Abstract
Here we present comparative data on the localization and identity of intracellular symbionts among the superfamily Lygaeoidea (Insecta: Hemiptera: Heteroptera: Pentatomomorpha). Five different lygaeoid species from the families Blissidae and Lygaeidae (sensu stricto; including the subfamilies Lygaeinae and Orsillinae) were analyzed. Fluorescence in situ hybridization (FISH) revealed that all the bugs studied possess paired bacteriomes that are differently shaped in the abdomen and harbor specific endosymbionts therein. The endosymbionts were also detected in female gonads and at the anterior poles of developing eggs, indicating vertical transmission of the endosymbionts via ovarial passage, in contrast to the posthatch symbiont transmission commonly found among pentatomoid bugs (Pentatomomorpha: Pentatomoidea). Phylogenetic analysis based on 16S rRNA and groEL genes showed that the endosymbionts of Ischnodemus sabuleti, Arocatus longiceps, Belonochilus numenius, Orsillus depressus, and Ortholomus punctipennis constitute at least four distinct clades in the Gammaproteobacteria. The endosymbiont phylogeny did not agree with the host phylogeny based on the mitochondrial cytochrome oxidase I (COI) gene, but there was a local cospeciating pattern within the subfamily Orsillinae. Meanwhile, the endosymbiont of Belonochilus numenius (Lygaeidae: Orsillinae), although harbored in paired bacteriomes as in other lygaeoid bugs of the related genera Nysius, Ortholomus, and Orsillus, was phylogenetically close to “Candidatus Rohrkolberia cinguli,” the endosymbiont of Chilacis typhae (Lygaeoidea: Artheneidae), suggesting an endosymbiont replacement in this lineage. The diverse endosymbionts and the differently shaped bacteriomes may reflect independent evolutionary origins of the endosymbiotic systems among lygaeoid bugs.
INTRODUCTION
The impressive diversity of insects would hardly be conceivable without beneficial bacterial symbionts associated with them. In particular, insects that feed exclusively on nutritionally restricted diets that are deficient in essential nutrients (e.g., amino acids or vitamins), such as woody materials, plant sap, seeds, vertebrate blood, or keratin materials, tend to possess obligate mutualistic symbionts (4, 5). In general, these symbiotic bacteria are either accommodated extracellularly in the gut cavity or harbored in specialized host cells called bacteriocytes or mycetocytes, which form symbiotic organs called bacteriomes or mycetomes. Especially the large insect group of the order Hemiptera stands out, with a large variety of symbiotic associations with microorganisms (3, 5). Most representatives of the five suborders Stenorrhyncha, Clypeorrhyncha, Archaeorrhyncha, Coleorrhyncha, and Heteroptera feed on nutritionally unbalanced plant xylem or phloem plant sap with their piercing-sucking mouth parts. Essential compounds, which the insect itself can neither synthesize nor obtain from its diet in sufficient quantities, are frequently supplied by the metabolic and biosynthetic capabilities of the symbionts. For example, essential amino acids deficient in plant sap are synthesized by Buchnera aphidicola in aphids and by Sulcia muelleri in spittlebugs and sharpshooters (33, 35).
