Abstract
Dimethylsulfoniopropionate (DMSP) is an organic sulfur compound that is rapidly metabolized by marine bacteria either by cleavage to dimethylsulfide (DMS) or demethylation to 3-methiolpropionate. The abundance and diversity of genes encoding bacterial DMS production (dddP) and demethylation (dmdA) were measured in the North Pacific subtropical gyre (NPSG) between May 2008 and February 2009 at Station ALOHA (22°45′N, 158°00′W) at two depths: 25 m and the deep chlorophyll maximum (DCM; ∼100 m). The highest abundance of dmdA genes was in May 2008 at 25 m, with ∼16.5% of cells harboring a gene in one of the eight subclades surveyed, while the highest abundance of dddP genes was in July 2008 at 25 m, with ∼2% of cells harboring a gene. The dmdA gene pool was consistently dominated by homologs from SAR11 subclades, which was supported by findings in metagenomic data sets derived from Station ALOHA. Expression of the SAR11 dmdA genes was low, with typical transcript:gene ratios between 1:350 and 1:1,400. The abundance of DMSP genes was statistically different between 25 m and the DCM and correlated with a number of environmental variables, including primary production, photosynthetically active radiation, particulate DMSP, and DMS concentrations. At 25 m, dddP abundance was positively correlated with pigments that are diagnostic of diatoms; at the DCM, dmdA abundance was positively correlated with temperature. Based on gene abundance, we hypothesize that SAR11 bacterioplankton dominate DMSP cycling in the oligotrophic NPSG, with lesser but consistent involvement of other members of the bacterioplankton community.
INTRODUCTION
Dimethylsulfoniopropionate (DMSP) is a common sulfur compound produced by phytoplankton for use as an osmolyte (40, 60). When released into seawater, DMSP is rapidly sequestered and degraded by members of the bacterioplankton community via two major metabolic pathways (28–30). The majority of DMSP (50 to 90%) is demethylated to 3-methiolpropionate (MMPA), ultimately producing sulfur and carbon intermediates which are incorporated into microbial biomass or further oxidized (29, 44). A competing metabolic pathway results in the production of dimethylsulfide (DMS) from DMSP (16, 21). DMS represents a major source of biogenic sulfur to the atmosphere, where oxidation products form cloud condensation nuclei and ultimately influence radiative backscatter (2, 33, 49).
Recent insights into the molecular mechanisms that drive bacterial DMSP degradation have provided an improved understanding of DMSP cycling at the genomic and transcriptional levels (5, 18, 44, 54, 56, 57). The identification of the DMSP demethylase gene (dmdA), which encodes the first step in the demethylation pathway, has enabled quantification of the gene in marine metagenomic surveys and revealed it to be taxonomically diverse and highly abundant (present in >50% of marine bacterioplankton) (19). To date, dmdA homologs, represented by 5 clades and 14 subclades, are known to be harbored by SAR11, roseobacters, Gammaproteobacteria, and SAR116 member “Candidatus Puniceispirillum marinum” IMCC1322 (19, 20, 62). While there is strong cohesion in dmdA-encoded amino acid sequences, there is extensive heterogeneity based on nucleotide sequences (19, 62). In comparison to dmdA, the genes involved in DMS production (dddD, dddL, dddP dddQ, dddY, and dddW, all of which mediate the same step of DMSP cleavage) are present in less than 10% of bacteria based on marine metagenomic surveys (9, 10, 19, 54–57). The most abundant ddd genes in bacterial taxa are dddP and dddQ, occurring in genomes of some roseobacters (19, 54, 55) and SAR116.
The objective of this study was to measure the distributions of genes diagnostic of DMSP degradation in the North Pacific subtropical gyre (NPSG), the world's largest biome with a surface area of 1 × 107 km2 (25). Station (Stn) ALOHA is located in the NPSG at 22°45′N, 158°00′W, and it represents the sampling site of the Hawaii Ocean Time-series (HOT) (23, 25), where a suite of biogeochemical and physical parameters are measured on a near-monthly basis to characterize the long-term biogeochemical cycling in this oligotrophic oceanic ecosystem. Here, we measured dmdA and dddP abundance and expression at two depths, at 25 m in the nutrient-depleted upper euphotic zone and at the persistent deep chlorophyll maximum (DCM), typically located at ∼100 m in the lower euphotic zone (24). Over a 10-month period, genetic and chemical analyses were conducted to identify the bacterial taxa and environmental factors that potentially influence DMSP fate in this ecosystem, including DMS and particulate DMSP (DMSPp) concentrations. Our results indicate that the targeted DMSP-degrading genes are abundant in surface waters of the NPSG, particularly those from the SAR11 clade, and that they show greater depth variability than temporal variability.
MATERIALS AND METHODS
Biogeochemical parameters.
Samples were collected at Stn ALOHA between May 2008 and February 2009 during HOT cruises 201 to 209. The core physical, chemical, and biological measurements were conducted as part of the HOT program as previously described (25, 26) and are available in the Hawaii Ocean Time-series Data Organization and Graphical System (HOT-DOGS; http://hahana.soest.hawaii.edu/hot/hot-dogs/interface.html). The mixed layer depth was calculated using the 0.125 potential density criterion (38). The average pH and salinity were 8.05 and 35.19, respectively, over the sampling period. Seawater concentrations of DMS and DMSP were quantified onboard using a cryogenic purge-and-trap technique followed by gas chromatography, as previously described (59). In brief, filtered seawater samples were sparged with helium and trapped in a sample loop maintained in liquid nitrogen. The sample loop was subsequently heated, and DMS was quantified using a gas chromatograph (Agilent 7890) equipped with a flame photometric detector (FPD) and a Chromasil 330 chromatography column. To measure particulate DMSP (DMSPp), a proxy for DMSP within phytoplankton, seawater samples were gently filtered through glass fiber filters, and the filters were exposed to NaOH (1 M) in gas-tight vials to hydrolyze DMSP to DMS, which was measured as described above.
Nucleic acid collection and extraction.
