Abstract
Ring-cleaving dioxygenases catalyze key reactions in the aerobic microbial degradation of aromatic compounds. Many pathways converge to catecholic intermediates, which are subject to ortho or meta cleavage by intradiol or extradiol dioxygenases, respectively. However, a number of degradation pathways proceed via noncatecholic hydroxy-substituted aromatic carboxylic acids like gentisate, salicylate, 1-hydroxy-2-naphthoate, or aminohydroxybenzoates. The ring-cleaving dioxygenases active toward these compounds belong to the cupin superfamily, which is characterized by a six-stranded β-barrel fold and conserved amino acid motifs that provide the 3His or 2- or 3His-1Glu ligand environment of a divalent metal ion. Most cupin-type ring cleavage dioxygenases use an FeII center for catalysis, and the proposed mechanism is very similar to that of the canonical (type I) extradiol dioxygenases. The metal ion is presumed to act as an electron conduit for single electron transfer from the metal-bound substrate anion to O2, resulting in activation of both substrates to radical species. The family of cupin-type dioxygenases also involves quercetinase (flavonol 2,4-dioxygenase), which opens up two C-C bonds of the heterocyclic ring of quercetin, a wide-spread plant flavonol. Remarkably, bacterial quercetinases are capable of using different divalent metal ions for catalysis, suggesting that the redox properties of the metal are relatively unimportant for the catalytic reaction. The major role of the active-site metal ion could be to correctly position the substrate and to stabilize transition states and intermediates rather than to mediate electron transfer. The tentative hypothesis that quercetinase catalysis involves direct electron transfer from metal-bound flavonolate to O2 is supported by model chemistry.
INTRODUCTION
Microorganisms have evolved a variety of aerobic as well as anaerobic pathways to degrade aromatic and heterocyclic compounds (40, 47, 48, 54). Since aromatic compounds, due to the delocalization of their π orbitals, are very stable, the key steps for degradation involve (i) activation of the aromatic ring by introduction of substituents and (ii) dearomatization. In aerobic microorganisms, activation of an aromatic substrate usually is achieved by hydroxylation reactions, and the critical dearomatization step is performed by ring-cleaving dioxygenases. Many pathways converge to catecholic substrates, which undergo cleavage either ortho to (between) the two hydroxyl substituents, catalyzed by intradiol dioxygenases, or meta (adjacent) to the hydroxyl substituents, catalyzed by extradiol dioxygenases. Ring-cleaving dioxygenases play important roles in the degradation of aromatic compounds by soil bacteria. They even can be key determinants of the fate of certain aromatic compounds in the environment, as in several instances their properties were shown to confine the specificity of a degradation pathway. For example, the presence of chloroaromatic compounds may prevent the degradation of methylaromatics, such as xylenes or cresols, via the meta cleavage pathway, because most catechol 2,3-dioxygenases are inactivated by 3-halocatechols (10, 69, 81). Meta cleavage also is the critical step in the degradation of polychlorinated biphenyls (PCBs), because the susceptibility of the enzyme to inactivation by ortho-substituted PCBs interferes with the degradation of other PCB congeners (28).
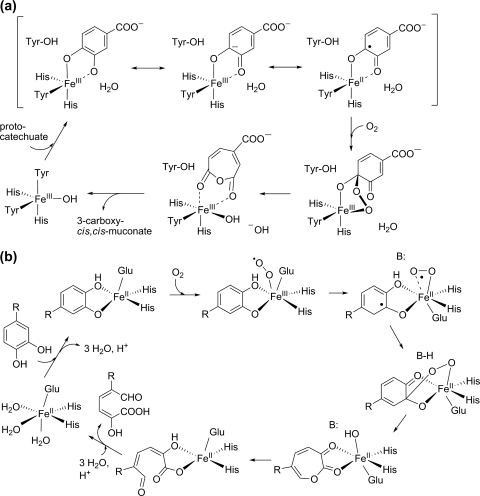
Enzymes of the intradiol dioxygenase family use a mononuclear FeIII center, coordinated to two tyrosine and two histidine ligands, for catalysis. Within this family, protocatechuate 3,4-dioxygenase has been studied most extensively. Crystal structures and X-ray absorption data revealed a ferric center in a trigonal bipyramidal geometry, with a hydroxide ligand completing the coordination sphere (31, 93, 94, 123). The substrate binds as a dianion, donating both its protons to the displaced hydroxide and tyrosyl ligands (36, 60, 96). Based on spectroscopic data and electronic structure calculations, it has been proposed that the iron-catecholate interaction introduces a semiquinonate radical character to the bound substrate, which reacts directly with dioxygen to form an alkylperoxo-FeIII intermediate. Rearrangement and O-O bond cleavage yield a cyclic anhydride and a metal-bound oxide or hydroxide, and the latter finally hydrolyzes the anhydride to yield the reaction product (30, 99) (Fig. 1a).
Fig 1.
Proposed reaction mechanisms for intradiol and extradiol ring-cleaving dioxygenases. (a) Mechanism of protocatechuate 3,4-dioxygenase (99). (b) Mechanism of homoprotocatechuate 2,3-dioxygenase (78, 83). R, -CH2COOH; B:, general base.
The canonical (type I) extradiol enzymes belong to the vicinal oxygen chelate (VOC) superfamily, which is characterized by paired βαβββ modules that provide a coordination environment for divalent metal ions. The types of reactions catalyzed by members of this superfamily are quite diverse and include isomerizations, epimerizations, nucleophilic substitutions, and oxidative C-C bond cleavage (5). Extradiol dioxygenases use an active-site FeII cofactor or, rarely, MnII for catalysis. The metal ion is coordinated by one glutamate and two histidine residues that occupy one face of a (pseudo)octahedral coordination sphere. The three adjacent coordination sites on the opposite face, usually occupied by easily displaceable solvent molecules, are available for the binding of substrates. This structural motif, termed the 2His-1carboxylate facial triad, is widespread among nonheme iron oxygenases (25, 73, 118). Whereas the intradiol enzymes appear to activate the organic substrate for electrophilic attack by O2, extradiol dioxygenases have been proposed to simultaneously activate substrate and O2 to form two metal-bound radical species (Fig. 1b). The reaction starts with bidentate binding of the substrate as catecholate monoanion to the active-site metal, displacing two or three water molecules (109, 124). Substrate binding dramatically increases the affinity of the metal center for O2 binding (4). In the ternary complex, the two substrates are electronically connected through the metal, facilitating electron transfer from catecholate to O2 to give a semiquinone-FeII-superoxide species. Such an activation of both substrates to radical species should allow rapid recombination to form an alkylperoxo intermediate. Subsequent rearrangement and O-O bond cleavage gives a seven-membered lactone intermediate and an FeII-bound hydroxide ion, which hydrolyzes the lactone to yield the 2-hydroxymuconate semialdehyde product (Fig. 1b) (44, 71, 72, 78, 83, 84).
A question pivotal to the understanding of the molecular mechanism of metal-dependent oxygenases refers to the role of the metal in oxygen activation. Interestingly, FeII-homoprotocatechuate 2,3-dioxygenase (Fe-HPCD) from Brevibacterium fuscum and the (closely related) Mn2+-dependent enzyme from Arthrobacter globiformis (126, 128) can each be prepared with the nonphysiological metal in the active site. Since the reduction potentials of iron and manganese differ by approximately 0.7 V in the absence of redox tuning by the protein, these enzymes were used to probe the relevance of the metal oxidation state in dioxygen activation. MnII- and FeII-HPCD had superimposable structures, suggesting that the difference in redox potential of the metals should be retained in the active site (37). Because the kinetic parameters of the “physiological” and “metal-swapped” enzymes for the organic substrate and for O2 were very similar, it has been discussed that oxygen activation and substrate oxidation steps can proceed without the requirement for an integral change in the metal redox state (37, 78). However, electron paramagnetic resonance (EPR) studies revealed the formation of a MnIII radical couple in Mn-HPCD (52), and recently, an FeIII-superoxo species and a [homoprotocatechuate semiquinone-FeIII-(hydro)peroxo] intermediate were trapped in a protein variant of HPCD (83, 84). These findings support the hypothesis that an FeIII-superoxide intermediate is formed in the catalytic cycle but has a very short lifetime. It seems that the metal center in HPCD and other extradiol dioxygenases, after having established the correct orientation of the two substrates, acts as a conduit for facile electron transfer from the catecholate to O2. Electron transfer from the divalent metal center to O2 is thought to elicit an immediate subsequent electron transfer from the bound substrate to the nascent trivalent metal center. The finding that MnII- and CoII-substituted HPCD are fully active or even hyperactive has been rationalized by the hypothesis that even though Mn2+ and, especially, Co2+ are poorer reducing agents than Fe2+, this is compensated by their MIII-superoxo intermediates acting as stronger oxidizing agents (44).
