Abstract
The increasing production of synthetic and natural poly(cis-1,4-isoprene) rubber leads to huge challenges in waste management. Only a few bacteria are known to degrade rubber, and little is known about the mechanism of microbial rubber degradation. The genome of Gordonia polyisoprenivorans strain VH2, which is one of the most effective rubber-degrading bacteria, was sequenced and annotated to elucidate the degradation pathway and other features of this actinomycete. The genome consists of a circular chromosome of 5,669,805 bp and a circular plasmid of 174,494 bp with average GC contents of 67.0% and 65.7%, respectively. It contains 5,110 putative protein-coding sequences, including many candidate genes responsible for rubber degradation and other biotechnically relevant pathways. Furthermore, we detected two homologues of a latex-clearing protein, which is supposed to be a key enzyme in rubber degradation. The deletion of these two genes for the first time revealed clear evidence that latex-clearing protein is essential for the microbial utilization of rubber. Based on the genome sequence, we predict a pathway for the microbial degradation of rubber which is supported by previous and current data on transposon mutagenesis, deletion mutants, applied comparative genomics, and literature search.
INTRODUCTION
The genus Gordonia is currently attracting increasing interest, due mainly to its ability to degrade a wide range of persistent compounds, such as environmental pollutants, xenobiotics, and hardly degradable natural polymers. Besides their bioremediation potential, strains of Gordonia have been isolated because of their anabolic and bioconversion capabilities (4). The diversity of compounds that can be degraded, transformed, and synthesized by Gordonia strains explains the rising interest in biotechnological utilization. Species of the genus have been isolated from many different habitats, such as soil, mangrove rhizospheres, estuaries, industrially contaminated habitats, wastewater, and wastewater treatment bioreactors, as well as from biofilters (1, 4). Some species have also been isolated from clinical specimens, which, in the majority of cases, were associated with infections in immunosuppressed humans (4, 39, 44). Until now, to the best of our knowledge, the only fully sequenced genome of this genus is that of G. bronchialis strain 3410T, which is a human pathogen (42).
Gordonia polyisoprenivorans strains were first isolated from soil of a rubber tree plantation and from fouled water inside a decayed automobile tire because of their capability to degrade natural (NR) and synthetic poly(cis-1,4-isoprene) (IR) rubber (53, 56). This capability is not widespread among bacteria and seems to be limited to Gram-positive actinomycetes (43), with only a few exceptions, like the Gram-negative Xanthomonas sp. strain 35Y (90). In contrast to bacteria that form translucent halos on latex-containing mineral agar (such as many Streptomyces strains), G. polyisoprenivorans belongs to the group of non-clear-zone-forming rubber-degrading bacteria. Members of this adhesively growing group of rubber degraders catabolize rubber more effectively than clear-zone-forming bacteria. Within this group, strains of G. polyisoprenivorans are deemed to be the most potent rubber-degrading bacteria (2, 13, 54, 72). G. polyisoprenivorans strain VH2, whose taxonomic classification has already been reported in detail (2), serves as a model organism for rubber degradation, due to its genetic accessibility (3) and efficient degradation of rubber (13). Up to now, most studies on microbial rubber degradation in the adhesively growing group of rubber degraders have been done with this strain.
A latex-clearing protein (Lcp), which has been considered a key enzyme in clear-zone-forming Streptomyces sp. strain K30 (73), was detected in G. polyisoprenivorans strain VH2 via PCR using degenerated oligonucleotides. Recombinant expression of the strain VH2 lcp gene (lcpVH2) enables non-clear-zone-forming Streptomyces lividans strain TK23 to form clear zones and aldehydes on latex overlay-agar plates. Furthermore, lcpVH2 transcripts in VH2 cultures grown on poly(cis-1,4-isoprene) as the sole carbon and energy source were detected by reverse transcription-PCR analysis, whereas no transcripts were detected after growth in the presence of sodium acetate. Nevertheless, a lcpVH2 disruption did not result in a negative rubber-degrading phenotype (21).
Furthermore, analysis of degradation products and selective inhibition of rubber degradation by the β-oxidation-specific inhibitor acrylic acid led to the conclusion that rubber intermediates are degraded via the β-oxidation pathway in bacteria (18). This assumption was also supported by transposon-induced mutants of G. polyisoprenivorans strain VH2. In the same studies, the involvement of an α-methylacyl-coenzyme A (CoA) racemase (Mcr) during rubber degradation was shown. Mutants with a disruption of the mcrVH2 gene lost the ability to metabolize poly(cis-1,4-isoprene) and related methyl-branched isoprenoid compounds (5, 9). Involvement of an extracellular superoxide dismutase (SodA) in rubber degradation was also demonstrated in this actinomycete (82).
Additionally, strains of G. polyisoprenivorans have been isolated from clinical specimens. There are case reports of septicemia, native valve endocarditis, and pneumonia with associated bacteremia caused by members of this species. All infected patients had fundamental health problems, were immunocompromised, and possessed a Hickman or Broviac catheter. The infections are believed to derive from these rubber-like-material-containing indwelling catheters (37, 45, 92). It is assumed that the ability of G. polyisoprenivorans strains to produce exopolysaccharides and mycolic acids enables them to form biofilms and to adhere to hydrophobic substrates, such as rubber and rubber-like polymer surfactants, e.g., on the intravascular devices (4, 92).
The increasing production of IR and NR for automobile tires and other applications leads to huge challenges in waste management. In order to recycle rubber-containing waste material in terms of new products, great efforts have been made to unravel the rubber degradation pathway. Nevertheless, only a little is known about the mechanism of microbial degradation. We sequenced and annotated the genome of G. polyisoprenivorans strain VH2 to shed light on rubber degradation and other features of this strain. In addition, we did comparative genomic studies, carried out transposon mutagenesis, and generated deletion mutants to obtain more information on poly(cis-1,4-isoprene) catabolism in G. polyisoprenivorans.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Strains of G. polyisoprenivorans and Escherichia coli used in this study are listed in Table 1. Cells of E. coli were cultivated aerobically at 37°C in lysogeny broth (LB) (78). Unless otherwise indicated, strains of G. polyisoprenivorans were cultivated aerobically at 30°C in Standard I (St-I) medium (Merck, Darmstadt, Germany).
Table 1.
List of strains, plasmids, and oligonucleotides used in this study
| Bacterial strains, plasmids, and oligonucleotides | Catalog no., sequence, or descriptiona | Source or referenceb |
|---|---|---|
| Strains | ||
| Gordonia polyisoprenivorans strain VH2 | DSM no. 44266; poly(cis-1,4-isoprene)-degrading wild type | 2 |
| G. polyisoprenivorans strain M71 | lcp1deletion mutant of G. polyisoprenivorans strain VH2; harbors kanamycin resistance cassette | This study |
| G. polyisoprenivorans strain M9 | lcp2 deletion mutant of G. polyisoprenivorans strain VH2; harbors apramycin resistance cassette | This study |
| G. polyisoprenivorans strain C15 | lcp1 and lcp2 deletion mutant of G. polyisoprenivorans strain VH2; harbors apramycin apramycin and kanamycin resistance cassette | This study |
| G. polyisoprenivorans strain TH15 | pMA5096 insertion upstream of lcp2 in deletion mutant of G. polyisoprenivorans strain M71; harbors kanamycin and apramycin resistance cassette | This study |
| G. polyisoprenivorans strain TH5 | pMA5096 insertion in a mammalian cell entry protein-encoding gene (RGOR02853) in lcp2 deletion mutant of G. polyisoprenivorans strain M71; harbors kanamycin and apramycin resistance cassette | This study |
| Escherichia coli XL10 Gold | Tetr Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac Hte [FrproAB lacIqZΔM15 Tn10 (Tetr) Amy Camr] | Stratagene |
| E. coli Top10 | F−araD139 Δ(ara leu)7697 ΔlacX74 galU galK rpsL (Strr) deoR ϕ80dlacZ ΔM15 endA1 nupG recA1 mcrA Δ(mrr hsdRMS mcrBC) | Invitrogen |
| Plasmids | ||
| pMA5096 | Apr; Aprar transposable plasmid comprising transposase Tn5096 | 9 |
| pSKsymΩKm | Apr; pSKsym (derivative of pBluescript SK− with a synthetic symmetrical multiple cloning site) harboring ΩKm in the SmaI site of the multiple cloning site | 65 |
| pBluescript SK− | Aprlac POZ′; T7 and T3 promoter | Stratagene |
| Oligonucleotides | ||
| P49fBamHI | AAAGGATCCCGTTGCACTGGTCGGTCAACC | This study |
| lcp1_A_BamHI | ||
| P938bHindIII | AAAAAGCTTTGTTGTGCTCCCGTCTATCGCC | This study |
| lcp1_B_HindIII | ||
| P2216fHindIII | AAAAAGCTTTGAATGGTGCGTTGCCGGG | This study |
| lcp1_C_HindIII | ||
| P2952bXbaI | AAATCTAGAATAGCGGGTCACGATTTCCACC | This study |
| lcp1_D_XbaI | ||
| lcp2_rew_KO_2 | GCGCCCAGCGAGGCACAGACG | This study |
| lcp2_fw_KO_1 | GTCGCCCGCCTGCACGCC | This study |
| apra_lcp2_3 | CTGACGGGTGCGCTGCTGCCACCGCGTCGCCCATCAA | This study |
| apra_lcp2_4 | TTGATGGGCGACGCGGTGGCAGCAGCGCACCCGTCAG | This study |
| lcp2_apra_5 | CACGGTGGGAATCTCGATGTCCAGCCCGGAGGGGTAAAC | This study |
| apra_lcp2_6 | GTTTACCCCTCCGGGCTGGACATCAGCATCGAGATTCCCACCGTG | This study |
For abbreviations used for genotypes, see reference 14a. DSM, German Collection of Microorganisms and Cell Cultures. Restriction sites used for cloning are underlined.
Stratagene, La Jolla, CA; Invitrogen, Carlsbad, CA.
For growth experiments, synthetic IR (CAS no. 104389-31-3; catalog no. 182141; Aldrich, Steinheim, Germany) was purified. IR (3% [wt/vol]) was dissolved in high-performance liquid chromatography (HPLC)-grade chloroform and subsequently precipitated by the slow addition of absolute ethanol while being stirred. The precipitated material was then dried. After a second dissolution in chloroform, the IR was precipitated by carefully pouring it into boiling water while stirring. The IR particles were then fully dried again. For growth experiments in liquid media, the IR was frozen in liquid nitrogen and then milled to a defined range of grain sizes (between 63 and 500 μm) according to the method of Warneke et al. (93). These grains were used to cultivate G. polyisoprenivorans and its derived mutants in liquid mineral salts medium (MSM) (80) containing 0.2% (wt/vol) IR as the sole carbon and energy source. This medium was inoculated with 5% (vol/vol) precultured cells, which were taken from liquid St-I cultures at the late exponential growth phase. The cells were washed two times with sterile liquid MSM before inoculation. Mutants were also characterized on MSM-IR sandwich agar plates according to the method of Banh et al. (9). Liquid cultures were incubated in Erlenmeyer flasks on a horizontal rotary shaker and agitated at 140 rpm. Solid media were prepared by the addition of 1.5% (wt/vol) agar-agar. For E. coli strains, antibiotics were applied as described by Sambrook et al. (78). For strains of G. polyisoprenivorans, 50 μg/ml of kanamycin and 50 μg/ml of apramycin were used.