Samples for DNA and RNA analysis were collected from 25 m and the DCM, which ranged from 110 to 140 m during the sampling period (see Table S1 in the supplemental material). Samples were obtained in triplicate from the same conductivity-temperature-depth (CTD) cast as the DMSP and DMS measurements and filtered through 25-mm-diameter 0.2-μm-pore-size Durapore filters. Approximately 1.8 liters of seawater was filtered for each DNA replicate and 1.4 to 1.9 liters for RNA replicates. For RNA samples, the filtration time averaged 15 min and was never >20 min. Immediately following filtration, filters were placed into 2-ml cryogenic vials, and 250 μl lysis buffer (20 mM Tris HCl, 2 mM EDTA, 1.2% Triton X, and 20 mg/ml lysozyme) was added to DNA filters and 250 μl RLT buffer (Qiagen, Valencia, CA) to RNA filters. All tubes, particularly RNA samples, were then immediately flash frozen in liquid nitrogen and subsequently stored at −80°C. DNA extractions were carried out with the DNeasy blood and tissue kit (Qiagen) following the pretreatment protocol for Gram-positive cells. RNA extractions were carried out using the RNeasy minikit (Qiagen). Immediately prior to extraction, an additional 600 μl of RLT buffer and 0.5 ml silica carbide beads (MO BIO Laboratories Inc., Carlsbad, CA) were added to the sample tube, which was placed on a bead beating vortex adapter for 5 min. The lysate was transferred to a new tube, and the rest of the extraction protocol was conducted as described in the RNeasy manual. Following extraction, RNA samples were DNase digested using the Turbo DNA-free kit (Ambion, Austin, TX) with double the enzyme volume. DNA and RNA samples used in quantitative PCR (qPCR) were quantified by Quant-iT PicoGreen and RiboGreen (Invitrogen Ltd., Carlsbad, CA) kits, respectively, using a TBS-380 fluorometer (Promega, Sunnyvale, CA) or for some RNA samples by a NanoDrop spectrophotometer (Thermo Fisher Scientific Inc., Wilmington, DE). Average DNA concentrations ranged from 0.2 ± 0.15 μg per liter at 25 m and 0.17 ± 0.12 μg per liter at the DCM. Average RNA concentrations ranged from 0.18 ± 0.17 μg per liter at 25 m and 0.05 ± 0.04 μg per liter at the DCM. The lower-concentration samples (<5 ng/μl) may have greater measurement error, particularly for NanoDrop quantification.
Quantitative and RT-qPCR.
The primer set sequences for dmdA subclades A/1, A/2, B/3, B/4, C/2, D/1, D/3, and E/2 primer and the corresponding annealing temperatures were described by Varaljay et al. (62), and the dddP primer set targeting group 1 was described by Levine et al. (32). The 16S rRNA BACT1369F and PROK1492R primers were from Suzuki et al. (53). As previously described (32), all quantitative PCRs (qPCRs) were run in duplicate in 25.0-μl volumes with iQ SYBR Green Supermix (Bio-Rad, Hercules, CA) and 300 nM final primer concentrations. qPCR product size and specificity were verified by agarose gel electrophoresis. For reverse transcription (RT)-qPCR assays, the 1× iScript one-step RT-PCR kit with SYBR green was used with specific priming cDNA reverse transcription, as specific priming is more sensitive than random hexamer priming. Additionally, for RT-qPCR, a 1.0-ng/μl final concentration of T4 gene 32 protein (Roche Applied Science, Indianapolis, IN) was added to decrease PCR inhibition (8, 32). For DNA and RNA samples, 3.0 μl and 5.0 μl of template were added to each reaction, respectively. All reactions were run on an iCycler iQ (Bio-Rad, Hercules, CA) with the following cycling conditions: initial denaturation at 95°C for 5 min, 40 cycles of 95°C for 30 s, specified annealing temperature (32, 62) for 30 s, extension at 72°C for 30 s, and then a final denaturation and annealing for 1 min each and a melt curve following each run. RT-qPCR cycling conditions were the same, except a 10-min 50°C reverse transcription step was included prior to the initial denaturation. The limit of detection was determined by (i) amplification above the lowest concentration of the standard curve (ranging from 5 to 300 copies per reaction across primer sets), (ii) specificity of melt curves, and (iii) minimal contamination (≥3 cycle difference) in the no-template or negative reverse transcriptase controls compared to the sample reactions. To normalize for any differences in extraction efficiency, all gene data were analyzed as a percentage of cells harboring the gene based on 16S rRNA qPCR (assuming 1.4 16S rRNA genes per cell as calculated from Stn ALOHA metagenomic 16S rRNA:recA estimates; see below).
All qPCR standards were constructed from TOPO TA (Invitrogen, Carlsbad, CA) plasmid clones with PCR gene product inserts. Product inserts and specificity of primer sets were previously verified for dmdA using 454 sequencing (as described in reference 62) and for dddP using Sanger sequencing (as described in reference 32). Five product inserts for 16S rRNA were sequenced from DNA obtained in this study, and one of these was used as a qPCR standard. Tenfold serially diluted standards were run on every DNA and RNA plate, with an average r2 of 0.997 and an efficiency of ∼94% across all standard curves.
Multidimensional scaling and statistical analyses.
The gene abundance and biogeochemical data sets were compared using multidimensional scaling (MDS) in R (42) using the vegan package and Bray-Curtis similarities (39). All gene data were normalized on a scale from 0 to 1 for the MDS analysis. Subclade B/4 was removed from analyses due to low abundance and nonspecific amplification. Three axes were chosen based on a significant decrease in stress; however, only the first and second axes, which contributed the most to the gene distribution variability (81% and 12%, respectively), were finally considered. The MDS output consisted of two matrices of similarity scores: (i) sample similarity scores based on the gene abundance patterns (n = 15) and (ii) gene similarity scores based on the sample patterns (n = 8). Both sets of scores were plotted on the MDS axes. Since these data did not appear normally distributed, nonparametric Spearman's rank correlations were determined between MDS axes and available environmental data, including depth, month, DMSPp, DMS, chlorophyll a (Chl a), photosynthetically available radiation (PAR), dissolved organic carbon (DOC), silicate, nitrate-nitrite, temperature, salinity, primary production, fucoxanthin, 19′-butanoyloxyfucoxanthin, 19′-hexanoyloxyfucoxanthin, picoeukaryote abundance, heterotrophic bacterial abundance, and cyanobacterial Synechococcus and Prochlorococcus abundance. Statistically significant correlations were determined using Student's t test on Spearman's correlation coefficient (ρ) at P values of <0.05. Differences in gene abundance and environmental parameter means between samples collected from 25 m and the DCM were verified using a two-sample t test at P values of <0.05. To negate the effects of autocorrelation of environmental data with depth, the samples were divided by depth, and intradepth patterns were identified based on rank correlations carried out between gene abundance and environmental variables using a significance level of P values of <0.05.
dmdA metagenomic and metatranscriptomic analyses.
