Abstract
Little is known about the gastric mucosal microbiota in healthy horses, and its role in gastric disease has not been critically examined. The present study used a combination of 16S rRNA bacterial tag-encoded pyrosequencing (bTEFAP) and fluorescence in situ hybridization (FISH) to characterize the composition and spatial distribution of selected gastric mucosal microbiota of healthy horses. Biopsy specimens of the squamous, glandular, antral, and any ulcerated mucosa were obtained from 6 healthy horses by gastroscopy and from 3 horses immediately postmortem. Pyrosequencing was performed on biopsy specimens from 6 of the horses and yielded 53,920 reads in total, with 631 to 4,345 reads in each region per horse. The microbiome segregated into two distinct clusters comprised of horses that were stabled, fed hay, and sampled at postmortem (cluster 1) and horses that were pastured on grass, fed hay, and biopsied gastroscopically after a 12-h fast (cluster 2). The types of bacteria obtained from different anatomic regions clustered by horse rather than region. The dominant bacteria in cluster 1 were Firmicutes (>83% reads/sample), mainly Streptococcus spp., Lactobacillus spp. and, Sarcina spp. Cluster 2 was more diverse, with predominantly Proteobacteria, Bacteroidetes, and Firmicutes, consisting of Actinobacillus spp. Moraxella spp., Prevotella spp., and Porphyromonas spp. Helicobacter sp. sequences were not identified in any of 53,920 reads. FISH (n = 9) revealed bacteria throughout the stomach in close apposition to the mucosa, with significantly more Streptococcus spp. present in the glandular region of the stomach. The equine stomach harbors an abundant and diverse mucosal microbiota that varies by individual.
INTRODUCTION
Historically, the stomach has been considered (based on microbial culture alone) a harsh environment colonized by a relatively small number of acid-tolerant bacteria such as Helicobacter spp. The clear association between Helicobacter pylori infection and development of gastric inflammation, ulceration, and cancer in humans has reinforced this notion (17, 37). However, the development of culture-independent molecular methods has dramatically changed our perception of the enteric microbiota (20, 25). A diverse and unique community of bacteria belonging to the phyla of the Proteobacteria, Firmicutes, Actinobacteria, and Bacteroidetes exists in the human stomach (11, 35). Fluorescence in situ hybridization (FISH) (with probes against bacterial 16S rRNA genes) has enabled characterization of the spatial distribution of gastric colonization in a variety of species (49, 57).
The most common equine gastric disease is gastric ulceration, while gastric impactions and neoplasia are reported infrequently (10, 12, 30, 56, 59). Gastric ulceration affects between 53 to 90% of adult horses and has been associated with colic, weight loss, and decreased performance (38, 61). Many (81%) outwardly healthy Thoroughbreds in race training also have ulcers (61). The specific cause of equine gastric ulcers is not fully elucidated, although several factors play a role. Increased gastric acidity coupled with decreased mucosal defense via a compromised “gastric mucosal barrier” is a major contributor. Reduced intragastric pH is attributed to increased secretion of hydrochloric acid plus bacterial fermentation of grain to volatile fatty acids (5–8, 43, 46, 47). Nearly 80% of ulcers in adult horses occur in the squamous region, usually close to the margo plicatus (9). Nonsteroidal anti-inflammatory drugs (NSAIDs) and corticosteroids contribute to ulceration of the glandular portion by altering the gastric mucosal barrier via prostaglandin inhibition. The antral mucosa at the entrance to the duodenum is susceptible, but ulceration of the fundic glandular region is much less common (44).
Relatively little is known about the gastric mucosal microbiota in healthy horses, and its role in gastric disease has not been critically examined using contemporary methodologies. The equine stomach is not sterile; bacteria have been visualized on the surfaces of the nonglandular and glandular equine stomach mucosae (4, 15, 28, 60, 62). Products of bacterial fermentation (such as lactic acid and volatile fatty acids) have been isolated from the equine gastric contents, and many anaerobes have been cultured. The anaerobes are comprised mostly of lactobacilli, streptococci, and lactic acid bacteria, of which specifically Lactobacillus salivarius, L. crispatus, L. reuteri, L. mucosae, L. delbrueckii, and L. agilis were identified using culture-independent technologies (3, 28, 62). Helicobacter-like organisms were seen in the gastric mucosae of 6/15 horses sampled (16), and Helicobacter-like DNA has been detected in the gastric mucosae of horses with and without ulcers (14). However, using FISH, gastric Helicobacter spp. were not identified in 36 horses with antral pathology (28). These contradictory findings indicate that further study is required to resolve the potential role of Helicobacter spp. in equine gastric ulcers.
The present study used a combination of 16S rRNA bacterial tag-encoded pyrosequencing (bTEFAP) and FISH to characterize the composition and spatial (anatomic) distribution of the gastric mucosal microbiota of healthy horses.
MATERIALS AND METHODS
Horses and gastric mucosal sample collection.
Gastric mucosal biopsy specimens were obtained from nine horses, and their management and signalment are shown in Table 1. None of the horses had clinical evidence of gastrointestinal disease or had received any nonsteroidal anti-inflammatory drugs, corticosteroids, or gastroprotectants within 1 month prior to gastric biopsy. The experiment was approved by the Cornell University Institutional Animal Care and Use Committee.
Table 1.
Summary of signalment, management, sample acquisition, and analyses for each horse
| Horse | Sampling date (mo/day/yr) | Age (yr)a | Breedb | Sexc | Management and sample acquisitiond | Gross ulcer severity grade | Gross ulcer no. score | Histopathology of abnormal mucosa | FISH | bTEFAP pyrosequencing |
|---|---|---|---|---|---|---|---|---|---|---|
| H1 | 5/23/06 | NR | NR | G | 1 | 0 | 0 | + | − | |
| H2 | 5/23/06 | NR | NR | G | 1 | 0 | 0 | + | + | |
| H3 | 5/31/06 | NR | NR | G | 1 | 2 | 1 | Ulcer | + | + |
| H35 | 6/14/06 | 7 | TB | M | 2 | 1 | 3 | Ulcer | + | + |
| H60 | 6/14/06 | 14 | WB | M | 2 | 0 | 0 | + | + | |
| H79 | 6/18/06 | 17 | TB | M | 2 | 1 | 2 | Parakeratosis | + | + |
| H86 | 6/14/06 | 15 | TB | M | 2 | 1 | 2 | Erosion | + | + |
| H131 | 1/30/07 | 9 | WB | G | 2 | 0 | 0 | + | − | |
| H158 | 1/30/07 | 8 | TB | G | 2 | 1 | 3 | Ulcer | + | − |
NR, not recorded.
TB, thoroughbred; WB, warm blood.
G, gelding; M, mare.
1, sampled postmortem, 5-mm punch biopsy specimen, nonfasted 12 h before sampling, nonpastured, exercise 30-min turnout/day in dirt paddock, subject to student lameness exams. 2, sampled antemortem, 2- to 3-mm gastroscopic biopsy specimen, fasted 12 h before sampling, pastured, not subject to student lameness exams.