DNA extraction and manipulations.
Oligonucleotides used in this study were synthesized by Eurofins MWG (Ebersberg, Germany) and are listed in Table 1. DNA from G. polyisoprenivorans and the derived mutants was isolated using a DNeasy blood and tissue kit (Qiagen, Hilden, Germany). Plasmid DNA was isolated using peqGOLD plasmid minipreparation kit I (Peqlab, Erlangen, Germany), and PCR fragments as well as restriction fragments were purified using a peqGOLD gel extraction kit I (Peqlab, Erlangen, Germany). PCR was done using Phusion high-fidelity DNA polymerase (Fermentas, Burlington, Ontario, Canada) or Biomix (Bioline, London, United Kingdom) according to the manufacturer's protocols. Restriction enzymes and T4 DNA ligase were obtained from Fermentas (Fermentas, Burlington, Ontario, Canada).
Genome sequencing, assembly, and gap closure.
A combination of Sanger sequencing and pyrosequencing was used for whole-genome sequencing of G. polyisoprenivorans strain VH2. Isolated DNA from strain VH2 was used to generate a 454 shotgun library according to the GS Rapid Library protocol (454 Life Sciences, Roche Applied Science, Branford, CT). The 454 DNA library was sequenced with the Genome Sequencer FLX system (454 Life Sciences, Roche Applied Science, Branford, CT) using titanium chemistry. A total of 495,486 shotgun reads were generated and assembled de novo into 92 large contigs (>500 bp) using Roche Newbler assembler software 2.0.1 FLX (454 Life Sciences, Roche Applied Science, Branford, CT). For the Sanger sequencing approach, a fosmid library was constructed according to the CopyControl fosmid library production kit manual (Epicentre Biotechnologies, Madison, WI, USA). Insert ends of 576 recombinant fosmids were sequenced with ABI 3730xl automated DNA sequencers (Life Technologies, Darmstadt, Germany), processed with Phred, and assembled using Phrap (http://www.phrap.org). Sequence editing was done by using GAP4 as part of the Staden software package (85), and final gap closure was performed by PCR and primer walking using a Bio-X-Act kit (Bioline, London, United Kingdom) and the 5 Prime Extender polymerase system (5 Prime GmbH, Hamburg, Germany) as described by the manufacturers.
Gene prediction, annotation, and analysis and comparative genomics.
Coding sequences (CDS) were predicted with YACOP (88), using the open reading frame (ORF) finders Glimmer, Critica, and Z-Curve. All CDS were manually curated and verified by using criteria such as the presence of a ribosome binding site, GC frame plot analysis, and comparison with sequences in the publicly available databases Swiss-Prot, Trembl, GenBank, COG, KEGG, ProDom, Pfam, Tigrfam, and Prosite, employing the annotation software tools ERGO (64) and Artemis (75). Prediction of twin arginine translocation (Tat) signal peptides was carried out using TatP 1.0 (11). For genome comparisons, the BiBag software tool (Bidirectional BLAST for the Identification of Bacterial Pan and Core Genomes; Göttingen Genomics Laboratory, Germany) was applied. The predicted proteome was clustered using the NCBI BLASTClust program. For this purpose, a minimum of 30% identity and 70% length coverage was chosen.
Plasmid and mutant construction.
To delete the first Lcp gene (lcp1), we amplified the flanking regions upstream and downstream of the gene, using the oligonucleotides P49fBamHI and P938bHindIII as well as P2216fHindIII and P2952bXbaI (Table 1), respectively, yielding fragments of 864 bp and 731 bp. The resulting fragments were purified and digested using BamHI/HindIII and HindIII/XbaI, respectively. A kanamycin resistance cassette (KmR) was excised from vector pSKsymΩKm (65) using the endonuclease HindIII and was subsequently purified. All three fragments were ligated into a BamHI/XbaI-digested pBluescript SK− (Stratagene, La Jolla, CA, USA) cloning vector. The resulting plasmids were transferred into E. coli cells (78), and selection was done on solid LB media containing ampicillin and kanamycin. The plasmid DNA was isolated and verified via digestion as well as sequencing. The linear construct (flankA/KmR/flankB) was used for transformation of electrocompetent G. polyisoprenivorans strain VH2 according to the method of Arenskötter et al. (3). The resulting transformants were plated onto solid St-I medium with kanamycin.
For deletion of the second Lcp gene (lcp2), a linear DNA deletion fragment was constructed via fusion PCR. For this process, total genomic DNA of G. polyisoprenivorans strain VH2 was used to amplify the flanking regions of lcp2. A flanking region upstream of lcp2 with a size of 914 bp was amplified using oligonucleotides lcp2_fw_KO_1 and apra_lcp2_3 (Table 1). A second fragment (880 bp) downstream of lcp2 was amplified using oligonucleotides apra_lcp2_6 and lcp2_rew_KO_2 (Table 1). An apramycin resistance cassette (ApraR) was amplified from the Tn5096 transposon-harboring vector pMA5096 (9) with oligonucleotides apra_lcp2_4 and lcp2_apra_5 (Table 1). Fusion PCR was carried out according to the method of Kuwayama et al. (52) using the purified fragments mentioned above and the oligonucleotides lcp2_fw_KO_1 and lcp2_rew_KO_2. The deletion fragment (flankA/ApraR/flankB) was purified and sequenced and was subsequently used for transformation of electrocompetent G. polyisoprenivorans strain VH2 as well as G. polyisoprenivorans strain VH2 Δlcp1ΩKmR according to the method of Arenskötter et al. (3). Transformants were plated onto solid St-I medium with either apramycin or apramycin and kanamycin, respectively.
All transformants of G. polyisoprenivorans were first screened via colony PCR using internal oligonucleotides of lcp1 and lcp2 as well as external oligonucleotides. The DNA of verified mutants was isolated, and the correct deletions were confirmed via PCR with oligonucleotides annealing outside the flanks as well as via sequencing.
Transposon mutagenesis was done using plasmid pMA5096 harboring transposon Tn5096. Screening and mapping of transposon insertions were carried out as described elsewhere (9).
Nucleotide sequence accession numbers.
The sequence of the complete and annotated genome of G. polyisoprenivorans strain VH2 is available under NCBI accession numbers CP003119 for the chromosome and CP003120 for the plasmid.
RESULTS AND DISCUSSION
General features.
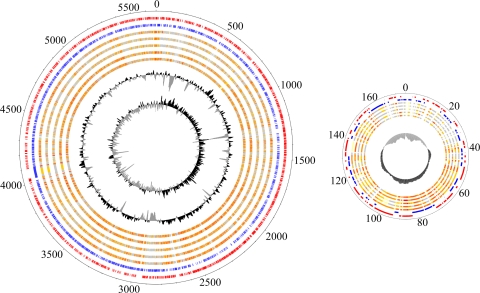
The genome of the actinomycete G. polyisoprenivorans strain VH2 consists of a chromosome of 5,669,805 bp and a plasmid (p174) of 174,494 bp with average GC contents of 67.0% and 65.7%, respectively (Table 2). Unlike the chromosomes in the genomes of other actinomycetes, such as Rhodococcus jostii strain RHA1 (59) and many strains of the genus Streptomyces (81), the chromosome is of a circular topology (Fig. 1). This topology was also described for the related actinomycetes G. bronchialis strain 3410T (42) and Nocardia farcinica strain IFM 10152 (41). The chromosome is predicted to contain 4,945 protein-coding sequences, whereas the plasmid is predicted to harbor 165. We could assign a putative function to 3,717 (72.7%) of the ORFs. In addition, the genome harbors 20 pseudogenes, 49 tRNA genes, and three copies of rRNA operons.
Table 2.
General features of the genome of G. polyisoprenivorans strain VH2
| Characteristic | Value |
|---|---|
| Size of genome (bp) | 5,844,299 |
| Size of chromosome (bp) | 5,669,805 |
| Size of plasmid (bp) | 174,494 |
| GC content (%) | 66.98 |
| No. of predicted protein-coding genes | 5,110 |
| No. of predicted proteins with putative function (%) | 3,717 (72.7) |
| No. of predicted proteins of unknown function (%) | 1,393 (27.3) |
| Coding density (%) | 89.56 |
| No. of RNA genes | 58 |
| No. of tRNA genes | 49 |
| No. of rRNA genes | 9 |
| No. of miscellaneous RNA genes | 1 |
| No. of pseudogenes | 20 |
Fig 1.
Visualization of the genome of G. polyisoprenivorans strain VH2 and its comparison to genomes of other rubber- and non-rubber-degrading bacteria. The chromosome is shown on the left and plasmid p174 on the right. Circle 1 shows positions in the chromosome and plasmid, respectively (in kb); circles 2 and 3 show proteins encoded on leading (red) and lagging (blue) strands of strain VH2; circle 4 shows the proteome of IR-degrading Nocardia farcinica strain IFM 10152; circle 5 shows the proteome of IR-degrading Actinosynnema mirum strain DSM 43827; circle 6 shows the proteome of IR-degrading Streptomyces flavogriseus strain ATCC 33331; circle 7 shows the proteome of non-IR-degrading Rhodococcus jostii strain RHA1; circle 8 shows the proteome of non-IR-degrading Mycobacterium smegmatis strain mc2155. In circles 4 to 8, similarities to proteins of strain VH2 are plotted in different colors (the darker the color, the higher the similarity). Circle 9 is a GC plot; circle 10 (chromosome only) is a GC skew. Comparative genomics was done using BiBag software.
For paralogue identification, the predicted proteome of strain VH2 was clustered. One thousand six hundred sixty-eight (32.6%) of the CDS were clustered into 461 paralogous families ranging from 2 to 78 members per family. We also compared the total numbers of proteins which were found to be part of the strikingly paralogous families in VH2 to the total numbers of these protein families found in the genomes of the pathogenic strains G. bronchialis DSM 43247 (42) and Mycobacterium tuberculosis H37Rv (26) as well as the harmless and typical soil bacterium Streptomyces avermitilis strain MA-4680 (40) (Table 3). The genome of VH2 harbors a high number of proteins associated with transport, including 78 members of the major facilitator superfamily (MFS) and 89 members of the ATP binding cassette (ABC) transporter. These numbers are much higher than those for the genomes of pathogenic bacteria, such as M. tuberculosis and G. bronchialis, and are more typical for soil bacteria, such as S. avermitilis. Furthermore, the genome also encodes many members of regulator families, such as LysR, GntR, IclR, and TetR. The high number of such transporters and regulators is important for the bacterium to adapt to various soil environments. On the other hand, the genome also possesses a large number of ORFs coding for proteins that are important for the intracellular growth of pathogenic bacteria like M. tuberculosis and G. bronchialis, such as short-chain dehydrogenases/reductases, acyl-CoA dehydrogenases, enoyl-CoA hydratase/isomerase family proteins, and esterase family proteins. The large number of proteins putatively involved in β-oxidation might be an explanation for the potent rubber-degrading phenotype of G. polyisoprenivorans and is discussed below. The composition of striking protein families found in the genome of strain VH2 is similar to that found in the genome of N. farcinica, which was isolated from a patient (41) and also shows adhesive growth on rubber.
Table 3.