Metagenomic and metatranscriptomic data sets from Stn ALOHA obtained on cruises HOT140 (11), HOT154 (11), HOT175 (41), HOT179 (14, 48), and HOT186 (51) and available in CAMERA (http://camera.calit2.net/) and NCBI's Short Read Archive (SRA; http://www.ncbi.nlm.nih.gov/sra) were mined for sequences representing dmdA, dddP, dddW, dddQ, dddY, dddD, and dddL and, for comparison, proteorhodopsin. Query sequences consisted of full-length protein sequences from cultured organisms (see accession numbers in Table S2 in the supplemental material). All functional gene BLASTs were carried out using tBLASTn, with an E-value cutoff of ≤10−4. To increase annotation confidence, all hits were compared to NCBI's RefSeq database in a BLASTx analysis using a bit score cutoff of ≥40 and manually verified for correct target function. For dmdA hits, an additional BLASTx analysis was done to identify clade affiliation (62), also using a bit score cutoff of ≥40, against a >3,000-member in-house DmdA (2,440 sequences) and GcvT (623 outgroup sequences) database consisting of cultured and metagenomic sequences from the GOS data set (46, 63). This was found to be the best approach for assigning short sequences (62), since a test using pplacer (37) with randomly trimmed 100-bp known dmdA or gcvT sequences misassigned 20% of the sequences (incorrectly placing dmdA sequences as gcvT or vice versa). The short sequences also limited the ability to accurately determine the percentage of sequences with matches to our qPCR primers.
To calculate the percentage of cells harboring DMSP-degrading genes, the ratio of 16S rRNA genes per recA was determined using Escherichia coli K-12 16S rRNA and recA genes (4, 19) as queries in BLASTn and tBLASTn, respectively, with an E-value cutoff of ≤10−4. Only those sequences with the correct target annotation and a reciprocal best hit with a bit score of ≥100 for 16S rRNA genes (BLASTn) and a bit score of ≥40 for RecA (BLASTx) were retained. In order to normalize for effects of gene size on the number of hits retrieved, 16S rRNA (1,542 bp) and recA (1,059 bp) genes were length normalized according to the method of Biers et al. (4), which resulted in an average ratio of 1.4 16S rRNA genes:1 recA gene. This ratio was used to calculate dmdA, dddP, and proteorhodopsin gene copy numbers as a percentage of cells, assuming a single copy of recA and 1.4 copies of 16S rRNA genes per cell. While multiple DMSP gene copies per cell could lead to inflated per-cell estimates, the SAR11 “Candidatus Pelagibacter ubique” HTCC7211 strain is the only marine isolate known to carry two dmdA gene copies (62).
RESULTS
HOT physical and biogeochemical data.
During the sampling period from May 2008 to February 2009, the depth of the surface mixed layer ranged from 22 to 114 m. With the exception of May 2008, seawater samples collected from 25 m were always located within the mixed layer (Fig. 1). The seawater temperatures at 25 m were 1 to 4°C higher than those in the DCM (see Fig. S1A in the supplemental material). The daily integrated flux of PAR at 25 m represented approximately 11% of surface irradiance, while the DCM was always situated below the 1% light level (see Fig. S1B in the supplemental material). Primary production was ∼13-fold higher at 25 m than at the DCM (see Fig. S1C in the supplemental material). Nitrate-nitrite concentrations were higher at the DCM (11 to 1,075 nmol liter−1) than at 25 m (2 to 6 nmol liter−1) (see Fig. S1D in the supplemental material).
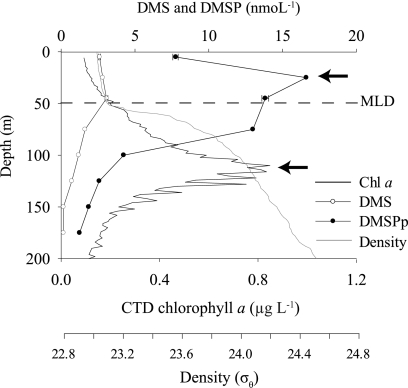
Fig 1.
Representative depth profile from August 2008 (HOT204) of DMSP-related biogeochemical data at Stn ALOHA. Peaks in the DMSPp concentration at 25 m and the Chl a concentration at the DCM are indicated by arrows. The mixed layer depth (MLD) is shown as a dotted line.
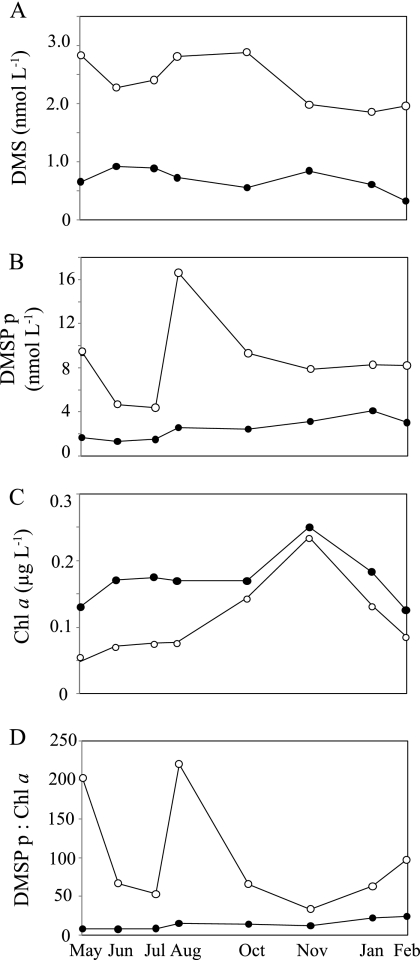
With respect to DMS(P) chemistry, DMS and DMSPp concentrations were highest in the upper 75 m of the water column and subsequently declined with depth to near-detection limits at 150 m (Fig. 1). For the discrete depths sampled for gene analysis, DMS and DMSPp concentrations were 2- to 6-fold higher at 25 m than at the DCM (Fig. 2A and B). Between May 2008 and February 2009, DMS concentrations at 25 m were highest from May to October (2.3 to 2.8 nmol liter−1) and lowest from November to February (1.9 to 2.0 nmol liter−1) (Fig. 2A). At the DCM, DMS concentrations were lower and less variable (0.3 to 0.9 nmol liter−1) (Fig. 2A). The concentration of DMSPp exceeded DMS concentrations at all sampled depths and dates (Fig. 2B). Chl a concentrations ranged from ∼1.5-fold to 3-fold higher at the DCM than at 25 m (Fig. 2C). The DMSPp:Chl a ratio (an indicator of the phytoplankton DMSP content per chlorophyll content) was up to 25-fold higher at 25 m (Fig. 2D), and the highest DMSPp:Chl a ratios occurred in May and August 2008 (Fig. 2D).
Fig 2.
DMSP-related biogeochemical properties at Stn ALOHA for 25 m (open symbols) and the DCM (closed symbols). Data were collected on HOT cruises 201 to 209.
dmdA and dddP abundances.
