of BrpA. In addition, we show that BrpA is required for optimal binding to salivary agglutinin. These results extend previous studies showing that BrpA plays a critical role in cell envelope biogenesis and cell envelope stress responses in S. mutans.
MATERIALS AND METHODS
Plasmids, bacterial strains, cell lines, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. S. mutans strains were maintained on brain heart infusion (BHI) medium. For biofilm formation, S. mutans was grown in modified biofilm medium (BM) with glucose (18 mM) and sucrose (2 mM) as supplemental carbon and energy sources (BMGS) (29, 42, 43). All solid media were prepared similarly, with inclusion of Bacto agar (Difco Laboratories, Franklin Lakes, NJ) at the level of 1.5% (wt/vol). When needed, erythromycin (10 μg/ml), kanamycin (1 mg/ml), and/or spectinomycin (1 mg/ml) was added. Unless otherwise stated, cells were grown at 37°C in an aerobic environment with 5% CO2. All Escherichia coli strains were grown aerobically in Luria-Bertani medium at 37°C with or without inclusion of kanamycin (40 μg/ml), ampicillin (100 μg/ml), spectinomycin (100 μg/ml), and/or erythromycin (300 μg/ml). Human coronary artery endothelial cells (HCAEC) were grown and maintained in endothelial cell basal medium 2 (EBM-2; Lonza) (2, 31).
Table 1.
Bacterial strains, plasmids, and primers used in the study
| Strain, plasmid, or primer | Major properties or DNA sequence(s) (5′ to 3′)a | Reference, source, or application |
|---|---|---|
| Strains | ||
| S. mutans UA159 | Wild type, serotype c | |
| S. mutans OMZ175 | Wild type, serotype f | 2 |
| S. mutans TW14 | UA159 derivative, brpA deficient, erythromycin resistant | 43 |
| S. mutans TW14D | UA159 derivative, brpA deficient, erythromycin resistant | 42 |
| S. mutans TW14K | UA159 derivative, brpA deficient, kanamycin resistant | This study |
| S. mutans TW230 | OMZ175 derivative, brpA deficient, erythromycin resistant | This study |
| E. coli DH10B | Cloning host; mcrA mcrBC mrr hsd | Invitrogen, Inc. |
| Plasmid | ||
| pFW5-luc | Integration vector containing a promoterless luciferase gene and a spectinomycin resistance marker | 22 |
| Primers | ||
| 5′ RACE Adapter | GCUGAUGGCGAUGAAUGAACACUGCGUUUGCUGGCUUUGAUGAAA | 5′ RACE |
| 5′ RACE Outer | GCTGATGGCGATGAATGAACACTG | 5′ RACE |
| 5′ RACE Inner | CGCGGATCCGAACACTGCGTTTGCTGGCTTTGATG | 5′ RACE |
| RACE brpA Rev | GTCAACTCCCATTAAGAGGATAC | 5′ RACE |
| brpA check Fw | GTTTACCTTAGGAGGAAACTGA | 5′ RACE |
| brpA SP1 | ACCTTTGGCAATTCCTTTTGTCA | Sequencing |
| brpA SP2 | AACTGACTTGACAGATAAAAACT | Sequencing |
| PbrpA | TTATGATGCTAGCAAGTCTCAAAGACA (forward), TCAGTTTCCTCCTCGAGTAAACATC (reverse) | Promoter-reporter fusion |
| SMU.409-brpA | AAGGCTGCCACTTTATCATTTGGATG (forward), AATCTTAATATCAAGCATATCCTGAA (reverse) | RT-PCR |
| brpA:erm | AGCTCAGATAAGGCTGAGCTCCTA (forward), AAACCGTCTTTCATGCCCATGTGCAT (reverse) | ΔbrpA:erm amplification |
| SMU.409P5 | TACAGCTAACTCTTCTGCAACACCATC (forward), ATTCGATAGGGATCCAAATGATAAAGTG (reverse) | 5′ fragment for polar insertion |
| SMU.409P3 | CACTTTATCATTTGGATCCCTATCGAAT (forward), ACGATACTTGCTGACACTGTCTAAAGCT (reverse) | 3′ fragment for polar insertion |
| SMU.246 | TCCTTCTTATGATTGGTGTT (forward), CTACTACTTCTTGACGGTAAT (reverse) | SMU.246 fragment, 135 bp |
| SMU.549 | GCAGTCTCTTACGATTATGG (forward), GCTACAACAGGAGGAACT (reverse) | SMU.549 fragment, 84 bp |
| SMU.599 | GTGCGACTACTATTCCTCAA (forward), TCTTCAACTTCTGCCAACT (reverse) | SMU.599 fragment, 82 bp |
| SMU.1677 | CTCATTATGGAAGTCTCAA (forward), AAGTAGGATGTTCAATCG (reverse) | SMU.1677 fragment, 121 bp |
| SecA | GTGCTTCCATTACCTATCA (forward), ATTCCTCTTCTTCTGTCTTC (reverse) | secA fragment, 87 bp |
| SecY | CAGGAAGTGTGGTTGTAA (forward), GCTTGAACGGATATTGAC (reverse) | secY fragment, 155 bp |
| BrpA | CGTGAGGTCATCAGCAAGGTC (forward), CGCTGTACCCCAAAAGTTTAGG (reverse) | brpA fragment, 148 bp |
Nucleotides underlined are restriction sites engineered for cloning.
DNA manipulation, transcriptional initiation site mapping, and construction of reporter fusions.
Standard recombinant DNA procedures were used (12, 37). All restriction and modifying enzymes were purchased from Invitrogen (Carlsbad, CA) or New England BioLabs (Ipswich, MA) and used as recommended by the suppliers. All primers (Table 1) were synthesized by Integrated DNA Technologies, Inc. (Iowa City, IA). RNA ligase-mediated rapid amplification of cDNA ends (RLM-RACE) (Ambion, Inc., Foster City, CA) was used to map the transcription initiation site (TIS) of brpA. Briefly, total RNA was prepared from early- (optical density at 600 nm [OD600] = 0.2) and late-exponential-phase (OD600 = 0.8) cultures grown in BHI using hot phenol (1, 42). The preparations were then treated with RNase-free DNase I (Ambion, Inc.), and RNA was retrieved using the Qiagen RNeasy purification kit (Qiagen, Inc., Valencia, CA). For cDNA synthesis, total RNA was treated with calf intestinal phosphatase and with tobacco acid pyrophosphatase by following the supplier's recommendations and then ligated to the supplied 5′ RACE adapter. cDNA was synthesized using iScript reverse transcriptase III (Invitrogen) and followed by a nested PCR using either the 5′ RACE outer primer or the 5′ RACE inner primer (Ambion, Inc.) and a brpA-specific reverse primer. The transcription initiation site (TIS) was determined by sequencing the resulting PCR amplicon.