Selection of protein families in G. polyisoprenivorans strain VH2 and homologous genes in related genomes
| Family | Rankinga | Incidence of homologues/percentage in genome of indicated bacterium |
||||
|---|---|---|---|---|---|---|
| G. polyisoprenivorans | G. bronchialis | S. avermitilis | M. tuberculosis | N. farcinica | ||
| Short-chain dehydrogenase/reductase | 1 | 86/1.68 | 58/1.24 | 99/1.29 | 64/1.6 | 95/1.6 |
| ABC transporter, ATP binding protein | 2 | 89/1.74 | 71/1.51 | 153/1.99 | 39/0.97 | 95/1.6 |
| Major facilitator superfamily | 3 | 78/1.53 | 49/1.04 | 92/1.2 | 20/0.5 | 64/1.08 |
| Acyl-CoA dehydrogenase | 4 | 47/0.92 | 41/0.87 | 31/0.4 | 32/0.8 | 48/0.81 |
| Aldehyde dehydrogenase | 6 | 28/0.55 | 17/0.36 | 25/0.33 | 10/0.25 | 24/0.4 |
| Enoyl-CoA reductase/hydratase | 7 | 26/0.51 | 26/0.55 | 19/0.25 | 24/0.6 | 28/0.47 |
| Two-component response regulator | 8 | 29/0.57 | 27/0.57 | 79/1.03 | 15/0.37 | 34/0.57 |
| Transcriptional regulator, LysR family | 10 | 27/0.53 | 16/0.34 | 32/0.42 | 4/0.1 | 22/0.37 |
| Transcriptional regulator, GntR family | 11 | 26/0.51 | 18/0.38 | 50/0.65 | 7/0.18 | 24/0.4 |
| Putative esterase | 12 | 20/0.39 | 11/0.23 | 1/0.01 | 8/0.2 | 15/0.25 |
| Thiolase | 17 | 13/0.25 | 10/0.21 | 12/0.16 | 10/0.25 | 13/0.22 |
| Transcriptional regulator, IclR family | 18 | 13/0.25 | 4/0.09 | 12/0.16 | 3/0.08 | 17/0.29 |
| Cytochrome P450 | 21 | 19/0.37 | 15/0.32 | 36/0.47 | 20/0.5 | 26/0.44 |
| YrbE protein | 22 | 10/0.2 | 8/0.17 | 2/0.03 | 8/0.2 | 12/0.2 |
| Transcriptional regulator, TetR family | 29 | 112/2.19 | 89/1.9 | 118/1.54 | 54/1.35 | 159/2.68 |
Ranking (according to size) of paralogous families in the genome of G. polyisoprenivorans strain VH2.
Virulence.
The genome of strain VH2 harbors five copies of the mammalian cell entry protein (mce1-5 locus), which are thought to be involved in virulence in M. tuberculosis. The latter harbors four such clusters in the genome. Several studies with M. tuberculosis indicated that some Mce clusters might play a role in the interaction with host cells as well as in uptake of the pathogen into the host (6, 25, 32). These proteins might therefore be one reason why strain VH2 could also be isolated from clinical specimens. However, the results of these studies are sometimes contradictory, and Mce clusters can also be found in the genomes of free-living, nonpathogenic bacteria (24). The role of one Mce cluster in G. polyisoprenivorans strain VH2 is discussed below in more detail. Furthermore, candidate genes were detected that might influence the virulence of strain VH2 by playing a role in forming surface-active compounds or in detoxification of reactive oxygen species (ROS) during oxidative attack of the host. A putative participation of these genes in rubber degradation is discussed below. The genome harbors some other ORFs with similarities to genes thought to be involved in the pathogenesis of M. tuberculosis and other actinobacteria. These ORFs encode, inter alia, putative secreted NlpC/P60 family proteins (GPOL_c16710, GPOL_c23360, GPOL_c28070, GPOL_c28080) with similarity to an invasion-associated protein of M. tuberculosis and PPE family proteins (GPOL_c32930, GPOL_c35060) which are thought to be of immunological importance in strains of Mycobacterium (26). ORFs with similarity to heparin binding hemagglutinin (GPOL_c09400, GPOL_c09490), which is required for mycobacterial extrapulmonary dissemination (66), are also located in the genome. The latter might be one explanation for why G. polyisoprenivorans was also shown to cause pneumonia in a patient (37). A phospholipase C (GPOL_c14800) involved in the pathogenesis of several bacteria, including M. tuberculosis (70), is also encoded in the genome of strain VH2. However, since all case reports of infections due to this species have in common that infected patients had fundamental health problems and were immunocompromised (37, 45, 92), G. polyisoprenivorans is clearly less pathogenic than M. tuberculosis.
Latex-clearing protein.
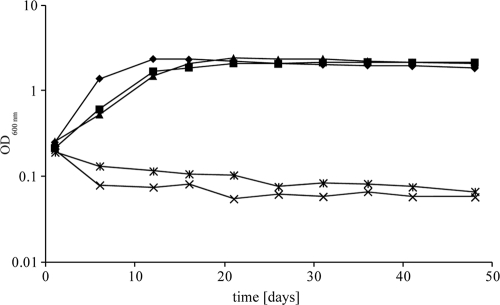
G. polyisoprenivorans strain VH2 was investigated mainly because of its ability to degrade rubber very effectively (2, 13, 54, 72). In recent studies, we identified a gene (GPOL_c48310) coding for a latex-clearing protein (lcp1VH2) in the genome of G. polyisoprenivorans strain VH2 by using degenerated primers. Heterologous expression of lcp1VH2 in the non-clear-zone-forming S. lividans strain TK23 enabled this bacterium to form clear zones and aldehydes on latex overlay-agar plates. Furthermore, after incubation of liquid medium with cells of this recombinant strain, the remaining polyisoprenoid molecules showed lower concentrations and molecular weights. Using reverse transcription-PCR analysis, we demonstrated that this gene is transcribed in the presence of rubber but not in the presence of sodium acetate (21). However, after this gene was deleted, the resulting mutant (G. polyisoprenivorans strain M71) showed growth behavior only slightly different from that of the wild type, as it grew only a little slower but reached the same cell density (Fig. 2). Therefore, we suspected that there is at least one or more homologue of lcp in the genome.
Fig 2.
Growth of G. polyisoprenivorans strain VH2 and lcp deletion mutants cultivated in mineral salt medium with 0.2% IR at 30°C (semilogarithmic plot; triple determination of growth). ♦, G. polyisoprenivorans strain VH2 (wild type); ■, G. polyisoprenivorans Δlcp1ΩKm (mutant strain M71); ▲, G. polyisoprenivorans Δlcp2ΩApra (mutant strain M9); ×, G. polyisoprenivorans Δlcp1ΩKm Δlcp2ΩApra (mutant strain C15);  , G. polyisoprenivorans Δlcp1ΩKm harboring pMA5096 upstream of lcp2 (mutant strain TH15).
, G. polyisoprenivorans Δlcp1ΩKm harboring pMA5096 upstream of lcp2 (mutant strain TH15).
In order to detect a putative second homologue of lcp, we carried out transposon mutagenesis in the G. polyisoprenivorans mutant strain M71. At the same time, we started to sequence the genome of the wild type. Almost simultaneously, we detected the second lcp (lcp2VH2) homologue in the genome. We identified a transposon-induced mutant, which had lost its ability to grow on IR as the sole source of carbon and energy (Fig. 2). The insertion occurred upstream of lcp2VH2, which was also found in the genome sequence (GPOL_174p00150) using BLAST search. No further lcp was detected in the genome. After deletion of lcp2VH2 in the lcp1VH2 deletion mutant strain M71, the resulting double mutant G. polyisoprenivorans strain C15 lost its ability to grow with IR as the sole carbon source (Fig. 2). A lcp2VH2 single-deletion mutant (G. polyisoprenivorans strain M9), which was also created, exhibited growth behavior on IR very similar to that of the lcp1VH2 single-deletion mutant strain M71. These observations demonstrated that either of the two Lcp-encoding genes allows adhesive growth of G. polyisoprenivorans strain VH2 on rubber. Since during rubber degradation a decrease in the number of cis-1,4 double bonds and the occurrence of ketone and aldehyde groups were observed when cells of strain VH2 were grown on NR latex gloves, an oxidative cleavage of the polymer at the cis-1,4 double bond is adopted (54). This kind of cleavage was also postulated for other adhesively and non-adhesively growing bacteria (18, 89). Since heterologous expression of lcp1VH2 enables S. lividans strain TK23 to form clear zones and aldehydes on latex overlay-agar plates, the remaining polyisoprenoid molecules are of lower polymer concentration and molecular weight (21), and a deletion of both Lcp-encoding genes of strain VH2 resulted in an IR-negative phenotype, we postulate that the lcp genes are responsible for this oxidative cleavage of rubber. The Lcp gene products comprise a Tat signal peptide at the N terminus. This postulate fits well with observations that were made for non-clear-zone-forming S. lividans strain TK23 heterologously expressing lcp1VH2 (21), and one could assume that Lcp is secreted by strain VH2. Furthermore, secretion of Lcp via the Tat pathway was already demonstrated in the clear-zone-forming Streptomyces sp. strain K30 (96).
The amino acid sequences of Lcp are highly conserved. Lcp1VH2 and Lcp2VH2 share over 60% identity to each other as well as to other already characterized Lcp proteins of adhesively growing rubber-degrading strains. Lcp of Streptomyces sp. strain K30 showed approximately 50% amino acid sequence identity to the Lcp sequences of strain VH2. Sequence similarity searches revealed that homologues are located in the genomes of strains belonging to the genus Gordonia, such as strains of G. westfalica, G. alkanivorans, and G. neofelifaecis, as well as in the genomes of other actinomycetes belonging to genera like Nocardia, Actinosynnema, Thermomonospora, Micromonospora, and Streptomyces. Only a few genomes harbor more than one lcp homologue. All these strains have in common that they grow in environments with access to numerous terpene-containing compounds, and many of them were studied because of their degradative properties (4, 30, 51, 55, 57). To the best of our knowledge, every actinomycete harboring one or more lcp homologue can also grow on rubber as the sole source of carbon and energy (21, 38, 73; unpublished data).
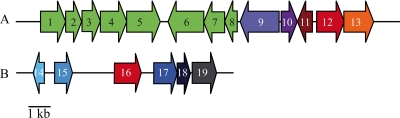
In the genome of G. polyisoprenivorans strain VH2, the gene encoding Lcp1VH2 is located downstream of a putative transcriptional regulator of the TetR family (Fig. 3). Such regulator genes are located next to an lcp gene in almost all genomes harboring one or more sequences homologous to lcp and therefore seem to play an important role in the regulation of lcp transcription. In addition, previous observations have shown that lcp1VH2 transcription is induced when strain VH2 is cultivated in the presence of poly(cis-1,4-isoprene) but not in the presence of acetate (21). This indicates that this putative regulator might function as a repressor for the expression of lcp and that it is inactivated directly or indirectly through molecules occurring during growth on rubber. Interestingly, a cluster of genes involved in the biosynthesis of terpenoids and carotenoids is located upstream of lcp1VH2 (Fig. 3). It might therefore be possible that Lcp also plays a role in the degradation of other extracellular poly- or oligoterpenes or terpene-containing compounds, like carotenoids, quinones with isoprenoid side chains, or steroids. The proximity of sequences homologous to lcp and genes involved in the biosynthesis of terpenoids and carotenoids is also detectable in the genomes of other actinomycetes, such as Streptosporangium roseum strain DSM 43021 and Actinosynnema mirum strain DSM 43827. Genes putatively involved in DNA replication, recombination, or reparation are located downstream of lcp1VH2. As shown in Fig. 3, the second lcp gene, detectable on the plasmid, is adjacent to an ORF encoding a hypothetical alpha/beta hydrolase-like protein (upstream) and an ORF encoding a putative lipoprotein of the ApbE family (downstream). Neither can be directly linked to microbial rubber degradation here.
