Abstract
The hypersensitive response and pathogenicity (hrp) genes of Dickeya dadantii 3937 encode a type III secretion system (T3SS) which is essential for its full virulence. Previous studies of the T3SS regulation in D. dadantii 3937 revealed that the expression of the hrp genes is regulated by a master regulator, HrpL, through the HrpX-HrpY-HrpS-HrpL and GacS-GacA-rsmB-RsmA pathways. In this work, we identified a novel regulator of the SlyA/MarR family, SlyA, which regulates hrp genes of the HrpL regulon in parallel with HrpL in D. dadantii. SlyA regulates the T3SS in a two-tier manner. It negatively regulates the expression of hrpL by downregulating hrpS and upregulating rsmA. Interestingly, concomitant with its downregulation of the hrpL, SlyA positively regulates the expression of hrpA and hrpN, two hrp genes located in the HrpL regulon. In contrast to Pectobacterium carotovorum, the expression of slyA is not controlled by ExpR and ExpI in D. dadantii 3937. We further show that SlyA is involved in controlling swimming motility and pellicle formation in D. dadantii 3937.
INTRODUCTION
Dickeya dadantii 3937 (formerly named Erwinia chrysanthemi 3937) is an enterobacterium that causes soft-rot, wilts, and dwarfing diseases in a wide range of plant species, including many economically important crops (22). It secretes a set of plant cell wall-degrading enzymes, such as pectinases, cellulases, polygalacturonases, and proteases, through the type I and type II secretion systems that together with the hrp-encoded type III secretion system (T3SS) are important for bacterial full virulence (13, 32, 40). Additionally, T3SS is required for pellicle formation and cell aggregation in D. dadantii 3937 (32, 37, 39).
Previous reports have described the genetic regulation of the group I T3SS genes in Pectobacterium carotovorum, Pseudomonas syringae, Dickeya dadantii, and Erwinia amylovora (27). In Dickeya dadantii, the expression of the hrp genes that encode the T3SS structural and functional proteins, such as hrpA, hrpN, and dspE, is controlled by the master regulator HrpL (24, 36). HrpL is an alternative sigma factor that binds to the hrp box in the promoter region of hrp genes to activate their expression (27, 36). Genes involved in T3SS are regulated by two sensory/regulatory pathways. First, the transcription of hrpL is regulated by the HrpX-HrpY-HrpS-HrpL pathway (40). HrpX and HrpY compose a two-component system (TCS). HrpX, a sensor histidine kinase, perceives environmental signals and in turn activates HrpY by phosphorylation (40, 41). HrpY is a response regulator which binds to the promoter region of hrpS to activate its expression (41). HrpS, an NtrC family enhancer protein, initiates the transcription of hrpL in an RpoN-dependent manner (27, 41). In addition to the transcriptional regulation, the amount of hrpL mRNA is controlled by the regulator of a secondary metabolism (Rsm) system through the GacS-GacA-rsmB-RsmA pathway (35, 43). The Rsm system in D. dadantii is composed of RsmA and rsmB. RsmA is a small RNA-binding protein that binds to hrpL mRNA and promotes its degradation. Alternatively, an untranslated regulatory small RNA, rsmB, binds to RsmA and neutralizes its negative regulatory effect by forming an inactive ribonucleoprotein complex (3–7, 35, 43). GacS/A is another TCS involved in the regulation of T3SS genes in D. dadantii 3937 (35). Under T3SS-inducing conditions, GacA positively regulates the transcription of hrpA, hrpN, and dspE through the Rsm posttranscriptional regulatory pathway by increasing the expression of rsmB, which thereby inhibits the degradation effect of RsmA on hrpL mRNA (35, 43).
SlyA is a member of the SlyA/MarR family transcriptional regulators, of which more than 130 SlyA protein homologues have been identified in bacteria and archaea (9, 23, 29, 31). SlyA/MarR regulators function as negative regulators of an array of genes involved in antibiotic resistance and virulence (1, 30). In Salmonella enterica serovar Typhimurium, SlyA regulates the expression of genes encoding the T3SS located on Salmonella pathogenicity islands 1 and 2 (SPI-1 and SPI-2) by directly controlling transcription of the TCS sensor kinase, SsrA (17, 23). In Pectobacterium carotovorum subsp. carotovorum, slyA was originally identified as a gene required for the production of pigment and the antibiotic carbapenem, whose function is similar to regulation of antibiotic and pigment (Rap) of Serratia marcescens (28). Recently, it was shown that SlyA contributes to the extracellular enzyme production and pellicle formation in soft-rot bacteria P. carotovorum subsp. carotovorum SCC3193 and D. dadantii (12, 26). In P. carotovorum subsp. carotovorum SCC3193, the expression of slyA is dependent on the quorum-sensing (QS) signal molecule N-acylhomoserine lactone (AHSL) synthesized by an AHSL synthase, ExpI (26). In the absence of AHSL, ExpR1 and ExpR2, two QS regulators, activate the expression of rsmA, which negatively regulates the expression of slyA (26).