To analyze the regulation of brpA expression, a promoterless luciferase gene (luc) was used as a reporter (23, 35). Briefly, the cognate brpA promoter region was amplified by PCR with primers PbrpA forward and PbrpA reverse. Following proper restriction digestions, the amplicon was cloned directly in front of the promoterless luc gene in the integration vector pFW11-luc (22), which also contains a Shine-Dalgarno sequence optimized for group A streptococci (35). Following confirmation of the correct sequence of the cloned element, the resulting construct, pFW11::pbrpA::luc, was introduced into S. mutans UA159 and TW14D and maintained on BHI agar containing 1 mg/ml spectinomycin. The expression of BrpA under different environmental conditions and cell envelope stressors was analyzed using a luciferase assay by following the protocol of Podbielski et al. (22, 35).
DNA microarray and real-time PCR analysis.
For DNA microarray analysis, total RNAs were extracted from early-exponential-phase (OD600 ≅ 0.3) cultures, treated with DNase I (Ambion, Inc.) to remove all DNA, and then retrieved with the RNeasy purification kit (Qiagen, Inc.) (42). Array analysis was performed by using the whole-genome S. mutans microarrays (version 2) that were obtained from The J. Craig Venter Institute (JCVI; http://pfgrc.jcvi.org) by following the protocols recommended by JCVI as described elsewhere (1, 42). Expression levels of selected genes identified by DNA microarray analysis were confirmed by real-time PCR procedures detailed elsewhere (Table 1) (5, 42).
Cell envelope antimicrobial susceptibility assays.
The susceptibility of S. mutans strains to antimicrobial agents was analyzed using microtiter plate-based assays as described previously (30, 40). Cell envelope antimicrobial agents tested included the antibiotics vancomycin (Sigma, St. Louis, MO), bacitracin (Sigma), and the β-lactam antibiotic penicillin G (Sigma), the bacteriocin nisin (Sigma), and the cell envelope active compounds sodium dodecyl sulfate (SDS) and chlorhexidine (Sigma). Briefly, 100 μl of properly diluted mid-exponential-phase cultures was added to 96-well plates containing BHI medium supplemented with 2-fold serial dilutions of the cell envelope antimicrobial agents. After 48 h, bacterial growth was measured spectrophotometrically using a Synergy 2 plate reader (BioTek, Inc.), and relative cell density percentages ([OD490 of cultures with antimicrobial agents/OD490 of the untreated cultures] × 100) were calculated. The MIC was defined as the lowest concentration at which the cultures did not grow to over 10% of the relative cell density. Minimal bactericidal concentration (MBC) assays were carried out using the MIC test plates. The MBC was determined as the lowest concentration for which fewer than 5 CFU were observed after 48 h when 20 μl of the cultures was plated on nonselective medium.
Biofilm formation and BIAcore assays.
Biofilm formation on 96-well plates precoated with salivary agglutinin was carried out as previously described (4, 42, 43). Interactions of S. mutans whole cells with salivary agglutinin were analyzed using BIAcore assays in which the receptor was immobilized on Pioneer F1 sensor chips (32).
Preparation of protein fractions and Western blot analysis.
Various fractions of proteins were prepared from BHI-grown early-exponential-phase (OD600 = 0.3) cultures of S. mutans (3, 41, 45). Briefly, whole-cell lysates were obtained by glass bead beating in SDS boiling buffer (60 mM Tris [pH 6.8], 10% glycerol, and 5% SDS). For surface-associated fractions, cells from 500-ml cultures were suspended in 25 ml of 0.2% N-dodecyl-N,N-dimethyl-3-ammonio-1-propanesulfonate (Zwittergent; Sigma) and incubated at 28°C with shaking at 80 rpm for 1 h. Following centrifugation, the supernatants were further concentrated using Amicon Ultra centrifugal filters (Millipore, Billerica, MA). In other cases, bacterial cells were suspended in 4% SDS and incubated at room temperature for 30 min. For cell-free fractions, cultural supernatants were precipitated by ammonium sulfate. For Western blot analysis, proteins (10 μg total) were separated using 7.5% SDS-PAGE, blotted onto Immobilon-FL membranes, and then probed with anti-P1 monoclonal antibodies (8, 41).
Bacterial invasion assay.
The impact of BrpA deficiency on the ability of S. mutans to invade host tissues was analyzed using primary human coronary artery endothelial cells as described elsewhere (2, 31). Briefly, overnight cultures were harvested by centrifugation at 14,000 × g for 5 min, and pellets were washed twice with phosphate-buffered saline (pH 7.2) and resuspended in endothelial cell basal medium 2 (EBM-2; Lonza). Aliquots (1 ml) of bacterial cells (with ∼5 × 107 CFU/ml) were mixed with HCAEC monolayers in 24-well plates for 2 hours. Following proper washes and additional incubation with gentamicin and penicillin G to eliminate extracellular bacterial cells, HCAE cells were lysed by osmotic shock, and serial dilutions of the lysates and bacterial cells released were plated on BHI agar in triplicate. The percentage of intracellular bacteria relative to the initial inoculum was calculated.
Wax worm infection model.
Galleria mellonella killing assays were performed by following the procedures described previously (21). Briefly, groups of 20 larvae, ranging from 200 to 300 mg in weight and with no signs of melanization, were randomly assigned. A 5-μl aliquot of properly diluted mid-exponential-phase (OD600 = 0.5) cultures of wild-type S. mutans or the BrpA-deficient mutant was injected into the hemocoel using a 10-μl Hamilton syringe (Hamilton Co., Reno, NV). Groups receiving heat-inactivated (10 min at 75°C) wild-type S. mutans or saline were used as controls. After injection, larvae were incubated at 37°C, and appearance (signs of melanization) and survival were recorded at selected intervals. Kaplan-Meier killing curves were plotted, and estimates of differences in survival were compared using a log rank test. A P value of ≤0.05 was considered significant. All data were analyzed with GraphPad Prism, version 4.0.
Microarray data accession number.
Microarray data have been deposited in NCBI (accession number GSE35349).
RESULTS
BrpA deficiency affects binding to immobilized salivary agglutinin.