Fig 3.
Location of lcp1, lcp2, and adjacent genes. (A) lcp1 and surrounding area (located on chromosome): 1, putative geranylgeranyl pyrophosphate synthase; 2, putative monooxygenase; 3, hypothetical protein; 4, glycosyl transferase family 2; 5, phytoene desaturase; 6, phytoene desaturase; 7, phytoene synthase; 8, isopentenyl-diphosphate delta-isomerase; 9, hypothetical protein; 10, putative protease; 11, putative transcriptional regulator, TetR family; 12, latex-clearing protein 1; 13, CinA domain-containing protein. (B) lcp2 and surrounding area (located on plasmid): 14, MarR family transcriptional regulator; 15, hypothetical protein; 16, latex-clearing protein 2; 17, ApbE family lipoprotein; 18, ferric reductase domain-containing protein; 19, NADH dehydrogenase.
Oxidation of aldehyde intermediates.
The low-molecular-weight aldehyde intermediates resulting from the oxidative cleavage of IR are further oxidized. In Streptomyces sp. strain K30, this step is thought to be catalyzed via a heterodimeric molybdenum-dependent hydroxylase encoded by oxiAB, which is located downstream of lcp. It enables the strain to oxidize these aldehyde intermediates (73), and it is transcribed during rubber degradation but not during growth on glucose (96). G. polyisoprenivorans does not possess oxiA and oxiB homologues. However, this heterodimeric hydroxylase is also missing in the genomes of other known rubber-degrading strains, no matter if they are adhesively growing (e.g., N. farcinica strain IFM 10152) or clear-zone forming (e.g., S. coelicolor strain A3 [2]). Which genes encode the enzyme(s) for this step cannot be clarified here, since the genome harbors a large number of putative candidates and experimental data must still be unraveled.
Degradation of rubber via the β-oxidation pathway.
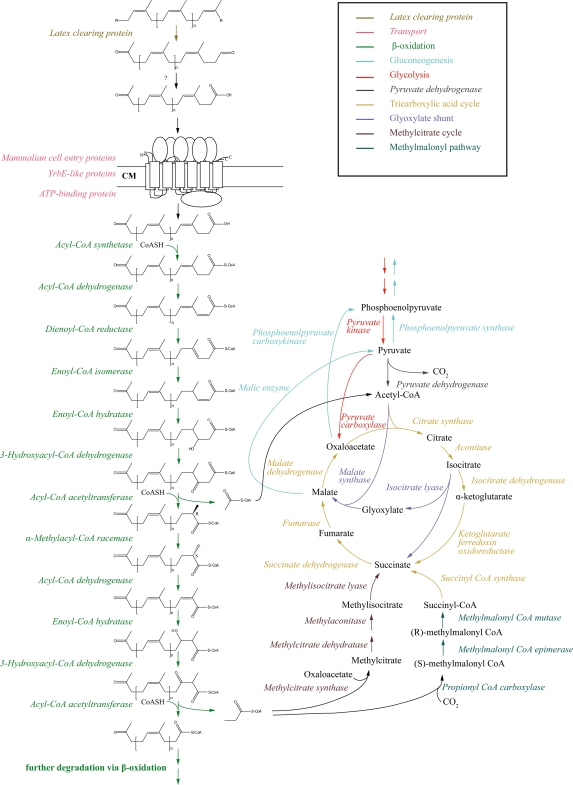
The resulting organic acids are most likely metabolized via β-oxidation for several reasons: (i) the occurrence of intermediates identified during rubber degradation by S. coelicolor A1 are explainable through degradation via β-oxidation; (ii) inhibition by acrylic acid, a β-oxidation-specific inhibitor, was observed in strain A1 (18) and other rubber-degrading strains (72); and (iii) β-oxidation is also involved in microbial degradation of branched-chain alkanes, such as isooctane (84), pristane (62), phytane (83), squalane, and squalene. The latter is also known to be metabolized by strain VH2 (14). In our proposed rubber degradation pathway, an acyl-CoA synthetase converts the acid to an acyl-CoA thioester (Fig. 4). In the genome of strain VH2, 37 candidate genes for this step were identified (Table 4). The resulting product is further catabolized by an acyl-CoA dehydrogenase, whereof we found 47 homologous ORFs in the genome sequence. Next, we propose two steps analogous to the degradation of polyunsaturated fatty acids in the rubber degradation pathway. First, the 2,4-dienoyl-CoA reductase (GPOL_c19120) catalyzes the reduction of double bonds at even-numbered positions, followed by an isomerization step. The latter reaction could be catalyzed by an enoyl-CoA isomerase. Within the genome of strain VH2, we found 26 candidates belonging to the enoyl-CoA hydratase/isomerase superfamily, which could also be involved in the following hydration step. In addition, we identified seven homologues of 3-hydroxyacyl-CoA dehydrogenases that could be responsible for the conversion of the hydroxyl derivate into the keto form. The last step of the first β-oxidation cycle is then catalyzed by the thiolase, for which we identified 12 homologous genes in the genome. In addition, the FadA/FadB β-oxidation complex (GPOL_c05460/GPOL_c05470) is encoded within the genome. The next step is most likely catalyzed by one of the two α-methylacyl-CoA racemases (GPOL_c25180, GPOL_c36450) located in the genome. In previous studies, one Mcr (GPOL_c36450) was found to be essential for strain VH2, since a disruption led to a total loss of the ability to utilize rubber for growth (5). This Mcr catalyzes the conversion of the (R)- into the (S)-stereoisomer. Only the (S)-isomer can serve as a substrate for the acyl-CoA dehydrogenase in the next β-oxidation cycle. Such an involvement of an Mcr was also suggested for the β-oxidation of methyl-branched alkanes in Mycobacterium sp. strain P101 (77). We propose successive cycles of β-oxidation, with propionyl-CoA and acetyl-CoA being consecutively released, as was also suggested, for example, for pristane (69). This considerable set of genes, which are putatively involved in β-oxidation, could be an explanation for why transposon-induced mutants, which are totally defective in rubber utilization and show transposon insertion in genes belonging to this pathway at the same time, did not occur. Other paralogous genes might be able to partially or even fully restore the function of a disrupted gene. However, in recent studies we identified mutants exhibiting impaired growth during cultivation on poly(cis-1,4-isoprene) as the sole carbon and energy source and confirming our proposed rubber degradation pathway via β-oxidation. In the transposon-induced mutant D6 (5), the insertion was mapped within the intergenic region of two genes encoding a thiolase (GPOL_c25440) and a short-chain dehydrogenase/reductase family protein (GPOL_c25450). Downstream of the latter gene, a putative acyl-CoA synthetase (GPOL_c25460) is located. The mutant shows a leaky phenotype during growth on liquid MSM with poly(cis-1,4-isoprene) as the sole carbon and energy source. In mutant strain C22 (5), the insertion of the transposon-harboring vector pMA5096 occurred in a putative acyl-CoA ligase (GPOL_c48500). Interestingly, sequencing and computational analyses revealed that this gene seems to be a pseudogene. However, the mutant shows reduced growth in liquid MSM supplemented with IR (5). Therefore, whether this ORF really represents a pseudogene must be questioned and experimentally investigated. In the transposon-induced mutant strain I45 (5), the insertion was mapped in a region putatively involved in the degradation of steroids like cholesterol (see below). The insertion of the transposon was detected upstream of an ORF (GPOL_c42810) which has some similarity to a steroid delta-isomerase. Upstream of this gene, we identified a cytochrome P450 monooxygenase, which is essential for R. jostii strain RHA1 to grow on 3-hydroxysterols, such as cholesterol (74). However, an acetyl-CoA acetyltransferase (GPOL_c42830) is located next to these genes. The expression of this gene could be affected due to polar effects and could therefore show impaired growth on MSM-IR sandwich agar plates (5).
Fig 4.
Predicted pathway of poly(cis-1,4-isoprene) rubber degradation by G. polyisoprenivorans strain VH2 based on annotated genome, transposon-induced mutants, comparative genomics, and literature search. The predicted transporter is visualized via a hypothetical arrangement based on that of Casali and Riley (24). CM, cytoplasmatic membrane; CoA and CoASH, coenzyme A.
Table 4.
ORFs in the genome of G. polyisoprenivorans strain VH2 potentially involved in degradation of rubber via the β-oxidation cycle
| Enzyme | Total no. | Locus tag(s) in genome of strain VH2a |
|---|---|---|
| Acyl-CoA synthetase | 37 | GPOL_c00650, GPOL_c00810, GPOL_c01490, GPOL_c02290, GPOL_c08760, GPOL_c08790, GPOL_c08990, GPOL_c10590, GPOL_c12010, GPOL_c15600, GPOL_c18870, GPOL_c23250, GPOL_c24340, GPOL_c24700, GPOL_c25330, GPOL_c25460, GPOL_c26980, GPOL_c28040, GPOL_c28930, GPOL_c31080, GPOL_c32520, GPOL_c36970, GPOL_c37880, GPOL_c38080, GPOL_c39560, GPOL_c40340, GPOL_c41480, GPOL_c42690, GPOL_c42730, GPOL_c42980, GPOL_c43120, GPOL_c43130, GPOL_c43210, GPOL_c47100, GPOL_c48960, GPOL_c48500, GPOL_c49330 |
| Acyl-CoA dehydrogenase | 47 | GPOL_c10560, GPOL_c39580, GPOL_c05310, GPOL_c06060, GPOL_c08610, GPOL_c08620, GPOL_c10630, GPOL_c11980, GPOL_c01410, GPOL_c14360, GPOL_c14370, GPOL_c14400, GPOL_c15560, GPOL_c16950, GPOL_c19020. GPOL_c21680, GPOL_c25260, GPOL_c25340, GPOL_c25500, GPOL_c31540, GPOL_c31730, GPOL_c31740, GPOL_c33130, GPOL_c33140, GPOL_c34390, GPOL_c36890, GPOL_c36920, GPOL_c37480, GPOL_c39570, GPOL_c40380, GPOL_c40390, GPOL_c40950, GPOL_c40960, GPOL_c40970, GPOL_c41350, GPOL_c41360, GPOL_c41520, GPOL_c41540, GPOL_c42440, GPOL_c42670, GPOL_c43200, GPOL_c45150, GPOL_c45280, GPOL_c45460, GPOL_c48400, GPOL_c49510, GPOL_c49930 |
| 2,4-Dienoyl-CoA reductase | 1 | GPOL_c19120 |
| Enoyl-CoA hydratase/isomerase | 26 | GPOL_c07190, GPOL_c09320, GPOL_c13550, GPOL_c16770, GPOL_c17840, GPOL_c25190, GPOL_c25210, GPOL_c25240, GPOL_c25250, GPOL_c28940, GPOL_c29800, GPOL_c30630, GPOL_c31570, GPOL_c36880, GPOL_c39120, GPOL_c40270, GPOL_c40940, GPOL_c41700, GPOL_c41720, GPOL_c41890, GPOL_c42740, GPOL_c43030, GPOL_c45480, GPOL_c46770, GPOL_c48490, GPOL_174p01070 |
| 3-Hydroxyacyl-CoA dehydrogenase | 7 | GPOL_c03320, GPOL_c06030, GPOL_c09390, GPOL_c24710, GPOL_c25200, GPOL_c40350, GPOL_c41710 |
| Thiolase | 12 | GPOL_c00880, GPOL_c05990, GPOL_c14950, GPOL_c18410, GPOL_c25220, GPOL_c40360, GPOL_c42830, GPOL_c43180, GPOL_c45290, GPOL_c38960, GPOL_c40330, GPOL_c42790 |
| α-Methylacyl-CoA racemase | 1 | GPOL_c36450 |
Underlining indicates evidence that the gene is involved in rubber degradation was detected in transposon-induced mutants showing impaired or no growth on rubber (5, 9); bold indicates that homologues can be found in the genomes of all reviewed rubber-degrading bacteria; italics indicate that disruption of α-methylacyl-CoA racemase resulted in a non-rubber-degrading phenotype (5).