Along with the known regulators reported in the HrpX-HrpY-HrpS-HrpL and GacS-GacA-rsmB-RsmA-HrpL pathways, we hypothesized that other unidentified regulators may play an essential role in T3SS regulation. In this study, slyA was identified as a T3SS regulator in a transposon mutagenesis screening. An slyA deletion mutant of D. dadantii 3937 was then constructed, and the regulatory effect of SlyA on T3SS gene expression was evaluated. We showed that in D. dadantii 3937, the regulation of SlyA on T3SS is through two different pathways: SlyA negatively regulates the expression of hrpL while it positively regulates the expression of HrpL regulon genes, hrpA and hrpN, in parallel with HrpL. Additionally, in contrast to P. carotovorum subsp. carotovorum, the regulation of slyA expression in D. dadantii 3937 is independent of the ExpI QS synthase. Finally, the effect of SlyA on pellicle formation and swimming motility was also investigated.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. D. dadantii strains were cultured in Luria-Bertani (LB) medium, mannitol-glutamate (MG) medium (33), or minimal medium (MM) (38) at 28°C. SOBG medium was used to evaluate pellicle formation (12, 40). Escherichia coli strains were cultured in LB medium at 37°C. Antibiotics were added at the following concentrations when needed: ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; streptomycin, 50 μg/ml; gentamicin, 10 μg/ml; and chloramphenicol, 50 μg/ml. Reporter plasmids pPhrpA, pPhrpN, pPhrpX, pPhrpY, and pPslyA (Table 1) were constructed by transcriptional fusion of the hrpA, hrpN, hrpX, hrpY, or slyA promoter regions to the gene encoding green fluorescent protein (gfp) in the promoter-probe vector, pPROBE-AT. The GFP reporter plasmids pPhrpS and pPhrpL were constructed previously (35). Primers used in this study are listed in Table S1 in the supplemental material. The overexpression plasmid pMLhrpL was constructed by cloning a 1.6-kb fragment containing hrpL into pML123; the nptII promoter in the plasmid permits constitutive expression of hrpL.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Characteristicsa | Reference or source |
|---|---|---|
| Strains | ||
| Escherichia. coli | ||
| EC100D+ | Epicentre Technologies | |
| S17-1λ pir/pMiniHimar RB1 | pMiniHimar RB1 transposon vector in S17-1λ pir; Kmr | 2 |
| Dickeya dadantii | ||
| 3937 | Wild type, Saintpaulia (African violet) isolate | N. Hugouvieux-Cotte-Pattat |
| WPP92 | hrpY::Kan; 3937 derivative; Kmr | 40 |
| WPP67 | hrpX:aadA; 3937 derivative; Spr Smr | 40 |
| Ech3049 | slyA::Kan; Himar RB1 transposon mutant; Kmr | This work |
| Ech4120 | slyA::Kan; Himar RB1 transposon mutant; Kmr | This work |
| Ech4380 | slyA::Kan; Himar RB1 transposon mutant; Kmr | This work |
| Ech4396 | slyA::Kan; Himar RB1 transposon mutant; Kmr | This work |
| Ech163 | slyA::Kan; deletion mutant; 3937 derivative; Kmr | This work |
| Ech164 | expI::Kan; deletion mutant; 3937 derivative; Kmr | This work |
| Ech165 | expR::Kan; deletion mutant; 3937 derivative; Kmr | This work |
| Ech166 | Chromosomal insertion of lacY-slyA-cm-prt in Ech163; Kmr Cmr | This work |
| Ech188 | hrpS::Kan; deletion mutant; 3937 derivative; Kmr | This work |
| Plasmids | ||
| pPROBE-AT | Promoter-GFP reporter plasmid; Apr | 21 |
| pKD4 | Template plasmid carrying Kan cassette; Kmr | 8 |
| pGEM-T easy | Cloning vector; lacZ; Apr | Promega Corp. |
| pPhrpS | hrpS promoter region cloned in pPROBE-AT; Apr | 34 |
| pPhrpL | hrpL promoter region cloned in pPROBE-AT; Apr | 34 |
| pPhrpA | hrpA promoter region cloned in pPROBE-AT; Apr | This work |
| pPhrpN | hrpN promoter region cloned in pPROBE-AT; Apr | This work |
| pPslyA | slyA promoter region cloned in pPROBE-AT; Apr | This work |
| pPhrpX | hrpX promoter region cloned in pPROBE-AT; Apr | This work |
| pPhrpY | hrpY promoter region cloned in pPROBE-AT; Apr | This work |
| pML123 | RSF1010-derived expression and lac-fusion broad-host-range vector, Gmr | 16 |
| pWM91 | Suicide vector; oriR6K mobRP4 lacZa (of pBluescript II) sacB; Sucr Apr | 20 |
| pWM91slyA::Kan | 3.7-kb fragment containing slyA::Kan in pWM91 | This work |
| pWM91expI::Kan | 3.0-kb fragment containing expI::Kan in pWM91 | This work |
| pWM91expR::Kan | 3.2-kb fragment containing expR::Kan in pWM91 | This work |
| pMLslyA | 1.5-kb slyA with native promoter region cloned in pML123; Gmr | This work |
| pMLhrpL | 1.6-kb hrpL with native promoter region cloned in pML123; Gmr | This work |
| pTCLS-Cm | 6.4-kb lacY-cm-prt region cloned in pGEM-T Easy | 15 |
| pTCLSslyA-Cm | 1.5-kb slyA with its promoter region cloned in pTCLS-Cm; Apr Cmr | This work |
Apr, ampicillin resistance; Cmr, chloramphenicol resistance; Kmr, kanamycin resistance; Gmr, gentamicin resistance; Spr, streptomycin resistance.
Transposon mutagenesis and fluorescence-activated cell sorting (FACS) screening.
To identify genes involved in regulation of hrpS in D. dadantii 3937, we performed a mutagenesis with the MiniHimar RB1 transposon (2). The transposon was mobilized into D. dadantii 3937 harboring GFP reporter plasmid pPhrpS by conjugation with E. coli S17-1 pir+ carrying pMiniHimar RB1. Transconjugants were selected on MG plates containing kanamycin and ampicillin. The resulting transposon mutants were grown in LB containing kanamycin and ampicillin at 28°C overnight, and cells were transferred to MM and grown for an additional 24 h. Bacterial cells were harvested by centrifugation, washed, and resuspended in phosphate-buffered saline (PBS) as previously described (35). The expression of hrpS was determined by measuring the GFP intensity using a Calibur flow cytometer (Becton Dickinson Biosciences, San Jose, CA), and the data were analyzed with Cell Quest software (BD Biosciences, San Jose, CA). To identify the genes disrupted by the transposon, the chromosomal DNA of the mutants was digested with BamHI, self-ligated, and transformed into E. coli EC100D+. Plasmid was isolated from kanamycin-resistant colonies and sequenced using the primers himar1 and 615 (2). Among the 11 mutants screened, four (Ech3049, Ech4120, Ech4380, and Ech4396) were found to have transposon insertions at different sites in the slyA gene.
Construction of mutants and functional complementation.
To construct the slyA mutant, fragments flanking the slyA gene were amplified from D. dadantii genomic DNA by PCR. A kanamycin cassette was also amplified using the plasmid pKD4 as a template (8). Primers for amplification of flanking regions and the kanamycin cassette were designed to incorporate restriction sites to facilitate cloning. DNA fragments slyA1 (flanks the 5′ end) and slyA2 (flanks the 3′ end) were digested with BamHI/EcoRI and HindIII/XhoI, respectively. The kanamycin cassette was digested with EcoRI and HindIII. The slyA1 and slyA2 fragments were ligated to the kanamycin cassette at EcoRI and HindIII sites, and this recombinant fragment was cloned into pWM91 at BamHI and XhoI sites to generate plasmid pWM91slyA::kan. The slyA mutant of D. dadantii was generated by biparental mating between the E. coli S17-1λ pir and D. dadantii 3937 as described previously (20). In this study, the expI and expR mutants were also constructed in a manner similar to that of the slyA mutant. These mutants were confirmed by PCR and DNA sequencing.
To complement the slyA mutant, a fragment containing the complete coding sequence for the slyA gene along with its native promoter was cloned to pTCLS-Cm between the lacY and cm loci at the NruI and XhoI sites. slyA was inserted into the chromosome of the D. dadantii strain at an intergenic region by allelic exchange as described previously (15).
RNA isolation and Northern blot analysis.