Eight dmdA subclades were selected for analysis with PCR primers, targeting ∼50% of known dmdA sequences (62), and copy numbers were normalized to 16S rRNA qPCR (based on a calculation of 1.4 16S rRNA genes per cell; see Materials and Methods). A higher proportion of bacterioplankton cells contained a dmdA gene from one of the 8 targeted subclades at 25 m (13% ± 2%) than at the DCM (6.5% ± 1%) throughout the sampling period (Fig. 3). The maximum frequency of dmdA-containing cells was 16.5%, occurring in May 2008 at 25 m. Like dmdA, a higher fraction of dddP-containing cells was observed at 25 m than at the DCM, although this gene was present at a consistently lower frequency than dmdA at every sampling occasion and depth. The maximum frequency of cells containing the targeted dddP subclade was 2.1%, occurring in July 2008 at 25 m (Fig. 3).
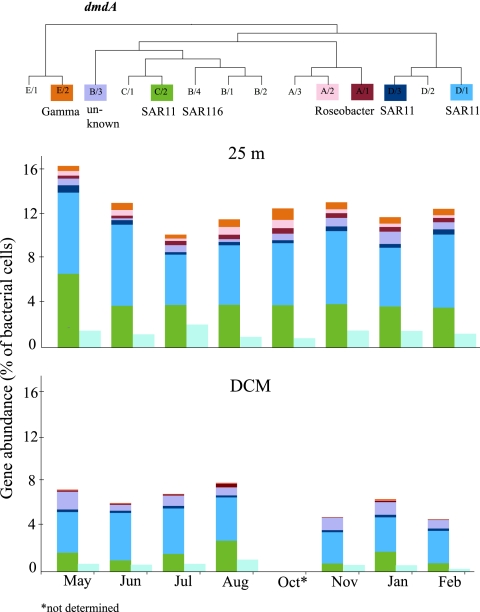
Fig 3.
Abundance of dmdA and dddP in Stn ALOHA bacterioplankton cells at two depths, 25 m (top bar graph) and the DCM (110 to 140 m) (bottom bar graph). dmdA abundances are shown in the multicolored bars, with the subclade color codes and phylogenetic relationships (20, 62) indicated at the top of the figure. dddP abundances are shown in the light blue bars. The relative gene abundances are calculated using the ratio of dmdA or dddP to 16S rRNA gene copies, assuming 1.4 copies per cell as calculated from HOT metagenomic data sets (see Materials and Methods).
The SAR11 subclades made up the greatest portion of dmdA genes, with subclades D/1 and C/2 responsible for 80% of the total dmdA genes measured. Together, these subclades were present in approximately 10% and 5% of cells at 25 m and the DCM, respectively (Fig. 3). dmdA genes from the Roseobacter subclade A/2 and Gammaproteobacteria subclade E/2 were particularly enriched in cells inhabiting surface waters, with an order of magnitude difference in their abundance relative to the DCM. All three SAR11 dmdA subclades, including the less abundant D/3, were most frequent in the bacterioplankton community at 25 m in May 2008, while the A/1, A/2, and E/2 subclades were most frequent at 25 m in October 2008. SAR116 dmdA subclade B/4 sequences were consistently below the detection limit.
MDS and statistical analyses.
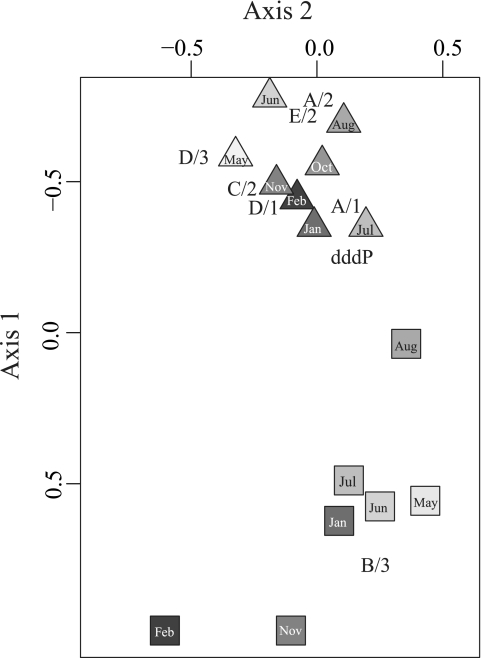
An MDS plot was used to explore dmdA and dddP profiles by depth and sampling occasion (Fig. 4). Together, the first two MDS axes represented ∼93% of the variability in abundance of measured DMSP-related genes. Samples were strongly separated by depth (Fig. 4). MDS scores for the individual genes indicated that surface subclades A/2 and E/2 (Roseobacter and Gammaproteobacteria) grouped together on the MDS plot (Fig. 4) and were highly correlated (ρ = 0.95, P < 0.001). Subclade B/3, which represents an unknown taxonomic group, was typically higher in abundance at the DCM (Fig. 4) and was negatively correlated with E/2 and D/1 (ρ < −0.56, P < 0.03) and not correlated with any other dmdA subclade or dddP.
Fig 4.
Multidimensional scaling (MDS) plot of dmdA and dddP gene abundance using R (vegan package and Bray-Curtis similarities). Triangles represent samples from 25 m, and squares represent samples from the DCM, with the sample month indicated within each symbol. The variabilities explained by axis 1 and axis 2 are 81% and 12%, respectively. Corresponding MDS scores for each dmdA subclade and dddP are also plotted.
To explore the factors that might be driving gene patterns, MDS axis scores were analyzed against biogeochemical parameters. MDS axis 1 correlated positively with depth (ρ = 0.87, P < 0.001) and less strongly but negatively with several other parameters that had the highest values at 25 m, including the DMSP:Chl a ratio, DMSPp concentration, DMS concentration, temperature, PAR, primary production rate, DOC, heterotrophic bacterial abundance, Prochlorococcus and Synechococcus abundance, and total DMSP gene counts (all ρ < −0.58, P < 0.05). In contrast, 19′-butanoyloxyfucoxanthin, 19′-hexanoyloxyfucoxanthin, nitrate-nitrite, Chl a, and silicate concentrations were positively correlated with MDS axis 1, being lower at 25 m than at the DCM (all ρ > 0.60, P < 0.05). MDS axis 2 was negatively correlated with fucoxanthin, a diagnostic pigment for diatoms (ρ = −0.51, P < 0.05), and with the DMSPp:Chl a ratio (i.e., an index of phytoplankton DMSP content per Chl a content; ρ = −0.52, P < 0.05), but this axis accounted for a smaller fraction of the variability (12%; Fig. 4).
To eliminate the strong influence of depth-related environmental signals that dominated axis 1 of the MDS plot, data were also analyzed independently for each depth. At 25 m, the overall dmdA gene abundance (all subclades combined) was not significantly correlated with any variable, although surface subclades A/2 and E/2 were negatively correlated with the diatom pigment fucoxanthin (both ρ < −0.85, P < 0.05), and subclade A/1 was positively correlated with Chl a (ρ = 0.93, P < 0.001). dddP gene counts were positively correlated with fucoxanthin (ρ = 0.78, P < 0.05). At the DCM, dmdA was positively correlated with temperature (ρ = 0.79, P < 0.05), while dddP was not significantly correlated with any of the environmental parameters measured.
dmdA gene expression.