In order to reduce the number of genes and find a minimal set of genes putatively involved in rubber degradation, we compared fully sequenced genomes of rubber-degrading strains using the BiBag software tool (Fig. 1). For this comparison, we used the genomes of adhesively growing N. farcinica strain IFM 10152, clear-zone-forming (unpublished data) A. mirum strain DSM 43827, and S. flavogriseus strain ATCC 33331. All these genomes harbor the genes carried on GPOL_c26980 and GPOL_c49330. These genes could be involved in activation of the molecules for further degradation via β-oxidation. The number of candidate genes involved in the following oxidation step was reduced to seven (GPOL_c06060, GPOL_c10630, GPOL_c11980, GPOL_c15560, GPOL_c36890, GPOL_c45280, GPOL_c45460). A 2,4-dienoyl-CoA reductase is present in all these genomes. Genes involved in the following steps of isomerization and hydration were reduced to five candidates (GPOL_c09320, GPOL_c30630, GPOL_c36880, GPOL_c41700, GPOL_174p01070). Furthermore, one gene probably involved in the next oxidation step (GPOL_c09390) and three candidates involved in thiolysis (GPOL_c05990, GPOL_c14950, GPOL_c18410) could be identified by comparing the genomes of the rubber-degrading strains. Genes encoding the FadA/FadB β-oxidation complex as well as Mcr are also located in all of these genomes. Interestingly, most of these genes can also be identified in the genomes of some non-rubber-degrading strains, such as R. jostii strain RHA1 and Mycobacterium smegmatis strain mc2155. In comparison to the proposed rubber degradation pathway shown in Fig. 4, both bacteria miss lcp homologues for the initial oxidative cleavage step only. This indicates that these genes encoding enzymes for β-oxidation might also be involved in the degradation of other, perhaps methyl-branched, isoprenoid compounds. A summary of ORFs putatively involved in the degradation of rubber via the β-oxidation cycle in strain VH2 is given in Table 4.
Central metabolism with respect to rubber degradation.
The genome sequence of G. polyisoprenivorans encodes central metabolic pathways, such as the Embden-Meyerhof-Parnas pathway, the pentose phosphate pathway, and the tricarboxylic acid cycle (TCC), for the enzymes of gluconeogenesis as well as the glyoxylate shunt and other anaplerotic enzymes. The glyoxylate shunt, for example, is, in addition to the β-oxidation cycle, required for growth on fatty acids, since it is essential for carbon anaplerosis in the TCC (47). Because reactions of the β-oxidation cycle of fatty acids are important during the catabolism of rubber (Fig. 4), the glyoxylate shunt should also play an essential role during utilization of rubber. The proposed pathway releases propionyl-CoA, which has to be further metabolized, in addition to acetyl-CoA. Strain VH2 grows on propionate as the sole source of carbon and energy (5). Its genome possesses genes encoding a methylcitrate synthase (GPOL_c17040), an aconitate hydratase (GPOL_c23340), a methylcitrate dehydratase (GPOL_c17020), and a methylisocitrate lyase (GPOL_c17030). It also exhibits genetic loci coding for biotin-dependent carboxylases. We detected two such carboxylase systems where the α- and β-subunit are located next to each other (GPOL_c11960/GPOL_c11970 and GPOL_c36900/GPOL_c36910), one α-subunit (GPOL_c17230), and four β-subunits (GPOL_c04190, GPOL_c17300, GPOL_c28880, GPOL_c38300) without a counterpart in the neighborhood. Some of the gene products probably play a role in generating precursors for fatty acid synthesis, and some are putatively involved in the methylmalonyl pathway. The carboxylase systems where the genes encoding the subunits are adjacent might be involved in the latter pathway, since they are located next to genes whose products are involved in the degradation of fatty acids (and perhaps rubber). Furthermore, genes coding for a methylmalonyl-CoA epimerase (GPOL_c18400) as well as a methylmalonyl-CoA mutase (GPOL_c23510/GPOL_c23520) are located in the genome. The methylmalonyl-CoA mutase-catalyzed step of the methylmalonyl pathway is vitamin B12 dependent (58). Interestingly, in recent studies we obtained transposon-induced mutants that were impaired in growth on rubber as the sole source of carbon and energy and in which the insertions were mapped in the neighborhood of genes putatively involved in cobalamin biosynthesis (9). Therefore, it might be possible that the methylmalonyl pathway plays an important role in rubber degradation. Remarkably, the methylmalonyl pathway is also present in rubber-degrading strains of N. farcinica, S. coelicolor, A. mirum, and S. flavogriseus, but the methylcitrate cycle is missing in these bacteria.
Transport of rubber degradation products into the cell.
Until now, nothing was known about the transport of rubber degradation products into the cell. During our screening of transposon-induced mutants generated in G. polyisoprenivorans M71 (see above), we detected a mutant showing a rubber-negative phenotype (data not shown) with the insertion of the transposon in a gene encoding an Mce protein (GPOL_c27400). This ORF is part of one of five Mce clusters located in the genome of G. polyisoprenivorans. All these Mce clusters consist of two genes encoding YrbE proteins, followed by six genes encoding Mce proteins (MceA-F). They can be found in all mycobacteria as well as in various other actinobacteria and are almost always clustered in the same manner (24). Mce clusters were first investigated in M. tuberculosis and are thought to be involved in the pathogenesis of this strain. In these studies, it was assumed that the operon-encoded proteins are membrane associated and that some of them might play a role in the uptake of M. tuberculosis inside mammalian cells (6, 25, 26, 32). However, these clusters were also found to be present in the genomes of free-living, nonpathogenic bacteria. Bioinformatic studies indicated that Mce clusters encode a novel subfamily of ABC uptake transporters, wherein the yrbE gene products function as permeases and the mce gene products as substrate binding proteins (24). Experimental evidence that these clusters encode uptake transporters was demonstrated for the mce4 locus of R. jostii strain RHA1 that encodes an ATP-dependent steroid uptake system (60). The ATPase, necessary for providing the required energy for this system, was predicted using a bioinformatics approach (24). A homologue to this predicted ATPase is also present in the genome of G. polyisoprenivorans (GPOL_c37790). We therefore propose a transport of the low-molecular-weight intermediates resulting from the oxidative cleavage of rubber outside the cell into the cytoplasm via an ATP-dependent, Mce protein-driven mechanism. The Mce proteins, which are predicted to be extracytoplasmatic or tethered to the membrane (24), could function as substrate-binding proteins and may mediate the movement of the intermediates across the cell wall.
Oxidative-stress response.
It is well known that rubber materials tend to auto-oxidize in the presence of atmospheric oxygen and ozone, leading to the formation of activated oxygen species. Furthermore, the oxidative cleavage of the double bond in the poly(cis-1,4-isoprene) backbone was shown via analysis of the degradation products from various bacterial cultures, and a cleavage of rubber involving radicals is conceivable (18, 72, 90). In recent studies, we showed a possible involvement of an extracellular superoxide dismutase (SodA; GPOL_c03380) in rubber degradation. SodA is believed to serve as a radical scavenger during degradation of poly(cis-1,4-isoprene), as the formation of SodA is induced during growth on rubber. In addition, a disruption of sodA in strain VH2 leads to reduced growth on rubber but not on succinate, for example, as the sole source of carbon and energy. As no secretory signal peptide was found in the amino acid sequence, two explanations for the extracellular occurrence during growth on rubber were discussed. One possibility is that SodA is released due to the long cultivation time by cell lysis in combination with a high level of SodA expression and enzyme stability. Another explanation is that an alternative SecA secretion pathway is responsible for the extracellular release (82), as was described for SodA in the pathogenic bacterium M. tuberculosis (19). In addition to one ORF encoding SecA (SecA1; GPOL_c34920) that is part of the general Sec pathway in all bacteria (28), an ORF encoding a second SecA (SecA2; GPOL_174p00700) is located on the plasmid of strain VH2. Auxiliary SecA-encoding genes have been described for only a few Gram-positive bacteria (71). In M. tuberculosis, SecA1 is the essential SecA and SecA2 is an accessory secretion factor involved in the export of a specific subset of proteins, such as SodA and KatG. Both enzymes in M. tuberculosis lack classical signal sequences (19), as is also found in the amino acid sequence of SodA from strain VH2. In addition, the catalase-peroxidase encoded by the plasmid of G. polyisoprenivorans (KatG; GPOL_174p01220) also lacks a putative leader peptide. SodA and KatG of strain VH2 show high similarity to SodA and KatG of M. tuberculosis (61% and 69%, respectively), which are involved in the detoxification of reactive oxygen that occurs during oxidative attack of the host (31, 61). Hence, it is quite conceivable that SodA and KatG from G. polyisoprenivorans are secreted via an accessory SecA2 system during growth on rubber or other polymers that are initially activated via O2 as a reactant to protect the cells against reactive oxygen species. No additional paralogues of the Sec pathway (such as SecY2) and accessory Sec proteins (such as Asp1 to Asp5) as described for Streptococcus gordonii (12, 87) are located in the genome of strain VH2. In addition to the iron- or manganese-dependent SodA, the genome of G. polyisoprenivorans encodes a copper- and zinc-dependent Sod (SodC; GPOL_c08150) containing a classical signal peptide sequence. This enzyme also plays a role in mycobacterial pathogenesis (67). Furthermore, in addition to katG, a KatA-encoding gene (GPOL_c46020) is located in the genome. The occurrence of putative peroxidases of the DyP-type peroxidase family (GPOL_c14220, GPOL_c44400, GPOL_c04890, GPOL_c045780), peroxiredoxins (GPOL_c18960, GPOL_c28800; AhpD, GPOL_c07030, GPOL_c21240; AhpC, GPOL_c21250, GPOL_c07010), dihydrolipoyl dehydrogenases (GPOL_c09230, GPOL_c14810, GPOL_c16800), and a dihydrolipoamide acyltransferase (DlaT; GPOL_c28340) was also detected. The latter enzymes also play an important role in protecting M. tuberculosis against oxidative and nitrosative stress as an immune response of the infected host (23, 48). To what extent these enzymes are involved in the protection of strain VH2 against ROS occurring during rubber degradation remains to be clarified.
Surface-active compounds.
Strains of Gordonia are known to produce surface-active compounds (SACs) (2, 4, 33, 34, 54, 86). SACs play an important role in biodegradation of water-insoluble and hydrophobic compounds such as n-alkanes (8). It is assumed that the occurrence of mycolic acids and the production of other SACs are significantly involved in rubber degradation by G. polyisoprenivorans and related strains. The surfactants are thought to be important for the formation of biofilms and for enabling the strains to get into direct contact with the solid and water-insoluble poly(cis-1,4-isoprene) during adhesive growth (4).