Cultures of D. dadantii strains grown overnight in LB broth were diluted 1:100 in MM and grown for 12 h at 28°C to induce expression of T3SS genes. Total RNA was isolated by TRI reagent (Sigma-Aldrich, St. Louis, MO) (34), and each sample was treated with 2 U of Turbo DNA-free DNase (Ambion, Austin, TX) for 1 h at 37°C. The concentration of RNA samples was determined using a spectrophotometer (ND-1000). For each sample, 10 to 15 μg of total RNA was electrophoresed on a 1.0% denaturing agarose gel. Hybridization probes of rsmB, hrpA, and rsmA were amplified by PCR using the primers listed in Table S1 in the supplemental material. Probes were labeled using a BrightStar psoralen-biotin kit (Ambion, Austin, TX). Hybridization and detection were performed by using the NorthernMax kit and the BrightStar BioDetect kit (Ambion, Austin, TX) according to the manufacturer's instructions. 16S rRNA was used as an internal control.
Real-time RT-PCR analysis.
Strains were grown overnight in LB broth. Cultures were diluted 1:100 in MM and grown for 12 h at 28°C. Total RNA was isolated from 2 ml of each sample using an RNeasy minikit (Qiagen Sciences, MD) and treated with a Turbo DNA-free DNase kit (Ambion, Austin, TX). cDNA was synthesized with 0.5 μg of treated RNA using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA). The Real Master Mix (5 PRIME, Gaithersburg, MD) was used to quantify the cDNA level of target genes. Real-time reverse transcription (RT)-PCR data were collected by the Opticon 2 system (Bio-Rad, Hercules, CA) as described previously (35) and analyzed using the relative expression software tool (25). The rplU was used as the endogenous control for data analysis (18, 43). Primers used for real-time PCR are listed in Table S1 in the supplemental material. Primer efficiencies were tested for all primers, all of which are within the 1.95-to-2.05 range.
Swimming motility and pellicle formation.
In the swimming motility assay, 10 μl of an overnight bacterial suspension (optical density at 600 nm [OD600] of 0.1) was inoculated in the center of an MG plate containing 0.3% agar (42). The plates were incubated at 28°C, and the diameter of radial growth was measured after 24 h. The experiment was repeated three times with three biological replicates in each experiment.
The pellicle-forming ability of bacterial strains was evaluated in SOBG medium as described before (12, 40). Briefly, overnight bacterial cultures in LB broth were inoculated to SOBG broth at a concentration of 106 CFU/ml. Cultures were incubated in glass test tubes without shaking at 28°C for 2 days to observe formation of the pellicle.
RESULTS
SlyA downregulates the expression of hrpS.
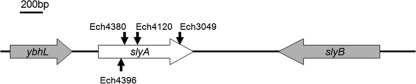
To identify novel T3SS regulators other than the previously identified TCS HrpX and HrpY that act upstream of hrpS, we employed a random transposon mutagenesis by introducing a MiniHimar RB1 transposon into D. dadantii 3937 carrying a plasmid-borne reporter, pPhrpS. pPhrpS contains an hrpS promoter-gfp transcriptional fusion which was used to identify transposon mutants with altered hrpS expression that can be detected using a flow cytometer to measure green fluorescent protein levels. Four mutants, Ech3049, Ech4120, Ech4380, and Ech4396, each with increased hrpS-gfp expression, had a transposon insertion in a 441-bp open reading frame (ORF) that encodes an SlyA protein (ASAP identifier [ID] 15312). Sequence analysis revealed that the transposons were inserted at bp position 392 (Ech3049), 177 (Ech4120), 122 (Ech4380), and 120 (Ech4396) of slyA (Fig. 1). The SlyA protein of D. dadantii 3937 showed 97% identity (99% similarity) to the SlyA protein of Dickeya zeae EC1 (NCBI accession no. ACC62398), 85% identity (94% similarity) to the SlyA protein of P. carotovorum subsp. carotovorum SCC3193 (NCBI accession no. ABY91292), and 75% identity (85% similarity) to the SlyA protein of Salmonella Typhimurium LT2 (NCBI accession no. NP460467). To confirm the increase in hrpS expression observed in the slyA transposon mutants, an slyA deletion mutant (Ech163) was constructed. Similar to the transposon mutants, dramatic increases of both promoter activity and mRNA level of hrpS were detected in Ech163 compared to those of the wild type (Table 2, Fig. 2A). The enhanced hrpS expression of Ech163 could be partially restored to the wild-type levels in Ech166, which contains an slyA gene integrated into a neutral chromosomal locus lacY-prt of Ech163 and expressed from its native promoter (Table 2).
Fig 1.
The slyA locus of D. dadantii 3937. The MiniHimar RB1 of four transposon mutants was inserted at bp 120 (Ech4396), 122 (Ech4380), 177 (Ech4120), and 392 (Ech3049) of the 441-bp nucleotide in the slyA ORF.
Table 2.
Promoter activity of hrpS in wild-type D. dadantii 3937 and slyA mutant Ech163 in minimal medium
| Gene promotera | Mean fluorescence intensityb |
|
|---|---|---|
| 12 h | 24 h | |
| 3937(pPhrpS) | 81.1 ± 3.2 | 82.9 ± 5.4 |
| Ech163(pPhrpS) | 185.7 ± 4.6* | 158.7 ± 5.0* |
| Ech166(pPhrpS) | 58.7 ± 1.1* | 66.3 ± 0.8 |
| 3937(pPhrpL) | 13.9 ± 1.1 | 14.6 ± 1.6 |
| Ech163(pPhrpL) | 24.2 ± 1.2* | 20.5 ± 2.0* |
| Ech166(pPhrpL) | 6.5 ± 0.1* | 9.5 ± 0.1* |
| 3937(pPhrpA) | 71.2 ± 8.3 | 88.1 ± 8.0 |
| Ech163(pPhrpA) | 24.3 ± 2.6* | 23.2 ± 6.3* |
| 3937(pPhrpN) | 44.5 ± 8.2 | 46.2 ± 8.0 |
| Ech163(pPhrpN) | 18.4 ± 3.8* | 12.1 ± 3.1* |
| 3937(pPROBE-AT) | 3.2 ± 0.3 | 3.4 ± 0.2 |
Ech163 is a deletion mutant of slyA in the D. dadantii 3937 background. Ech166 contains the slyA gene with its native promoter integrated into a neutral chromosomal locus, lacY-prt, of Ech163.
GFP intensity was determined on gated populations of bacterial cells by flow cytometry. Values of mean fluorescence intensity (MFI) are an average GFP fluorescence intensity of total bacterial cells with standard deviations (SD). Similar results have been observed in two individual experiments, and the results of one experiment have been shown here. Three replicates were used in this experiment. Asterisks indicate statistically significant differences in GFP MFI between the wild-type strain (3937) and the slyA mutant or the slyA complemented strain (P < 0.01, Student's t test).