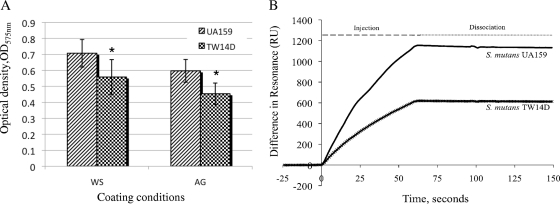
Binding to salivary agglutinin and other glycoproteins, primarily through the multifunctional adhesin P1 (also called antigen I/II, PAc, or SpaP), is considered to be a major mechanism used by S. mutans to colonize the tooth surface (15, 19, 25, 27). On 96-well plates precoated with whole saliva and purified salivary agglutinin (4), wild-type S. mutans UA159 formed robust biofilms after 24 h, consistent with previous findings (4). Relative to UA159, however, biofilm accumulation by the BrpA-deficient mutant TW14D was significantly lower (P < 0.05) (Fig. 1A). We also used BIAcore assays to analyze the impact of BrpA deficiency on P1-mediated adherence and biofilm formation. Affinity-purified, high-molecular-weight salivary glycoprotein agglutinin was immobilized on a Pioneer F1 sensor chip, and interaction of S. mutans with immobilized agglutinin was measured by BIAcore, a proven technique for assessment of salivary-agglutinin-mediated adherence (32). Compared to the wild type, the capacity of salivary-agglutinin-mediated whole-cell-receptor interactions in the mutant lacking BrpA was decreased by more than 57%, with the average resonance signal at 938.95 (±102.45) resonance units (RU) for the wild-type UA159 strain and 413.6 (±186.7) RU for the mutant TW14D strain (P < 0.01) (Fig. 1B).
Fig 1.
Biofilm formation (A) and BIAcore assays (B). (A) S. mutans UA159 and TW14D were grown on 96-well plates that were precoated with unstimulated whole saliva (WS) or affinity-purified salivary agglutinin (AG). Data show the average densities (± standard deviations [error bars]) of 24-hour biofilms from more than three independent sets of experiments, with an asterisk indicating a significant difference between UA159 and TW14D under the conditions specified (P < 0.05). (B) P1-mediated S. mutans whole-cell interactions with salivary agglutinin were measured using BIAcore assays. Salivary agglutinin was immobilized on an F1 chip surface. S. mutans UA159 and the BrpA-deficient mutant TW14D were injected for 60 s. S. mutans UA159 yielded an average resonance signal of 938.95 resonance units (RU), while TW14D had an average resonance signal of 413.6 RU. Results indicate that BrpA deficiency affects P1-mediated whole-cell adhesin-receptor interactions. The panel shows representatives of two independent experiments.
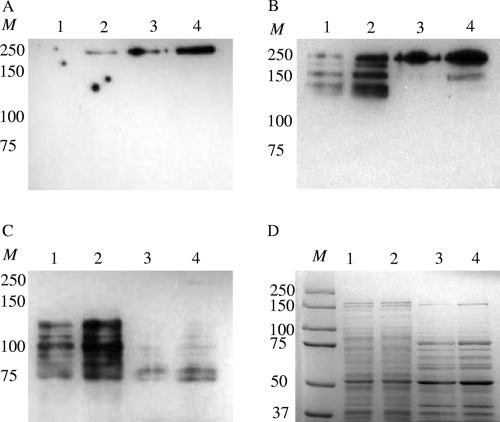
Western blot analysis was then carried out to further examine the levels of P1 in whole-cell lysates, cell-free fractions, and surface-associated fractions from UA159 and TW14D using monoclonal antibodies (MAbs) against P1 as probes (3, 8, 41, 45). When probed with MAb 6-8C, which reacts to the C terminus of P1 (8), a single band with a molecular mass of around 200 kDa was apparent in the surface-associated fractions of both UA159 and TW14D (Fig. 2A). In comparison, however, the density of this reactive band in TW14D was over 2-fold higher than that of the one in UA159. A similar band was also detected in the whole-cell lysate of TW14D but was not detectable in UA159. When probed with MAb 4-10A, which recognizes the A-P stalk, one major band with a molecular mass of around 200 kDa was apparent in both TW14 and UA159 (Fig. 2B), with the density of the band in TW14D being more than 4-fold higher than that of the band found in UA159. Besides these, multiple bands with molecular masses of around 150 kDa were apparent in the whole-cell lysates, but again these bands were more than 9-fold denser in TW14D than those in UA159. With MAb 3-8D, which recognizes the A region of the P1, used as a probe, multiple bands with similar molecular masses of around 100 kDa were identified in the whole-cell lysates of both UA159 and TW14D (Fig. 2C). In a comparison, the densities of these bands were about 6-fold higher in the mutant than in the wild type. Multiple bands reactive to MAb 3-8D were also seen in the surface-associated fractions, but unlike those from the whole-cell lysates, these bands were mostly around 75 kDa.
Fig 2.
Western blot (A to C) and SDS-PAGE (D) analysis of P1 in wild-type S. mutans UA159 (lanes 1 and 3) and the BrpA-deficient mutant TW14D (lanes 2 and 4). Proteins (10 μg total) of whole-cell lysates (lanes 1 and 2) and surface-associated fractions (lanes 3 and 4) were separated using 7.5% SDS-PAGE (D), blotted onto a polyvinylidene fluoride (PVDF) nitrocellulose membrane, and then probed with anti-P1 monoclonal antibodies (MAbs). Panel A shows results when probed with MAb 6-8C, which recognizes the C terminus of P1. A single band with a molecular mass of around 200 kDa was apparent, and the density of this band in TW14D was more than 2-fold higher than that of the one in UA159. Panels B and C show results when probed with MAb 4-10A and MAB 3-8D, which recognize the A-P stalk and the alanine-rich region of P1, respectively, but both are shown to react to truncated peptides (8). Multiple bands with molecular masses of around 150 (B) and 100 (C) kDa were identified in both UA159 and TW14D, but the densities of these bands were significantly higher in TW14D than in UA159. M, molecular weight markers.
BrpA-deficient mutants are more sensitive to cell envelope-active antimicrobials.