The genome of G. polyisoprenivorans exhibits many ORFs predicted to be involved in biosynthesis, modification, assembly, and transport of lipopolysaccharides and/or exopolysaccharides putatively involved in biofilm formation. Especially in a specific 68-kb region (bp 1266000 to bp 1334000), many candidate genes were detected playing a significant role in forming high-molecular-weight SACs. Within this region, most genes (rmlABC) were found coding for proteins responsible for the synthesis of dTDP-l-rhamnose and are involved in the biosynthesis of precursors of lipopolysaccharides, for example, in Xanthomonas campestris pv. campestris ATCC 33913 (rmlABCD) (46). Gene rmlD is located outside this region in the genome of strain VH2 (GPOL_c17430). A putative glucose-1-phosphate cytidylyltransferase as well as an UDP-glucose 6-dehydrogenase found inside this region also seem to play a role in forming precursors for synthesis or modification of high-molecular-weight polymers. Furthermore, enzymes believed to transfer sugar to a lipid carrier, many putative glycosyl transferases, and genes involved in polymerization as well as export of polysaccharides are located in this region. High-molecular-weight SACs are important during stabilization of oil-in-water emulsions and play an important role in biofilm formation.
The synthesis of mycolic acids is also closely associated with biofilm formation in, for example, M. smegmatis (63), and these SACs are known to be responsible for the adhesive properties of coryneform bacteria (10). G. polyisoprenivorans strain VH2 is known to produce mycolic acids with overall numbers of carbon atoms of 60, 62, and 64 (2). It is assumed that the hydrophobicity of the cell surface affected by these SACs plays an essential role in adhesive growth on rubber (4). Homologues to almost all genes responsible for mycolic acid biosynthesis in M. tuberculosis (17) were also found in the genome of strain VH2. Nevertheless, in comparison to the genome of M. tuberculosis, two differences could be observed in the fatty acid synthase II (FAS-II) system of G. polyisoprenivorans. First, strain VH2 exhibits only one 3-oxoacyl-[acyl-carrier-protein] synthase (kasA; GPOL_c28870) in the genome, whereas M. tuberculosis harbors two Kas genes (kasA and kasB). In the latter, KasA and KasB catalyze the initiation of subsequent rounds of acyl extension by the FAS-II system (79). However, in M. smegmatis, kasA seems to be essential, since it could not be deleted whereas kasB could (16). Previous studies indicated that both enzymes have a discrete set of substrates and that KasB is responsible for the full extension of mycolic acids in Mycobacterium strains, since mutants with disrupted kasB produce meromycolate chains that are shorter than those produced by the wild type (17). This might be an explanation for why strain VH2 produces mycolic acids that are shorter than mycobacterial mycolic acids (60 to 90 carbon atoms). A prevalence of shorter-chain-length mycolic acids was observed in M. smegmatis during biofilm formation. In this study, it was proposed that KasA and the unusual and nonessential chaperone GroEL1 play an important role in building up a biofilm-specific FAS-II system (63). Since strain VH2 possesses three GroEL paralogues (the supposed housekeeping chaperone GroEL2 [GPOL_c16000], an adopted nonessential GroEL1 [GPOL_c08950], and a more distantly related GroEL3 [GPOL_c34350]), the involvement of such a biofilm-specific system during biofilm formation on rubber is conceivable. Furthermore, the absence of kasB might be one explanation for why G. polyisoprenivorans is less infectious than M. tuberculosis, since KasB plays an important role in mycobacterial virulence (15, 35). The second difference in the FAS-II system of VH2 compared to the well-studied mycobacterial FAS-II system is found in the gene composition responsible for β-hydroxyacyl-acyl carrier protein (ACP) dehydratase activity. In M. tuberculosis, an operon comprising three genes (hadA, hacB, hadC) is involved in this enzyme activity. Previous studies pointed out that HadB forms heterodimers with either HadA or HadC. The resulting HadAB and HadBC heterodimers show distinct chain length specificity. HadBC has a preference for longer substrates than HadAB (22, 76). However, the genome of strain VH2 does not exhibit such an operon but possesses an ORF putatively encoding such a β-hydroxyacyl-ACP dehydratase (GPOL_c11830). Within GPOL_c11830, orthologues of hadA and hadB are fused in one gene. This phenomenon was also described for N. farcinica and was deemed to be responsible for the formation of smaller meromycolic chains in this strain (76). This is also conceivable for strain VH2.
Plasmid.
The plasmid p174 harbors 165 ORFs, 67.9% of which we could assign to a putative function. Except for the second lcp and a gene encoding an enoyl-CoA hydratase (see above), the plasmid does not exhibit genes that can be associated with the proposed rubber degradation pathway. p174 is considerably different from the plasmid pKB1, which was shown to be essential for rubber degradation by G. westfalica (20). However, the plasmid also harbors genes that might play a role in genetic information storage and processing (including many transposases) as well as in metabolism (such as uptake and utilization of amino acids and carbon metabolism). ORFs possibly encoding proteins that are involved in heavy metal resistance could also be identified on p174. This, inter alia, includes a heavy metal-translocating P-type ATPase (GPOL_174p01290).
Further metabolic capabilities.
Strains of Gordonia have so far been investigated mainly because of their biodegradation and bioremediation capabilities (4). The genome of strain VH2 gives the impression of high metabolic potential. In addition to many genes involved in the catabolism of fatty acids, it harbors numerous determinants for the catabolism of various aromatic compounds, such as putative ring-cleavage dioxygenases (GPOL_c41080, GPOL_c41220), a putative manganese-dependent 2,3-dihydroxybiphenyl 1,2-dioxygenase (GPOL_c33190), and putative ring-hydroxylating dioxygenases (GPOL_c41100, GPOL_c41200). In addition to many other monooxygenase systems, the genome harbors a cluster putatively coding for a toluene-4-monooxygenase (GPOL_c08420, GPOL_c08470) with similarity to the cluster found in Pseudomonas mendocina strain KR1. This multicomponent system has broad substrate specificity in strain KR1 (94, 95), and it might therefore play a role not only in the initial hydroxylation of toluene in strain VH2 but also in the degradation of xenobiotics. A ring-hydroxylating dioxygenase system also putatively involved in aromatic compound metabolism or xenobiotic degradation is located elsewhere (benABC; GPOL_c38890, GPOL_c38910). The genome also possesses 19 putative cytochrome P450 monooxygenases, which might play a role in the survival of the strain in human bodies or be involved in the degradation of xenobiotics, lipids, or steroidal compounds. For the degradation of steroid compounds, such as cholesterol and other steroids with long carbon side chains, strain VH2 harbors two putative cholesterol oxidases (GPOL_c16080, GPOL_c38540) that are also found in the genome of G. cholesterolivorans strain DSM 45229. GPOL_c38540 has the highest similarity to ChoOx-2, which is essential for growth on cholesterol in strain DSM 45229 (29). It also exhibits homologous sequences to all genes assigned to the cholesterol degradation pathway in R. jostii and M. tuberculosis, including the essential hsaC (GPOL_c41080), supAB (GPOL_c42630, GPOL_c42640), and mce4 genes (GPOL_c42570, GPOL_c42620) (91), which are part of the steroid uptake system (60). Furthermore, the ability to grow on cholesterol is widespread within the genus Gordonia (29). Thus, strain VH2 might utilize sterols like cholesterol. Another example of the strain's high metabolic potential is a putative propane degradation pathway involving one of the 17 putative flavin-containing monooxygenases located in the genome. Herein, all genes responsible for propane utilization as described for Gordonia sp. strain TY-5, involving a novel Baeyer-Villiger monooxygenase (BVMO)-dependent acetone metabolism, are also present in strain VH2. Propane is first oxidized via monooxygenase-mediated subterminal oxidation to 2-propanol (GPOL_c34280, GPOL_c34310). The further oxidation of 2-propanol to acetone is then catalyzed by NAD+-dependent secondary alcohol dehydrogenases (GPOL_c34340, GPOL_c06040, GPOL_c02950) (49). Acetone is oxidized through a BVMO (GPOL_c02620) to methyl acetate, which is then hydrolyzed by an esterase (GPOL_c02630) to acetic acid and methanol (50). Because of the very high identities of the amino acid sequences (average identity, 98.7%; the amino acid sequences of the alcohol dehydrogenases of strain TY-5 are 100% identical to that of strain VH2), one can assume that strain VH2 also possesses this pathway. All responsible genes are clustered as described for strain TY-5.
G. polyisoprenivorans strain VH2 shows strong orange pigmentation of the colonies after exposure to light. This indicates the production of significant amounts of carotenoids (4, 9). Concerning the production of carotenoids, we identified transposon-induced mutants in previous studies that showed a phenotype different from that of the wild type (9). One mutant (34-07) exhibits a carotenoid-negative, white phenotype, and the transposon was mapped in a phytoene desaturase (GPOL_c48250), which might play a role in desaturation of colorless phytoene to neurosporene and lycopene. This ORF is part of a cluster involved in the synthesis of isoprenoids and carotenoids found upstream of lcp1 (Fig. 3). In the genome of another mutant (38-33) showing a bright yellow phenotype, a transposon insertion was detected next to an ORF showing similarity to a spheroidene monooxygenase (crtA; GPOL_c48210) also located within this cluster. The protein is normally involved in later steps of carotenoid biosynthesis, such as the conversion of spirilloxanthin to 2-ketospirilloxanthin or of spheroidene to spheroidenone (36, 68). Interestingly, no sequences homologous to hydroxyneurosporene synthase (crtC) or hydroxyneurosporene-O-methyltransferase (crtF) were detected inside the genome. However, an ORF putatively encoding a methoxyneurosporene dehydrogenase (crtD; GPOL_c48240) is located downstream of crtI. Genes for a putative lycopene cyclase (crtL; GPOL_c23090) as well as a putative β-carotene ketolase (crtO; GPOL_c27870) are found elsewhere. This might indicate that strain VH2 synthesizes carotenoids like canthaxanthin, shown to be present in G. jacobaea strain MV-1 (27). However, a concrete statement on the carotenoids produced in the mevalonate-independent-pathway-possessing strain VH2 cannot be done here and requires further analysis.
Conclusions.
The genome sequence of G. polyisoprenivorans strain VH2 together with generated transposon-induced mutants, deletion mutants, and literature search allowed us to predict a pathway for the degradation of IR. First, by deleting both lcp homologues found in the genome, we showed the importance of this enzyme for rubber degradation, as the double-deletion mutant was not able to grow on IR, in contrast to the single-deletion mutants. From these results and previous studies done on lcp1 of this organism (and others), we conclude that Lcp participates in the initial step of rubber degradation. Another transposon-induced mutant, which could no longer grow on IR, showed the involvement of an Mce cluster that seems to be involved in the transport of the oligomers generated outside the cell. Our proposal of a further degradation of the cleavage products similar to the degradation of fatty acids is consistent with mutants of strain VH2 generated in the past (5, 9) and is supported by analysis of the genome sequence. The genome harbors an extensive number of putative genes involved in β-oxidation. By using a comparative approach, we limited the number to a minimal set of candidate genes involved in the rubber degradation pathway.
In addition to genes involved in the utilization of rubber, the genome harbors many genes involved in degradation, bioconversion, and synthesis of other interesting compounds. The study presented here offers a powerful tool for further studies on the bioremediation and biotechnical utilization of rubber-containing waste material, which is becoming an increasing environmental problem.