Postmortem gastric biopsy specimens were obtained from three (of the nine total) nonfasted horses within 30 min of euthanasia using sterile 6-mm punch biopsy instruments. Biopsies from each horse and each stomach region were done using a new biopsy instrument to avoid potential carryover of microbial DNA. These three horses had been used for student lameness examinations, including perineural and joint anesthesia plus diagnostic imaging. For 2 weeks prior to euthanasia, these horses were stabled and received only free-choice hay (Table 1).
Gastroscopic biopsy specimens were obtained from the other six horses, which were kept at pasture and supplemented with hay (Table 1). The biopsy specimens were obtained after a 12-h fast and sedation with detomidine (Dormosedan; Pfizer Animal Health, Exton, PA) using a 3-m gastroscope (Olympus CV-100 video endoscope) and 3-mm sterile biopsy forceps (Olympus of America, Melville, New York). Separate sterilized biopsy forceps were used on each horse. Forceps were decontaminated between stomach regions by serial washes in 6.15% sodium hypochlorite bleach and sterile 0.9% saline to reduce the possibility of microbial DNA carryover between the individual samples taken from a horse. The endoscope was sterilized between horses with 55% glutaraldehyde.
Macroscopic evaluation of the gastric mucosa was performed at postmortem examination or during endoscopy, and a number and severity score (36) were assigned to any gastric ulcers present (Table 1). Gastric biopsy specimens were obtained from the squamous, glandular, and antral regions and from any ulcerated mucosae of each horse. All horses had two biopsy specimens per region placed in formalin for FISH, fixed overnight, and paraffin embedded for standard histopathology; two other biopsy specimens per region were placed in tissue lysis (ATL) buffer (Qiagen Inc., Valencia, CA) for DNA extraction.
Histopathology.
Formalin-fixed paraffin-embedded sections of gastric mucosa stained with hematoxylin and eosin (H&E) and Masson's Trichrome were “blindly” evaluated for the presence or absence of ulceration and gastritis by one pathologist (S.P.M.). The presence of bacteria in Gram- and modified-Steiner-stained sections was qualitatively evaluated.
Pyrosequencing.
DNA was extracted from mucosal biopsy specimens of all regions of the stomach from six horses (H2, H3, H35, H60, H79, and H86) (Table 1) using the QIAamp tissue kit (Qiagen Inc., Valencia, CA) according to the manufacturer's instructions. Three horses (H1, H131, and H158) did not have bTEFAP performed due to inadequate or lost samples. The data processing was performed as described previously, with minor alterations (26, 31, 52). The bTEFAP sequencing was performed on the GS FLX Titanium sequencing platform (Roche Applied Science, Indianapolis, IN). Primers 28F (5′GAGTTTGATCNTGGCTCAG) and 519r (5′GTNTTACNGCGGCKGCTG) were used to sequence an approximately 500-bp fragment of the 16S rRNA gene (Research and Testing Laboratory, Lubbock, TX). Raw data from bTEFAP were screened and trimmed based upon Q20 Phred-based quality scoring and binned into individual biopsy sample collections. Sequence collections were depleted of chimeras using B2C2 (Research and Testing Laboratory, Lubbock, TX). The resulting files were then depleted of short reads (<200 bp), tags, primers, nonbacterial ribosome sequences (mitochondrial, plastid, etc.), sequences with degenerate bases, and sequences with >6 homopolymer stretches. The bacterial taxa were then identified using BLASTn comparison to a curated high-quality 16S rRNA gene database derived from NCBI (http://www.ncbi.nlm.nih.gov/). Relative percentages of bacteria at each taxonomic level were determined for each individual biopsy sample. Data were also compiled at each individual taxonomic level according to the NCBI taxonomy criteria as described previously (18, 19).
Bacterial diversity indices were determined for on trimmed, nonribosomal sequence-depleted, chimera-depleted, high-quality reads as described previously (1). Multiple-sequence alignment was done using MUSCLE (21) (with parameters -maxiters 1, -diags1, and -sv). Based on the alignment, a distance matrix was constructed using DNAdist from PHYLIP version 3.6 with default parameters (23). These pairwise distances served as input to DOTUR (51) for clustering the sequences into operational taxonomic units (OTUs) of defined sequence similarity ranging from 0% to 20% dissimilarity. From the literature, we can expect that 0% and even 3% dissimilarity in sequences generated from pyrosequencing (based upon rarefaction) will provide dramatic overestimation of the phylotypes present in a sample (32, 50). At 5% dissimilarity (roughly genus-level classification), we expect to obtain a more accurate estimation of comparative diversity present across the samples. Although such estimates have limitations, the comparison among samples that were treated using similar procedures was felt to be valid, yet noting that some overestimation or underestimation is likely. The clusters based upon dissimilarity of 3% and 5% served as OTUs for generating predictive rarefaction models and for making calculations with the richness (diversity) estimators ACE and Chao1 (13). Correlation between the numbers of OTUs and estimated number of OTUs was calculated in the R package (http://www.R-project.org).
A hierarchical cluster analysis was performed using the PVClust software (55) available in the R package (http://www.R-project.org). The data used for the cluster analysis were the relative-abundance data on the genus level, using both the correlation distance option and the average cluster method. Approximately unbiased P values as computed by the PVClust software (55) were reported to determine the significance of the clusters found. Principal-component analyses (PCA) and principal-coordinate analysis (PCoA) were performed in the ape package version 2.8 (48) in the R package. The PCoA was based on a Bray-Curtis dissimilarity matrix produced in the vegan package (version 1.17-12, Oksanen et al. [http://CRAN.R-project.org/package=vegan]). To determine the significance of the effects of horse and stomach region on the microbiota, a permutation test for constrained correspondence analysis was performed in the vegan package.
To determine which phylotypes were most abundant within a sample, representatives of the five most abundant reads were compared to type strains in the RDP database (version 10.19; http://rdp.cme.msu.edu/). The choice to limit the analyses to the five most abundant reads was arbitrary and was made to limit the amount of data analyzed. To get a better understanding of the phylogenetic placement of these sequences, we also performed phylogenetic comparisons of these representatives of the five most abundant reads to type strains in the RDP database. Reads were aligned to the type strains of selected groups using the existing RDP alignment. Phylogenetic inferences were performed in RaxML version 7.0.4 (53) with 1,000 rapid bootstrap replicates to infer robustness of the individual clades. The most abundant reads were extracted from the data sets using tools available in the RDP pyrosequencing pipeline (http://pyro.cme.msu.edu/).
FISH.
The number and spatial distribution of mucosal bacteria in all nine horses and gastric regions (Table 1) were evaluated by fluorescence in situ hybridization (FISH) with labeled oligonucleotide probes directed against bacterial 16S rRNA as previously described (29). Sections were hybridized with a probe that recognizes the vast majority of bacteria (EUB-338, GCTGCCTCCCGTAGGAGT labeled with 6-carboxyfluorescein [6-FAM]) in combination with a probe directed against Streptococcus spp. (STREP, CACTCTCCCCTTCTGCAC labeled with Cy3) (58). There also was a probe directed against Lactobacillus spp. [LAB-158, GGTATTAGCA(C/T)CTGTTTCCA labeled with Cy3] (24). The specificity of hybridization was controlled by the combined application of an irrelevant probe (non-EUB338, ACTCCTACGGGAGGCAGC) and inclusion of control slides spotted with a broad spectrum of bacteria (29). Hybridized samples were washed in phosphate-buffered saline (PBS), air dried, and mounted with the ProLong antifade kit. Sections were examined with an Olympus BX51 epifluorescence microscope and images captured with an Olympus DP-70 camera and analysis system (Olympus America, PA). The numbers of bacteria located in the free mucus, adherent mucus, superficial epithelium, glandular epithelium, and glandular mucus or that were invasive were determined using counts from 10 representative fields of view (magnification, ×60) and expressed as bacteria/mm2. The same person (A.J.B.) performed all bacterial localization and counting.