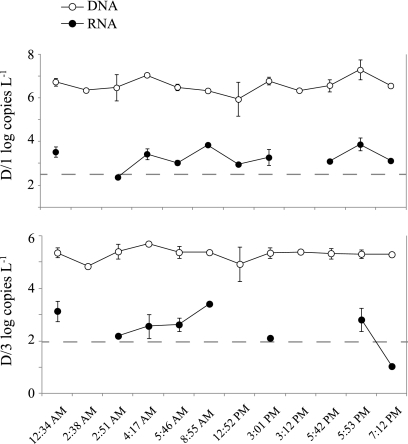
Select samples from three dates (August 2008, October 2008, and January 2009) and both depths (25 m and the DCM) were analyzed for dmdA transcript levels (SAR11 D/1 and D/3 subclades only), but most were at or below the detection limit. In the October 2008 samples from 25 m, however, both D/1 and D/3 subclades had measurable expression. Average transcript-gene ratios were 1:350 for D/3 and 1:1,400 for D/1 in samples for which transcription could be accurately quantified.
Metagenomic data set analysis.
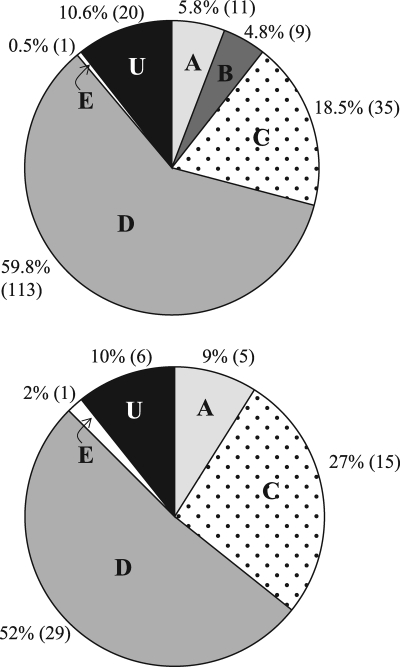
A homology search of dmdA sequences against available metagenomic and metatranscriptomic data sets from Stn ALOHA (11, 14, 41, 48, 51) resulted in hits to clade A (Roseobacter), clades C and D (SAR11), and clade E (Gammaproteobacteria), with the majority of sequences having best matches to the two SAR11 clades (>75%; Fig. 5). From estimates of recA abundance in the metagenomic data sets and assuming a single copy of dmdA and recA per cell, the percentage of cells carrying a dmdA gene was ∼40%, 3-fold higher than the qPCR-based average of ∼13% for the 8 primer sets combined. The abundance of cells harboring a dddP gene in the HOT metagenomes was 4%, while our qPCR estimate averaged 1.4%. The percentage of cells harboring a dddD or a dddQ was ∼3% or 1.6%, respectively. Reads with significant homology to dddW, dddY, and dddL sequences were not detected in the Stn ALOHA metagenomes or metatranscriptomes.
Fig 5.
Relative abundance of dmdA clades in two metagenomic (top) and three metatranscriptomic (bottom) data sets collected on multiple HOT cruises at Stn ALOHA (11, 14, 41, 48, 51). The five clades (A, B, C, D, and E) plus U (which designates unclassified dmdA sequences) are shown as percentages of total dmdA hits. The number of hits to each clade is shown in parentheses.
DISCUSSION
The goals of this study were to determine whether bacterioplankton DMSP genes in the NPSG covary with physical or chemical parameters that indicate the environmental conditions conducive for DMSP-relevant microbial processes and whether variations in taxonomic affiliations of genes over time or space could signal shifts in the dominant bacterial taxa mediating DMSP cycling. Previously, both primary production and solar radiation have been hypothesized to influence DMSP and DMS production. The former is thought to track with rates of biosynthesis of DMSP by phytoplankton cells (3, 36). The latter assumes a role for DMSP in cellular scavenging of reactive oxygen species (50, 52, 61), and indeed solar radiation levels have been linked to increased assimilation of DMSP by microorganisms in the surface mixed layer of the NPSG (12). In this study, primary productivity and solar radiation may also have driven the consistently higher abundance of bacterioplankton cells harboring one of the dmdA subclades or dddP at 25 m compared to the DCM (Fig. 3), but many other depth-related parameters also showed strong vertical structure at Stn ALOHA (11, 22, 23), including DMS, DMSPp, Chl a, dissolved organic carbon concentrations, DMSP:Chl a ratios, and temperature. Because of autocorrelation with depth, these could not be individually resolved in this study.
At Stn ALOHA, non-DMSP-producing Prochlorococcus species are the dominant phytoplankton (6, 7, 65), with DMSP-producing species such as diatoms and prymnesiophytes present in lower abundance (1, 31, 47). The pigment-related correlations emerging for some DMSP-degrading genes at 25 m (see Fig. S2 in the supplemental material), however, might indicate ecological interactions between DMSP-degrading bacteria and one of the DMSP-producing phytoplankton groups, such as the positive correlation between dddP gene abundance (known to be harbored by Roseobacter [54] and SAR116 cells thus far) and fucoxanthin, a pigment diagnostic of diatom cells. The DMSPp:Chl a ratio is considered an indicator of the fractional importance of DMSP in the available organic carbon pool (27, 45) and is expected to track with the abundance of high DMSP-producing phytoplankton species within a given light regime. However, on the two sampling occasions during which the DMSPp:Chl a ratio was 3-fold higher than average (Fig. 2; 25 m in May and August 2008), there were no obvious changes in composition or abundance of the DMSP gene pool targeted with the qPCR primer sets (Fig. 3).
SAR11 clades (particularly clade D/1) dominated the dmdA pool at every depth and station in this study, similar to what was found over 36 ocean surface waters surveyed in the 2007 GOS data set (19) and consistent with the recognized abundance of SAR11 in the Stn ALOHA bacterioplankton (13). Two other dmdA clades, Roseobacter clade A/2 and Gammaproteobacteria clade E/2, were largely confined to the upper mixed layer, while subclade B/3 (taxonomic affiliation unknown) was primarily a DCM indicator. These patterns of taxonomic affiliations of DMSP-related genes at Stn ALOHA were not correlated to DMSP or DMS pool sizes at either 25 m or the DCM but might nonetheless signal differences in the dominant pathways or processing rates. For example, SAR11 bacteria rely on reduced sulfur for growth (58) and are capable only of DMSP demethylation, based on the genome sequences available thus far, while some roseobacters are able to both demethylate and cleave DMSP (16, 19, 35). These overall findings are also consistent with a study of the upper 60 m of another oligotrophic ocean gyre, the Sargasso Sea, in which SAR11 dmdA genes dominate year-round (32).