ACKNOWLEDGMENTS
The authors gratefully acknowledge financial support from the German Federal Ministry of Education and Research (BMBF) in the framework of the funding measure GenoMik-Plus (FKZ 0313751E, FKZ 0313751A, FKZ 0313807C).
We thank A. Wollherr for providing the BiBag software tool.
Footnotes
Published ahead of print 10 February 2012
REFERENCES
- 1. Alvarez LA, Exton DA, Timmis KN, Suggett DJ, McGenity TJ. 2009. Characterization of marine isoprene-degrading communities. Environ. Microbiol. 11:3280–3291 [DOI] [PubMed] [Google Scholar]
- 2. Arenskötter M, et al. 2001. Taxonomic characterization of two rubber degrading bacteria belonging to the species Gordonia polyisoprenivorans and analysis of hyper variable regions of 16S rDNA sequences. FEMS Microbiol. Lett. 205:277–282 [DOI] [PubMed] [Google Scholar]
- 3. Arenskötter M, Baumeister D, Kalscheuer R, Steinbüchel A. 2003. Identification and application of plasmids suitable for transfer of foreign DNA to members of the genus Gordonia. Appl. Environ. Microbiol. 69:4971–4974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Arenskötter M, Bröker D, Steinbüchel A. 2004. Biology of the metabolically diverse genus Gordonia. Appl. Environ. Microbiol. 70:3195–3204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Arenskötter Q, Heller J, Dietz D, Arenskötter M, Steinbüchel A. 2008. Cloning and characterization of α-methylacyl coenzyme A racemase from Gordonia polyisoprenivorans VH2. Appl. Environ. Microbiol. 74:7085–7089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Arruda S, Bomfim G, Knights R, Huima-Byron T, Riley LW. 1993. Cloning of an M. tuberculosis DNA fragment associated with entry and survival inside cells. Science 261:1454–1457 [DOI] [PubMed] [Google Scholar]
- 7. Reference deleted.
- 8. Banat IM, et al. 2010. Microbial biosurfactants production, applications and future potential. Appl. Microbiol. Biotechnol. 87:427–444 [DOI] [PubMed] [Google Scholar]
- 9. Banh Q, Arenskötter M, Steinbüchel A. 2005. Establishment of Tn5096-based transposon mutagenesis in Gordonia polyisoprenivorans. Appl. Environ. Microbiol. 71:5077–5084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bendinger B, Rijnaarts HH, Altendorf K, Zehnder AJ. 1993. Physicochemical cell surface and adhesive properties of coryneform bacteria related to the presence and chain length of mycolic acids. Appl. Environ. Microbiol. 59:3973–3977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bendtsen JD, Nielsen H, Widdick D, Palmer T, Brunak S. 2005. Prediction of twin-arginine signal peptides. BMC Bioinformatics 6:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bensing BA, Sullam PM. 2002. An accessory sec locus of Streptococcus gordonii is required for export of the surface protein GspB and for normal levels of binding to human platelets. Mol. Microbiol. 44:1081–1094 [DOI] [PubMed] [Google Scholar]
- 13. Berekaa MM, Linos A, Reichelt R, Keller U, Steinbüchel A. 2000. Effect of pretreatment of rubber material on its biodegradability by various rubber degrading bacteria. FEMS Microbiol. Lett. 184:199–206 [DOI] [PubMed] [Google Scholar]
- 14. Berekaa MM, Steinbüchel A. 2000. Microbial degradation of the multiply branched alkane 2,6,10,15,19, 23-hexamethyltetracosane (squalane) by Mycobacterium fortuitum and Mycobacterium ratisbonense. Appl. Environ. Microbiol. 66:4462–4467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14a. Berlyn MKB, Low KB, Rudd KE. 1996. Linkage map of Escherichia coli K-12, p 1715–1902 In Neidhardt FC, et al. (ed), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed American Society for Microbiology, Washington, Dc [Google Scholar]
- 15. Bhatt A, et al. 2007. Deletion of kasB in Mycobacterium tuberculosis causes loss of acid-fastness and subclinical latent tuberculosis in immunocompetent mice. Proc. Natl. Acad. Sci. U. S. A. 104:5157–5162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bhatt A, Kremer L, Dai AZ, Sacchettini JC, Jacobs WR., Jr 2005. Conditional depletion of KasA, a key enzyme of mycolic acid biosynthesis, leads to mycobacterial cell lysis. J. Bacteriol. 187:7596–7606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bhatt A, Molle V, Besra GS, Jacobs WR, Jr, Kremer L. 2007. The Mycobacterium tuberculosis FAS-II condensing enzymes: their role in mycolic acid biosynthesis, acid-fastness, pathogenesis and in future drug development. Mol. Microbiol. 64:1442–1454 [DOI] [PubMed] [Google Scholar]
- 18. Bode HB, Kerkhoff K, Jendrossek D. 2001. Bacterial degradation of natural and synthetic rubber. Biomacromolecules 2:295–303 [DOI] [PubMed] [Google Scholar]
- 19. Braunstein M, Espinosa BJ, Chan J, Belisle JT, Jacobs WR., Jr 2003. SecA2 functions in the secretion of superoxide dismutase A and in the virulence of Mycobacterium tuberculosis. Mol. Microbiol. 48:453–464 [DOI] [PubMed] [Google Scholar]
- 20. Bröker D, Arenskötter M, Legatzki A, Nies DH, Steinbüchel A. 2004. Characterization of the 101-kilobase-pair megaplasmid pKB1, isolated from the rubber-degrading bacterium Gordonia westfalica Kb1. J. Bacteriol. 186:212–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bröker D, Dietz D, Arenskötter M, Steinbüchel A. 2008. The genomes of the non-clearing-zone-forming and natural-rubber-degrading species Gordonia polyisoprenivorans and Gordonia westfalica harbor genes expressing Lcp activity in Streptomyces strains. Appl. Environ. Microbiol. 74:2288–2297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brown AK, et al. 2007. Identification of the dehydratase component of the mycobacterial mycolic acid-synthesizing fatty acid synthase-II complex. Microbiology 153:4166–4173 [DOI] [PubMed] [Google Scholar]
- 23. Bryk R, Lima CD, Erdjument-Bromage H, Tempst P, Nathan C. 2002. Metabolic enzymes of mycobacteria linked to antioxidant defense by a thioredoxin-like protein. Science 295:1073–1077 [DOI] [PubMed] [Google Scholar]
- 24. Casali N, Riley LW. 2007. A phylogenomic analysis of the Actinomycetales mce operons. BMC Genomics 8:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chitale S, et al. 2001. Recombinant Mycobacterium tuberculosis protein associated with mammalian cell entry. Cell. Microbiol. 3:247–254 [DOI] [PubMed] [Google Scholar]
- 26. Cole ST, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537–544 [DOI] [PubMed] [Google Scholar]
- 27. de Miguel T, Sieiro C, Poza M, Villa TG. 2000. Isolation and taxonomic study of a new canthaxanthin-containing bacterium, Gordonia jacobaea MV-1 sp. nov. Int. Microbiol. 3:107–111 [PubMed] [Google Scholar]
- 28. Driessen AJ, Nouwen N. 2008. Protein translocation across the bacterial cytoplasmic membrane. Annu. Rev. Biochem. 77:643–667 [DOI] [PubMed] [Google Scholar]
- 29. Drzyzga O, Fernández de las Heras L, Morales V, Navarro Llorens JM, Perera J. 2011. Cholesterol degradation by Gordonia cholesterolivorans. Appl. Environ. Microbiol. 77:4802–4810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E. (ed). 2006. Prokaryotes, 3rd ed, vol 3 Springer Science & Business Media LLC, New York, NY [Google Scholar]
- 31. Edwards KM, et al. 2001. Iron-cofactored superoxide dismutase inhibits host responses to Mycobacterium tuberculosis. Am. J. Respir. Crit. Care Med. 164:2213–2219 [DOI] [PubMed] [Google Scholar]
- 32. El-Shazly S, Ahmad S, Mustafa AS, Al-Attiyah R, Krajci D. 2007. Internalization by HeLa cells of latex beads coated with mammalian cell entry (Mce) proteins encoded by the mce3 operon of Mycobacterium tuberculosis. J. Med. Microbiol. 56:1145–1151 [DOI] [PubMed] [Google Scholar]
- 33. Franzetti A, Bestetti G, Caredda P, La Colla P, Tamburini E. 2008. Surface-active compounds and their role in the access to hydrocarbons in Gordonia strains. FEMS Microbiol. Ecol. 63:238–248 [DOI] [PubMed] [Google Scholar]
- 34. Fusconi R, Godinho MJ. 2002. Screening for exopolysaccharide-producing bacteria from sub-tropical polluted groundwater. Braz. J. Biol. 62:363–369 [DOI] [PubMed] [Google Scholar]
- 35. Gao LY, et al. 2003. Requirement for kasB in Mycobacterium mycolic acid biosynthesis, cell wall impermeability and intracellular survival: implications for therapy. Mol. Microbiol. 49:1547–1563 [DOI] [PubMed] [Google Scholar]
- 36. Garcia-Asua G, Lang HP, Cogdell RJ, Hunter CN. 1998. Carotenoid diversity: a modular role for the phytoene desaturase step. Trends Plant Sci. 3:445–449 [Google Scholar]
- 37. Gupta M, Prasad D, Khara HS, Alcid D. 2010. A rubber-degrading organism growing from a human body. Int. J. Infect. Dis. 14:e75–e76 [DOI] [PubMed] [Google Scholar]
- 38. Ibrahim EM, Arenskötter M, Luftmann H, Steinbüchel A. 2006. Identification of poly(cis-1,4-isoprene) degradation intermediates during growth of moderately thermophilic actinomycetes on rubber and cloning of a functional lcp homologue from Nocardia farcinica strain E1. Appl. Environ. Microbiol. 72:3375–3382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Iida S, et al. 2005. Gordonia otitidis sp. nov., isolated from a patient with external otitis. Int. J. Syst. Evol. Microbiol. 55:1871–1876 [DOI] [PubMed] [Google Scholar]
- 40. Ikeda H, et al. 2003. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 21:526–531 [DOI] [PubMed] [Google Scholar]
- 41. Ishikawa J, et al. 2004. The complete genomic sequence of Nocardia farcinica IFM 10152. Proc. Natl. Acad. Sci. U. S. A. 101:14925–14930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ivanova N, et al. 2010. Complete genome sequence of Gordonia bronchialis type strain (3410). Stand. Genomic Sci. 2:19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jendrossek D, Tomasi G, Kroppenstedt RM. 1997. Bacterial degradation of natural rubber: a privilege of actinomycetes? FEMS Microbiol. Lett. 150:179–188 [DOI] [PubMed] [Google Scholar]
- 44. Kageyama A, et al. 2006. Gordonia araii sp. nov. and Gordonia effusa sp. nov., isolated from patients in Japan. Int. J. Syst. Evol. Microbiol. 56:1817–1821 [DOI] [PubMed] [Google Scholar]
- 45. Kempf VA, et al. 2004. Gordonia polyisoprenivorans septicemia in a bone marrow transplant patient. Eur. J. Clin. Microbiol. Infect. Dis. 23:226–228 [DOI] [PubMed] [Google Scholar]
- 46. Köplin R, Wang G, Hötte B, Priefer UB, Pühler A. 1993. A 3.9-kb DNA region of Xanthomonas campestris pv. campestris that is necessary for lipopolysaccharide production encodes a set of enzymes involved in the synthesis of dTDP-rhamnose. J. Bacteriol. 175:7786–7792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kornberg HL, Krebs HA. 1957. Synthesis of cell constituents from C2-units by a modified tricarboxylic acid cycle. Nature 179:988–991 [DOI] [PubMed] [Google Scholar]