Many biochemical and structural studies on intradiol and type I extradiol dioxygenases have contributed to a rather detailed understanding of these enzymes, which have been the subject of several comprehensive reviews (14–17, 25, 73, 78, 125). Genomic and metagenomic studies provided insight into their phylogenetic distribution, diversity, and abundance in the environment (13, 120, 127). However, in a number of bacterial degradation pathways, intermediates may be converted to gentisate, i.e., a para-diol, rather than to a catecholic compound. Other metabolic pathways involve monohydroxylated aromatic carboxylic acids that directly undergo dioxygenolytic ring cleavage reactions. Enzymes catalyzing these reactions have been referred to as “type III extradiol dioxygenases,” even though most substrates lack the diol character. Another group of particular ring-cleaving dioxygenases open up two C-C-bonds of the flavonol heterocycle, releasing the hydroxy-substituted carbon atom at the C-3 position as carbon monoxide. These ring-cleavage enzymes active toward noncatecholic substrates, which are the main focus of this review, belong to the cupin superfamily of proteins (Table 1).
Table 1.
Biochemically or structurally characterized microbial ring cleavage dioxygenases with a cupin fold
| Dioxygenase | Source | Domain structure, quaternary structure | Metal dependence | Ligand(s) to the metal iona | Reference(s) |
|---|---|---|---|---|---|
| Gentisate 1,2-dioxygenase | Pseudomonas acidovorans ATCC 17438, P. testosteroni ATCC 49249 | Bicupin, homotetramer | Fe2+ | NS | 55, 56 |
| Escherichia coli O157:H7 | Bicupin, homotetramer | Fe2+ | 3His, 3H2O | 1 | |
| Silicibacter pomeroyi DSS-3 | Bicupin, homotetramer | Fe2+ | 3His | 20 | |
| Salicylate 1,2-dioxygenase | Pseudaminobacter salicylatoxidans BN12 | Bicupin, homotetramer | Fe2+ | 3His | 39, 59, 82 |
| 1-Hydroxy-2-naphthoate 1,2-dioxygenase | Nocardioides sp. KP7 | Bicupin, homohexamer | Fe2+ | NS | 62 |
| 3-Hydroxyanthranilate 3,4-dioxygenase | Saccharomyces cerevisiae | Monocupin, homodimer | Fe2+ | 2His, 1Glu (bidentate), 2H2O | 74, 76 |
| Ralstonia metallidurans | Monocupin, homodimer | Fe2+ | 2His, 1Glu (bidentate), 2H2O | 130 | |
| 4-Amino-3-hydroxybenzoate 2,3-dioxygenase | Bordetella sp. 10d | Monocupin, homodimer | Fe2+ | NS | 88, 119 |
| Flavonol 2,4-dioxygenase (quercetinase) | Aspergillus japonicus | Bicupin, homodimer | Cu2+ | 3His, 1H2O (major form); 3His, 1Glu, 1H2O; (minor form) | 49, 70, 113, 114 |
| Bacillus subtilis | Bicupin, homodimer | Mn2+ > Co2+ > Fe2+, Ni2+, Cu2+ | 3His, 1Glu, 1H2O | 51, 107 | |
| Streptomyces sp. FLA | Monocupin, homodimer | Ni2+ > Co2+ > Mn2+, Fe2+ | NS | 85, 86 |
NS, not specified.
THE CUPIN SUPERFAMILY
The cupin superfamily is characterized by a conserved six-stranded β-barrel fold (“cupa” is the Latin term for small barrel). Most family members either comprise a single cupin domain (“monocupins”) or have a duplicated domain structure (“bicupins”) (Fig. 2). Cupins are found in all kingdoms of life. The superfamily is functionally highly diverse and includes nonenzymatic proteins, such as plant seed storage proteins, transcription regulators, and stress-related proteins, and a wide variety of enzymes. Enzymatic cupins involve isomerases and epimerases, decarboxylases, and many oxygenases, with the 2-oxoglutarate-dependent dioxygenases as the largest subset (for reviews, see references 21, 33–35, and 68).
Fig 2.
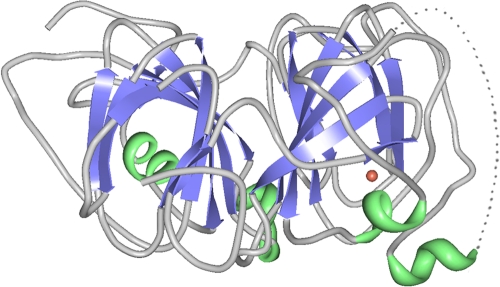
Ribbon diagram of the monomer of quercetinase of A. japonicus, illustrating the two barrels of the bicupin fold. The dotted line indicates a flexible region of the structure (residues 154 to 169) (49). The copper atom located in the N-terminal domain is shown as a red sphere. The native protein is a homodimer. The figure was generated from PDB code 1JUH using the RCSB Viewer RCSB PDB Protein Workshop 3.9 (87).
The cupin domain comprises two conserved amino acid motifs with the consensus sequences G(X)5HXH(X)3-4E(X)6G (motif 1) and G(X)5-7PXG(X)2H(X)3N (motif 2). Each motif corresponds to two β strands separated by another two strands with an intervening loop. The two histidine residues and the glutamate residue in motif 1, together with the conserved histidine residue in motif 2, can act as ligands for the binding of a divalent metal ion. This ligand environment has been observed in many cupin structures, such as germin (129), oxalate decarboxylase (2), flavonol 2,4-dioxygenases (49, 51), and acireductone dioxygenase (19, 27, 29, 64, 102). The majority of the enzymatic cupins use Fe2+ or Mn2+ as an active-site metal ion, but other members contain Cu2+, Zn2+, Co2+, or Ni2+. The type of metal ion and its ligand environment within the cupin fold likely are major determinants for the reaction chemistry at the active site. The metal centers usually are hexa-coordinated, adopting an octahedral (bipyramidal) or distorted octahedral geometry, with two water molecules completing the coordination sphere. Some members, however, contain a penta-coordinated metal site (32, 49, 51, 113). Interestingly, some cupin subfamilies do not contain all four of the conserved metal-binding residues. A key feature of the thiol dioxygenase family, for example, is that the glutamate residue in motif 1 is replaced by a cysteine in mammalian cysteine dioxygenases and by a glycine in bacterial thiol dioxygenases (111, 116). Other enzymes with a “reduction” of the 3His-1Glu motif are the gentisate 1,2-dioxygenases (GDOs) and 3-hydroxyanthranilate 3,4-dioxygenases (HADs) (see below). Most remarkably, the metal binding site in polyketide cyclase RemF, a monocupin protein, contains an octahedral zinc site, with four histidine residues as protein ligands to the Zn2+ ion (110).
Many ring cleavage dioxygenases within the cupin superfamily use a mononuclear FeII center for catalysis. Interestingly, however, flavonol-cleaving 2,4-dioxygenases from different sources exhibit different metal selectivities or even are cambialistic enzymes, capable of using several divalent metal ions as a cofactor. Such “metal promiscuity” raises the question of the role of the metal center in catalysis and the mechanistic relatedness of cupin dioxygenases and classical catechol dioxygenases.