The average dmdA and dddP frequencies obtained by qPCR analysis were systematically lower than those estimated from the HOT179 metagenomic data, but this is attributable to the requirement for highly conserved nucleotide sequences for qPCR primer design and, as a consequence, the fact that the eight qPCR primer sets used here targeted only about half of the currently known dmdA sequences from the 2007 GOS metagenome (62). Furthermore, dmdA groups not represented in the GOS 2007 may be present at Stn ALOHA, particularly since the DCM habitat was not sampled in the GOS (46). Given the constraints of environmental primer design, the qPCR and metagenomic estimates were in good agreement. Both the HOT179 metagenome and qPCR data similarly found that most dmdA genes in NPSG surface waters were associated with SAR11 clades (∼75% for the HOT179 and 80% for qPCR). In addition, both methods found an order of magnitude difference in dmdA and dddP abundance (13:1 for the HOT179 metagenome and 9:1 for qPCR), which may be a common feature of marine environments (19, 32, 44, 54). Field studies indicate that ∼80% of bacterially metabolized DMSP is processed through the demethylation pathway and only ∼20% is cleaved to DMS (29). Evolutionary pressure for marine bacteria to maintain the ability to demethylate DMSP could explain the consistently high and stable dmdA gene frequencies found in ocean bacteria, including those at Stn ALOHA. Transcript levels in NPSG surface waters, which should better correlate with the conditions under which the gene products are ecologically advantageous, were also quantified, but only the October samples had expression levels above the limit of detection for dmdA subclades D/1 and D/3.
Poor transcript detection for DMSP-related genes may be due in part to the relatively low RNA concentrations recovered from Stn ALOHA (many RNA yields were below 100 ng liter−1), or detection may appear low if the DNA pool is artificially inflated by detrital material (17, 66). However, there is also evidence that dmdA has inherently low expression relative to other bacterial genes at Stn ALOHA, since low transcript abundance was also found in the HOT179 metatranscriptomic data. For example, the expression ratio in the HOT179 sequence libraries (calculated as % representation in the cDNA library/% representation in the DNA library, according to Frias-Lopez et al. [14], which is a relative expression ratio) was 0.11, whereas the expression ratio calculated in the same way for proteorhodopsin, another high-frequency SAR11-dominated gene in the bacterioplankton DNA, was 100-fold higher (see Fig. S3 in the supplemental material). Since a diel survey conducted during the October cruise did not find significant variation in transcript abundance across 8 to 10 time points over 48 h (Fig. 6), low dmdA expression levels cannot simply be attributed to inopportune sampling times. However, storage of undegraded DMSP by bacterioplankton for use as an osmolyte (43), short mRNA half-lives, or a highly stable or efficient dmdA protein product might uncouple instantaneous gene transcription rates from DMSP turnover rates. Low expression levels for dmdA have been detected in a number of other marine systems using qPCR and metatranscriptomics (15, 32, 64). Understanding the relationship between gene frequency, transcript abundance, protein levels, and biogeochemical rates is a crucial challenge for future research.
Fig 6.
Gene and transcript copies at Stn ALOHA over a diel cycle for October at 25 m (HOT205) for SAR11 dmdA subclades D/1 (top) and D/3 (bottom). Vertical bars are standard deviations for duplicate or triplicate biological samples. Dotted lines are the detection limits. RNA samples were not collected at the 2:38 AM or 3:12 PM sampling times, and expression could not be quantified for D/1 at the 2:51 AM sampling time or for D/3 at the 12:52, 5:42, or 7:12 PM sampling times.
Overall, dmdA and dddP gene pools targeted by our suite of primers at Stn ALOHA showed greater variation between the surface mixed layer and the DCM than they did within either depth throughout the 10-month study period (Fig. 3), in agreement with the strong vertical structure but low seasonality of the NPSG (34). We propose that the high and relatively invariant inventory of bacterial DMSP genes in the NPSG indicates strong evolutionary pressure on bacterioplankton to maintain this capability and that DMSP degradation is not the purview of specialized bacteria. Based on the composition of the DMSP gene pool, SAR11 bacterioplankton dominate DMSP cycling in the upper ocean of the oligotrophic NPSG throughout the year, with lesser but consistent involvement of members of the Roseobacter and Gammaproteobacteria taxa.
Supplementary Material
ACKNOWLEDGMENTS
We thank the captain and crew of the R/V Kilo Moana and the Hawaii Ocean Time-series (HOT) for availability of environmental data and M. Church for additional DNA samples collected during 2008 for qPCR verification. N. Levine, H. Luo, R. Newton, and S. Holland provided helpful discussions and advice on statistical analyses.
This work was supported by grants from the Gordon and Betty Moore Foundation, NSF (EnGen award OCE0724017), and the NSF-supported Center for Microbial Oceanography: Research and Education (C-MORE) (EF0424599).