Nonparametric comparisons were used on the quantitative counts of bacteria/mm2 (derived from FISH) for each stomach region as follows. Friedman's test was used to compare the localization (“depth”) of bacteria within the mucosae of the various stomach regions and for comparison of the number of each type of bacteria in the different regions (squamous, antral, and glandular), all controlled for horse. Significance was set at a P value of ≤0.05 but with a Bonferroni correction (for three stomach regions or for three categories of bacteria), considering any test-wise (2-sided) P value of ≤0.0167 significant. All analyses were performed using Statistix 8 v.8.0 (Analytical Software, Tallahassee, FL).
Accession numbers.
Collection and sequence information has been deposited at MG-RAST (http://metagenomics.anl.gov/) under accession numbers 4468640.3 to 4468664.3.
RESULTS
Gastric morphology.
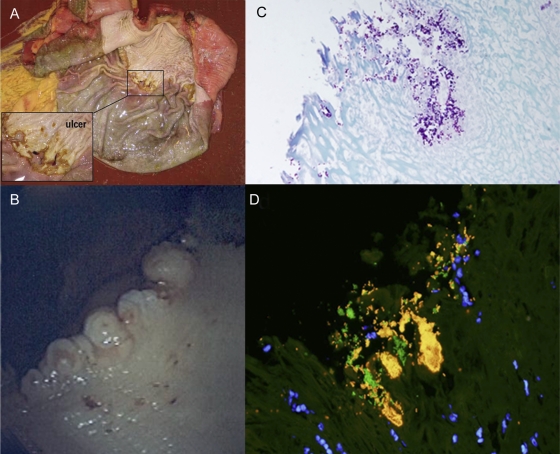
The stomach was macroscopically abnormal in five out of nine horses, with one horse (H3) having a grade 2 ulcer present in the squamous portion adjacent to the margo plicatus, along the greater curvature, at necropsy (Table 1; Fig. 1A). On histopathology, this grade 2 ulcer had marked hyperplasia and parakeratosis with moderate neutrophilic infiltrate and abundant adherent bacteria. The other four horses with macroscopic lesions had multiple (grade 2 or 3 for the number score) small pinpoint (grade 1 for the severity score) ulcers about 2 to 3 mm in diameter in the squamous mucosa adjacent to the margo plicatus at the cardiac region (Fig. 1B). Histopathologically, the pinpoint ulcers seen endoscopically were confirmed to be ulcers in only three horses (H3, H35, and H158) (Fig. 1C and D). One horse (H79) was normal with parakeratosis of the mucosa at this site, while the other (H86) had a mild erosion (Table 1). Endoscopic biopsy specimens taken from the squamous region in five horses were too small (consisting only of superficial parakeratosis or epidermis) for objective evaluation. The gastric mucosa revealed bacteria on all sections that were predominantly Gram-positive cocci, and bacterial colonization appeared to be particularly dense in ulcerated mucosa (Fig. 1C and D).
Fig 1.
The equine stomach during gross postmortem and endoscopic examinations followed by a closer investigation with histopathology and FISH. (A) Postmortem examination of H3, with a grade 2 ulcer in the squamous portion of the stomach near the margo plicatus. (B) Endoscopic view of the cardiac region (H158) of the stomach, with small pinpoint ulcers (grade 1) at the junction of the squamous and glandular regions. (C) Gram stain of ulcerated mucosa (H158) shows colonization by Gram-positive cocci and rods (magnification, ×40). (D) FISH with probes directed against bacteria in general (EUB338, 6-FAM) and Streptococcus spp. (STREP, Cy-3), showing dense colonization of the ulcerated mucosa by Streptococcus spp. (bright yellow) and fewer other bacteria (green). DAPI (4′,6′-diamidino-2-phenylindole)-stained nuclei appear blue. Original magnification, ×60.
Cluster analysis of the equine gastric mucosal microbiome based on relative abundance of bacterial genera shows two distinct clusters.
Pyrosequencing yielded 72,283 total sequences; after quality control, 53,920 sequences were utilized for analysis, with a range from 641 to 5,345 reads in each sample in the six horses analyzed (Table 2). No significant correlation (Pearson's correlation, P > 0.35) was found between the number of reads per sample, the number of OTUs, and the estimated number of OTUs, and hence we conclude that read coverage per sample does not influence estimation of phylotype diversity and richness.
Table 2.
Numbers of reads, observed OTUs, and estimated OTUs (ACE and Chao1 estimators) at the 3 and 5% sequence divergence levels
| Samplea | No. of reads/sample | No. of OTUs |
|||||
|---|---|---|---|---|---|---|---|
| Observed |
Estimated |
||||||
| ACE |
Chao1 |
||||||
| 3% | 5% | 3% | 5% | 3% | 5% | ||
| H2-A | 3,093 | 125 | 105 | 148 | 122 | 147 | 121 |
| H2-G | 4,150 | 126 | 107 | 166 | 128 | 163 | 120 |
| H2-SQ | 2,079 | 113 | 100 | 132 | 114 | 151 | 131 |
| H3-A | 1,278 | 94 | 81 | 126 | 120 | 118 | 108 |
| H3-G | 1,491 | 77 | 62 | 80 | 54 | 74 | 52 |
| H3-SQ | 4,248 | 62 | 46 | 59 | 42 | 57 | 40 |
| H3-U | 4,345 | 69 | 48 | 81 | 53 | 96 | 71 |
| H35-A | 1,613 | 122 | 103 | 129 | 113 | 131 | 116 |
| H35-G | 2,300 | 175 | 140 | 200 | 157 | 204 | 160 |
| H35-SQ | 3,124 | 226 | 174 | 282 | 216 | 287 | 215 |
| H35-U | 1,783 | 133 | 109 | 148 | 117 | 146 | 120 |
| H60-A | 712 | 89 | 80 | 77 | 65 | 81 | 64 |
| H60-G | 1,778 | 156 | 136 | 172 | 149 | 170 | 149 |
| H60-SQ | 3,033 | 192 | 149 | 235 | 179 | 232 | 174 |
| H79-A | 1,576 | 197 | 159 | 236 | 181 | 247 | 188 |
| H79-G | 2,441 | 210 | 165 | 253 | 195 | 243 | 194 |
| H79-PK | 2,008 | 189 | 155 | 235 | 185 | 230 | 178 |
| H79-SQ | 2,183 | 209 | 161 | 240 | 175 | 239 | 176 |
| H86-A | 641 | 80 | 70 | 69 | 62 | 67 | 60 |
| H86-E | 1,611 | 121 | 97 | 136 | 105 | 131 | 101 |
| H86-G | 963 | 106 | 87 | 110 | 88 | 104 | 93 |
| H86-SQ | 787 | 93 | 78 | 100 | 80 | 101 | 80 |
A, antral; G, glandular; SQ, squamous; U, ulcer; PK, parakeratosis; E, erosion.