GENTISATE 1,2-DIOXYGENASE AND RELATED ENZYMES
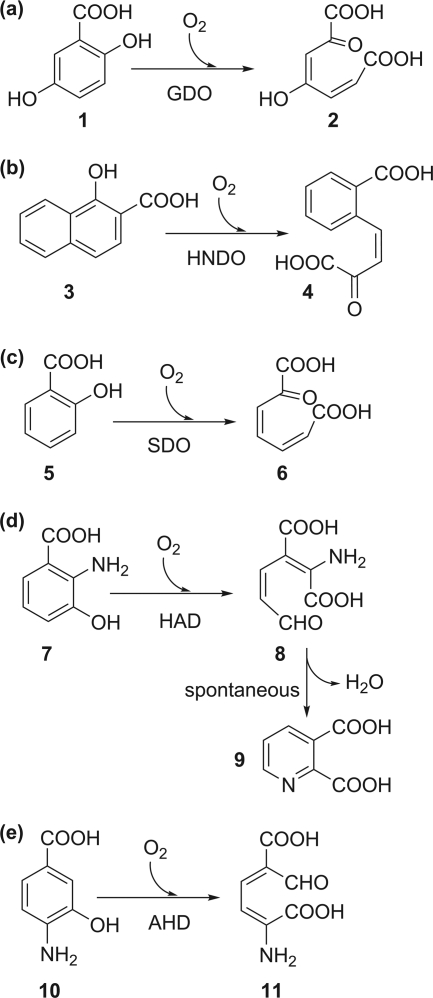
Gentisate (2,5-dihydroxybenzoate) is formed from 3-chloro- and 3-hydroxybenzoate or from salicylate in the bacterial degradation of many simple as well as complex aromatic compounds (for a compilation of pertinent pathways, see the University of Minnesota Biocatalysis/Biodegradation [UM-BB] Database, http://umbbd.msi.umn.edu/ [50]). Gentisate 1,2-dioxygenase (GDO), as well as 1-hydroxy-2-naphthoate and salicylate 1,2-dioxygenases (HNDO and SDO, respectively), cleave their substrates between the adjacent carbon atoms carrying a carboxylate and a hydroxyl substituent (Fig. 3a to c). The GDOs isolated from two Pseudomonas species are both Fe2+-dependent enzymes with a high specificity for gentisate, with 5-aminosalicylate as a poor alternative substrate. Salicylate is not converted but acts as a competitive inhibitor (55, 56). All GDOs characterized to date belong to the bicupin family (1, 20). Each bicupin subunit of the homotetrameric enzyme from Escherichia coli contains a mononuclear iron center coordinated to three His ligands, leaving three solvent-occupied sites on the iron available for interactions with substrate (1). The conserved glutamate of the cupin consensus motif 1 is replaced by alanine (or another hydrophobic or a polar residue) in virtually all confirmed and predicted GDOs. This substitution has been discussed to possibly represent an adaptation for binding of both the aromatic carboxylate and O2 to the metal. Earlier EPR studies on GDO from Pseudomonas testosteroni provided evidence that O2 and gentisate indeed are coordinated simultaneously to the active-site iron (56). Based on spectroscopic data and supported by structural data, the catalytic cycle has been proposed to start with bidentate coordination of the substrate via its hydroxyl and carboxylate groups to FeII, which activates the FeII center for subsequent binding of O2. Polarization of electron density from the aromatic ring toward the iron-bound O2 in the ternary complex primes O2 for attack at carbon 1 of the bound gentisate to form an alkylperoxo intermediate. Cleavage of the O-O bond and insertion of one oxygen atom into the ring, promoted by ketonization of the hydroxyl substituent at C-5, would generate a cyclic lactone that may be hydrolyzed by the hydroxide that after O2 cleavage remained at the iron (1, 20, 56). The mechanism proposed for GDO is very similar to that of the type I extradiol dioxygenases (cf. Fig. 1b).
Fig 3.
Reactions of cupin-type dioxygenases which catalyze the cleavage of an aromatic C-C bond. (a) Gentisate 1,2-dioxygenase (GDO); (b) 1-hydroxy-2-naphthoate 1,2-dioxygenase (HNDO), (c) salicylate 1,2-dioxygenase (SDO); (d) 3-hydroxyanthranilate 3,4-dioxygenase (HAD); (e) 4-amino-3-hydroxybenzoate 2,3-dioxygenase (AHD). 1, Gentisic acid; 2, maleylpyruvic acid; 3, 1-hydroxy-2-naphthoic acid; 4, (3Z)-4-(2-carboxyphenyl)-2-oxobut-3-enoic acid; 5, salicylic acid; 6, 2-oxohepta-3,5-dienedioic acid; 7, 3-hydroxyanthranilic acid; 8, 2-amino-3-carboxymuconic semialdehyde; 9, quinolinic acid; 10, 4-amino-3-hydroxybenzoic acid; 11, 2-amino-5-carboxymuconic semialdehyde.
1-Hydroxy-2-naphthoate is an intermediate in the microbial degradation of phenanthrene and chrysene (53, 62, 90). Salicylate can be formed in the degradation of naphthalene and derivatives, polycyclic aromatic hydrocarbons, dibenzofuran, and the organophosphorous insecticide isocarbophos, as described in the UM-BB database (http://umbbd.msi.umn.edu/) (50). 1-Hydroxy-2-naphthoate 1,2-dioxygenase (HNDO) from the phenanthrene-degrading strain Nocardioides sp. strain KP7 and salicylate 1,2-dioxygenase (SDO) from the naphthalenesulfonate-degrading bacterium Pseudoaminobacter salicylatoxidans BN12 exhibit significant sequence similarity to GDOs (39, 59, 82). However, gentisate and salicylate were neither substrates nor competitive inhibitors of HNDO, suggesting stringent adaptation of this enzyme to its physiological substrate (62). In contrast, SDO from P. salicylatoxidans BN12 has a remarkably broad specificity. The highest activity was observed with gentisate, and the gentisate analogue 5-aminosalicylate, as well as 1-hydroxy-2-naphthoate, was also converted with higher activity than salicylate. SDO therefore seems to be basically a GDO that has relaxed its specificity toward the conversion of monohydroxylated substrates (39, 59, 82). Like E. coli GDO, SDO is a homotetramer of bicupin subunits, with one 3His-coordinated FeII site per subunit.
A 5-nitrosalicylate dioxygenase, which is distantly related to salicylate and gentisate 1,2-dioxygenases, was recently identified in the 5-nitroanthranilic acid-degrading bacterium Bradyrhizobium sp. strain JS329. The enzyme also catalyzes the oxidation of 5-chlorosalicylate, whereas conversion of 5-hydroxyanthranilate, 4-nitrocatechol, or 4-chlorocatechol was not observed (104, 105).
AMINOHYDROXYBENZOATE DIOXYGENASES
3-Hydroxyanthranilate 3,4-dioxygenase (HAD) is involved in tryptophan catabolism and seems to be conserved from bacteria to humans (18, 22, 32, 74, 75, 79, 89). 2-Amino-3-carboxymuconic 6-semialdehyde, the product of the HAD reaction, may either undergo further degradation via 2-aminomuconate 6-semialdehyde or spontaneously cyclize to quinolinate (23) (Fig. 3d). In the mammalian central nervous system, quinolinate binds to the N-methyl-d-aspartate receptor and thus has neurotoxic capacity (108, 117), but on the other hand, quinolinate is also a source for nicotinate mononucleotide, required for the biosynthesis of the NAD cofactors. HAD therefore may have a biosynthetic role in both eukaryotes and bacteria (74, 75). However, utilization of 2-nitrobenzoate by Pseudomonas fluorescens strain KU-7 (58, 89) and Arthrobacter protophormiae RKJ100 (97) proceeds via 3-hydroxyanthranilate and 2-amino-3-carboxymuconic 6-semialdehyde, with the latter subsequently undergoing decarboxylation and oxidation to 2-amino-cis,cis-muconate (89). Quite unusually, Geobacillus thermodenitrificans NG80-2 degrades anthranilate via 3-hydroxyanthranilate and subsequent 3,4-dioxygenolytic ring cleavage (79). In these cases, ring cleavage serves for the utilization of an aromatic compound as carbon and energy source rather than for quinolinate formation.
All HAD proteins characterized until now depend on Fe2+ for activity (18, 32, 89, 130). HADs from Ralstonia metallidurans and from yeast are homodimeric proteins, each monocupin subunit harboring a catalytic FeII site and a rubredoxin-like (FeS4) center of unknown function located at the C-terminal periphery of the molecule (76, 130). The arrangement of the two cupin barrels in the homodimer resembles bicupin structures (130). In contrast to the bacterial HAD, the bovine and human proteins are monomeric bicupins lacking the rubredoxin-like center, with only the N-terminal cupin domain containing an active site (32). Despite these structural differences, the active-site residues considered relevant for the catalytic reaction appear to be fully conserved in the prokaryotic and eukaryotic proteins, suggesting the same catalytic mechanism. In all HAD proteins, the first conserved histidine of the consensus sequence of motif 1 is substituted, resulting in a 2His-1Glu coordination sphere. However, in contrast to the 2His-1Glu facial triad of the type I extradiol dioxygenases, the glutamate binds in a bidentate manner (130) (Fig. 4). The crystal structures of the yeast and human enzymes and of HAD from R. metallidurans suggest a distorted octahedral coordination geometry with Fe2+ bound to two His ligands, bidentate Glu, and two ligands modeled as water molecules (76, 130). 3-Hydroxyanthranilate, possibly in its phenolate form, binds as a bidentate ligand, presumably displacing one water molecule and the carbonyl oxygen of the Glu ligand. A conserved glutamate, which is not part of the first coordination sphere, was proposed to be involved in deprotonation of the substrate's hydroxy group. Structures with bound inhibitor and NO or O2 suggest a direct interaction of the dioxygen molecule with the metal ion. The proposed reaction mechanism is analogous to that of the type I extradiol dioxygenases (24, 76, 130) (Fig. 4).
Fig 4.
Proposed mechanism for the reaction catalyzed by 3-hydroxyanthranilate 3,4-dioxygenase (HAD) (76). B:, general base.
Another Fe2+-containing ring-cleaving enzyme active toward a carboxy-substituted o-aminophenol is 4-amino-3-hydroxybenzoate 2,3-dioxygenase (AHD) from Bordetella sp. strain 10d (Fig. 3e). Its amino acid sequence shows 24 to 29% identity to those of the HAD proteins; however, Pseudomonas HAD and Bordetella AHD are highly specific for their respective physiological substrates (88, 89, 119). Active-site residues which in HADs are involved in metal binding and deprotonation of the hydroxy group of 3-hydroxyanthranilate are conserved in AHD, tentatively suggesting a similar catalytic mechanism.