Previously, it was shown that BrpA deficiency in S. mutans caused elevations in autolysis and reductions in viability, with more dead cells and cell debris in biofilms in the mutant than in the wild-type strain (13, 42). To analyze whether BrpA in S. mutans affects cell envelope integrity, the MIC and MBC against several cell envelope antibacterial agents were measured using microtiter plate-based assays. As shown in Table 2, the BrpA-deficient mutant had a decreased ability to survive the treatment of several different antimicrobial agents compared to that of the wild-type strain under the same conditions. TW14D had 1.8- and 2-fold reductions in MICs to nisin and bacitracin, respectively, compared to those of the wild type. Similar trends were also detected with vancomycin, penicillin G, d-cycloserine, SDS, and triclosan, although the differences between these two strains were not statistically significant. When MBCs were analyzed, the deficient mutant had ≥1.5-fold reductions in sensitivity to nisin, chlorhexidine, and SDS compared to those of the wild type. Slight, but not statistically significant, decreases were also seen with bacitracin, vancomycin, and triclosan.
Table 2.
Effect of BrpA deficiency on susceptibility to cell envelope antimicrobialsa
| Strain | MIC and MBC (μg/ml) for each antimicrobial |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lipid II inhibitors |
Non-lipid II inhibitors |
Cell membrane-disrupting agents |
||||||||||||||
| Van |
Nis |
Bac |
Pen |
d-Cyc |
Chl |
SDS |
Tri |
|||||||||
| MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | |
| UA159 | 0.75 | 1.25 | 17.5 | 27.5 | 100 | 1,600 | 0.04 | 0.075 | 300 | 11,000 | 1.5 | 6 | 40 | 60 | 60 | 75 |
| TW14D | 0.50 | 1 | 10b | 15b | 50c | 1,400 | 0.03 | 0.075 | 250 | 11,000 | 1.5 | 3c | 30 | 40b | 50 | 70 |
Van, vancomycin; Nis, nisin; Bac, bacitracin; Pen, penicillin G; d-Cyc, d-Cycloserine; Chl, chlorhexidine; SDS, sodium dodecyl sulfate; Tri, triclosan.
Reduction in MIC and/or MBC of more than 1.5-fold compared to UA159.
Reduction in MIC and/or MBC of more than 2-fold compared to UA159.
BrpA deficiency affects virulence in a Galleria mellonella model.
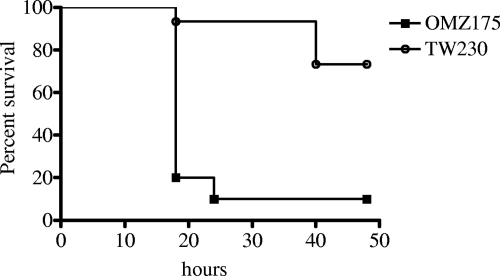
Recent studies have shown that certain strains of S. mutans, such as OMZ175 (serotype f), are highly invasive and consequently may play a significant role in development of certain systemic diseases, such as infective endocarditis (2, 31). In contrast, UA159, a commonly used laboratory strain, possesses only limited capacity to invade endothelial cell lines (2). To create a BrpA-deficient mutant of OMZ175, PCRs were conducted with genomic DNA from TW14D as the template (Table 1) (42). The resulting amplicon, containing DNA fragments flanking brpA and an erythromycin resistance element (Ermr), was used to replace the brpA-coding sequence in S. mutans OMZ175, and mutants were selected on BHI with erythromycin and further confirmed by DNA sequencing. When analyzed by invasion assays using human coronary artery endothelial cells (HCAEC) (2), the BrpA-deficient mutant TW230 had a slightly reduced invasion efficiency compared to that of OMZ175, with average invasion rates of 0.18% for TW230 and 0.49% for OMZ175 (P = 0.089). When tested in the G. mellonella (wax worm) virulence model (21), the survival rate of worms receiving the BrpA-deficient mutants was significantly (P < 0.01) higher than that of those receiving strain OMZ175 (Fig. 3). Not surprisingly, considering the poor virulence of strain UA159 in this model, no major differences (P > 0.05) were observed when TW14D and UA159 were compared in the wax worms (data not shown).
Fig 3.
Killing of G. mellonella larvae infected with wild-type S. mutans and the corresponding BrpA-deficient mutants at 37°C. Survival (Kaplan-Meier) plots of S. mutans OMZ175 (solid squares) and the BrpA-deficient mutant TW230 (open circles), injected at 1 × 107 CFU/larva, are shown. There was no killing of larvae injected with saline and minimum killing of larvae injected with heat-killed S. mutans UA159 cells (data not shown). The experiments were repeated three times, and the data presented here are results representative of a typical experiment. Compared to the wild-type parent strain, OMZ175, the BrpA-deficient mutant TW230 showed attenuated virulence (P ≤ 0.01). No significant differences were measured between UA159 and TW14D (data not shown).
brpA is cotranscribed with SMU.409.
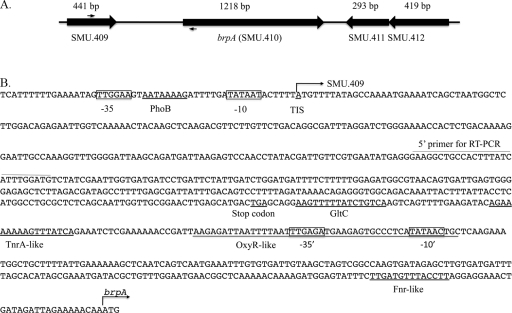
The brpA gene (SMU.410) in S. mutans is flanked by downstream SMU.411 and upstream SMU.409 (Fig. 4), which encode a streptococcus-specific hypothetical protein and a putative bacterial ATPase/GTPase (www.oralgen.lanl.gov), respectively. The open reading frames in SMU.409 and brpA are arranged in the same orientation, while SMU.411 and brpA are transcribed in opposite directions. To map the promoter region of brpA, the TIS was examined using 5′ RACE with total RNA extracted from BHI-grown cultures. Results of the 5′ RACE PCR showed that multiple brpA transcripts existed (data not shown). Sequence analysis of the major cDNA product revealed that the major TIS of brpA was 774 bp upstream of the translational start codon ATG (Fig. 4), suggesting that SMU.409 and brpA are cotranscribed under the conditions studied. Reverse transcription-PCR (RT-PCR) with total RNA extracted from BHI-grown planktonic cultures and 3-day-old biofilms grown on BMGS confirmed that brpA was cotranscribed with the upstream gene SMU.409 (Fig. 5A) under both of the conditions tested. Insertion of a polar kanamycin resistance element, ΩKm (34), at SMU.409 also caused a reduction in brpA transcription of more than 25-fold, as shown by real-time PCR with total RNA preparations of early-exponential-phase (OD600 = 0.25) cultures of the insertional mutant and its parent strain UA159 (data not shown). Similar results were also obtained with Western blot analysis (data not shown).