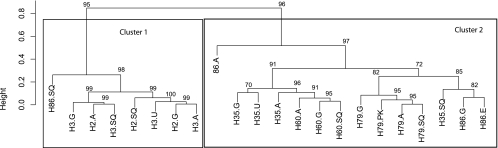
Analyses of the relative abundances of individual bacterial genera showed two distinct clusters with a significant approximately unbiased P value (P > 0.95) (Fig. 2); (i) cluster 1 consisted of samples from horses H2, H3, and H86, and cluster 2 consisted of samples from horses H35, H60, H79, and H86. These clusters segregated by groups which differed by sampling method (fasted and nonfasted; specimens obtained endoscopically and postmortem) and management practice, with differences including but not limited to feeding practices, pasture turnout, exercise, and socializing. Specific known differences are listed in Table 1. The microbiomes did not cluster by gastric region; however, 12 out of 22 samples clustered by horse, indicating more similarity in the relative abundance and taxonomic composition of the microbiotas of different stomach regions within an individual horse than between different horses. While there was generally no significant (P < 0.95) approximately unbiased P value supporting this clustering by horse, a constrained correspondence analysis (CCA) further confirmed that the horse from which a sample is derived has a significant effect (analysis of variance [ANOVA]-like permutation test, P < 0.001) on the microbial community, while the effect of stomach region is not significant (ANOVA-like permutation test, P = 0.561). Samples taken from horse H86 did not cluster exclusively in one of the two clusters; samples from the glandular, antral, and ulcerated regions segregated throughout cluster 2, whereas the squamous region of H86 segregated into cluster 1. Differences in bacterial diversity between cluster 1 (horses H2 and H3) and cluster 2 (horses H35, H60, H79, and H86) were most obvious at the genus (5% divergence) level: the average number of OTUs at the 5% divergence level for cluster 1 was 78 OTUs, whereas it was 128 OTUs in cluster 2. A similar pattern was seen at the species level (3% divergence level). The ACE and Chaol diversity estimators showed similar patterns of a higher microbial diversity in cluster 2 than in cluster 1. The estimated diversity was generally between 3 and 25% higher than the observed number of OTUs. This suggests that the real diversity of OTUs of the individual samples is potentially higher than the observed diversity.
Fig 2.
Hierarchical cluster analysis of samples of the equine gastric mucosa based on relative abundance of sequence reads on the bacterial genus level. The two main clusters found in this analysis coincide with differences in management (feeding, exercise, housing, etc.) and sampling method. Horses sampled immediately postmortem (H2 and H3) and fed hay formed cluster 1, whereas horses biopsied endoscopically after a 12-hour fast (H35, H60, and H79) and fed hay and grass formed cluster 2. The exception was horse H86, which appeared in both clusters. An individual horse had a distinct equine gastric mucosa microbiota without differences between regions. Numbers above the nodes are approximate unbiased values. Labels indicate horse identification number along with stomach region (Sq, squamous; G, glandular; A, antral; U, ulcer; E, erosion; PK, superficial parakeratosis).
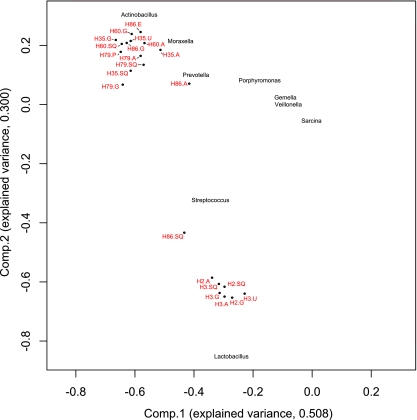
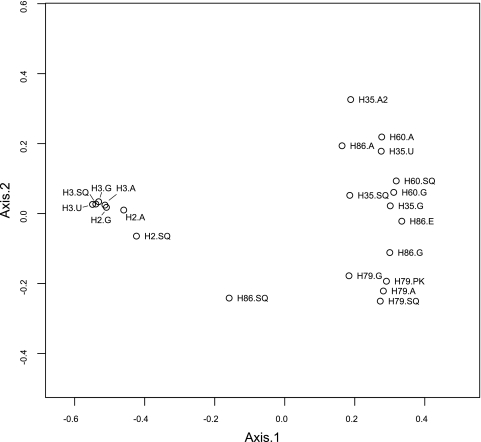
Principal-component analysis of the relative abundances of individual bacterial genera showed that the variation in the data (>80%) could be explained by the first two principal components (Fig. 3). This analysis also confirmed the subdivision of the samples into two main clusters, a cluster consisting of samples obtained from horses H2 and H3 (cluster 1) and a cluster containing samples obtained from horses H35, H60, and H79 and all but one sample from horse H86 (cluster 2). The microbiota of the sample obtained from the squamous region of horse H86 is more similar to those of samples of cluster 1. A limited number of genera were responsible for the variance explained by components 1 and 2; high abundances of Actinobacillus, Moraxella, Prevotella, Porphyromonas, and, to a lesser extent, Gemella and Veillonella were associated with cluster 2, while Lactobacillus, Streptococcus, and Sarcina were associated with cluster 1. The remainder of the principal-component analysis explained a variance in the data of only 7% or less, and none of these components could be associated with factors that were not correlated to the subdivision of cluster 1 and 2, such as ulcer grade. Principal-coordinate analyses of the relative abundance data on the genus level revealed the same subdivision of the samples into two clusters (Fig. 4). In addition, this analysis also showed that samples taken from different regions in an individual horses are more similar to each other than they are to samples from other horses, with the exception of horse H86.
Fig 3.
Principal-component analysis plot of the horse stomach samples (dots) based on the relative abundance of bacterial genera per horse. Labels indicate horse identification number and stomach region (SQ, squamous; G, glandular; A, antral; U, ulcer; E, erosion; P, superficial parakeratosis). Names of the bacterial genera with the highest loadings have been placed in the plot according to their correlation to the first two components.
Fig 4.
Principal-coordinate ordination plot of the samples from the horses, based on a Bray-Curtis matrix derived from the relative abundance of bacterial genera per horse. Labels indicate horse identification number and stomach region (SQ, squamous; G, glandular; A, antral; U, ulcer; E, erosion; PK, = superficial parakeratosis).
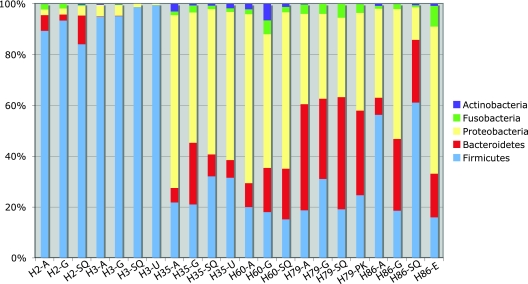
Comparison of the microbiotas on the phylum level (Fig. 5) further shows that cluster 1 and cluster 2 (as found in the cluster analysis based on data on the genus level) differed in the composition and diversity of their microbiotas. The dominant phyla in cluster 1 were Firmicutes (>83% reads/sample), whereas cluster 2 was more diverse, with three main phyla: Proteobacteria (31 to 68% of the reads per sample), Bacteroidetes (6 to 44% of reads per sample), and Firmicutes (15 to 42% of reads per sample).