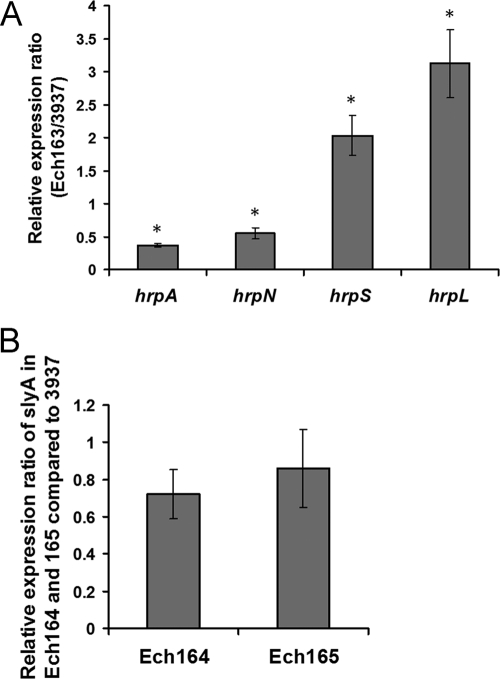
Fig 2.
(A) Expression ratios of hrpA, hrpN, hrpS, and hrpL mRNAs in slyA mutant Ech163 compared to those of D. dadantii 3937. (B) The relative mRNA levels of slyA in the expI mutant Ech164 and the expR mutant Ech165 compared to that of D. dadantii 3937. Bacterial strains were grown in minimal medium for 12 h. Total RNA was isolated, and the mRNA of target genes was measured by real-time RT-PCR. Asterisks indicate statistically significant differences in mRNA levels of the mutants compared to that of the wild type (P < 0.05). Similar results were observed in two independent experiments.
SlyA and HrpXY regulate the expression of hrpS independently.
The expression of hrpS in D. dadantii 3937 is dependent on the TCS HrpX/HrpY, with HrpY directly activating expression of hrpS (40, 41). In this study, we showed that the expression of hrpS is also controlled by SlyA. To determine whether the HrpX/HrpY signaling cascade regulates the expression of hrpS through SlyA, the promoter activity of slyA was measured in D. dadantii 3937, an hrpX mutant (WPP67), and an hrpY mutant (WPP92). Similar expression levels of slyA were observed among the wild type, WPP67, and WPP92 (Table 3). This result suggests that the expression of slyA is not influenced by HrpX and HrpY. SlyA regulates T3SS expression by controlling the transcription of the TCS kinase SsrA in S. Typhimurium (17, 23). We also investigated the effect of SlyA on expression of the hrpX and hrpY genes. The GFP intensity of Ech163 carrying the reporters pPhrpX and pPhrpY, which contain transcriptional fusions of hrpX and hrpY promoters with the gfp gene, respectively, was measured by flow cytometry. Compared with the wild-type strain, the slyA mutant exhibited no difference in either hrpX or hrpY expression (Table 3). This indicates that SlyA does not regulate the expression of hrpX and hrpY.
Table 3.
Promoter activities of hrpX and hrpY in D. dadantii 3937 and Ech163 and of slyA in 3937, WPP67, and WPP92
| Gene promotera | Mean fluorescence intensityb |
||
|---|---|---|---|
| 6 h | 12 h | 24 h | |
| 3937(pPslyA) | 119.5 ± 7.3 | 131.7 ± 3.3 | 128.1 ± 2.7 |
| WPP67(pPslyA) | 119.1 ± 2.4 | 133.7 ± 0.8 | 140.7 ± 0.8 |
| WPP92(pPslyA) | 127.5 ± 5.4 | 128.5 ± 0.5 | 134.3 ± 5.3 |
| 3937(pPROBE-AT) | 2.6 ± 0.5 | 2.1 ± 0.1 | 3.4 ± 0.6 |
| 3937(pPhrpX) | 10.8 ± 0.5 | 14.3 ± 0.8 | 14.8 ± 0.7 |
| Ech163(pPhrpX) | 11.0 ± 0.2 | 12.0 ± 0.4 | 13.5 ± 0.9 |
| 3937(pPhrpY) | 25.4 ± 1.0 | 35.2 ± 1.1 | 34.3 ± 0.9 |
| Ech163(pPhrpY) | 24.8 ± 0.6 | 29.5 ± 0.8 | 28.6 ± 0.6 |
| 3937(pPROBE-AT) | 2.3 ± 0.1 | 2.6 ± 0.3 | 4.4 ± 0.6 |
WPP67 and WPP92 are hrpX and hrpY mutants of D. dadantii 3937, respectively. Ech163 is the slyA mutant of D. dadantii 3937.
Similar results have been observed in two individual experiments with three biological replicates, and the results of one experiment have been shown here. No significant difference in slyA promoter activity was observed among the wild-type strain (3937), WPP67, WPP92, and Ech163 (P > 0.05, Student's t test).
SlyA negatively regulates hrpL at transcriptional and posttranscriptional levels.
In D. dadantii 3937, HrpS initiates the transcription of hrpL (40). As discovered from the transposon mutagenesis and the deletion of slyA, SlyA downregulates hrpS expression. To determine whether SlyA also regulates hrpL, the expression level of hrpL was evaluated in the wild type and slyA mutant Ech163 by promoter activity assay and quantitative RT-PCR (qRT-PCR). Compared with that of the wild type, an approximate 2-fold increase in hrpL promoter activity was observed in Ech163 (Table 2). The increased hrpL expression observed in Ech163 could be partially restored to the wild-type level in an slyA complementation strain, Ech166 (Table 2). In addition, analysis by qRT-PCR showed a 3-fold increase in the hrpL mRNA in Ech163 (P ≤ 0.01) compared to the wild type (Fig. 2A).
In D. dadantii 3937, in addition to the transcriptional regulation by HrpS, the hrpL mRNA level is also controlled through the GacS-GacA-rsmB-RsmA-HrpL regulatory pathway at the posttranscriptional level. To further explore the role of this pathway, the expression levels of rsmA and rsmB in wild type and Ech163 were analyzed by Northern blot analysis. Compared to the wild type, a reduction in rsmA mRNA was detected in Ech163 and was restored to near-wild-type levels in the Ech166 complemented strain (Fig. 3A). Similar amounts of rsmB RNA were observed between the wild type and Ech163 (Fig. 3B). RsmA is a small-RNA-binding protein that acts by reducing the half-life of the hrpL mRNA. These results suggest that the increased hrpL mRNA level observed in Ech163 is derived not only from the increased hrpS expression but also from the decreased rsmA expression. Overall, SlyA negatively regulates hrpL by downregulating hrpS and upregulating rsmA.
Fig 3.
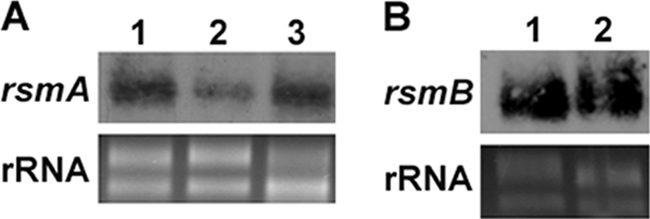
(A) Northern blot analysis of rsmA in D. dadantii 3937 (lane 1), slyA mutant Ech163 (lane 2), and slyA chromosomal complementation strain Ech166 (lane 3). (B) Northern blot analysis of rsmB mRNA in D. dadantii 3937 (lane 1) and Ech163 (lane 2). Similar results were observed in two independent experiments.