Most of 42,300 described species of the suborder Heteroptera (13) feed on plant sap as well, but seed-sucking, blood-sucking, and predacious feeding habits are also found. In plant-sucking stinkbugs of the superfamily Pentatomoidea (Heteroptera: Pentatomomorpha), most species of the families Acanthosomatidae, Cydnidae, Parastrachiidae, Pentatomidae, Plataspidae, and Scutelleridae harbor specific bacterial symbionts, which belong to distinct lineages in the Gammaproteobacteria, extracellularly in separated sections of the posterior midgut called crypts or ceca (8, 9, 17, 18, 20, 24, 29, 40, 44). Typically, the symbionts harbored in the midgut crypts are transmitted vertically by postnatal transmission mechanisms such as egg smearing. The importance of these specific symbionts has been experimentally demonstrated, as aposymbiotic insects exhibit retarded growth, mortality, and/or sterility (1, 14, 16, 24, 36, 41). Comparable sac- or tube-like outgrowths are found in bugs of the families Berytidae, Blissidae, Cymidae, Pachygronthidae, and Rhyparochromidae (Heteroptera: Pentatomomorpha: Lygaeoidea) (12, 13) and also in the species of the families Coreidae and Alydidae (Heteroptera: Pentatomomorpha: Coreoidea) (23, 25). However, in contrast to stinkbugs of the superfamily Pentatomoidea, most representatives of lygaeoid and coreoid stinkbugs are associated with betaproteobacterial Burkholderia symbionts in the midgut appendages (23), which are not vertically transmitted but are acquired from the environment by nymphal insects every generation (22).
On the other hand, some species of the superfamily Lygaeoidea possess neither midgut appendages nor Burkholderia symbionts. These lygaeoid species, which are often associated with specific host plants, are characterized by the presence of different types of bacteriomes for harboring specific endosymbionts, which was first described by Schneider (45) in detail using light microscopy. Later, Carayon (6) and Cobben (7) described bacteriome-associated endosymbionts from Arocatus species (Lygaeoidea: Lygaeidae: Lygaeinae) and Orsillus depressus (Lygaeoidea: Lygaeidae: Orsillinae), respectively. All of this histological work on the symbiotic systems in lygaeoid bugs was summarized by Pericart (39).
Recently, several bacteriome-associated symbiotic systems of lygaeoid bugs were analyzed using molecular methods (27, 28, 32). In Kleidocerys resedae and Kleidocerys ericae (Lygaeoidea: Lygaeidae: Ischnorhynchinae), their endosymbiont, “Candidatus Kleidoceria schneideri” (Gammaproteobacteria), is harbored in a single red, raspberry-shaped bacteriome, which is closely associated with but not connected to the midgut (27). In Nysius spp. (Lygaeoidea: Lygaeidae: Orsillinae), their endosymbiont “Candidatus Schneideria nysicola” is localized in paired red bacteriomes associated with the gonads (32). In Chilacis typhae (Lygaeoidea: Artheneidae), the endosymbiotic system is distinct from those in Kleidocerys spp. and Nysius spp.: a “bacteriocytic belt” exists at the anterior end of the midgut, which consists of circularly arranged, strongly enlarged midgut epithelial cells whose cytoplasm is full of a specific endosymbiont belonging to the Gammaproteobacteria (28). In all these cases, the endosymbionts are vertically transmitted via transovarial passages, which is typical of intracellular symbiotic associations (35).
In this study, we describe the endosymbiotic bacteria from five additional lygaeoid species: Ischnodemus sabuleti (Lygaeoidea: Blissidae), Arocatus longiceps (Lygaeoidea: Lygaeidae: Lygaeinae), and Belonochilus numenius, Orsillus depressus, and Ortholomus punctipennis (all Lygaeoidea: Lygaeidae: Orsillinae). All these species are monophagous, living on a specific host plant: I. sabuleti is associated predominantly with Glyceria (Poaceae), A. longiceps and B. numenius live on seed balls of sycamore trees (Platanus); O. depressus feeds primarily on Juniperus and Cupressus species, and O. punctipennis is associated predominantly with Potentilla (Rosaceae) (49). Notably, A. longiceps, B. numenius, and O. depressus have been spreading in Central Europe in recent years (42). B. numenius, originating from North America, was first detected on the European continent in 2008 (26, 31).
Our histological and microbiological inspections unveiled a striking diversity in the structure and localization of the bacteriomes and also in phylogenetic affinity of the endosymbionts among these lygaeoid bugs.
MATERIALS AND METHODS
Sampling and histology.