FLAVONOL 2,4-DIOXYGENASES (QUERCETINASES)
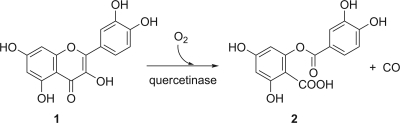
Flavonols are polyphenolic compounds synthesized by numerous higher plants, in most cases as O-glycosides (63). Major plant flavonols are quercetin (3,5,7,3′,4′-pentahydroxyflavone), kaempferol (3,5,7,4′-tetrahydroxyflavone), myricetin (3,5,7,3′,4′,5′-hexahydroxyflavone), and isorhamnetin (3,5,7,4′-tetrahydroxy-3′-methoxyflavone). Quercetin is found in considerable amounts in many vegetables and fruits, such as onions, broccoli, and apples, and thus is the major flavonol in the human diet (38, 100, 103). Quercetin exhibits antioxidative, antiinflammatory, and vasodilating effects and has been proposed to be a potential anticancer agent (38, 67, 100). It also exhibits antibacterial activity, which is at least partially due to inhibition of DNA gyrase B (26, 101). Due to release from rotting plant material and as constituents of leaf, seed, and root exudates (57, 66, 115), quercetin conjugates are widespread in soil, and various filamentous fungi and bacteria have been described to detoxify or to degrade quercetin (reviewed in reference 121). The initial step of the aerobic metabolism is a 2,4-dioxygenolytic ring cleavage to form 2-protocatechuoylphloroglucinol carboxylic acid and carbon monoxide, catalyzed by quercetinase (Fig. 5).
Fig 5.
Reaction catalyzed by flavonol 2,4-dioxygenase (quercetinase). 1, Quercetin (3,5,7,3′,4′-pentahydroxyflavone); 2, protocatechuoylphloroglucinol carboxylic acid.
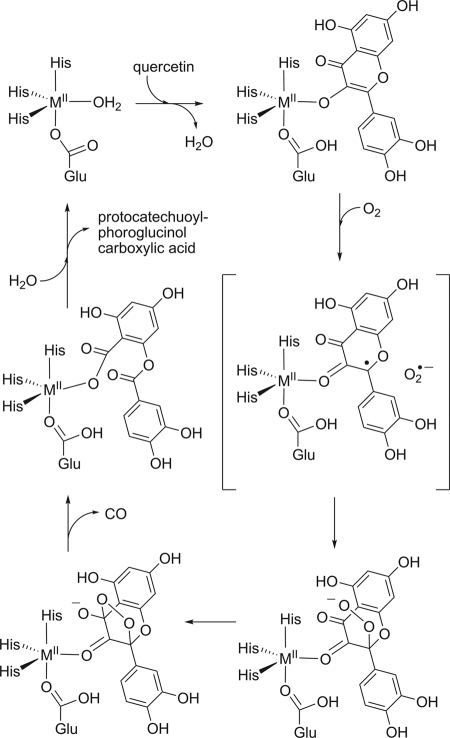
Fungal quercetinases are extracellular glycoproteins with a bicupin fold. The reactivity of quercetinases from Aspergillus japonicus, other Aspergillus species, and Penicillium olsonii depends on a mononuclear CuII center (49, 61, 70, 95, 122). In the crystal structure of quercetinase of A. japonicus, the copper ion is mainly coordinated by three His residues and a water molecule in a distorted tetrahedral geometry; in a minor form, the metal is penta-coordinated by three His, a glutamate, and an aquo ligand in a trigonal bipyramidal geometry. The glutamate ligand has been proposed to act as a general base to generate the flavonolate anion, which binds in a monodentate fashion through its O-3 atom to the copper site (49, 70, 113, 114). A flavonoxy radical-CuI valence tautomer, arising from the flow of one electron from the flavonolate to the copper, was hypothesized to be the active species that can react with dioxygen. This proposed mechanism of substrate activation shares similarity with that of the intradiol catechol dioxygenases (113).
Whereas fungal quercetinases appear to exclusively utilize a Cu2+ ion for catalysis, other quercetinases are cambialistic, i.e., function with several or even a variety of metal ions. Quercetinase of Bacillus subtilis, at least when produced in Escherichia coli, is able to exchange its active-site metal ion while retaining catalytic activity. The recombinant enzyme is most active with Mn2+, whereas nickel is a poor cofactor (9, 11, 107). The crystal structure of the FeII isoform shows that three His residues and one Glu residue provide the protein-based ligands for the Fe2+ ion (51). For the Bacillus quercetinase, Schaab et al. (107) suggested a mechanism similar to that of the extradiol dioxygenases, involving a [quercetin radical-MII-superoxo] intermediate. However, EPR studies on the Fe form of Bacillus quercetinase indicated that enzyme-bound quercetin shields the FeII cofactor from interactions with the O2 mimic nitric oxide (51), tentatively suggesting that the reaction catalyzed by Bacillus (Fe-)quercetinase may proceed without direct interaction of dioxygen and metal ion.
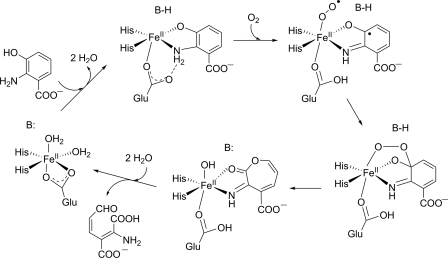
A most interesting case of a cambialistic quercetinase is the enzyme from Streptomyces sp. strain FLA which, in contrast to the Bacillus enzyme, has a monocupin fold and is most active with Ni2+ as a cofactor. A nickel preference is very unusual for oxygenases. Co2+, Fe2+, and Mn2+ can replace Ni2+ and also support catalytic activity to some extent (85, 86). Given that quercetinases are evolutionarily related and catalyze the same reaction, their variability in metal selectivity is surprising. It is also interesting to note that the metal ions that afford catalytically active enzyme have standard reduction potentials that span a range of more than 1.5 V (Table 2), suggesting that the redox properties of the bound metal are relatively unimportant for the reaction. However, a mechanism which involves initial electron transfer from the divalent metal to O2, as proposed for the extradiol dioxygenases, requires that a MIII-superoxo state is thermodynamically accessible (44). On the other hand, a mechanism as discussed for copper quercetinase, with the bound substrate acting as the initial electron donor to the divalent metal center, requires accessibility of the MI state. EPR spectra of the Co form of Streptomyces quercetinase were indicative of a high-spin CoII species in a trigonal-bipyramidal or tetrahedral coordination geometry (85). Studies with model CoII complexes suggested that a 3N/1O ligand environment stabilizes the CoII state (106). Moreover, NiII centers in ligand environments dominated by O and N donors (in contrast to thiolate-ligated NiII sites) were proposed to be redox inert (80). Thus, for the CoII and, especially, the NiII center of quercetinases, the accessibility of redox states other than MII is a matter of debate. These considerations have led to the suggestion that the MII-flavonolate complex is oxidized by an outer-sphere electron transfer to dioxygen, generating the superoxide anion radical along with the MII-[flavonoxy radical] without valence change of the metal (Fig. 6). However, experimental evidence for intermediates of the reaction catalyzed by bacterial quercetinases is relatively scarce compared to the wealth of information on the classical intradiol and extradiol dioxygenases.
Table 2.
Standard reduction potentials of selected transition metal cations
Fig 6.
Hypothetical reaction mechanism of bacterial quercetinases.
The hypothesis of direct electron transfer from the metal-bound quercetin anion to O2 is also inspired by the intrinsic chemical reactivity of flavonolates toward dioxygen. Organic model reactions showed that basic conditions and the presence of dioxygen are sufficient for 2,4-dioxygenolytic cleavage of flavonols, demonstrating that a metal catalyst is not necessarily required for the reaction to occur. Aprotic conditions increase the rate of base-catalyzed dioxygenolysis (6, 8, 65, 91, 92). Interestingly, recent studies on model systems suggested that the presence of a carboxylate coligand on CuII-flavonolate complexes induces monodentate binding of flavonolate to the metal, resulting in high electron density on the C-2 atom of the flavonolate, which possibly facilitates the direct single electron transfer from the activated flavonolate to dioxygen (98). Thus, the major role of the divalent metal ion in the active site of quercetinases could be to correctly position the substrate and to stabilize transition states and intermediates rather than to mediate electron transfer.
In the context of a possible nonredox role of the metal centers of quercetinases, it is interesting to note that oxygenases active toward 3-hydroxy-4(1H)-quinolones, which, like quercetinase, catalyze a 2,4-dioxygenolytic ring cleavage reaction, neither require nor contain a metal ion for catalysis (41, 45, 46, 112). For these enzymes, which have an α/β-hydrolase fold (45, 112), and also for other cofactor-independent oxygenases from different fold families, base-catalyzed abstraction of a proton from the organic substrate seems to be the common initial step in catalysis. Subsequent catalytic steps are thought to be triggered by the intrinsic reactivity of the bound carbanion and, presumably, also by the ability of the organic anions to form resonance-stabilized radicals upon single-electron oxidation (42, 43). The biochemistry of cofactor-independent oxygenases supports the notion that the oxygenation of (carb)anionic substrates does not necessarily require a redox-active metal center.