Fig 4.
Schematic diagram of the brpA-flanking region (A) and analysis of the brpA promoter region (B). Panel A shows the genetic organization of the regions flanking brpA, with locus name and size labeled under and above the arrows, respectively. Approximate positions of the primers used for RT-PCR are shown with solid arrows above SMU.409 and under brpA. Panel B highlights the promoter region of brpA, which includes the coding sequence of SMU.409, with the translation initiation sites indicated by arrows above the start codons of SMU.409 and brpA, respectively. As determined by 5′ RACE, the transcription initiation site (TIS, underlined) of brpA was 774 bp upstream of its translation start site, which overlaps the translation start site of SMU.409 as annotated by NCBI (www.oralgen.lanl.gov). Computer-based analysis using BPROM, a bacterial sigma70 promoter recognition program, revealed two sets of putative −35 and −10 sites (boxed) and binding sites for putative transcriptional regulators (underlined). Further analysis using Virtual Footprint, a program especially designed to analyze transcription factor binding sites, also identified sequences with high similarity to binding sites for transcriptional factors GltC, TnrA, and Fnr (underlined).
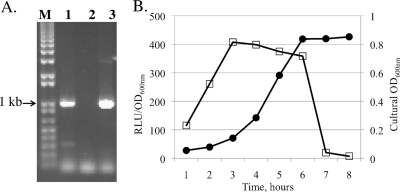
Fig 5.
RT-PCR analysis (A) and luciferase activity assays (B). (A) RT-PCR analysis was carried out using total RNA isolated from mid-exponential-phase (OD600 = 0.5) cultures. Lanes M, 1, 2, and 3 contain DNA markers, RT-PCR products, controls with no reverse transcriptase, and PCR products with genomic DNA as the template, respectively. Similar results were obtained with total RNA extracted from 3-day-old biofilms (data not shown). (B) For the reporter gene fusion study, the full-length brpA promoter region was cloned in front of a promoterless firefly luciferase gene, and the resulting promoter-reporter fusion was integrated into the S. mutans genome. Cells carrying the reporter fusion were collected at different times during growth and assayed for luciferase activity by mixing 200 μl of whole cells with 50 μl of 1 mM d-luciferin (pH 6.0). Data show reporter luciferase activity (open squares) in relative light units (RLU) calibrated with the optical density (OD600) of the cultures (solid circles) at each time point.
Expression of BrpA is regulated in response to environmental conditions.
In cultures grown in BHI broth, luciferase expression from the full brpA promoter (a 1,119-bp fragment) was measured at its maximum during early exponential phase (OD600 = 0.3) (Fig. 5B), consistent with our earlier study with Northern blotting (44). Considering the fact that mutants lacking BrpA had significant defects in their abilities to survive low pH and hydrogen peroxide challenge (42), cells of early-exponential-phase cultures carrying the reporter fusion were treated with hydrogen peroxide and methyl viologen (Paraquat; Sigma) in the growth medium for 90 min. Results showed that, relative to the untreated controls, the level of luciferase activity in cells treated with hydrogen peroxide and methyl viologen was increased significantly (Table 3). Such increases appeared to be concentration dependent when the amount used was within a certain threshold (see Fig. S1 in the supplemental material). However, beyond the threshold, luciferase activity was decreased when more hydrogen peroxide and methyl viologen were used. Similar results were also obtained with cells treated with various cell envelope antimicrobials at subinhibitory concentrations (Tables 2 and 3), with the most significant impact measured with chlorhexidine, a chemical commonly used in dental care products and in prevention of tooth decay. Efforts were also made to evaluate the impact of pH on BrpA expression by incubating the cells carrying the reporter fusion in BHI broth adjusted to different pH values, but the results were inconclusive, probably due to the impact of low pH on the luciferase enzyme (data not shown). To circumvent this problem, a study is under way using chloramphenicol acetyltransferase as a reporter under controlled conditions in a chemostat.
Table 3.
Luciferase expression in response to oxidative stress and cell envelope-active antimicrobial agents
| Stimulus or stressor (concn) | Luciferase activity ratio (RLU)a |
|---|---|
| Hydrogen peroxide (0.4 mM) | 2.09 ± 0.33d |
| Methyl viologen (10 mM) | 2.07 ± 0.55c |
| SDS (8 μg/ml) | 2.51 ± 0.50d |
| Chlorhexidine (1.5 μg/ml) | 3.03 ± 0.42d |
| Vancomycin (0.75 μg/ml) | 1.41 ± 0.03b |
| Bacitracin (20 μg/ml) | 2.24 ± 0.13b |
| Nisin (10 μg/ml) | 2.25 ± 0.47c |
| d-Cycloserine (20 μg/ml) | 1.62 ± 0.31c |
| Penicillin G (0.04 μg/ml) | 1.66 ± 0.35b |
Data are the ratios (average ± standard deviation) of the luciferase activity (in RLU) at the conditions specified to that of controls that received an equal amount of solvent instead of the stressor indicated.
Difference between the groups at the significance level of P < 0.05.
Difference between the groups at the significance level of P < 0.01.
Difference between the groups at the significance level of P < 0.001.
BrpA deficiency causes substantial alterations in the transcriptional profiles of the deficient mutant.
In consideration of the fact that BrpA expression is at its maximum during early exponential phase, as shown by Northern blotting (44) and reporter gene fusion assays, we carried out another DNA microarray analysis using total RNA from early-exponential-phase cultures (OD600 = 0.3). It was found that 92 genes were upregulated and 90 downregulated by a factor of at least 1.5-fold in TW14D (P ≤ 0.001) (see Tables S1 and S2 in the supplemental material). At a P level of <0.01, 176 additional genes were found to be differentially expressed in TW14D, with 77 genes upregulated and 90 downregulated (data not shown). Based on the descriptions and putative functions of the genes identified at the significance level of a P value of <0.001, BrpA deficiency affects almost every aspect of cellular physiology as well as virulence properties, including amino acid biosynthesis (10), carbohydrates and energy metabolism (18), nucleic acids and DNA metabolism (10), transcriptional regulation (9), ABC transporters (29), molecular chaperones and other cellular processes (13), and hypothetical and conserved hypothetical proteins (50). The breadth of impact of BrpA deficiency on the transcriptional profile of the deficient mutant is similar to what was observed previously with mid-exponential-phase cells (42). However, comparison of the two transcriptional profiles revealed that only a small number of genes, which include recA (for recombinant protein RecA), gtfD (for glucosyltransferase D), wapA (for surface-associated protein WapA), groEL (for molecular chaperone GroEL), and sod (for Mn-dependent superoxide dismutase [SOD]), were consistently up- or downregulated in both early- and mid-exponential-phase cultures (see Tables S1 and S2 in the supplemental material) (42). In addition, the magnitude of alterations in gene expression was more dramatic in cells of the early exponential phase than in the mid-exponential-phase cultures.