SlyA upregulates the transcription of hrp genes in the HrpL regulon in spite of its downregulation of hrpL.
To further investigate whether SlyA affects the expression of genes in the HrpL regulon, the expression of hrpA and hrpN was measured in D. dadantii 3937 and Ech163 by measuring promoter activity. Surprisingly, a reduction in hrpA and hrpN expression was observed in Ech163 (Table 2). A decrease in the mRNA level of hrpA and hrpN was also confirmed in Ech163 in comparison to that of the wild-type strain by qRT-PCR (Fig. 2A). Finally, the expression of hrpA was partially complemented in Ech166 (Fig. 4A). These results suggest that SlyA exerts two different modes of regulation on the T3SS in D. dadantii 3937: it negatively regulates the expression of hrpS and hrpL, whereas it positively regulates the expression of HrpL regulon genes, such as hrpA and hrpN.
Fig 4.
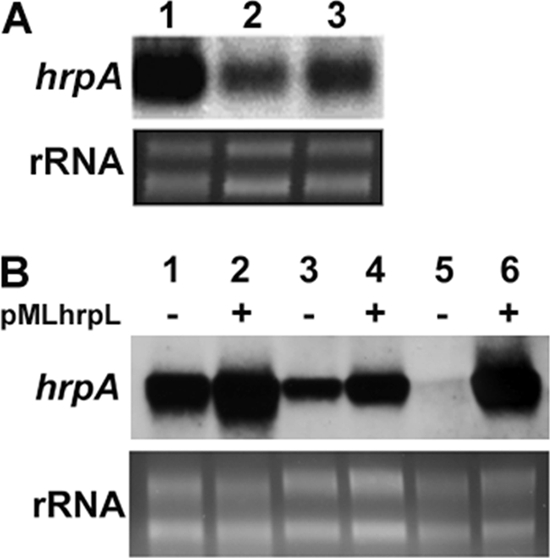
(A) Northern blot analysis of hrpA in wild-type D. dadantii 3937 (lane 1), slyA mutant Ech163 (lane 2), and Ech166, the chromosomal complemented strain of Ech163 (lane 3). (B) Northern blot analysis of hrpA mRNA in D. dadantii 3937 carrying empty vector pML123 (lane 1), D. dadantii 3937 carrying hrpL overexpression plasmid pMLhrpL (lane 2), slyA mutant Ech163 carrying pML123 (lane3), Ech163 carrying pMLhrpL (lane 4), hrpS mutant Ech188 carrying pML123 (lane 5), and Ech188 carrying pMLhrpL (lane 6). Similar results were observed in two independent experiments.
SlyA controls the HrpL regulon genes in parallel with the T3SS master regulator HrpL.
In the slyA mutant, the expression of hrpA and hrpN is reduced even though hrpL expression is induced, which suggests that SlyA regulates HrpL regulon genes independent of HrpL. To test this hypothesis, the hrpA mRNA levels were compared in D. dadantii 3937, the slyA mutant Ech163, and the hrpS mutant Ech188 in the presence and absence of an hrpL overexpressing plasmid pMLhrpL (Fig. 4B). When hrpL is overexpressed in D. dadantii 3937, a dramatic induction of hrpA expression was observed compared to that of the wild type carrying the empty vector pML123 (Fig. 4B, lanes 1 and 2). HrpS controls hrpA expression through its regulation of hrpL. As expected, the hrpS mutant Ech188 was deficient in hrpA expression (Fig. 4B, lane 5). By introducing pMLhrpL into Ech188, the expression of hrpA was restored to a level similar to that of the wild type carrying pMLhrpL (Fig. 4B, lane 6). The hrpA expression was greatly reduced in the slyA mutant, which indicates that SlyA positively regulates hrpA (Fig. 4B, lane 3). However, in the slyA mutant Ech163 with pMLhrpL, hrpA expression was unable to be restored to the same level as that in Ech188 with pMLhrpL (Fig. 4B, lanes 4 and 6). In fact, the hrpA expression in Ech163 carrying pMLhrpL (lane 4) was slightly lower than that of D. dadantii 3937 with the empty vector (lane 1). Similar results were observed by Northern blot analysis using an hrpN probe (data not shown). These results suggest that the regulation of SlyA on the HrpL regulon genes, such as hrpA and hrpN, is in parallel with the T3SS master regulator HrpL.
slyA is not controlled by ExpR or ExpI in D. dadantii 3937.
In P. carotovorum subsp. carotovorum SCC3193, the expression of slyA is dependent on the concentration of AHSL, a QS signal molecule (26). A mutation in expI, a gene encoding the AHSL synthase, leads to a reduction in slyA expression. This is due to the fact that in the absence of AHSL, ExpR1 and ExpR2 activate the expression of rsmA, which in turn negatively regulates slyA expression. However, in the presence of the AHSL, the expression of rsmA in P. carotovorum subsp. carotovorum SCC3193 is fully repressed (26). The D. dadantii 3937 genome contains expI (ASAP ID 19415) and expR (ASAP ID 19414), encoding an AHL synthase and an AHSL responsive regulator, respectively (10, 14). The D. dadantii genome also contains a putative LuxR regulator (ASAP ID 15900) with low similarity (23% identity) to ExpR. Besides expI, no other homologue of the AHSL synthase was found in the D. dadantii 3937 genome. To determine whether QS controls the expression of T3SS by regulating slyA in D. dadantii 3937, deletion mutants of expI (Ech164) and expR (Ech165) were generated, and the expression of rsmA, slyA, and hrpA was measured in D. dadantii 3937, Ech164, and Ech165. In contrast to that in P. carotovorum subsp. carotovorum SCC3193, similar levels of rsmA RNA were observed among the wild type, the expI mutant (Ech164), and the expR mutant (Ech165) (Fig. 5A). qRT-PCR analysis showed no significant differences in slyA mRNA levels in Ech164 (P = 0.55) or Ech165 (P = 0.983) compared to that in the wild-type strain (Fig. 2B). In addition, similar levels of hrpA mRNA were observed in the expI and expR mutants and the wild type when examined by Northern blot analysis (Fig. 5B). Given that HrpY is essential for the expression of hrpA, an hrpY mutant (WPP92) was included as a control (Fig. 5B). These results indicate that unlike P. carotovorum subsp. carotovorum SCC3193, ExpI and ExpR are not involved in regulating the expression of slyA or the T3SS genes in D. dadantii 3937.
Fig 5.