Fig 5.
Top five phyla found in the gastric mucosae of 6 healthy horses by bTEFAP. The top five phyla represented were Firmicutes, Bacteroidetes, Proteobacteria, Fusobacteria, and Actinobacteria. Cluster 1 horses (H2 and H3) had less diversity, with a predominance of Firmicutes (>82%), whereas horses in cluster 2 (H35, H60, H79, and H86) had a more diverse population consisting of higher percentages of Proteobacteria and Bacteroidetes, as well as Firmicutes. The horse identification number and stomach region (SQ, squamous; G, glandular; A, antral; U, ulcer; E, erosion; PK, superficial parakeratosis) are along the x axis.
A phylogenetic analysis of the five most abundant reads showed further differences in the microbial composition of samples within a cluster and within individual samples (Table 3 gives an overview of the single most abundant read types per sample). The most abundant read types in horses 35, 60, 79, and 86 could be phylogenetically assigned to Eubacterium, Actinobacillus, Moraxella, Acinetobacter, and Veillonella species. Two of three samples from horse H2 were found in a sister group to the 16S sequences of the type strains of Lactobacillus jensenii and L. fornicalis (bootstrap value [BS] of 77). In contrast, the most abundant read type in the squamous region of horse H2 was found in a clade with the type strains of Sarcina maxima and S. ventriculi (BS of 90). The most abundant read type found in two samples (antrum and glandular) from horse H3 are phylogenetically close to the type strain of the horse-associated species Lactobacillus hayakitensis and are found in a moderately supported cluster (BS of 77) together with the type strain in the phylogenetic analysis. The most abundant read type in the squamous region of horse H3 was a Streptococcus species; however, these reads did not cluster with a specific type strain. The most dominant read type in the ulcerated region of horse H3 was found in a well-supported (BS of 87) cluster with another horse-associated species, Lactobacillus equigenerosi (22, 42). Although Helicobacter spp. have been reported to be present in the horse stomach, (14, 16), none of the 53,920 reads matched this genus, indicating the absence of Helicobacter spp. in our samples.
Table 3.
Most abundant species or phylotype in individual samples (values indicate the percentage of the total number of reads)
| Phylotype or speciesb | % of total no. of reads for samplea: |
|||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2A | 2G | 2SQ | 3A | 3G | 3SQ | 3U | 35A | 35G | 35SQ | 35U | 60A | 60G | 60SQ | 79A | 79G | 79SQ | 79PK | 86A | 86G | 86SQ | 86E | |
| Sister group to L. jensenii and L. fornicalis | 19 | 29 | ||||||||||||||||||||
| L. hayakitensis | 19 | 20 | ||||||||||||||||||||
| L. equigenerosi | 18 | |||||||||||||||||||||
| Streptococcus sp. | 24 | |||||||||||||||||||||
| Sarcina sp. | 13 | |||||||||||||||||||||
| Eubacterium sp. | 14 | |||||||||||||||||||||
| Actinobacillus sp. | 19 | 6 | ||||||||||||||||||||
| Moraxella sp. | 9 | 18 | 11 | 6 | 10 | 10 | 8 | 11 | 19 | 22 | ||||||||||||
| Acinetobacter sp. | 7 | |||||||||||||||||||||
| Veillonella sp. | 10 | |||||||||||||||||||||
Samples are designated by horse number and gastric region (A, antral; G, glandular; SQ, squamous; U, ulcer; PK, parakeratosis; E, erosion).
Assignment of the most abundant read type to a species or phylotype was done using a phylogenetic approach; reads were aligned to existing 16S alignments of type sequences obtained from the RDP database, and phylogenetic similarity was inferred by a maximum-likelihood analysis (using RaxML version 7.0.4 [53]).
FISH demonstrates gastric colonization by mucosal bacteria.
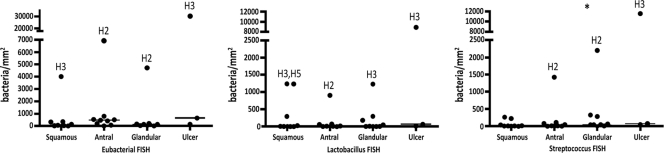
FISH analysis was performed with probes directed against all bacteria (eubacterial, EUB-338), Lactobacillus spp. (LAB-158), and Streptococcus spp. (STREP). An abundant mucosal microbiota was present in all regions of the gastric mucosa examined (Fig. 6). For all nine horses, the glandular region had significantly more Streptococcus bacteria (P = 0.0098) than the squamous and antral regions (Fig. 7). However, neither Lactobacillus spp. (P = 0.31) nor total bacteria (P = 0.034) differed by region. Horse H3 (with the grade 2 ulcer) had the most bacteria at the ulcer site (30,530 bacteria/mm2) (Fig. 7). When looking at specific localization (“depth”) of bacteria within the mucosa, in all regions and for the Streptococcus and eubacterial probes, significantly more bacteria were located in the tightly adherent mucus, followed by the free mucus and the superficial epithelium (P ≤ 0.015 for all by Friedman's test). The same was true for the glandular region and ulcers for the Lactobacillus probe (P ≤ 0.014 for both), but the P values for the squamous (P = 0.055) and antral (P = 0.025) regions indicated no significant variation in Lactobacillus numbers by “depth.” Deeply invasive bacteria were restricted to the ulcerated mucosa.
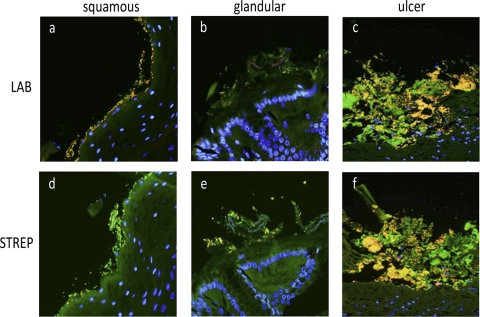
Fig 6.
FISH shows abundant bacteria in close apposition to the equine gastric mucosa. Histologic sections from the squamous (a and d), glandular (b and e), and ulcerated (c and f) mucosa were examined using labeled oligonucleotide probes directed against bacteria in general (EUB-338, 6-FAM, green) in combination with probes directed against Lactobacillus spp. (a, b, and c) (Cy-3) or Streptococcus spp. (d, e, and f) (Cy-3). In healthy squamous mucosa, Lactobacillus spp. (a) (yellow) and Streptococcus spp. (d) (yellow) are in close apposition to the epithelium. In the glandular region, both Lactobacillus spp. (b) (yellow) and Streptococcus spp. (e) (yellow) colonize the mucosa but appear to be present in smaller amounts than in the squamous region. Ulcerated mucosa from H3 (c and f) contains an abundant mixed bacterial population that includes Streptococcus spp. (f) (yellow), Lactobacillus spp. (c) (yellow), and other bacterial species (c and f) (green). Nuclei are blue (DAPI). Original magnification, ×600.
Fig 7.