Footnotes
Published ahead of print 10 February 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Andersen RA, Bidigare RR, Keller MD, Latasa M. 1996. A comparison of HPLC pigment signatures and electron microscopic observations for oligotrophic waters of the North Atlantic and Pacific Oceans. Deep Sea Res. 43:517–537 [Google Scholar]
- 2. Andreae MO, Crutzen PJ. 1997. Atmospheric aerosols: biogeochemical sources and role in atmospheric chemistry. Science 276:1052–1058 [Google Scholar]
- 3. Bell TG, Poulton AJ, Malin G. 2010. Strong linkages between dimethylsulphoniopropionate (DMSP) and phytoplankton community physiology in a large subtropical and tropical Atlantic Ocean data set. Global Biogeochem. Cycles 24:1–12 [Google Scholar]
- 4. Biers EJ, Sun S, Howard EC. 2009. Prokaryotic genomes and diversity in surface ocean waters: interrogating the global ocean sampling metagenome. Appl. Environ. Microbiol. 75:2221–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bürgmann H, et al. 2007. Transcriptional response of Silicibacter pomeroyi DSS-3 to dimethylsulfoniopropionate (DMSP). Environ. Microbiol. 9:2742–2755 [DOI] [PubMed] [Google Scholar]
- 6. Campbell L, Nolla HA, Vaulot D. 1994. The importance of Prochlorococcus to community structure in the central North Pacific Ocean. Limnol. Oceanogr. 39:954–961 [Google Scholar]
- 7. Campbell L, Vaulot D. 1993. Photosynthetic picoplankton community structure in the subtropical North Pacific Ocean near Hawaii (Station ALOHA). Deep Sea Res. 40:2043–2060 [Google Scholar]
- 8. Chandler DP, Wagnon CA, Bolton H., Jr 1998. Reverse transcriptase (RT) inhibition of PCR at low concentrations of template and its implications for quantitative RT-PCR. Appl. Environ. Microbiol. 64:669–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Curson ARJ, Rogers R, Todd JD, Brearley CA, Johnston AWB. 2008. Molecular genetic analysis of a dimethylsulfoniopropionate lyase that liberates the climate-changing gas dimethylsulfide in several marine α-proteobacteria and Rhodobacter sphaeroides. Environ. Microbiol. 10:757–767 [DOI] [PubMed] [Google Scholar]
- 10. Curson ARJ, Sullivan MJ, Todd JD, Johnston AWB. 2011. DddY, a periplasmic dimethylsulfoniopropionate lyase found in taxonomically diverse species of Proteobacteria. ISME J. 5:1191–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. DeLong EF, et al. 2006. Community genomics among stratified microbial assemblages in the ocean's interior. Science 311:496–503 [DOI] [PubMed] [Google Scholar]
- 12. Del Valle DA, Kiene RP, Karl DM. Effect of visible light on dimethylsulfoniopropionate assimilation and conversion to dimethylsulfide in the North Pacific Subtropical Gyre. Aquat. Microb. Ecol., in press [Google Scholar]
- 13. Eiler A, Hayakawa DH, Church MJ, Karl DM, Rappé MS. 2009. Dynamics of the SAR11 bacterioplankton lineage in relation to environmental conditions in the oligotrophic North Pacific Subtropical Gyre. Environ. Microbiol. 11:2291–2300 [DOI] [PubMed] [Google Scholar]
- 14. Frias-Lopez J, et al. 2008. Microbial community gene expression in ocean surface waters. P. Natl. Acad. Sci. U. S. A. 105:3805–3810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gifford SM, Sharma S, Rinta-Kanto JM, Moran MA. 2011. Quantitative analysis of a deeply sequenced marine microbial metatranscriptome. ISME J. 5:461–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gonzalez JM, Kiene RP, Moran MA. 1999. Transformation of sulfur compounds by an abundant lineage of marine bacteria in the α-subclass of the class Proteobacteria. Appl. Environ. Microbiol. 65:3810–3819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Holm-Hansen O, Sutcliffe WH, Jr, Sharp J. 1968. Measurement of deoxyribonucleic acid in the ocean and its ecological significance. Limnol. Oceanogr. 13:507–514 [Google Scholar]
- 18. Howard EC, et al. 2006. Bacterial taxa that limit sulfur flux from the ocean. Science 314:649–652 [DOI] [PubMed] [Google Scholar]
- 19. Howard EC, Sun S, Biers EJ, Moran MA. 2008. Abundant and diverse bacteria involved in DMSP degradation in marine surface waters. Environ. Microbiol. 10:2397–2410 [DOI] [PubMed] [Google Scholar]
- 20. Howard EC, et al. 2011. Changes in dimethylsulfoniopropionate demethylase gene assemblages in response to an induced phytoplankton bloom. Appl. Environ. Microbiol. 77:524–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Johnston AWB, et al. 2008. Molecular diversity of bacterial production of the climate-changing gas, dimethyl sulphide, a molecule that impinges on local and global symbioses. J. Exp. Bot. 59:1059–1067 [DOI] [PubMed] [Google Scholar]
- 22. Karl DM. 2007. Microbial oceanography: paradigms, processes and promises. Nat. Rev. Microbiol. 5:759–769 [DOI] [PubMed] [Google Scholar]
- 23. Karl DM. 1999. A sea of change: biogeochemical variability in the North Pacific Subtropical Gyre. Ecosystems 2:181–214 [Google Scholar]
- 24. Karl DM, Bidigare RR, Letelier RM. 2002. Sustained and aperiodic variability in organic matter production and phototrophic microbial community structure in the North Pacific Subtropical Gyre, p 222–264 In Williams PJ, Thomas DN, Reynolds CS. (ed), Phytoplankton productivity: carbon assimilation in marine and freshwater ecosystems. Blackwell Science Ltd., Ames, IA [Google Scholar]
- 25. Karl DM, Lukas R. 1996. The Hawaii Ocean Time-series (HOT) program: background, rationale and field implementation. Deep Sea Res. 43:129–156 [Google Scholar]
- 26. Karl DM, Winn CD, Hebel DVW, Letelier R. 1990. Hawaii Ocean Time-series program. Field and laboratory protocols. School of Ocean and Earth Science and Technology, University of Hawaii, Honolulu, HI [Google Scholar]
- 27. Kiene RP, Linn LJ. 2000. Distribution and turnover of dissolved DMSP and its relationship with bacterial production and dimethylsulfide in the Gulf of Mexico. Limnol. Oceanogr. 45:849–861 [Google Scholar]
- 28. Kiene RP, Linn LJ, Bruton JA. 2000. New and important roles for DMSP in marine microbial communities. J. Sea Res. 43:209–224 [Google Scholar]
- 29. Kiene RP, Linn LJ, Gonzalez J, Moran MA, Bruton JA. 1999. Dimethylsulfoniopropionate and methanethiol are important precursors of methionine and protein-sulfur in marine bacterioplankton. Appl. Environ. Microbiol. 65:4549–4558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ledyard KM, Dacey JWH. 1996. Microbial cycling of DMSP and DMS in coastal and oligotrophic seawater. Limnol. Oceanogr. 41:33–40 [Google Scholar]
- 31. Letelier R, et al. 1993. Temporal variability of phytoplankton community structure based on pigment analysis. Limnol. Oceanogr. 38:1420–1437 [Google Scholar]
- 32. Levine NM, et al. 2012. Environmental, biochemical, and genetic drivers of DMSP degradation and DMS production in the Sargasso Sea. Environ. Microbiol. doi:10.1111/j.1462-2920.2012.02700.x [DOI] [PubMed] [Google Scholar]