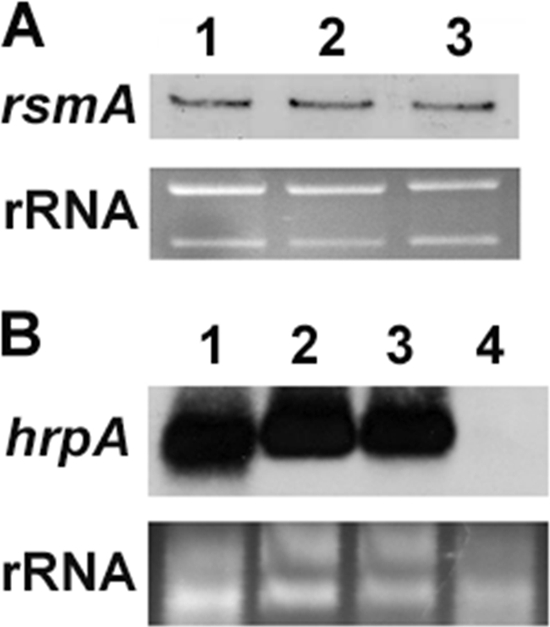
(A) Northern blot analysis of rsmA mRNA in D. dadantii 3937 (lane 1), the expI mutant Ech164 (lane 2), and the expR mutant Ech165 (lane 3). (B) Northern blot analysis of hrpA in D. dadantii 3937 (lane 1), expI mutant Ech164 (lane 2), expR mutant Ech165 (lane3), and hrpY mutant WPP92 (lane 4). Similar results were observed in two independent experiments.
SlyA controls bacterial swimming motility and pellicle formation.
Motility contributes to bacterial virulence, whereas biofilm and pellicle formation aids bacterial survival in a variety of environments. In D. dadantii 3937, the T3SS is required for pellicle formation (12, 40). Compared to wild-type D. dadantii 3937, increased swimming motility was observed in Ech163, and this phenotype could be complemented in the complementation strain Ech166 (Fig. 6A). Additionally, pellicle formation was completely suppressed in Ech163 compared to that in the wild type, and the mutant phenotype could be restored to the wild-type level in Ech166 (Fig. 6B).
Fig 6.
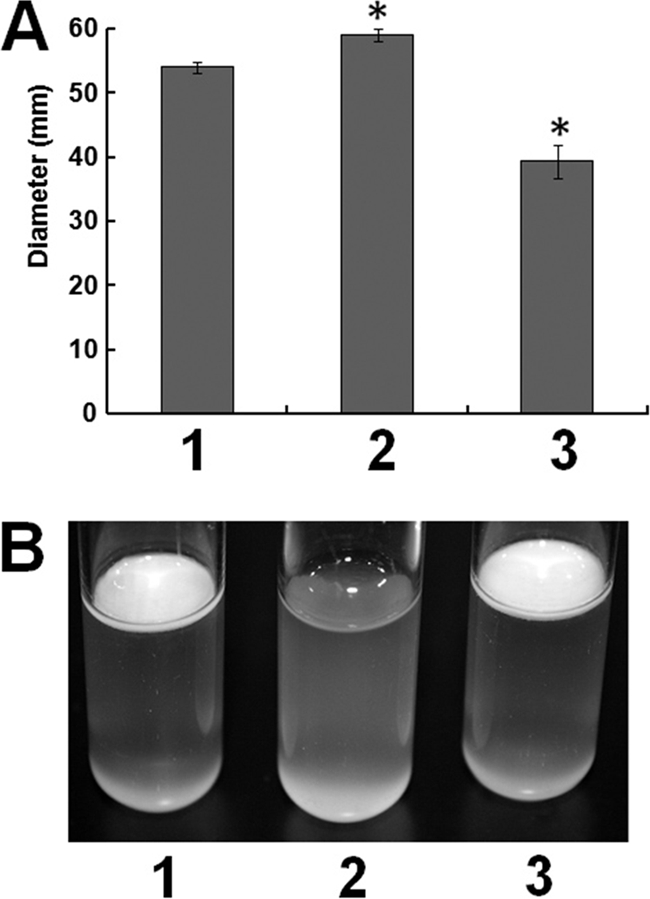
(A) Swimming motility of D. dadantii 3937 (lane 1), slyA mutant Ech163 (lane 2), and Ech166 (lane 3), a slyA chromosomal complementation strain of Ech163. (B) Pellicle formation of D. dadantii 3937 (lane 1), Ech163 (lane 2), and Ech166 (lane 3). Similar results were observed in two independent experiments with two biological replicates in each experiment.
DISCUSSION
In this study, a novel regulator, SlyA, which displays a complex and intricate regulation of the T3SS in D. dadantii 3937 (Fig. 7), was identified and characterized. Previous reports have described that the T3SS structural and effector genes are regulated by two major regulatory pathways, HrpX/HrpY-HrpS-HrpL and GacS/GacA-rsmB-RsmA pathways. Our work demonstrates that, in contrast to HrpX/HrpY, SlyA regulates hrpS negatively (Table 3). In addition, SlyA positively controls RsmA, a posttranscriptional regulator of hrpL (Fig. 3A). The negative regulation on hrpS and positive regulation on rsmA leads to a decrease in hrpL expression (Fig. 2A and Table 2). Interestingly, although SlyA represses the hrpL expression, the deletion of slyA leads to reduced expressions of two T3SS genes in the HrpL regulon, hrpA and hrpN (Fig. 2A and Table 2). These results suggest that SlyA plays an opposing role in T3SS regulation: it acts as a negative regulator of hrpS and hrpL but serves as a positive regulator of HrpL regulon genes, such as hrpA and hrpN. This also implies that along with the well-studied HrpX-HrpY-HrpS-HrpL and GacA-GacS-RsmA-rsmB-HrpL pathways, an additional regulatory pathway, SlyA-HrpA/HrpN, regulates T3SS in D. dadantii 3937 (Fig. 7). Finally, hrpA expression was completely shut down when HrpL was absent in an hrpS mutant, but a reduced level of hrpA expression was observed in the slyA mutant (Fig. 4B, lanes 3 and 5). This suggests that HrpL plays a greater role in controlling the HrpL regulon genes than SlyA. In this study, the expression of hrpS, hrpL, and hrpA was partially complemented when the slyA gene was inserted into a neutral chromosomal site of Ech163. The partially complemented phenotype in the mutant may be due to temporal and spatial differences in the placement of the complemented copy of slyA in the intergenic region.
Fig 7.
The regulation of SlyA on the T3SS expression and extracellular enzyme production. SlyA negatively regulates the transcription of hrpS, through an HrpX/HrpY-independent pathway. It also upregulates the expression of rsmA. The influence of SlyA on hrpS and rsmA leads to a downregulation of hrpL. However, SlyA upregulates the expression of the HrpL regulon genes, such as hrpA and hrpN, in parallel with HrpL. The lines with bars and arrows represent negative and positive regulations, respectively.
HrpL is the master regulator of the T3SS genes encoding the basic structural and functional components of the T3SS. By binding to a consensus sequence called the hrp box, HrpL initiates the transcription of the HrpL regulon genes. Our data suggest that SlyA regulates HrpL regulon genes in parallel with HrpL. Reports have been published describing novel T3SS regulators of the group I T3SS; however, the influence of these regulators on the HrpL regulon genes are mediated through the regulation of HrpL, either transcriptionally or posttranscriptionally (22, 42, 43). To our knowledge, this is the first report describing a transcriptional regulator that controls HrpL regulon genes in an HrpL-independent manner (Fig. 7).