DISCUSSION
The cell envelope is of essential importance for growth, cell division, interaction with the environment, and antimicrobial resistance. Previous studies have shown that BrpA, a paralogue of the LCP family of cell wall-associated transcriptional attenuators, strongly influences S. mutans biofilm formation and survival against low pH and reactive oxygen species (42, 43). Strains lacking BrpA also displayed increased autolysis rates and decreased viability, suggesting a role for BrpA in regulation of cell envelope biogenesis or homeostasis (13, 42, 43). In this study, BrpA deficiency was shown to significantly weaken the ability of S. mutans to survive cell envelope stresses induced by cell envelope-targeting antimicrobials (Table 2). Among the antimicrobial agents tested, the most significant influences on MIC were measured with nisin and bacitracin, two antibiotics that interfere with lipid II cycling, blocking peptidoglycan and cell wall biosynthesis (9). The most significant impacts on MBC were seen with chlorhexidine and SDS, two chemicals commonly used in oral health care products that compromise membrane integrity. These results provide further support for a role for BrpA in regulation of cell envelope biogenesis or maintenance by S. mutans, consistent with the roles of certain LCP paralogues in other bacterial species (18, 20, 33, 36, 39).
The bacterial cell wall is a repeating, three-dimensional polymer known as peptidoglycan or murein that consists of a linear, alternating N-acetylmuramic acid (MurNAc) and N-acetylglucosamine (GlcNAc) motif, cross-linked via peptides appended to MurNAc. Of the genes altered as a result of BrpA deficiency in TW14D, several were found to encode proteins with potential roles in peptidoglycan biosynthesis (Table 4). Among them are SMU.246 for a phospho-MurNAc-pentapeptide transferase (RgpG), SMU.549 for an undecaprenyl-PP-MurNAc-pentapeptide-UDP-GlcNAc transferase (MurG), SMU.599 for a d-alanine-d-alanine ligase (DdlA), and SMU.1677 for a UDP-MurNAc-tripeptide synthetase (MurE). While the exact role of these gene products in S. mutans cellular physiology awaits further investigation, downregulation of genes involved in peptidoglycan synthesis would have an impact on cell envelope biogenesis, likely leading to defects in wall integrity. Such a defect would be consistent with the weakened resistance to cell envelope antimicrobials, reduced viability, and increased autolysis observed for BrpA-deficient mutants (13, 43). In support of a role in cell envelope biogenesis, the expression of a luciferase reporter fusion under the direction of a brpA promoter was also upregulated in response to cell envelope stresses induced by exposure to subinhibitory concentrations of antimicrobial agents that target the cell envelope (Table 3). Defects in cell envelope integrity would likely result in vulnerability of the bacterial cells to environmental insults and therefore can partly explain the weakened acid and oxidative stress responses of the BrpA-deficient mutants (42).
Table 4.
Selected genes identified by DNA microarray analysis
| Unique IDa | Description/putative functionb | Array ratioc | qPCR ratioc | P value |
|---|---|---|---|---|
| SMU.246 | Glycosyltransferase N-acetylglucosaminyltransferase, RgpG | −2.6 | −4.0 | 5.9E-3 |
| SMU.549 | Undecaprenyl-PP-MurNAc-pentapeptide-UDPGlcNAc GlcNAc transferase, MurG | −2.7 | −2.0 | 3.6E-3 |
| SMU.599 | d-Alanine-d-alanine ligase, DdlA | −1.8 | −3.2 | 7.1E-3 |
| SMU.1677 | UDP-N-acetylmuramoylananine-d-glutamate-2,6-diaminopimelate ligase, UDP-MurNac-tripeptide synthetase, MurE | −13.1 | −2.7 | 3.6E-3 |
| SMU.1689 | d-Alanyl carrier protein, DltC | 3.4 | ND | 6.9E-3 |
| SMU.1691 | d-Alanine-d-alanyl carrier protein ligase, DltA | 3.4 | ND | 3.7E-5 |
| SMU.1838 | Preprotein translocase subunit SecA | −1.9 | −2.0 | 6.6E-3 |
| SMU.1948 | Preprotein translocase subunit SecE | −2.8 | ND | 2.1E-3 |
| SMU.2006 | Preprotein translocase SecY protein | 3.9 | 1.9 | 1.6E-5 |
ID, identification.
Descriptions and putative functions of the identified genes are based upon the published S. mutans database.
Levels of expression in the BrpA-deficient mutant relative to those of the wild type, as shown by DNA microarray analysis (array ratio) and real-time PCR (quantitative PCR [qPCR]), with negative numbers indicating downregulation. ND, not done.
P1, a cell wall-anchored adhesin, is considered a key contributor to S. mutans colonization of the tooth surface (7, 15). P1 mediates the adherence through interactions with high-molecular-weight salivary agglutinin in the enamel pellicle. Both biofilm formation assays and BIAcore analysis showed that BrpA affects the ability of S. mutans to interact with salivary agglutinin (Fig. 1). As shown by Western blotting, however, the level of P1 expression was increased by more than 2-fold as a result of BrpA deficiency (Fig. 2A). When analyzed by DNA microarrays, several genes encoding components of the Sec translocase were also found altered in TW14D. These included secA, secE, and secY, encoding the ATP-dependent motor of the translocation machinery, SecA, and the translocon pore components SecE and SecY, respectively. Both secA and secE were downregulated by more than 2-fold, while secY was upregulated by more than 2-fold (Table 4). The Sec secretion system participates in translocation of polypeptides across, or integration into, the cytoplasmic membrane (46). Alteration in expression of individual members of the Sec translocon complex, as well as global defects in cell envelope integrity, will likely influence the function of the translocation/secretion machinery. As a result of altered Sec function, the P1 adhesin may be compromised in conformation, stability, and/or distribution on the surface. Therefore, the increased expression could be a compensatory response to such a compromise, but the underlying mechanism awaits further investigation. In addition, the disproportional increases in density of the lower-molecular-mass bands in TW14D that were reactive to MAbs 3-8D and 4-10A, which are shown to recognize truncated peptides (8), suggest that the stability of P1 may be reduced in the brpA mutant (Fig. 2B and C). Therefore, decreased stability and/or misfolding of P1 may underlie the reduced binding to salivary agglutinin by strain TW14D. These in vitro results also suggest that BrpA deficiency may affect bacterial adherence and biofilm initiation by S. mutans in the oral cavity as well.