- 48. Koshkin A, Knudsen GM, Ortiz de Montellano PR. 2004. Intermolecular interactions in the AhpC/AhpD antioxidant defense system of Mycobacterium tuberculosis. Arch. Biochem. Biophys. 427:41–47 [DOI] [PubMed] [Google Scholar]
- 49. Kotani T, Yamamoto T, Yurimoto H, Sakai Y, Kato N. 2003. Propane monooxygenase and NAD+-dependent secondary alcohol dehydrogenase in propane metabolism by Gordonia sp. strain TY-5. J. Bacteriol. 185:7120–7128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kotani T, Yurimoto H, Kato N, Sakai Y. 2007. Novel acetone metabolism in a propane-utilizing bacterium, Gordonia sp. strain TY-5. J. Bacteriol. 189:886–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kummer C, Schumann P, Stackebrandt E. 1999. Gordonia alkanivorans sp. nov., isolated from tar-contaminated soil. Int. J. Syst. Bacteriol. 49:1513–1522 [DOI] [PubMed] [Google Scholar]
- 52. Kuwayama H, et al. 2002. PCR-mediated generation of a gene disruption construct without the use of DNA ligase and plasmid vectors. Nucleic Acids Res. 30:E2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Linos A, Steinbüchel A. 1998. Microbial degradation of natural and synthetic rubbers by novel bacteria belonging to the genus Gordona. Kautsch. Gummi Kunstst. 51:496–499 [Google Scholar]
- 54. Linos A, et al. 2000. Biodegradation of cis-1,4-polyisoprene rubbers by distinct actinomycetes: microbial strategies and detailed surface analysis. Appl. Environ. Microbiol. 66:1639–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Linos A, et al. 2002. Gordonia westfalica sp. nov., a novel rubber-degrading actinomycete. Int. J. Syst. Evol. Microbiol. 52:1133–1139 [DOI] [PubMed] [Google Scholar]
- 56. Linos A, Steinbüchel A, Spröer C, Kroppenstedt RM. 1999. Gordonia polyisoprenivorans sp. nov., a rubber-degrading actinomycete isolated from an automobile tyre. Int. J. Syst. Bacteriol. 49:1785–1791 [DOI] [PubMed] [Google Scholar]
- 57. Liu Y, et al. 2011. Gordonia neofelifaecis sp. nov., a cholesterol side-chain-cleaving actinomycete isolated from the faeces of Neofelis nebulosa. Int. J. Syst. Evol. Microbiol. 61:165–169 [DOI] [PubMed] [Google Scholar]
- 58. Mancia F, et al. 1996. How coenzyme B12 radicals are generated: the crystal structure of methylmalonyl-coenzyme A mutase at 2 Å resolution. Structure 4:339–350 [DOI] [PubMed] [Google Scholar]
- 59. McLeod MP, et al. 2006. The complete genome of Rhodococcus sp. RHA1 provides insights into a catabolic powerhouse. Proc. Natl. Acad. Sci. U. S. A. 103:15582–15587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mohn WW, et al. 2008. The actinobacterial mce4 locus encodes a steroid transporter. J. Biol. Chem. 283:35368–35374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ng VH, Cox JS, Sousa AO, MacMicking JD, McKinney JD. 2004. Role of KatG catalase-peroxidase in mycobacterial pathogenesis: countering the phagocyte oxidative burst. Mol. Microbiol. 52:1291–1302 [DOI] [PubMed] [Google Scholar]
- 62. Nhi-Cong LT, Mikolasch A, Klenk H-P, Schauer F. 2009. Degradation of the multiple branched alkane 2,6,10,14-tetramethyl-pentadecane (pristane) in Rhodococcus ruber and Mycobacterium neoaurum. Int. Biodeter. Biodegrad. 63:201–207 [Google Scholar]
- 63. Ojha A, et al. 2005. GroEL1: a dedicated chaperone involved in mycolic acid biosynthesis during biofilm formation in mycobacteria. Cell 123:861–873 [DOI] [PubMed] [Google Scholar]
- 64. Overbeek R, et al. 2003. The ERGO genome analysis and discovery system. Nucleic Acids Res. 31:164–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Overhage J, Priefert H, Rabenhorst J, Steinbüchel A. 1999. Biotransformation of eugenol to vanillin by a mutant of Pseudomonas sp. strain HR199 constructed by disruption of the vanillin dehydrogenase (vdh) gene. Appl. Microbiol. Biotechnol. 52:820–828 [DOI] [PubMed] [Google Scholar]
- 66. Pethe K, et al. 2001. The heparin-binding haemagglutinin of M. tuberculosis is required for extrapulmonary dissemination. Nature 412:190–194 [DOI] [PubMed] [Google Scholar]
- 67. Piddington DL, et al. 2001. Cu,Zn superoxide dismutase of Mycobacterium tuberculosis contributes to survival in activated macrophages that are generating an oxidative burst. Infect. Immun. 69:4980–4987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Pinta V, et al. 2003. Characterization of unusual hydroxy- and ketocarotenoids in Rubrivivax gelatinosus: involvement of enzyme CrtF or CrtA. Arch. Microbiol. 179:354–362 [DOI] [PubMed] [Google Scholar]
- 69. Pirnik MP, Atlas RM, Bartha R. 1974. Hydrocarbon metabolism by Brevibacterium erythrogenes: normal and branched alkanes. J. Bacteriol. 119:868–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Raynaud C, et al. 2002. Phospholipases C are involved in the virulence of Mycobacterium tuberculosis. Mol. Microbiol. 45:203–217 [DOI] [PubMed] [Google Scholar]
- 71. Rigel NW, Braunstein M. 2008. A new twist on an old pathway—accessory secretion systems. Mol. Microbiol. 69:291–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Rose K, Steinbüchel A. 2005. Biodegradation of natural rubber and related compounds: recent insights into a hardly understood catabolic capability of microorganisms. Appl. Environ. Microbiol. 71:2803–2812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Rose K, Tenberge KB, Steinbüchel A. 2005. Identification and characterization of genes from Streptomyces sp. strain K30 responsible for clear zone formation on natural rubber latex and poly(cis-1,4-isoprene) rubber degradation. Biomacromolecules 6:180–188 [DOI] [PubMed] [Google Scholar]
- 74. Rosłoniec KZ, et al. 2009. Cytochrome P450 125 (CYP125) catalyses C26-hydroxylation to initiate sterol side-chain degradation in Rhodococcus jostii RHA1. Mol. Microbiol. 74:1031–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Rutherford K, et al. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944–945 [DOI] [PubMed] [Google Scholar]
- 76. Sacco E, et al. 2007. The missing piece of the type II fatty acid synthase system from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 104:14628–14633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Sakai Y, et al. 2004. Role of α-methylacyl coenzyme A racemase in the degradation of methyl-branched alkanes by Mycobacterium sp. strain P101. J. Bacteriol. 186:7214–7220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 79. Schaeffer ML, et al. 2001. Purification and biochemical characterization of the Mycobacterium tuberculosis beta-ketoacyl-acyl carrier protein synthases KasA and KasB. J. Biol. Chem. 276:47029–47037 [DOI] [PubMed] [Google Scholar]
- 80. Schlegel HG, Kaltwasser H, Gottschalk G. 1961. Ein submersverfahren zur kultur wasserstoffoxidierender bakterien: wachstumsphysiologische untersuchungen. Arch. Mikrobiol. 38:209–222 [PubMed] [Google Scholar]
- 81. Schrempf H. 2006. The family Streptomycetaceae. Part II: Molecular biology, p 605–622 In Dworkin M. (ed), Prokaryotes, 3rd ed, vol 3 Springer Science & Business Media LLC, New York, NY [Google Scholar]
- 82. Schulte C, et al. 2008. Possible involvement of an extracellular superoxide dismutase (SodA) as a radical scavenger in poly(cis-1,4-isoprene) degradation. Appl. Environ. Microbiol. 74:7643–7653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Silva RA, Grossi V, Alvarez HM. 2007. Biodegradation of phytane (2,6,10,14-tetramethylhexadecane) and accumulation of related isoprenoid wax esters by Mycobacterium ratisbonense strain SD4 under nitrogen-starved conditions. FEMS Microbiol. Lett. 272:220–228 [DOI] [PubMed] [Google Scholar]
- 84. Solano-Serena F, Marchal R, Heiss S, Vandecasteele JP. 2004. Degradation of isooctane by Mycobacterium austroafricanum IFP 2173: growth and catabolic pathway. J. Appl. Microbiol. 97:629–639 [DOI] [PubMed] [Google Scholar]
- 85. Staden R, Beal KF, Bonfield JK. 2000. The Staden package, 1998. Methods Mol. Biol. 132:115–130 [DOI] [PubMed] [Google Scholar]
- 86. Ta-Chen L, Chang JS, Young CC. 2008. Exopolysaccharides produced by Gordonia alkanivorans enhance bacterial degradation activity for diesel. Biotechnol. Lett. 30:1201–1206 [DOI] [PubMed] [Google Scholar]
- 87. Takamatsu D, Bensing BA, Sullam PM. 2005. Two additional components of the accessory Sec system mediating export of the Streptococcus gordonii platelet-binding protein GspB. J. Bacteriol. 187:3878–3883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Tech M, Merkl R. 2003. YACOP: enhanced gene prediction obtained by a combination of existing methods. In Silico Biol. 3:441–451 [PubMed] [Google Scholar]
- 89. Tsuchii A, Suzuki T, Takeda K. 1985. Microbial degradation of natural rubber vulcanizates. Appl. Environ. Microbiol. 50:965–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Tsuchii A, Takeda K. 1990. Rubber-degrading enzyme from a bacterial culture. Appl. Environ. Microbiol. 56:269–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Van der Geize R, et al. 2007. A gene cluster encoding cholesterol catabolism in a soil actinomycete provides insight into Mycobacterium tuberculosis survival in macrophages. Proc. Natl. Acad. Sci. U. S. A. 104:1947–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Verma P, et al. 2006. Native valve endocarditis due to Gordonia polyisoprenivorans: case report and review of literature of bloodstream infections caused by Gordonia species. J. Clin. Microbiol. 44:1905–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Warneke S, Arenskötter M, Tenberge KB, Steinbüchel A. 2007. Bacterial degradation of poly(trans-1,4-isoprene) (gutta percha). Microbiology 153:347–356 [DOI] [PubMed] [Google Scholar]
- 94. Yen KM, Karl MR. 1992. Identification of a new gene, tmoF, in the Pseudomonas mendocina KR1 gene cluster encoding toluene-4-monooxygenase. J. Bacteriol. 174:7253–7261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Yen KM, et al. 1991. Cloning and characterization of a Pseudomonas mendocina KR1 gene cluster encoding toluene-4-monooxygenase. J. Bacteriol. 173:5315–5327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Yikmis M, et al. 2008. Secretion and transcriptional regulation of the latex-clearing protein, Lcp, by the rubber-degrading bacterium Streptomyces sp. strain K30. Appl. Environ. Microbiol. 74:5373–5382 [DOI] [PMC free article] [PubMed] [Google Scholar]