Quorum sensing (QS) is a common mechanism of gene regulation among Gram-negative bacteria in which individual cells produce and respond to special AHSL signal molecules (11). The SlyA homologue has recently been described as a QS target in P. carotovorum subsp. carotovorum SCC3193 (26). An increase in slyA expression was observed in an expI mutant of P. carotovorum subsp. carotovorum SCC3193 (26). In this regulatory pathway, RsmA negatively regulates the expression of slyA, and rsmA expression is positively regulated by two QS regulators, ExpR1 and ExpR2, in the absence of AHSL (26). The SlyA protein of D. dadantii 3937 was annotated based on high similarity to SlyA of P. carotovorum. However, our result showed that inactivation of expI or expR of D. dadantii 3937 had little effect on the expression of slyA. This suggests that the expression of slyA may be controlled dissimilarly among different bacteria. For example, SlyA is controlled in an AHSL-independent manner in P. carotovorum subsp. carotovorum strain ATTn10 (19). A previous report demonstrated that the expression of hrpN of D. dadantii 3937 is independent of the QS signal N-(3-oxo-hexanoyl)-l-homoserine lactone (OHHL) (22). Similarly, our results show that in D. dadantii 3937, the expression of hrpA and hrpN is not under the control of ExpI and ExpR.
Cell-cell aggregation is an important process for bacteria switching from a single-cell mode to a multicellular community (39). In D. dadantii 3937, the T3SS genes are required for pellicle formation (12, 40). Our study demonstrated that SlyA is required for the pellicle formation, which may be due to the regulatory effect of SlyA on hrpA and hrpN expression.
SlyA homologues represent a growing family of novel bacterial regulatory proteins which play important roles in the global regulation of diverse physiological processes in animal and plant pathogens (28). In soft-rot bacteria, a reduction in extracellular enzymes and pellicle formation was observed in an slyA mutant of P. carotovorum subsp. carotovorum Scc3139 and D. dadantii (12, 26). In this study, SlyA also shows a global regulatory effect to control other biological characteristics, such as T3SS, motility, and pellicle formation. Our results indicate that SlyA upregulates the expression of hrpA and hrpN, which is independent of the known HrpS-HrpL and posttranscriptional Rsm-mediated regulatory pathways.
Supplementary Material
ACKNOWLEDGMENTS
This work is dedicated to Noel T. Keen.
We thank Daad Saffarini and Amy Charkowski for providing the bacterial strains S17-1λ pir/pMiniHimar, WPP67, and WPP92 and Nicole Perna of the University of Wisconsin—Madison for providing access to the annotated D. dadantii genome sequences (https://asap.ahabs.wisc.edu/asap/ASAP1.htm). We thank Eulandria Biddle for critical discussions and reading of the manuscript and Tracy Doyle for assistance in constructing the slyA mutant.
This project is supported by grants from the National Science Foundation (award no. EF-0332163), the Research Growth Initiative of the University of Wisconsin—Milwaukee, and the Catalyst Grant Program of the UWM Research Foundation.
Footnotes
Published ahead of print 20 January 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Alekshun MN, Kim YS, Levy SB. 2000. Mutational analysis of MarR, the negative regulator of marRAB expression in Escherichia coli, suggests the presence of two regions required for DNA binding. Mol. Microbiol. 35:1394–1404 [DOI] [PubMed] [Google Scholar]
- 2. Bouhenni R, Gehrke A, Saffarini D. 2005. Identification of genes involved in cytochrome c biogenesis in Shewanella oneidensis, using a modified mariner transposon. Appl. Environ. Microbiol. 71:4935–4937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chatterjee A, Cui Y, Liu Y, Dumenyo CK, Chatterjee AK. 1995. Inactivation of rsmA leads to overproduction of extracellular pectinases, cellulases, and proteases in Erwinia carotovora subsp. carotovora in the absence of the starvation/cell density-sensing signal, N-(3-oxohexanoyl)-l-homoserine lactone. Appl. Environ. Microbiol. 61:1959–1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cui Y, Chatterjee A, Liu Y, Dumenyo CK, Chatterjee AK. 1995. Identification of a global repressor gene, rsmA, of Erwinia carotovora subsp. carotovora that controls extracellular enzymes, N-(3-oxohexanoyl)-l-homoserine lactone, and pathogenicity in soft-rotting Erwinia spp. J. Bacteriol. 177:5108–5115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cui Y, Chatterjee A, Yang H, Chatterjee AK. 2008. Regulatory network controlling extracellular proteins in Erwinia carotovora subsp. carotovora: FlhDC, the master regulator of flagellar genes, activates rsmB regulatory RNA production by affecting gacA and hexA (lrhA) expression. J. Bacteriol. 190:4610–4623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cui Y, Madi L, Mukherjee A, Dumenyo CK, Chatterjee AK. 1996. The RsmA− mutants of Erwinia carotovora subsp. carotovora strain Ecc71 overexpress hrpNEcc and elicit a hypersensitive reaction-like response in tobacco leaves. Mol. Plant Microbe Interact. 9:565–573 [DOI] [PubMed] [Google Scholar]
- 7. Cui Y, Mukherjee A, Dumenyo CK, Liu Y, Chatterjee AK. 1999. rsmC of the soft-rotting bacterium Erwinia carotovora subsp. carotovora negatively controls extracellular enzyme and harpin(Ecc) production and virulence by modulating levels of regulatory RNA (rsmB) and RNA-binding protein (RsmA). J. Bacteriol. 181:6042–6052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ferrandiz MJ, Bishop K, Williams P, Withers H. 2005. HosA, a member of the SlyA family, regulates motility in enteropathogenic Escherichia coli. Infect. Immun. 73:1684–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Glasner JD, et al. 2011. Genome sequence of the plant pathogenic bacterium Dickeya dadantii 3937. J. Bacteriol. 193:2076–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gray KM, Garey JR. 2001. The evolution of bacterial LuxI and LuxR quorum sensing regulators. Microbiology 147:2379–2387 [DOI] [PubMed] [Google Scholar]
- 12. Haque MM, Kabir MS, Aini LQ, Hirata H, Tsuyumu S. 2009. SlyA, a MarR family transcriptional regulator, is essential for virulence in Dickeya dadantii 3937. J. Bacteriol. 191:5409–5418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hugouvieux-Cotte-Pattat N, Condemine G, Nasser W, Reverchon S. 1996. Regulation of pectinolysis in Erwinia chrysanthemi. Annu. Rev. Microbiol. 50:213–257 [DOI] [PubMed] [Google Scholar]