CONCLUSIONS
Gentisate, salicylate, 1-hydroxy-2-naphthoate, 3-hydroxyanthranilate, and 4-amino-3-hydroxybenzoate occur as intermediates in the bacterial degradation of substituted benzoates, naphthalene and derivatives, and some polycyclic aromatic hydrocarbons. Ring-cleaving dioxygenases active toward these noncatecholic compounds belong to the cupin superfamily and utilize a mononuclear FeII center for catalysis. Their catalytic strategy has been proposed to involve a one-electron transfer to dioxygen, possibly via transient formation of an FeIII-O2 · − intermediate, as observed in type I extradiol dioxygenases (83, 84), which are members of the VOC superfamily (5). However, it should be emphasized that compared to the thoroughly studied extradiol dioxygenase reaction mechanism, much less experimental evidence is available on the catalytic mechanism of the cupin-type ring-cleaving dioxygenases.
Both the cupin β-barrel fold and the paired βαβββ modules of the VOC superfamily proteins provide a scaffold for a metal coordination environment with two or three readily accessible coordination sites, and both folds afford a remarkable breadth of catalytic diversity (5, 33–35, 68). With respect to the dioxygenases that catalyze the cleavage of aromatic C-C bonds, it seems likely that convergent evolution of the Fe2+-dependent enzymes with a cupin scaffold and the extradiol dioxygenases of the VOC family has led to a similar catalytic mechanism.
The 3His- or 2- or 3His-1Glu Fe2+ binding motif of the cupin dioxygenases can be considered a variation of the 2His-1carboxylate facial triad motif, which is common not only in the type I extradiol dioxygenases but also in many other nonheme FeII-containing oxygenases of different fold families (25, 73, 118). However, the cupin-type motif apparently is less selective with regard to the divalent metal ion utilized than the 2His-1carboxylate center. Metal “promiscuity” in particular seems to be a distinctive feature of the few bacterial quercetinases identified until now. The enzymes from Bacillus subtilis and Streptomyces sp. can utilize several divalent metal ions for catalysis, but it is interesting to note that the Streptomyces enzyme is most active with Ni2+, whereas the Bacillus enzyme prefers Mn2+. In contrast to the cambialistic bacterial enzymes, fungal quercetinases seem to depend on Cu2+. With respect to the biochemistry of quercetinases, a number of questions remain to be answered. The molecular basis for the observed differences in metal specificity or promiscuity and the mechanistic implications of distinct metal preferences are at present unclear. It seems that monodentate (instead of bidentate) binding of the organic substrate as a flavonolate anion is a key step in catalysis, but the mode of dioxygen binding and activation is not well understood. In particular, the question of whether or not the metal ion of prokaryotic quercetinases mediates electron transfer is still under discussion. It is tempting to speculate that in the cambialistic enzymes, which utilize a range of divalent metal ions, the metal has a nonredox role in catalysis.
ACKNOWLEDGMENTS
The financial support of this work on oxygenases by the Deutsche Forschungsgemeinschaft (grants FE 383/15-1 and FE 383/18-1) is gratefully acknowledged.
I thank Dimitrios Nianios for the preparation of Fig. 2.
Footnotes
Published ahead of print 27 January 2012
REFERENCES
- 1. Adams MA, Singh VK, Keller BO, Jia Z. 2006. Structural and biochemical characterization of gentisate 1,2-dioxygenase from Escherichia coli O157:H7. Mol. Microbiol. 61:1469–1484 [DOI] [PubMed] [Google Scholar]
- 2. Anand R, Dorrestein PC, Kinsland C, Begley TP, Ealick SE. 2002. Structure of oxalate decarboxylase from Bacillus subtilis at 1.75 Å resolution. Biochemistry 41:7659–7669 [DOI] [PubMed] [Google Scholar]
- 3. Antelman MS, Harris FJ. 1982. The encyclopedia of chemical electrode potentials. Plenum, London, United Kingdom [Google Scholar]
- 4. Arciero DM, Orville AM, Lipscomb JD. 1985. [17O]water and nitric oxide binding by protocatechuate 4,5-dioxygenase and catechol 2,3-dioxygenase. Evidence for binding of exogenous ligands to the active site Fe2+ of extradiol dioxygenases. J. Biol. Chem. 260:14035–14044 [PubMed] [Google Scholar]
- 5. Armstrong RN. 2000. Mechanistic diversity in a metalloenzyme superfamily. Biochemistry 39:13625–13632 [DOI] [PubMed] [Google Scholar]
- 6. Balogh-Hergovich E, Speier G. 2001. Kinetics and mechanism of the base-catalyzed oxygenation of flavonol in DMSO-H2O solution. J. Org. Chem. 66:7974–7978 [DOI] [PubMed] [Google Scholar]
- 7. Bard AJ, Parsons R, Jordan J. 1985. Standard potentials in aqueous solution. Marcel Dekker, New York, NY [Google Scholar]
- 8. Barhacs L, Kaizer J, Speier G. 2000. Kinetics and mechanism of the oxygenation of potassium flavonolate. Evidence for an electron transfer mechanism. J. Org. Chem. 65:3449–3452 [DOI] [PubMed] [Google Scholar]
- 9. Barney BM, Schaab MR, LoBrutto R, Francisco WA. 2004. Evidence for a new metal in a known active site: purification and characterization of an iron-containing quercetin 2,3-dioxygenase from Bacillus subtilis. Protein Expr. Purif. 35:131–141 [DOI] [PubMed] [Google Scholar]
- 10. Bartels I, Knackmuss H-J, Reineke W. 1984. Suicide inactivation of catechol 2,3-dioxygenase from Pseudomonas putida mt-2 by 3-halocatechols. Appl. Environ. Microbiol. 47:500–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bowater L, Fairhurst SA, Just VJ, Bornemann S. 2004. Bacillus subtilis YxaG is a novel Fe-containing quercetin 2,3-dioxygenase. FEBS Lett. 557:45–48 [DOI] [PubMed] [Google Scholar]
- 12. Bratsch SG. 1989. Standard electrode potentials and temperature coefficients in water at 298.15 K. J. Phys. Chem. Ref. Data 18:1–21 [Google Scholar]
- 13. Brennerova MV, Josefiova J, Brenner V, Pieper DH, Junca H. 2009. Metagenomics reveals diversity and abundance of meta-cleavage pathways in microbial communities from soil highly contaminated with jet fuel under air-sparging bioremediation. Environ. Microbiol. 11:2216–2227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Broderick JB. 1999. Catechol dioxygenases. Essays Biochem. 34:173–189 [DOI] [PubMed] [Google Scholar]
- 15. Bugg TDH. 2003. Dioxygenase enzymes: catalytic mechanisms and chemical models. Tetrahedron 59:7075–7101 [Google Scholar]
- 16. Bugg TDH, Lin G. 2001. Solving the riddle of the intradiol and extradiol catechol dioxygenases: how do enzymes control hydroperoxide rearrangements? Chem. Commun. (Camb.) 35:941–952 [Google Scholar]
- 17. Bugg TDH, Ramaswamy S. 2008. Non-heme iron-dependent dioxygenases: unravelling catalytic mechanisms for complex enzymatic oxidations. Curr. Opin. Chem. Biol. 12:134–140 [DOI] [PubMed] [Google Scholar]
- 18. Calderone V, Trabucco M, Menin V, Negro A, Zanotti G. 2002. Cloning of human 3-hydroxyanthranilic acid dioxygenase in Escherichia coli: characterization of the purified enzyme and its in vitro inhibition by Zn2+. Biochim. Biophys. Acta 1596:283–292 [DOI] [PubMed] [Google Scholar]
- 19. Chai SC, et al. 2008. Characterization of metal binding in the active sites of acireductone dioxygenase isoforms from Klebsiella ATCC 8724. Biochemistry 47:2428–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen J, et al. 2008. Crystal structure and mutagenic analysis of GDOsp, a gentisate 1,2-dioxygenase from Silicibacter pomeroyi. Protein Sci. 17:1362–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Clifton IJ, et al. 2006. Structural studies on 2-oxoglutarate oxygenases and related double-stranded β-helix fold proteins. J. Inorg. Biochem. 100:644–669 [DOI] [PubMed] [Google Scholar]