Healthy horses generally have an abundance of bacteria of <1,000 bacteria/mm2 in all gastric mucosal samples by as determined by FISH (n = 9). FISH was done with probes directed against bacteria in general (eubacteria), Streptococcus spp., and Lactobacillus spp. A few horses (H2, H3, and H35) had outliers as indicated. Horse 3's grade 2 ulcer had an abundance of bacteria (30,529 bacteria/mm2). All other horses had small pinpoint ulcers, with total numbers of bacteria similar to those in other regions of the stomach. Each symbol represents a horse. There were significantly more Streptococcus bacteria in the glandular mucosa than in the squamous and antral mucosae (P = 0.0098), as indicated by the asterisk.
DISCUSSION
Little is known about the gastric mucosal microbiota in healthy horses, and its role in gastric disease has not previously been examined. Only 30% of the fecal microbiota in people is considered cultivable, and advances in molecular microbiology have revealed important variation in the microbiota between different gastrointestinal segments and in luminal contents versus mucosa in healthy individuals (20, 25, 27, 33). In the present study, we used a combination of culture-independent approaches, 16S rRNA bacterial tag-encoded pyrosequencing (bTEFAP) and FISH to characterize the composition and spatial distribution of the gastric mucosal microbiota of healthy horses. The results of our analyses show that (i) Firmicutes, Proteobacteria, and Bacteroidetes are the dominant bacterial phyla in the equine gastric mucosa in this set of horses, (ii) based on the taxonomic composition of the gastric microbiota, this set of horses can be subdivided into two groups, (iii) certain species of Lactobacillus, which have been previously associated with equine fecal matter, are also present in the gastric mucosae of the horses, (iv) in particular Lactobacillaceae and Streptococcaceae form a distinct association with the gastric mucosa, and (v) Helicobacter is not a abundant taxon in the microbiota of healthy horses.
Firmicutes, Proteobacteria, and Bacteroidetes are the dominant bacterial phyla in the equine gastric mucosa.
This culture-independent approach enabled us to identify Firmicutes, Proteobacteria, and Bacteroidetes as the dominant bacterial phyla in the equine gastric mucosa in this set of horses. These phyla were also identified as the dominant phyla in the human gastric environment (11) and mammalian gut microbiota (34), suggesting that these phyla represent the dominant bacterial phyla in the mammalian gastrointestinal tract. While the human stomach environment seems to be dominated by Proteobacteria (11), the dominant phylum seems to differ by sample and horse, with Firmicutes being the most dominant phylum in all samples from horses H2 and H3 and in samples from the squamous and antral regions of horse H86, while Proteobacteria were dominant in samples from horses H35 and H60 and the glandular and erosion samples from horse H86 (Fig. 3). Bacteroidetes, while not necessarily being the dominant phylum, made up a large proportion of the microbiota found in horse H79. The composition of the gastric mucosal microbiota therefore seems to differ substantially per horse.
Based on the taxonomic composition of the gastric microbiota, gastric samples from horses can be subdivided into two groups.
Cluster analyses and factorial analyses (both principal-component analysis and principal-coordinate analysis) of the composition of the samples at the genus level show that the samples cluster into two groups: (i) a group (cluster 1) consisting of samples dominated by Firmicutes, in particular the genera Lactobacillus, Streptococcus, and Sarcina, and (ii) a group (cluster 2) consisting of samples dominated by either Proteobacteria (in particular Actinobacillus and Moraxella) or Bacteroidetes (Prevotella and Porphyromonas). We found that this subdivision overlapped remarkably well with the way the samples were obtained from the horses and the husbandry of the horses (the only exception being horse H86). For example, horses in cluster 1 were stabled, fed hay, and sampled postmortem, while horses in cluster 2 were pastured on grass, fed hay, and biopsied gastroscopically after a 12-h fast. It is important to note that these two groups of horses had many other differences, such as housing, socialization, exercise, and others, that may have contributed to the differences in the bacterial communities present (Table 1 gives an overview). De Fombelle et al. (15) commented that the stomach ecosystem seemed the most affected by the composition of the last pelleted meal ingested, and hence nutrition and the 12-h fast maybe the main reasons for the differences seen between these two groups of horses. The 12-h fast needed to visualize the stomach by endoscopy may alter bacterial types and numbers because of a lack of substrate available for microorganisms as well as alterations in environmental conditions, specifically a decrease in gastric pH (2, 39, 45, 54). The human stomach microbiota had bacterial diversity similar to that of our gastroscopically sampled horses and was also studied by taking gastroscopic biopsy specimens after a period of fasting (11). In essence, our two nonfasted horses sampled immediately postmortem demonstrate a unique data set and bacterial community, with a predominance of Firmicutes, although it is unknown how much the gastric microbiota changes in the short time from euthanasia to retrieval of the stomach biopsy specimens. Whether these two horses represent the normal equine gastric environment and microbiota better than the four horses sampled endoscopically after a 12-h fast is yet to be determined. Another factor that could have contributed to the difference observed between the two clusters is the fact that the abundance of lactic acid bacteria can be heavily skewed by differences in PCR conditions and methods of DNA isolation (40). While the primary goal of this study was to determine the microbial diversity in normal and abnormal equine gastric mucosae in healthy horses, the overwhelming difference in the composition of the microbiota of samples in cluster 1 versus cluster 2 suggests that other factors that were not accounted for in this study may affect the composition of the microbiota. Future studies should therefore take factors such as husbandry, sex, and age into account in the study design.
Certain species of Lactobacillus, which have been previously associated with equine fecal matter, are also present in the gastric mucosae of the horses.
Phylogenetic analyses of the most dominant read types found among the different samples showed that the sequences obtained by 16S rRNA bacterial tag-encoded pyrosequencing (bTEFAP) were phylogenetically informative enough to identify reads assigned to Lactobacillus to species, while reads assigned to other genera (for instance, Moraxella) were not. We found one group of reads (the most dominant read type in the antral and glandular samples from horse 2) to be a sister group of L. jensenii and L. fornicalis, which may represent a taxonomically uncharacterized species. In addition to this group, we found reads that clustered with the type strains of L. hayakitensis and L. equigenerosi, two distinct Lactobacillus species that were reported to be the predominant lactobacilli in the intestinal microbiota of healthy thoroughbred horses (41). The observation that these species are also abundant in the gastric mucosae of healthy horses suggests that these species may also form a predominant component of the local microbiota of the gastric mucosa.
Lactobacillaceae and Streptococcaceae form a distinct association with the gastric mucosa.
FISH delivered additional evidence that at least some part of the microbiota observed by pyrosequencing is part of the local microbiota of the equine gastric mucosa and does not present an amalgam of transitional bacteria that have entered the gastric environment through a nasal pharyngeal route. Lactobacillus- and Streptococcus-specific probes showed the majority of the bacteria being tightly adhered to the mucosal surface and to any ulcerated mucosa. Similar observations of lactobacilli closely adhered to the squamous epithelium of the horse stomach were made by Yuki et al. (62). They identified these lactobacilli as L. salivarius, L. crispatus, L. reuteri, and L. agilis by 16S rRNA gene sequence analysis and DNA-DNA hybridization. Yuki et al. (62) further found that these bacteria adhered to horse epithelial cells but not to rat epithelial cells, suggesting host specificity of these Lactobacillus strains. The study by Yuki et al. and the results of our FISH experiments combined with the pyrosequencing results thus suggest that the gastric mucosa is inhabited by highly host-specific lactobacilli and streptococci.