Adults and nymphs of I. sabuleti, A. longiceps, B. numenius, O. depressus, and O. punctipennis were collected from their host plants (Table 1). These bugs were brought alive to the laboratory and dissected. The isolated tissues were subjected to either histology, fluorescence in situ hybridization (FISH), or microbial characterization. Before fixation in 4% paraformaldehyde overnight, the hemelytra were removed from the insects. The fixed bugs were washed in 0.5× phosphate-buffered saline and 48% (vol/vol) ethanol, dehydrated serially in ethanol (70%, 90%, and [twice] 100%), and embedded in Unicryl (Plano GmbH, Germany). Serial sections (2 μm) were cut using a Leica Jung RM2035 rotary microtome (Leica Instruments GmbH, Wetzlar, Germany), mounted on epoxy-coated glass slides, and subjected to FISH. Symbiont cultivation experiments were performed with insects that had been surface sterilized by ethanol with a subsequent external flame treatment. Cultivability of the symbiotic bacteria was tested by plating tissue extract containing bacteria on two standard microbiological media, brain heart infusion (BHI) and lysogeny broth (LB) media (Merck).
Table 1.
Samples of lygaeoid bugs used in this studya
| Taxon | Locality | Date (yr or yr/mo/day) | No. and sexb | Host plant | Symbiotic organ |
|---|---|---|---|---|---|
| Family Blissidae | |||||
| Ischnodemus sabuletic | Bayreuth, Bavaria, Germany | 2009–2011 | Multitudinous | Glyceria (Poaceae) | Paired bacteriome (tubular) |
| Erlangen, Bavaria, Germany | 10/05/26 | 25 M, 13 F | |||
| Schweinfurt, Bavaria, Germany | 09/06/17 | 14 M, 15 F | |||
| Mistelgau, Bavaria, Germany | 2009–2011 | Multitudinous | |||
| Bautzen, Saxony, Germany | 10/05/04 | 35 M, 27 F, 51 larvae | |||
| Family Lygaeidae | |||||
| Subfamily Lygaeinae | |||||
| Arocatus longicepsd | Schweinfurt, Bavaria, Germany | 2009–2011 | Multitudinous | Platanus (Platanaceae) | Paired bacteriome (three part) |
| Erlangen, Bavaria, Germany | 10/02/16 | 43 M, 52 F | |||
| Cecina, Toskana, Italy | 10/05/05 | 18 M, 12 F, 5 larvae | |||
| Subfamily Orsillinae | |||||
| Belonochilus numeniuse | Cecina, Toskana, Italy | 10/05/05 | 3 M, 4 F, 38 larvae | Platanus (Platanaceae) | Paired bacteriome |
| L'Escalet, France | 11/09/09 | 1 M, 1 F | |||
| Vienna, Austria | 11/09/13 | 7 M, 6 F | |||
| Orsillus depressusf | Horb am Neckar, Swabia, Germany | 10/07/30 | 3 M, 5 F | Thuja, Juniperus (Cupressaceae) | |
| Künzelsau, Swabia, Germany | 11/07/04 | 1 M, 7 F, 17 larvae | |||
| Ingelfingen, Swabia, Germany | 11/07/04 | 2 M, 1 F, 3 larvae | |||
| Ortholomus punctipennisg | Bayreuth, Bavaria, Germany | 2009–2011 | 4 M, 9 F | Potentilla (Rosaceae) | |
| Erlangen, Bavaria, Germany | 11/06/25 | 3 M |
DNA samples from different species were subjected to cloning, RFLP typing, and clone sequencing.
M, male; F, female.
Ten samples; 45 clones by RFLP; 6 clones sequenced.
Ten samples; 40 clones by RFLP; 6 clones sequenced.
Five samples; 38 clones by RFLP; 8 clones sequenced.
Five samples; 40 clones by RFLP; 6 clones sequenced.
Four samples; 40 clones by RFLP; 6 clones sequenced.
DNA extraction, cloning, and sequencing.