- 33. Lovelock JE, Maggs RJ, Rasmussen RA. 1972. Atmospheric dimethyl sulphide and the natural sulphur cycle. Nature 237:452–453 [Google Scholar]
- 34. Malmstrom RR, et al. 2010. Temporal dynamics of Prochlorococcus ecotypes in the Atlantic and Pacific oceans. ISME J. 4:1252–1264 [DOI] [PubMed] [Google Scholar]
- 35. Malmstrom RR, Kiene RP, Cottrell MT, Kirchman DL. 2004. Contribution of SAR11 bacteria to dissolved dimethylsulfoniopropionate and amino acid uptake in the North Atlantic Ocean. Appl. Environ. Microbiol. 70:4129–4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Matrai P, Vernet M, Wassman P. 2007. Relating temporal and spatial patterns of DMSP in the Barents Sea to phytoplankton biomass and productivity. J. Marine Syst. 67:83–101 [Google Scholar]
- 37. Matsen EA, Kodner RB, Armbrust EV. 2010. pplacer: linear time maximum-likelihood and Bayesian phylogenetic placement of sequences onto a fixed reference tree. BMC Bioinformatics 11:538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Monterey GI, Levitus S. 1997. Seasonal variability of mixed layer depth for the world ocean. NOAA NESDIS Atlas 14:5 [Google Scholar]
- 39. Oksanen J, et al. 2009. vegan: community ecology package. R package version 1.15-4. http://CRAN.R-project.org/package=vegan
- 40. Pichereau V, Pocard J-A, Hamelin J, Blanco C, Bernard T. 1998. Differential effects of dimethylsulfoniopropionate, dimethylsulfonioacetate, and other S-methylated compounds on the growth of Sinorhizobium meliloti at low and high osmolarities. Appl. Environ. Microbiol. 64:1420–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Poretsky RS, et al. 2009. Comparative day/night metatranscriptomic analysis of microbial communities in the North Pacific Subtropical Gyre. Environ. Microbiol. 11:1358–1375 [DOI] [PubMed] [Google Scholar]
- 42. R Development Core Team 2011. R: a language and environment for statistical computing. R foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org [Google Scholar]
- 43. Reisch CR, Moran MA, Whitman WB. 2008. Dimethylsulfoniopropionate-dependent demethylase (DmdA) from Pelagibacter ubique and Silicibacter pomeroyi. J. Bacteriol. 190:8018–8024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Reisch CR, et al. 2011. Novel pathway for assimilation of dimethylsulphoniopropionate widespread in marine bacteria. Nature 473:208–211 [DOI] [PubMed] [Google Scholar]
- 45. Rinta-Kanto JM, Sun S, Sharma S, Kiene RP, Moran MA. 2012. Bacterial community transcription patterns during a marine phytoplankton bloom. Environ. Microbiol. 14:228–239 [DOI] [PubMed] [Google Scholar]
- 46. Rusch DB, et al. 2007. The Sorcerer II global ocean sampling expedition: Northwest Atlantic through Eastern Tropical Pacific. PLoS Biol. 5:e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Scharek R, Latasa M, Karl DM, Bidigare RR. 1999. Temporal variations in diatom abundance and downward vertical flux in the oligotrophic North Pacific Gyre. Deep Sea Res. 46:1051–1075 [Google Scholar]
- 48. Shi Y, Tyson GW, DeLong EF. 2009. Metatranscriptomics reveals unique microbial small RNAs in the ocean's water column. Nature 459:266–269 [DOI] [PubMed] [Google Scholar]
- 49. Simo R. 2001. Production of atmospheric sulfur by oceanic plankton: biogeochemical, ecological and evolutionary links. Trends Ecol. Evol. 16:287–294 [DOI] [PubMed] [Google Scholar]
- 50. Slezak D, Kiene R, Toole D, Simó R, Kieber D. 2007. Effects of solar radiation on the fate of dissolved DMSP and conversion to DMS in seawater. Aquat. Sci. 69:377–393 [Google Scholar]
- 51. Stewart FJ, Ottesen EA, DeLong EF. 2010. Development and quantitative analyses of a universal rRNA-subtraction protocol for microbial metatranscriptomics. ISME J. 4:896–907 [DOI] [PubMed] [Google Scholar]
- 52. Sunda W, Kieber DJ, Kiene RP, Huntsman S. 2002. An antioxidant function for DMSP and DMS in marine algae. Nature 418:317–320 [DOI] [PubMed] [Google Scholar]
- 53. Suzuki MT, Taylor LT, DeLong EF. 2000. Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Appl. Environ. Microbiol. 66:4605–4614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Todd JD, Curson AJ, Dupont CL, Nicholson P, Johnston AWB. 2009. The dddP gene, encoding a novel enzyme that converts dimethylsulfoniopropionate into dimethyl sulfide, is widespread in ocean metagenomes and marine bacteria and also occurs in some Ascomycete fungi. Environ. Microbiol. 11:1624–1625 [DOI] [PubMed] [Google Scholar]
- 55. Todd JD, et al. 2011. DddQ, a novel, cupin-containing, dimethylsulfoniopropionate lyase in marine roseobacters and in uncultured marine bacteria. Environ. Microbiol. 13:427–438 [DOI] [PubMed] [Google Scholar]
- 56. Todd JD, Kirkwood M, Newton-Payne S, Johnston AWB. 2012. DddW, a third DMSP lyase in a model Roseobacter marine bacterium, Ruegeria pomeroyi DSS-3. ISME J. 6:223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Todd JD, et al. 2007. Structural and regulatory genes required to make the gas dimethyl sulfide in bacteria. Science 315:666–669 [DOI] [PubMed] [Google Scholar]
- 58. Tripp HJ, et al. 2008. SAR11 marine bacteria require exogenous reduced sulphur for growth. Nature 452:741–744 [DOI] [PubMed] [Google Scholar]
- 59. Turner SM, Malin G, Bågander LE, Leck C. 1990. Interlaboratory calibration and sample analysis of dimethyl sulphide in water. Mar. Chem. 29:47–62 [Google Scholar]
- 60. Vairavamurthy A, Andreae MO, Iverson RL. 1985. Biosynthesis of dimethylsulfide and dimethylpropiothetin by Hymenomonas carterae in relation to sulfur source and salinity variations. Limnol. Oceanogr. 30:59–70 [Google Scholar]
- 61. Vallina SM, Simo R. 2007. Strong relationship between DMS and the solar radiation dose over the global surface ocean. Science 315:506–508 [DOI] [PubMed] [Google Scholar]
- 62. Varaljay VA, Howard EC, Sun S, Moran MA. 2010. Deep sequencing of a dimethylsulfoniopropionate-degrading gene (dmdA) by using PCR primer pairs designed on the basis of marine metagenomic data. Appl. Environ. Microbiol. 76:609–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Venter JC, et al. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66–74 [DOI] [PubMed] [Google Scholar]
- 64. Vila-Costa M, et al. 2010. Transcriptomic analysis of a marine bacterial community enriched with dimethylsulfoniopropionate. ISME J. 4:1410–1420 [DOI] [PubMed] [Google Scholar]
- 65. Vila-Costa M, et al. 2006. Dimethylsulfoniopropionate uptake by marine phytoplankton. Science 314:652–654 [DOI] [PubMed] [Google Scholar]
- 66. Winn CD, Karl DM. 1986. Diel nucleic acid synthesis and particulate DNA concentrations: conflicts with division rate estimates by DNA accumulation. Limnol. Oceanogr. 31:637–645 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.