Helicobacter is not an abundant taxon in the microbiota of healthy horses.
The microbiota found in abnormal (e.g., ulcerated or eroded) regions of the gastric mucosa did not differ from that in normal regions, nor could we find any specific groups of bacteria among the abundant groups that were specific for ulcerated regions. An important finding was the absence of Helicobacter sequences in over 53,920 reads from healthy horses. On a whole-horse basis, none of the six healthy horses were carriers of Helicobacter spp.; however, we acknowledge that the upper 95% confidence limit on 0/9 would allow for as high as 37% carrier prevalence. There has only been weak historical evidence of Helicobacter infection in association with ulcers in horses (14, 16), and Helicobacter spp. were not identified in 36 horses with antral pathology using FISH (28). Taken as a whole, these studies indicate that Helicobacter spp. are probably not a common asymptomatic inhabitant of the equine stomach, and their role in gastric ulceration in the horse is yet to be determined.
This study establishes the presence of a rich and diverse mucosa-associated bacterial microbiota in the stomachs of healthy horses, providing a starting point for research on the function of the native microbiota in the equine gastrointestinal tract.
ACKNOWLEDGMENTS
We thank M. Baumgart and S. Janecsko for their assistance with the laboratory work and especially thank Paula Sharp and Francis Davis for their technical assistance.
J. Parker was supported by the Cornell Leadership Program and the Wellcome Trust, and R. L. Rosenthal was supported by a Research Apprenticeship in Biological Sciences at Cornell University. This project was funded by the U.S. Equestrian Federation, Inc.
Footnotes
Published ahead of print 3 February 2012
REFERENCES
- 1. Acosta-Martinez V, Dowd SE, Sun Y, Allen V. 2009. Tag-encoded pyrosequencing analysis of bacterial diversity in a single soil type as affected by management and land use. Soil Biol. Biochem. 4:2762–2770 [Google Scholar]
- 2. Alexander F. 1972. Certain aspects of the physiology and pharmacology of the horse's digestive tract. Equine Vet. J. 4:166–169 [Google Scholar]
- 3. Al Jassim RAM, Scott T, Trebbin AL, Trott D, Pollitt CC. 2005. The genetic diversity of lactic acid producing bacteria in the equine gastrointestinal tract. FEMS Microbiol. Lett. 248:75–81 [DOI] [PubMed] [Google Scholar]
- 4. Al Jassim RAM. 2006. Supplementary feeding of horses with processed sorghum grains and oats. Anim. Feed Sci. Tech. 125:33–44 [Google Scholar]
- 5. Andrews FM, Buchanan BR, Elliot SB, Clairday NA, Edwards LH. 2005. Gastric ulcers in horses. J. Anim. Sci. 83:E18–E21 [Google Scholar]
- 6. Andrews FM, et al. 2008. In vitro effects of hydrochloric and lactic acids on bioelectric properties of equine gastric squamous mucosa. Equine Vet. J. 40:301–305 [DOI] [PubMed] [Google Scholar]
- 7. Andrews FM, Buchanan BR, Smith SH, Elliott SB, Saxton AM. 2006. In vitro effects of hydrochloric acid and various concentrations of acetic, propionic, butyric, or valeric acids on bioelectric properties of equine gastric squamous mucosa. Am. J. Vet. Res. 67:1873–1882 [DOI] [PubMed] [Google Scholar]
- 8. Argenzio RA. 1999. Comparative pathophysiology of nonglandular ulcer disease: a review of experimental studies. Equine Vet. J. Suppl. 29:19–23 [DOI] [PubMed] [Google Scholar]
- 9. Begg LM, O'Sullivan CB. 2003. The prevalence and distribution of gastric ulceration in 345 racehorses. Aust. Vet. J. 81:199–201 [DOI] [PubMed] [Google Scholar]
- 10. Bell RJ, Mogg TD, Kingston JK. 2007. Equine gastric ulcer syndrome in adult horses: a review. N. Z. Vet. J. 55:1–12 [DOI] [PubMed] [Google Scholar]
- 11. Bik EM, et al. 2006. Molecular analysis of the bacterial microbiota in the human stomach. Proc. Natl. Acad. Sci. U. S. A. 103:732–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Buchanan BR, Andrews FM. 2003. Treatment and prevention of equine gastric ulcer syndrome. Vet. Clin. North Am. Equine Pract. 19:575–597 [DOI] [PubMed] [Google Scholar]
- 13. Chao A, Bunge J. 2002. Estimating the number of species in a stochastic abundance model. Biometrics 58:531–539 [DOI] [PubMed] [Google Scholar]
- 14. Contreras M, et al. 2007. Detection of Helicobacter-like DNA in the gastric mucosa of Thoroughbred horses. Lett. Appl. Microbiol. 45:553–557 [DOI] [PubMed] [Google Scholar]
- 15. de Fombelle A, et al. 2003. Characterization of the microbial and biochemical profile of the different segments of the digestive tract in horses given two distinct diets. Anim. Sci. 77:293–304 [Google Scholar]
- 16. Dimola S, Caruso ML. 1999. Helicobacter pylori in animals affecting the human habitat through the food chain. Anticancer Res. 19:3889–3894 [PubMed] [Google Scholar]
- 17. Dorer MS, Talarico S, Salama NR. 2009. Helicobacter pylori's unconventional role in health and disease. PLoS Pathog. 5:e1000544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dowd SE, et al. 2008. Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP). BMC Microbiol. 8:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dowd SE, et al. 2008. Polymicrobial nature of chronic diabetic foot ulcer biofilm infections determined using bacterial tag encoded FLX amplicon pyrosequencing (bTEFAP). PLoS One 3:e3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Eckburg PB, et al. 2005. Diversity of the human intestinal microbial flora. Science 308:1635–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Endo A, Roos S, Satoh E, Morita H, Okada S. 2008. Lactobacillus equigenerosi sp. nov., a coccoid species isolated from faeces of Thoroughbred racehorses. Int. J. Syst. Evol. Microbiol. 58:914–918 [DOI] [PubMed] [Google Scholar]
- 23. Felsenstein J. 1989. PHYLIP—Phylogeny Inference Package (version 3.2). Cladistics 5:164–166 [Google Scholar]
- 24. Franks AH, et al. 1998. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 64:3336–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Goodman AL, et al. 2011. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc. Natl. Acad. Sci. U. S. A. 108:6252–6257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Handl S, Dowd SE, Garcia-Mazcorro JF, Steiner JM, Suchodolski JS. 2011. Massive parallel 16S rRNA gene pyrosequencing reveals highly diverse fecal bacterial and fungal communities in healthy dogs and cats. FEMS Microbiol. Ecol. 76:301–310 [DOI] [PubMed] [Google Scholar]
- 27. Hayashi H, Sakamoto M, Benno Y. 2002. Phylogenetic analysis of the human gut microbiota using 16S rDNA clone libraries and strictly anaerobic culture-based methods. Microbiol. Immunol. 46:535–548 [DOI] [PubMed] [Google Scholar]