Previously, Northern blotting showed that transcription of brpA was maximal during early exponential phase (OD600≈ 0.3) and that deficiency of LuxS dramatically decreased brpA transcription, indicating that expression of brpA is regulated in response to environmental conditions and by LuxS-mediated quorum sensing (44). In this study, we used luciferase reporter gene fusion assays to show that the expression of BrpA is strongly dependent on growth phase, with maximal activity measured during early exponential phase (Fig. 5B). These results again suggest that environmental conditions and cell density play an important role in the regulation of BrpA expression. Differences in environmental conditions, such as pH and concentration of ROS, and cell density may, in part, account for some of the discrepancies observed between the two transcriptional profiles for the early- and mid-exponential-phase cultures (see Tables S1 and S2 in the supplemental material) (42). However, whether BrpA affects different group of genes in response to environmental stimuli awaits further investigation.
Both 5′ RACE and RT-PCR showed that under the conditions studied, the major transcript was a product of cotranscription of brpA with its upstream locus SMU.409 (Fig. 4 and 5). Consistently, both real-time PCR and Western blot analysis (data not shown) showed that a polar insertion at SMU.409 resulted in a dramatic reduction of BrpA expression. However, we have previously shown that possession in trans of the brpA-coding sequence plus a 344-bp fragment upstream of its start codon was able to partially complement the deficient mutant TW14 in an acid tolerance response (42). The luciferase reporter fusion with a fragment of 683 bp upstream of brpA also showed promoter activity in this intergenic region, although it is much weaker than the full-length (1,119-bp) promoter (data not shown). Computer-based analysis of this intergenic region using BPROM, a bacterial sigma70 promoter recognition program, and Virtual Footprint, a program especially designed to analyze transcription factor binding sites, also revealed putative −10 (TATAAc) and −35 (TTGAgA) sites and regions with high similarity to binding sites for several putative transcriptional regulators (Fig. 4). These results further suggest that transcription of brpA may be initiated at different sites under different environmental conditions. A study is under way to dissect the underlying mechanisms, including the cis- and trans-acting elements involved in regulation of brpA expression. SMU.409 encodes a putative ATPase/GTPase. While the exact role of SMU.409 in regulation of S. mutans cellular physiology and brpA expression is still under investigation, the close association of this gene with brpA suggests its likely involvement in BrpA-regulated cell envelope biogenesis/homeostasis.
In summary, the results presented here further support that S. mutans BrpA is involved in the regulation of cell envelope biogenesis/maintenance and that deficiency of BrpA affects the fitness of the deficient mutants and decreases the virulence of OMZ175, a highly invasive strain in a wax worm model. Current efforts are directed to further investigation of the underlying mechanisms.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIDCR grant DE19452 to Z.T.W. and in part by the South Louisiana Institute of Infectious Disease Research.
We thank Fengxia (Felicia) Qi at the University of Oklahoma Health Sciences Center for her kind gift of the integration vector pFW11-luc and James H. Miller for his assistance with invasion and wax worm infection assays.
Footnotes
Published ahead of print 10 February 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Abranches J, Candella M, Wen TZ, Baker HV, Burne RA. 2006. Different roles of EIIABMan and EIIGlc in the regulation of energy metabolism, biofilm development, and competence in Streptococcus mutans. J. Bacteriol. 188:3748–3756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Abranches J, et al. 2009. Invasion of human coronary artery endothelial cells by Streptococcus mutans OMZ175. Oral Microbiol. Immunol. 24:141–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ahn SJ, Burne RA. 2006. The atlA operon of Streptococcus mutans: role in autolysin maturation and cell surface biogenesis. J. Bacteriol. 188:6877–6888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ahn SJ, Wen ZT, Brady LJ, Burne RA. 2008. Characteristics of biofilm formation by Streptococcus mutans in the presence of saliva. Infect. Immun. 76:4259–4268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ahn SJ, Wen ZT, Burne RA. 2006. Multilevel control of competence development and stress tolerance in Streptococcus mutans UA159. Infect. Immun. 74:1631–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Biswas I, Drake L, Erkina D, Biswas S. 2008. Involvement of sensor kinases in the stress tolerance response of Streptococcus mutans. J. Bacteriol. 190:68–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brady LJ, et al. 2010. The changing faces of Streptococcus antigen I/II polypeptide family adhesins. Mol. Microbiol. 77:276–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brady LJ, Piacentini DA, Crowley PJ, Bleiweis AS. 1991. Identification of monoclonal antibody-binding domains within antigen P1 of Streptococcus mutans and cross-reactivity with related surface antigens of oral streptococci. Infect. Immun. 59:4425–4435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Breukink E, de Kruijff B. 2006. Lipid II as a target for antibiotics. Nat. Rev. Drug Discov. 5:321–332 [DOI] [PubMed] [Google Scholar]
- 10. Burne RA. 1998. Oral streptococci… products of their environment. J. Dent. Res. 77:445–452 [DOI] [PubMed] [Google Scholar]
- 11. Burne RA, et al. 2011. Functional genomics of Streptococcus mutans, p 185–204 In Kolenbrander PE. (ed), Oral microbial communities: genomic inquires and interspecies communication. ASM Press, Washington, DC [Google Scholar]
- 12. Burne RA, Wen ZT, Chen YM, Penders JEC. 1999. Regulation of expression of the fructan hydrolase gene of Streptococcus mutans GS-5 by induction and carbon catabolite repression. J. Bacteriol. 181:2863–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chatfield CH, Koo H, Quivey RG., Jr 2005. The putative autolysin regulator LytR in Streptococcus mutans plays a role in cell division and is growth-phase regulated. Microbiology 151:625–631 [DOI] [PubMed] [Google Scholar]
- 14. Cieslewicz MJ, Kasper DL, Wang Y, Wessels MR. 2001. Functional analysis in type Ia group B Streptococcus of a cluster of genes involved in extracellular polysaccharide production by diverse species of streptococci. J. Biol. Chem. 276:139–146 [DOI] [PubMed] [Google Scholar]