- 22. Colabroy KL, Begley TP. 2005. Tryptophan catabolism: identification and characterization of a new degradative pathway. J. Bacteriol. 187:7866–7869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Colabroy KL, Begley TP. 2005. The pyridine ring of NAD is formed by a nonenzymatic pericyclic reaction. J. Am. Chem. Soc. 127:840–841 [DOI] [PubMed] [Google Scholar]
- 24. Colabroy KL, et al. 2005. The mechanism of inactivation of 3-hydroxyanthranilate 3,4-dioxygenase by 4-chloro-3-hydroxyanthranilate. Biochemistry 44:7623–7631 [DOI] [PubMed] [Google Scholar]
- 25. Costas M, Mehn MP, Jensen MP, Que L., Jr 2004. Dioxygen activation at mononuclear nonheme iron active sites: enzymes, models, and intermediates. Chem. Rev. 104:939–986 [DOI] [PubMed] [Google Scholar]
- 26. Cushnie TP, Lamb AJ. 2005. Antimicrobial activity of flavonoids. Int. J. Antimicrob. Agents 26:343–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dai Y, Pochapsky TC, Abeles RH. 2001. Mechanistic studies of two dioxygenases in the methionine salvage pathway of Klebsiella pneumoniae. Biochemistry 40:6379–6387 [DOI] [PubMed] [Google Scholar]
- 28. Dai SD, et al. 2002. Identification and analysis of a bottleneck in PCB biodegradation. Nat. Struct. Biol. 9:934–939 [DOI] [PubMed] [Google Scholar]
- 29. Dai Y, Wensink PC, Abeles RH. 1999. One protein, two enzymes. J. Biol. Chem. 274:1193–1195 [DOI] [PubMed] [Google Scholar]
- 30. Davis MI, et al. 2002. Spectroscopic and electronic structure studies of protocatechuate 3,4-dioxygenase: nature of tyrosinate-Fe(III) bonds and their contribution to reactivity. J. Am. Chem. Soc. 124:602–614 [DOI] [PubMed] [Google Scholar]
- 31. Davis MI, et al. 1999. Spectroscopic investigation of reduced protocatechuate 3,4-dioxygenase: charge-induced alterations in the active site iron coordination environment. Inorg. Chem. 38:3676–3683 [DOI] [PubMed] [Google Scholar]
- 32. Dilovic I, Gliubich F, Malpeli G, Zanotti G, Matkovic-Calogovic D. 2009. Crystal structure of bovine 3-hydroxyanthranilate 3,4-dioxygenase. Biopolymers 91:1189–1195 [DOI] [PubMed] [Google Scholar]
- 33. Dunwell JM, Culham A, Carter CE, Sosa-Aguirre CR, Goodenough PW. 2001. Evolution of functional diversity in the cupin superfamily. Trends Biochem. Sci. 26:740–746 [DOI] [PubMed] [Google Scholar]
- 34. Dunwell JM, Khuri S, Gane PJ. 2000. Microbial relatives of the seed storage proteins of higher plants: conservation of structure and diversification of function during evolution of the cupin superfamily. Microbiol. Mol. Biol. Rev. 64:153–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dunwell JM, Purvis A, Khuri S. 2004. Cupins: the most functionally diverse protein superfamily? Phytochemistry 65:7–17 [DOI] [PubMed] [Google Scholar]
- 36. Elgren TE, et al. 1997. Crystal structure and resonance Raman studies of protocatechuate 3,4-dioxygenase complexed with 3,4-dihydroxyphenylacetate. Biochemistry 36:11504–11513 [DOI] [PubMed] [Google Scholar]
- 37. Emerson JP, Kovaleva EG, Farquhar ER, Lipscomb JD, Que L., Jr 2008. Swapping metals in Fe- and Mn-dependent dioxygenases: evidence for oxygen activation without a change in metal redox state. Proc. Natl. Acad. Sci. U. S. A. 105:7347–7352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Erlund I. 2004. Review of the flavonoids quercetin, hesperetin, and naringenin. Dietary sources, bioactivities, bioavailability, and epidemiology. Nutr. Res. 24:851–874 [Google Scholar]
- 39. Ferraroni M, et al. 2012. Crystal structures of salicylate 1,2-dioxygenase-substrates adducts: a step towards the comprehension of the structural basis for substrate selection in class III ring cleaving dioxygenases. J. Struct. Biol. 177:431–438 [DOI] [PubMed] [Google Scholar]
- 40. Fetzner S. 1998. Bacterial degradation of pyridine, indole, quinoline, and their derivatives under different redox conditions. Appl. Microbiol. Biotechnol. 49:237–250 [Google Scholar]
- 41. Fetzner S. 2002. Oxygenases without requirement for cofactors or metal ions. Appl. Microbiol. Biotechnol. 60:243–257 [DOI] [PubMed] [Google Scholar]
- 42. Fetzner S. 2007. Cofactor-independent oxygenases go it alone. Nat. Chem. Biol. 3:374–375 [DOI] [PubMed] [Google Scholar]
- 43. Fetzner S, Steiner RA. 2010. Cofactor-independent oxidases and oxygenases. Appl. Microbiol. Biotechnol. 86:791–804 [DOI] [PubMed] [Google Scholar]
- 44. Fielding AJ, Kovaleva EG, Farquhar ER, Lipscomb JD, Que L., Jr 2011. A hyperactive cobalt-substituted extradiol-cleaving catechol dioxygenase. J. Biol. Inorg. Chem. 16:341–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fischer F, Künne S, Fetzner S. 1999. Bacterial 2,4-dioxygenases: new members of the α/β hydrolase-fold superfamily of enzymes functionally related to serine hydrolases. J. Bacteriol. 181:5725–5733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Frerichs-Deeken U, Ranguelova K, Kappl R, Hüttermann J, Fetzner S. 2004. Dioxygenases without requirement for cofactors and their chemical model reaction: compulsory order ternary complex mechanism of 1H-3-hydroxy-4-oxoquinaldine 2,4-dioxygenase involving general base catalysis by histidine 251 and single-electron oxidation of the substrate dianion. Biochemistry 43:14485–14499 [DOI] [PubMed] [Google Scholar]
- 47. Fuchs G. 2008. Anaerobic metabolism of aromatic compounds. Ann. N. Y. Acad. Sci. 1125:82–99 [DOI] [PubMed] [Google Scholar]
- 48. Fuchs G, Boll M, Heider J. 2011. Microbial degradation of aromatic compounds—from one strategy to four. Nat. Rev. Microbiol. 9:803–816 [DOI] [PubMed] [Google Scholar]
- 49. Fusetti F, et al. 2002. Crystal structure of the copper-containing quercetin 2,3-dioxygenase from Aspergillus japonicus. Structure 10:259–268 [DOI] [PubMed] [Google Scholar]
- 50. Gao J, Ellis LBM, Wackett LP. 2010. The University of Minnesota Biocatalysis/Biodegradation database: improving public access. Nucleic Acids Res. 38:D488–D491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gopal B, Madan LL, Betz SF, Kossiakoff AA. 2005. The crystal structure of a quercetin 2,3-dioxygenase from Bacillus subtilis suggests modulation of enzyme activity by a change in the metal ion at the active site (s). Biochemistry 44:193–201 [DOI] [PubMed] [Google Scholar]
- 52. Gunderson WA, et al. 2008. Electron paramagnetic resonance detection of intermediates in the enzymatic cycle of an extradiol dioxygenase. J. Am. Chem. Soc. 130:14465–14467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hadibarata T, Tachibana S, Itoh K. 2009. Biodegradation of chrysene, an aromatic hydrocarbon by Polyporus sp. S133 in liquid medium. J. Hazard. Mater. 164:911–917 [DOI] [PubMed] [Google Scholar]
- 54. Haritash AK, Kaushik CP. 2009. Biodegradation aspects of polycyclic aromatic hydrocarbons (PAHs): a review. J. Hazard. Mater. 169:1–15 [DOI] [PubMed] [Google Scholar]
- 55. Harpel MR, Lipscomb JD. 1990. Gentisate 1,2-dioxygenase from Pseudomonas. Purification, characterization, and comparison of the enzymes from Pseudomonas testosteroni and Pseudomonas acidovorans. J. Biol. Chem. 265:6301–6311 [PubMed] [Google Scholar]
- 56. Harpel MR, Lipscomb JD. 1990. Gentisate 1,2-dioxygenase from Pseudomonas. Substrate coordination to active site Fe2+ and mechanism of turnover. J. Biol. Chem. 265:22187–22196 [PubMed] [Google Scholar]
- 57. Hartwig UA, Joseph CM, Phillips DA. 1991. Flavonoids released naturally from alfalfa seeds enhance growth rate of Rhizobium meliloti. Plant Physiol. 95:797–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hasegawa Y, et al. 2000. A novel degradative pathway of 2-nitrobenzoate via 3-hydroxyanthranilate in Pseudomonas fluorescens strain KU-7. FEMS Microbiol. Lett. 190:185–190 [DOI] [PubMed] [Google Scholar]