- 28. Husted L, Jensen TK, Olsen SN, Molbak L. 2010. Examination of equine glandular stomach lesions for bacteria, including Helicobacter spp by fluorescence in situ hybridisation. BMC Microbiol. 10:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Janeczko S, et al. 2008. The relationship of mucosal bacteria to duodenal histopathology, cytokine mRNA, and clinical disease activity in cats with inflammatory bowel disease. Vet. Microbiol. 128:178–193 [DOI] [PubMed] [Google Scholar]
- 30. Kellam LL, Johnson PJ, Kramer J, Keegan KG. 2000. Gastric impaction and obstruction of the small intestine associated with persimmon phytobezoar in a horse. J. Am. Vet. Med. Assoc. 216:1279–1281 [DOI] [PubMed] [Google Scholar]
- 31. Kellermayer R, et al. 2011. Colonic mucosal DNA methylation, immune response, and microbiome patterns in Toll-like receptor 2-knockout mice. FASEB J. 25:1449–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lemos LN, Fulthorpe RR, Triplett EW, Roesch LF. 2011. Rethinking microbial diversity analysis in the high throughput sequencing era. J. Microbiol. Methods 86:42–51 [DOI] [PubMed] [Google Scholar]
- 33. Lepage P, et al. 2005. Biodiversity of the mucosa-associated microbiota is stable along the distal digestive tract in healthy individuals and patients with IBD. Inflamm. Bowel Dis. 11:473–480 [DOI] [PubMed] [Google Scholar]
- 34. Ley RE, et al. 2008. Evolution of mammals and their gut microbes. Science 320:1647–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li XX, et al. 2009. Bacterial microbiota profiling in gastritis without Helicobacter pylori infection or non-steroidal anti-inflammatory drug use. PLoS One 4:e7985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. MacAllister CG, Andrews FM, Deegan E, Ruoff W, Olovson SG. 1997. A scoring system for gastric ulcers in the horse. Equine Vet. J. 29:430–433 [DOI] [PubMed] [Google Scholar]
- 37. Marshall BJ, Warren JR. 1984. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet i:1311–1315 [DOI] [PubMed] [Google Scholar]
- 38. McClure SR, Glickman LT, Glickman NW. 1999. Prevalence of gastric ulcers in show horses. J. Am. Vet. Med. Assoc. 215:1130–1133 [PubMed] [Google Scholar]
- 39. Merritt AM. 1999. Normal equine gastroduodenal secretion and motility. Equine Vet. J. Suppl. 29:7–13 [DOI] [PubMed] [Google Scholar]
- 40. Morgan JL, Darling AE, Eisen JA. 2010. Metagenomic sequencing of an in vitro-simulated microbial community. PLoS One 5:e10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Morita H, et al. 2009. Lactobacillus hayakitensis, L. equigenerosi and L. equi, predominant lactobacilli in the intestinal flora of healthy Thoroughbreds. Anim. Sci. J. 80:339–346 [DOI] [PubMed] [Google Scholar]
- 42. Morita H, et al. 2007. Lactobacillus hayakitensis sp. nov., isolated from intestines of healthy Thoroughbreds. Int. J. Syst. Evol. Microbiol. 57:2836–2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Murray MJ, Eichorn ES, Jeffrey SC. 2001. Histological characteristics of induced acute peptic injury in equine gastric squamous epithelium. Equine Vet. J. 33:554–560 [DOI] [PubMed] [Google Scholar]
- 44. Murray MJ, Nout YS, Ward DL. 2001. Endoscopic findings of the gastric antrum and pylorus in horses: 162 cases (1996-2000). J. Vet. Intern. Med. 15:401–406 [PubMed] [Google Scholar]
- 45. Murray MJ, Schusser GF. 1993. Measurement of 24-h gastric pH using an indwelling pH electrode in horses unfed, fed and treated with ranitidine. Equine Vet. J. 25:417–421 [DOI] [PubMed] [Google Scholar]
- 46. Nadeau JA, et al. 2003. Effects of hydrochloric, acetic, butyric, and propionic acids on pathogenesis of ulcers in the nonglandular portion of the stomach of horses. Am. J. Vet. Res. 64:404–412 [DOI] [PubMed] [Google Scholar]
- 47. Nadeau JA, et al. 2003. Effects of hydrochloric, valeric, and other volatile fatty acids on pathogenesis of ulcers in the nonglandular portion of the stomach of horses. Am. J. Vet. Res. 64:413–417 [DOI] [PubMed] [Google Scholar]
- 48. Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20:289–290 [DOI] [PubMed] [Google Scholar]
- 49. Priestnall SL, et al. 2004. Evaluation of “Helicobacter heilmannii” subtypes in the gastric mucosas of cats and dogs. J. Clin. Microbiol. 42:2144–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Roesch LF, et al. 2007. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 1:283–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schloss PD, Handelsman J. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Smith DM, et al. 2010. Evaluation of the bacterial diversity of pressure ulcers using bTEFAP pyrosequencing. BMC Med. Genomics 3:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690 [DOI] [PubMed] [Google Scholar]
- 54. Stick JA, Robinson NE, Krehbiel JD. 1981. Acid-base and electrolyte alterations associated with salivary loss in the pony. Am. J. Vet. Res. 42:733–737 [PubMed] [Google Scholar]
- 55. Suzuki R, Shimodaira H. 2006. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22:1540–1542 [DOI] [PubMed] [Google Scholar]
- 56. Taylor SD, Haldorson GJ, Vaughan B, Pusterla N. 2009. Gastric neoplasia in horses. J. Vet. Intern. Med. 23:1097–1102 [DOI] [PubMed] [Google Scholar]
- 57. Trebesius K, Adler K, Vieth M, Stolte M, Haas R. 2001. Specific detection and prevalence of Helicobacter heilmannii-like organisms in the human gastric mucosa by fluorescent in situ hybridization and partial 16S ribosomal DNA sequencing. J. Clin. Microbiol. 39:1510–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Trebesius K, et al. 2000. Culture independent and rapid identification of bacterial pathogens in necrotising fasciitis and streptococcal toxic shock syndrome by fluorescence in situ hybridisation. Med. Microbiol. Immunol. 188:169–175 [DOI] [PubMed] [Google Scholar]
- 59. Vainio K, Sykes BW, Blikslager AT. 2011. Primary gastric impaction in horses: a retrospective study of 20 cases (2005-2008). Equine Vet. Educ. 23:186–190 [Google Scholar]
- 60. Varloud M, Fonty G, Roussel A, Guyonvarch A, Julliand V. 2007. Postprandial kinetics of some biotic and abiotic characteristics of the gastric ecosystem of horses fed a pelleted concentrate meal. J. Anim. Sci. 85:2508–2516 [DOI] [PubMed] [Google Scholar]
- 61. Vatistas NJ, et al. 1999. Cross-sectional study of gastric ulcers of the squamous mucosa in Thoroughbred racehorses. Equine Vet. J. Suppl. 31:34–39 [DOI] [PubMed] [Google Scholar]
- 62. Yuki N, et al. 2000. Colonization of the stratified squamous epithelium of the nonsecreting area of horse stomach by lactobacilli. Appl. Environ. Microbiol. 66:5030–5034 [DOI] [PMC free article] [PubMed] [Google Scholar]