- 15. Crowley PJ, Brady LJ, Michalek SM, Bleiweis AS. 1999. Virulence of a spaP mutant of Streptococcus mutans in a gnotobiotic rat model. Infect. Immun. 67:1201–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hanson BR, Lowe BA, Neely MN. 2011. Membrane topology and DNA-binding ability of the streptococcal CpsA protein. J. Bacteriol. 193:411–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hubscher J, Luthy L, Berger-Bachi B, Stutzmann Meier P. 2008. Phylogenetic distribution and membrane topology of the LytR-CpsA-Psr protein family. BMC Genomics 9:617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hubscher J, et al. 2009. MsrR contributes to cell surface characteristics and virulence in Staphylococcus aureus. FEMS Microbiol. Lett. 295:251–260 [DOI] [PubMed] [Google Scholar]
- 19. Jenkinson HF, Lamont RJ. 2005. Oral microbial communities in sickness and in health. Trends Microbiol. 13:589–595 [DOI] [PubMed] [Google Scholar]
- 20. Johnsborg O, Havarstein LS. 2009. Pneumococcal LytR, a protein from the LytR-CpsA-Psr family, is essential for normal septum formation in Streptococcus pneumoniae. J. Bacteriol. 191:5859–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kajfasz JK, et al. 2010. Two Spx proteins modulate stress tolerance, survival, and virulence in Streptococcus mutans. J. Bacteriol. 192:2546–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kreth J, Merritt J, Shi W, Qi F. 2005. Co-ordinated bacteriocin production and competence development: a possible mechanism for taking up DNA from neighbouring species. Mol. Microbiol. 57:392–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kreth J, Merritt J, Shi W, Qi F. 2005. Competition and coexistence between Streptococcus mutans and Streptococcus sanguinis in the dental biofilm. J. Bacteriol. 187:7193–7203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kuramitsu HK, He X, Lux R, Anderson MH, Shi W. 2007. Interspecies interactions within oral microbial communities. Microbiol. Mol. Biol. Rev. 71:653–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lamont RJ, Demuth DR, Davis CA, Malamud D, Rosan B. 1991. Salivary-agglutinin-mediated adherence of Streptococcus mutans to early plaque bacteria. Infect. Immun. 59:3446–3450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lazarevic V, Margot P, Soldo B, Karamata D. 1992. Sequencing and analysis of the Bacillus subtilis lytRABC divergon: a regulatory unit encompassing the structural genes of the N-acetylmuramoyl-l-alanine amidase and its modifier. J. Gen. Microbiol. 138:1949–1961 [DOI] [PubMed] [Google Scholar]
- 27. Lee SF, Progulske-Fox A, Bleiweis AS. 1988. Molecular cloning and expression of a Streptococcus mutans major surface protein antigen, P1 (I/II), in Escherichia coli. Infect. Immun. 56:2114–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lemos JA, Burne RA. 2008. A model of efficiency: stress tolerance by Streptococcus mutans. Microbiology 154:3247–3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Loo CY, Corliss DA, Ganeshkumar N. 2000. Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J. Bacteriol. 182:1374–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McBain AJ, Ledder RG, Sreenivasan P, Gilbert P. 2004. Selection for high-level resistance by chronic triclosan exposure is not universal. J. Antimicrob. Chemother. 53:772–777 [DOI] [PubMed] [Google Scholar]
- 31. Nakano K, Fujita K, Nishimura K, Nomura R, Ooshima T. 2005. Contribution of biofilm regulatory protein A of Streptococcus mutans to systemic virulence. Microbes Infect. 7:1246–1255 [DOI] [PubMed] [Google Scholar]
- 32. Oli MW, McArthur WP, Brady LJ. 2006. A whole cell BIAcore assay to evaluate P1-mediated adherence of Streptococcus mutans to human salivary agglutinin and inhibition by specific antibodies. J. Microbiol. Methods 65:503–511 [DOI] [PubMed] [Google Scholar]
- 33. Over B, et al. 2011. LytR-CpsA-Psr proteins in Staphylococcus aureus display partial functional redundancy and the deletion of all three severely impairs septum placement and cell separation. FEMS Microbiol. Lett. 320:142–151 [DOI] [PubMed] [Google Scholar]
- 34. Perez-Casal J, Caparon MG, Scott JR. 1991. Mry, a trans-acting positive regulator of the M protein gene of Streptococcus pyogenes with similarity to the receptor proteins of two-component regulatory systems. J. Bacteriol. 173:2617–2624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Podbielski A, Woischnik M, Leonard BA, Schmidt KH. 1999. Characterization of nra, a global negative regulator gene in group A streptococci. Mol. Microbiol. 31:1051–1064 [DOI] [PubMed] [Google Scholar]
- 36. Rossi J, Bischoff M, Wada A, Berger-Bachi B. 2003. MsrR, a putative cell envelope-associated element involved in Staphylococcus aureus sarA attenuation. Antimicrob. Agents Chemother. 47:2558–2564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 38. Senadheera D, et al. 2009. Inactivation of VicK affects acid production and acid survival of Streptococcus mutans. J. Bacteriol. 191:6415–6424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Steidl R, et al. 2008. Staphylococcus aureus cell wall stress stimulon gene-lacZ fusion strains: potential for use in screening for cell wall-active antimicrobials. Antimicrob. Agents Chemother. 52:2923–2925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Suntharalingam P, Senadheera MD, Mair RW, Levesque CM, Cvitkovitch DG. 2009. The LiaFSR system regulates the cell envelope stress response in Streptococcus mutans. J. Bacteriol. 191:2973–2984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wen TZ, Suntharaligham P, Cvitkovitch DG, Burne RA. 2005. Trigger factor in Streptococcus mutans is involved in stress tolerance, competence development, and biofilm formation. Infect. Immun. 73:219–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wen ZT, Baker HV, Burne RA. 2006. Influence of BrpA on critical virulence attributes of Streptococcus mutans. J. Bacteriol. 188:2983–2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wen ZT, Burne RA. 2002. Functional genomics approach to identifying genes required for biofilm development by Streptococcus mutans. Appl. Environ. Microbiol. 68:1196–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wen ZT, Burne RA. 2004. LuxS-mediated signaling in Streptococcus mutans is involved in regulation of acid and oxidative stress tolerance and biofilm formation. J. Bacteriol. 186:2682–2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wilkins JC, Beighton D, Homer KA. 2003. Effect of acidic pH on expression of surface-associated proteins of Streptococcus oralis. Appl. Environ. Microbiol. 69:5290–5296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zimmer J, Nam Y, Rapoport TA. 2008. Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature 455:936–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.