- 59. Hintner J-P, Reemtsma T, Stolz A. 2004. Biochemical and molecular characterization of a ring fission dioxygenase with the ability to oxidize (substituted) salicylate(s) from Pseudaminobacter salicylatoxidans. J. Biol. Chem. 279:37250–37260 [DOI] [PubMed] [Google Scholar]
- 60. Horsman GP, et al. 2005. Spectroscopic studies of the anaerobic enzyme-substrate complex of catechol 1,2-dioxygenase. J. Am. Chem. Soc. 127:16882–16891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hund HK, et al. 1999. Flavonol 2,4-dioxygenase from Aspergillus niger DSM 821, a type 2 CuII-containing glycoprotein. Eur. J. Biochem. 263:871–878 [DOI] [PubMed] [Google Scholar]
- 62. Iwabuchi T, Harayama S. 1998. Biochemical and molecular characterization of 1-hydroxy-2-naphthoate dioxygenase from Nocardioides sp. KP7. J. Biol. Chem. 273:8332–8336 [DOI] [PubMed] [Google Scholar]
- 63. Iwashina T. 2000. The structure and distribution of the flavonoids in plants. J. Plant Res. 113:287–299 [Google Scholar]
- 64. Ju T, et al. 2006. One protein, two enzymes revisited: a structural entropy switch interconverts the two isoforms of acireductone dioxygenase. J. Mol. Biol. 363:823–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kaizer J, Balogh-Hergovich E, Czaun M, Csay T, Speier G. 2006. Redox and nonredox metal assisted model systems with relevance to flavonol and 3-hydroxyquinolin-4(1H)-one 2,4-dioxygenase. Coord. Chem. Rev. 250:2222–2233 [Google Scholar]
- 66. Kalinova J, Vrchotova N, Triska J. 2007. Exudation of allelopathic substances in buckwheat (Fagopyrum esculentum Moench). J. Agric. Food Chem. 55:6453–6459 [DOI] [PubMed] [Google Scholar]
- 67. Kandaswami C, Middleton E., Jr 1994. Free radical scavenging and antioxidant activity of plant flavonoids. Adv. Exp. Med. Biol. 366:351–376 [DOI] [PubMed] [Google Scholar]
- 68. Khuri S, Bakker FT, Dunwell JM. 2001. Phylogeny, function, and evolution of the cupins, a structurally conserved, functionally diverse superfamily of proteins. Mol. Biol. Evol. 18:593–605 [DOI] [PubMed] [Google Scholar]
- 69. Klecka GM, Gibson DT. 1981. Inhibition of catechol 2,3-dioxygenase from Pseudomonas putida by 3-chlorocatechol. Appl. Environ. Microbiol. 41:1159–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kooter IM, et al. 2002. EPR characterization of the mononuclear Cu-containing Aspergillus japonicus quercetin 2,3-dioxygenase reveals dramatic changes upon anaerobic binding of substrates. Eur. J. Biochem. 269:2971–2979 [DOI] [PubMed] [Google Scholar]
- 71. Kovaleva EG, Lipscomb JD. 2007. Crystal structures of Fe2+ dioxygenase superoxo, alkylperoxo, and bound product intermediates. Science 316:453–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kovaleva EG, Lipscomb JD. 2008. Intermediate in the O–O bond cleavage reaction of an extradiol dioxygenase. Biochemistry 47:11168–11170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kovaleva EG, Lipscomb JD. 2008. Versatility of biological non-heme Fe(II) centers in oxygen activation reactions. Nat. Chem. Biol. 4:186–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kucharczyk R, Zagulski M, Rytka J, Herbert CJ. 1998. The yeast gene YJR025c encodes a 3-hydroxyanthranilic acid dioxygenase and is involved in nicotinic acid biosynthesis. FEBS Lett. 424:127–130 [DOI] [PubMed] [Google Scholar]
- 75. Kurnasov O, et al. 2003. NAD biosynthesis: identification of the tryptophan to quinolinate pathway in bacteria. Chem. Biol. 10:1195–1204 [DOI] [PubMed] [Google Scholar]
- 76. Li X, et al. 2006. Crystal structure of 3-hydroxyanthranilic acid 3,4-dioxygenase from Saccharomyces cerevisiae: a special subgroup of the type III extradiol dioxygenases. Protein Sci. 15:761–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Lide DR. 1999. Handbook of chemistry and physics, 80th ed CRC Press, Boca Raton, FL [Google Scholar]
- 78. Lipscomb JD. 2008. Mechanism of extradiol aromatic ring-cleaving dioxygenases. Curr. Opin. Struct. Biol. 18:644–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Liu X, et al. 2010. Characterization of the anthranilate degradation pathway in Geobacillus thermodenitrificans NG80-2. Microbiology 156:589–595 [DOI] [PubMed] [Google Scholar]
- 80. Maroney MJ. 1999. Structure/function relationships in nickel metallobiochemistry. Curr. Opin. Chem. Biol. 3:188–199 [DOI] [PubMed] [Google Scholar]
- 81. Mars AE, Kingma J, Kaschabek SR, Reineke W, Janssen DB. 1999. Conversion of 3-chlorocatechol by various catechol 2,3-dioxygenases and sequence analysis of the chlorocatechol dioxygenase region of Pseudomonas putida GJ31. J. Bacteriol. 181:1309–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Matera I, et al. 2008. Salicylate 1,2-dioxygenase from Pseudaminobacter salicylatoxidans: crystal structure of a peculiar ring-cleaving dioxygenase. J. Mol. Biol. 380:856–868 [DOI] [PubMed] [Google Scholar]
- 83. Mbughuni MM, et al. 2010. Trapping and spectroscopic characterization of an FeIII-superoxo intermediate from a nonheme mononuclear iron-containing enzyme. Proc. Natl. Acad. Sci. U. S. A. 107:16788–16793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Mbughuni MM, et al. 2011. Oxy intermediates of homoprotocatechuate 2,3-dioxygenase: facile electron transfer between substrates. Biochemistry 50:10262–10274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Merkens H, Kappl R, Jakob RP, Schmid FX, Fetzner S. 2008. Quercetinase QueD of Streptomyces sp. FLA, a monocupin dioxygenase with preference for nickel and cobalt. Biochemistry 47:12185–12196 [DOI] [PubMed] [Google Scholar]
- 86. Merkens H, Sielker S, Rose K, Fetzner S. 2007. A new monocupin quercetinase of Streptomyces sp. FLA: identification and heterologous expression of the queD gene and activity of the recombinant enzyme towards different flavonols. Arch. Microbiol. 187:475–487 [DOI] [PubMed] [Google Scholar]
- 87. Moreland JL, Gramada A, Buzko OV, Zhang Q, Bourne PE. 2005. The Molecular Biology Toolkit (MBT): a modular platform for developing molecular visualization applications. BMC Bioinformatics 6:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Murakami S, Sawami Y, Takenaka S, Aoki K. 2004. Cloning of a gene encoding 4-amino-3-hydroxybenzoate 2,3-dioxygenase from Bordetella sp. 10d. Biochem. Biophys. Res. Commun. 314:489–494 [DOI] [PubMed] [Google Scholar]
- 89. Muraki T, Taki M, Hasegawa Y, Iwaki H, Lau PCK. 2003. Prokaryotic homologs of the eukaryotic 3-hydroxyanthranilate 3,4-dioxygenase and 2-amino-3-carboxymuconate-6-semialdehyde decarboxylase in the 2-nitrobenzoate degradation pathway of Pseudomonas fluorescens strain KU-7. Appl. Environ. Microbiol. 69:1564–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Nayak AS, Sanganal SK, Mudde SK, Oblesha A, Karegoudar TB. 2011. A catabolic pathway for the degradation of chrysene by Pseudoxanthomonas sp. PNK-04. FEMS Microbiol. Lett. 320:128–134 [DOI] [PubMed] [Google Scholar]
- 91. Nishinaga A, Matsuura T. 1973. Base-catalyzed autoxidation of 3,4′-dihydroxyflavone. J. Chem. Soc. Chem. Commun. (Camb.) 1973:9–10 [Google Scholar]
- 92. Nishinaga A, Tojo T, Tomita H, Matsuura T. 1979. Base-catalyzed oxygenolysis of 3-hydroxyflavones. J. Chem. Soc. Perkin I. 1979:2511–2516 [Google Scholar]
- 93. Ohlendorf DH, Lipscomb JD, Weber PC. 1988. Structure and assembly of protocatechuate 3,4-dioxygenase. Nature 336:403–405 [DOI] [PubMed] [Google Scholar]
- 94. Ohlendorf DH, Orville AM, Lipscomb JD. 1994. Structure of protocatechuate 3,4-dioxygenase from Pseudomonas aeruginosa at 2.15 Å resolution. J. Mol. Biol. 244:586–608 [DOI] [PubMed] [Google Scholar]