- 14. Hussain MB, et al. 2008. The acyl-homoserine lactone-type quorum-sensing system modulates cell motility and virulence of Erwinia chrysanthemi pv. zeae. J. Bacteriol. 190:1045–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jahn CE, Willis DK, Charkowski AO. 2008. The flagellar sigma factor fliA is required for Dickeya dadantii virulence. Mol. Plant Microbe Interact. 21:1431–1442 [DOI] [PubMed] [Google Scholar]
- 16. Labes M, Puhler A, Simon R. 1990. A new family of RSF1010-derived expression and lac-fusion broad-host-range vectors for Gram-negative bacteria. Gene 89:37–46 [DOI] [PubMed] [Google Scholar]
- 17. Linehan SA, Rytkonen A, Yu XJ, Liu M, Holden DW. 2005. SlyA regulates function of Salmonella pathogenicity island 2 (SPI-2) and expression of SPI-2-associated genes. Infect. Immun. 73:4354–4362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mah TF, et al. 2003. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 426:306–310 [DOI] [PubMed] [Google Scholar]
- 19. McGowan SJ, et al. 2005. Carbapenem antibiotic biosynthesis in Erwinia carotovora is regulated by physiological and genetic factors modulating the quorum sensing-dependent control pathway. Mol. Microbiol. 55:526–545 [DOI] [PubMed] [Google Scholar]
- 20. Metcalf WW, et al. 1996. Conditionally replicative and conjugative plasmids carrying lacZ alpha for cloning, mutagenesis, and allele replacement in bacteria. Plasmid 35:1–13 [DOI] [PubMed] [Google Scholar]
- 21. Miller WG, Leveau JH, Lindow SE. 2000. Improved gfp and inaZ broad-host-range promoter-probe vectors. Mol. Plant Microbe Interact. 13:1243–1250 [DOI] [PubMed] [Google Scholar]
- 22. Nasser W, Reverchon S, Vedel R, Boccara M. 2005. PecS and PecT coregulate the synthesis of HrpN and pectate lyases, two virulence determinants in Erwinia chrysanthemi 3937. Mol. Plant Microbe Interact. 18:1205–1214 [DOI] [PubMed] [Google Scholar]
- 23. Okada N, et al. 2007. Identification of amino acid residues of Salmonella SlyA that are critical for transcriptional regulation. Microbiology 153:548–560 [DOI] [PubMed] [Google Scholar]
- 24. Peng Q, et al. 2006. Population behavior analysis of dspE and pelD regulation in Erwinia chrysanthemi 3937. Mol. Plant Microbe Interact. 19:451–457 [DOI] [PubMed] [Google Scholar]
- 25. Pfaffl MW, Horgan GW, Dempfle L. 2002. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sjoblom S, Harjunpaa H, Brader G, Palva ET. 2008. A novel plant ferredoxin-like protein and the regulator Hor are quorum-sensing targets in the plant pathogen Erwinia carotovora. Mol. Plant Microbe Interact. 21:967–978 [DOI] [PubMed] [Google Scholar]
- 27. Tang X, Xiao Y, Zhou JM. 2006. Regulation of the type III secretion system in phytopathogenic bacteria. Mol. Plant Microbe Interact. 19:1159–1166 [DOI] [PubMed] [Google Scholar]
- 28. Thomson NR, et al. 1997. The rap and hor proteins of Erwinia, Serratia, and Yersinia: a novel subgroup in a growing superfamily of proteins regulating diverse physiological processes in bacterial pathogens. Mol. Microbiol. 26:531–544 [DOI] [PubMed] [Google Scholar]
- 29. Thomson NR, Crow MA, McGowan SJ, Cox A, Salmond GP. 2000. Biosynthesis of carbapenem antibiotic and prodigiosin pigment in Serratia is under quorum sensing control. Mol. Microbiol. 36:539–556 [DOI] [PubMed] [Google Scholar]
- 30. Wilkinson SP, Grove A. 2006. Ligand-responsive transcriptional regulation by members of the MarR family of winged helix proteins. Curr. Issues Mol. Biol. 8:51–62 [PubMed] [Google Scholar]
- 31. Wu RY, et al. 2003. Crystal structure of Enterococcus faecalis SlyA-like transcriptional factor. J. Biol. Chem. 278:20240–20244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yang CH, et al. 2002. hrp genes of Erwinia chrysanthemi 3937 are important virulence factors. Mol. Plant Microbe Interact. 15:472–480 [DOI] [PubMed] [Google Scholar]
- 33. Yang CH, Menge JA, Cooksey DA. 1994. Mutations affecting hyphal colonization and pyoverdine production in pseudomonads antagonistic toward Phytophthora parasitica. Appl. Environ. Microbiol. 60:473–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yang S, et al. 2008. Type III secretion system genes of Dickeya dadantii 3937 are induced by plant phenolic acids. PLoS One 3:e2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yang S, et al. 2008. Dynamic regulation of GacA in type III secretion, pectinase gene expression, pellicle formation, and pathogenicity of Dickeya dadantii (Erwinia chrysanthemi 3937). Mol. Plant Microbe Interact. 21:133–142 [DOI] [PubMed] [Google Scholar]
- 36. Yang S, et al. 2010. Genome-wide identification of HrpL-regulated genes in the necrotrophic phytopathogen Dickeya dadantii 3937. PLoS One 5:e13472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yang S, et al. 2004. Genome-wide identification of plant-upregulated genes of Erwinia chrysanthemi 3937 using a GFP-based IVET leaf array. Mol. Plant Microbe Interact. 17:999–1008 [DOI] [PubMed] [Google Scholar]
- 38. Yang S, et al. 2007. Global effect of indole-3-acetic acid biosynthesis on multiple virulence factors of Erwinia chrysanthemi 3937. Appl. Environ. Microbiol. 73:1079–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yap MN, Rojas CM, Yang CH, Charkowski AO. 2006. Harpin mediates cell aggregation in Erwinia chrysanthemi 3937. J. Bacteriol. 188:2280–2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yap MN, Yang CH, Barak JD, Jahn CE, Charkowski AO. 2005. The Erwinia chrysanthemi type III secretion system is required for multicellular behavior. J. Bacteriol. 187:639–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yap MN, Yang CH, Charkowski AO. 2008. The response regulator HrpY of Dickeya dadantii 3937 regulates virulence genes not linked to the hrp cluster. Mol. Plant Microbe Interact. 21:304–314 [DOI] [PubMed] [Google Scholar]
- 42. Yi X, Yamazaki A, Biddle E, Zeng Q, Yang CH. 2010. Genetic analysis of two phosphodiesterases reveals cyclic diguanylate regulation of virulence factors in Dickeya dadantii. Mol. Microbiol. 77:787–800 [DOI] [PubMed] [Google Scholar]
- 43. Zeng Q, Ibekwe AM, Biddle E, Yang CH. 2010. Regulatory mechanisms of exoribonuclease PNPase and regulatory small RNA on T3SS of Dickeya dadantii. Mol. Plant Microbe Interact. 23:1345–1355 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.