- 95. Oka T, Simpson FJ, Krishnamurty HG. 1972. Degradation of rutin by Aspergillus flavus. Studies on specificity, inhibition, and possible reaction mechanism of quercetinase. Can. J. Microbiol. 18:493–508 [DOI] [PubMed] [Google Scholar]
- 96. Orville AM, Lipscomb JD, Ohlendorf DH. 1997. Crystal structures of substrate and substrate analog complexes of protocatechuate 3,4-dioxygenase: endogenous Fe3+ ligand displacement in response to substrate binding. Biochemistry 36:10052–10066 [DOI] [PubMed] [Google Scholar]
- 97. Pandey G, Paul D, Jain RK. 2003. Branching of o-nitrobenzoate degradation pathway in Arthrobacter protophormiae RKJ100: identification of new intermediates. FEMS Microbiol. Lett. 229:231–236 [DOI] [PubMed] [Google Scholar]
- 98. Pap JS, Kaizer J, Speier G. 2010. Model systems for the CO-releasing flavonol 2,4-dioxygenase enzyme. Coord. Chem. Rev. 254:781–793 [Google Scholar]
- 99. Pau MY, Davis MI, Orville AM, Lipscomb JD, Solomon EI. 2007. Spectroscopic and electronic structure study of the enzyme-substrate complex of intradiol dioxygenases: substrate activation by a high-spin ferric non-heme iron site. J. Am. Chem. Soc. 129:1944–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Pietta PG. 2000. Flavonoids as antioxidants. J. Nat. Prod. 63:1035–1042 [DOI] [PubMed] [Google Scholar]
- 101. Plaper A, et al. 2003. Characterization of quercetin binding site on DNA gyrase. Biochem. Biophys. Res. Commun. 306:530–536 [DOI] [PubMed] [Google Scholar]
- 102. Pochapsky TC, et al. 2002. Modeling and experiment yields the structure of acireductone dioxygenase from Klebsiella pneumoniae. Nat. Struct. Biol. 9:966–972 [DOI] [PubMed] [Google Scholar]
- 103. Prior RL. 2003. Fruits and vegetables in the prevention of cellular oxidative damage. Am. J. Clin. Nutr. 78:570–578 [DOI] [PubMed] [Google Scholar]
- 104. Qu Y, Spain JC. 2010. Biodegradation of 5-nitroanthranilic acid by Bradyrhizobium sp. strain JS329. Appl. Environ. Microbiol. 76:1417–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Qu Y, Spain JC. 2011. Molecular and biochemical characterization of the 5-nitroanthranilic acid degradation pathway in Bradyrhizobium sp. strain JS329. J. Bacteriol. 193:3057–3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Scarpellini M, Wu AJ, Kampf JW, Pecoraro VL. 2005. Corroborative models of the cobalt(II) inhibited Fe/Mn superoxide dismutases. Inorg. Chem. 44:5001–5010 [DOI] [PubMed] [Google Scholar]
- 107. Schaab MR, Barney BM, Francisco WA. 2006. Kinetic and spectroscopic studies on the quercetin 2,3-dioxygenase from Bacillus subtilis. Biochemistry 45:1009–1016 [DOI] [PubMed] [Google Scholar]
- 108. Schwarcz R, Whetsell WO, Jr, Mangano RM. 1983. Quinolinic acid: an endogenous metabolite that produces axon-sparing lesions in rat brain. Science 219:316–318 [DOI] [PubMed] [Google Scholar]
- 109. Shu L, et al. 1995. X-ray absorption spectroscopic studies of the Fe(II) active site of catechol 2,3-dioxygenase. Implications for the extradiol cleavage mechanism. Biochemistry 34:6649–6659 [DOI] [PubMed] [Google Scholar]
- 110. Silvennoinen L, Sandalova T, Schneider G. 2009. The polyketide cyclase RemF from Streptomyces resistomycificus contains an unusual octahedral zinc binding site. FEMS Lett. 583:2917–2921 [DOI] [PubMed] [Google Scholar]
- 111. Simmons CR, et al. 2006. Crystal structure of mammalian cysteine dioxygenase. A novel mononuclear iron center for cysteine thiol oxidation. J. Biol. Chem. 281:18723–18733 [DOI] [PubMed] [Google Scholar]
- 112. Steiner RA, Janßen HJ, Roversi P, Oakley AJ, Fetzner S. 2010. Structural basis for cofactor-independent dioxygenation of N-heteroaromatic compounds at the α/β hydrolase fold. Proc. Natl. Acad. Sci. U. S. A. 107:657–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Steiner RA, Kalk KH, Dijkstra BW. 2002. Anaerobic enzyme-substrate structures provide insight into the reaction mechanism of the copper-dependent quercetin 2,3-dioxygenase. Proc. Natl. Acad. Sci. U. S. A. 99:16625–16630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Steiner RA, Kooter IM, Dijkstra BW. 2002. Functional analysis of the copper-dependent quercetin 2,3-dioxygenase. 1. Ligand-induced coordination changes probed by X-ray crystallography: inhibition, ordering effect, and mechanistic insights. Biochemistry 41:7955–7962 [DOI] [PubMed] [Google Scholar]
- 115. Stevens JF, et al. 1999. Leaf surface flavonoids of Chrysothamnus. Phytochemistry 51:771–780 [Google Scholar]
- 116. Stipanuk MH, Simmons CR, Karplus PA, Dominy JE., Jr 2011. Thiol dioxygenases: unique families of cupin proteins. Amino Acids 41:91–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Stone TW, Perkins MN. 1981. Quinolinic acid: a potent endogenous excitant at amino acid receptors in CNS. Eur. J. Pharmacol. 72:411–412 [DOI] [PubMed] [Google Scholar]
- 118. Straganz GD, Nidetzky B. 2006. Variations of the 2-His-1-carboxylate theme in mononuclear non-heme FeII oxygenases. ChemBioChem 7:1536–1548 [DOI] [PubMed] [Google Scholar]
- 119. Takenaka S, Asami T, Orii C, Murakami S, Aoki K. 2002. A novel meta-cleavage dioxygenase that cleaves a carboxyl-group-substituted 2-aminophenol. Eur. J. Biochem. 269:5871–5877 [DOI] [PubMed] [Google Scholar]
- 120. Tancsics A, et al. 2010. Investigation of catechol 2,3-dioxygenase and 16S rRNA gene diversity in hypoxic, petroleum hydrocarbon contaminated groundwater. Syst. Appl. Microbiol. 33:398–406 [DOI] [PubMed] [Google Scholar]
- 121. Tranchimand S, Brouant P, Iacazio G. 2010. The rutin catabolic pathway with special emphasis on quercetinase. Biodegradation 21:833–859 [DOI] [PubMed] [Google Scholar]
- 122. Tranchimand S, et al. 2008. Biochemical and molecular characterization of a quercetinase from Penicillium olsonii. Biochimie 90:781–789 [DOI] [PubMed] [Google Scholar]
- 123. True AE, Orville AM, Pearce LL, Lipscomb JD, Que L., Jr 1990. An EXAFS study of the interaction of substrate with the ferric active site of protocatechuate 3,4-dioxygenase. Biochemistry 29:10847–10854 [DOI] [PubMed] [Google Scholar]
- 124. Vaillancourt FH, et al. 2002. Definitive evidence for monoanionic binding of 2,3-dihydroxybiphenyl to 2,3-dihydroxybiphenyl 1,2-dioxygenase from UV resonance Raman spectroscopy, UV/Vis absorption spectroscopy, and crystallography. J. Am. Chem. Soc. 124:2485–2496 [DOI] [PubMed] [Google Scholar]
- 125. Vaillancourt FH, Bolin JT, Eltis LD. 2006. The ins and outs of ring-cleaving dioxygenases. Crit. Rev. Biochem. Mol. Biol. 41:241–267 [DOI] [PubMed] [Google Scholar]
- 126. Vetting MW, Wackett LP, Que L, Jr, Lipscomb JD, Ohlendorf DH. 2004. Crystallographic comparison of manganese- and iron-dependent homoprotocatechuate 2,3-dioxygenases. J. Bacteriol. 186:1945–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Vilchez-Vargas R, Junca H, Pieper DH. 2010. Metabolic networks, microbial ecology and “omics” technologies: towards understanding in situ biodegradation processes. Environ. Microbiol. 12:3089–3104 [DOI] [PubMed] [Google Scholar]
- 128. Whiting AK, Boldt YR, Hendrich MP, Wackett LP, Que L., Jr 1996. Manganese(II)-dependent extradiol-cleaving catechol dioxygenase from Arthrobacter globiformis CM-2. Biochemistry 35:160–170 [DOI] [PubMed] [Google Scholar]
- 129. Woo E-J, Dunwell JM, Goodenough PW, Marvier AC, Pickersgill RW. 2000. Germin is a manganese containing homohexamer with oxalate oxidase and superoxide dismutase activities. Nat. Struct. Biol. 7:1036–1040 [DOI] [PubMed] [Google Scholar]
- 130. Zhang Y, Colabroy KL, Begley TP, Ealick SE. 2005. Structural studies on 3-hydroxyanthranilate 3,4-dioxygenase: the catalytic mechanism of a complex oxidation involved in NAD biosynthesis. Biochemistry 44:7632–7643 [DOI] [PubMed] [Google Scholar]