DNA of the dissected bacteriome was extracted using a Qiagen DNeasy tissue kit (Qiagen GmbH, Hilden, Germany) following the protocol for animal tissue. The eubacterial 16S rRNA gene was PCR amplified using the universal primers 07F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1507R (5′-TACCTTGTTACGACTTCAC-3′) (30). A 1.65-kb segment of the bacterial groEL gene (which encodes the 60-kDa heat shock protein GroEL) was amplified with the primers Gro-F2 (5′-ATGGCAGCTAAAGAMGTAAAATTYGG-3′) and Gro-R2 (5′-TTACATCATRCCRCCCAT-3′) (27). The insect mitochondrial cytochrome oxidase I (COI) gene was amplified and sequenced with the primers mtD4_F (5′-TACAATTTATCGCCTAAACTTCAGCC-3′) and Nancy_R (5′-CCCGGTAAAATTAAA ATATAAACTTC-3′) (46).
All PCRs were performed on a Biometra thermal cycler with the following program: an initial denaturing step at 94°C for 3 min, followed by 34 cycles of 94°C for 30 s, 50°C for 2 min, and 72°C for 1 min. A final extension step of 72°C for 10 min was included. PCR products of the expected sizes were cloned using the CloneJET PCR cloning kit (Fermentas). Suitable clones for sequencing were selected by restriction fragment length polymorphism (RFLP). Inserts were digested with restriction endonucleases RsaI and HhaI. Plasmids containing the DNA inserts of the expected sizes were sequenced with the pJET1.2 forward and pJET1.2 reverse sequencing primers (Fermentas).
FISH.
The following probes were used for specific endosymbiont detection: Ischno500 [5′-(Cy3)-TTATTTACATTATTATTTTCCTCCC-3′] and the two helping probes Ischno500H1 [5′-AGGTAACGTCAGATAATAATG-3′] and Ischno500H2 [5′-CCCTACCGAAAGTGCTTTACA-3′] for the I. sabuleti endosymbiont, Belono500 [5′-(Cy3)-TTGCTGCTTTCCTCATCGCT-3′] for the B. numenius endosymbiont, Aro450 [5′-(Cy3)-ACGCTATCGCCTTCCTCCCC-3′] for the A. longiceps endosymbiont, Orsdep1350 [5′-(Cy3)-TGTACTTTTTGAGGTTGGCTTAATC-3′] for the O. depressus endosymbiont, and Ortpun1350 [5′-(Cy3)-TGTAGTTTGTGAGGTTGGCTTGCTC-3′] for the O. punctipennis endosymbiont. The tissue sections were incubated with a hybridization buffer (20 mM Tris-HCl [pH 8.0], 0.9 M NaCl, 0.01% sodium dodecyl sulfate [SDS], 20% formamide) containing 10 pmol/ml each of the fluorescent probes, kept at 46°C for 90 min, rinsed with a washing buffer (20 mM Tris-HCl [pH 8.0], 450 mM NaCl, 0.01% SDS), mounted with an antibleaching solution (Vectashield mounting medium; Vector Laboratories, Peterborough, United Kingdom), and viewed under a fluorescence microscope.
Electron microscopy.
Dissected tissues were fixed in 2.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.3) for 1 h, embedded in a 2% agarose gel, and fixed again in 2.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.3) overnight. The tissue was washed in 0.1 M cacodylate buffer for 20 min three times. Following postfixation in 2% osmium tetroxide for 2 h, the sample was washed and stained en bloc in 2% uranyl acetate for 90 min. After fixation, the tissue was dehydrated serially in ethanol (30%, 50%, 70%, 95%, and [three times] 100%), transferred to propylene oxide, and embedded in Epon. Ultrathin sections (70 nm) were cut using a diamond knife (Micro-Star, Huntsville, TX) on a Leica Ultracut UCT microtome (Leica Microsystems, Vienna, Austria). Ultrathin sections were mounted on pioloform-coated copper grids and stained with saturated uranyl acetate, followed by lead citrate. The sections were viewed using a Zeiss CEM 902 A transmission electron microscope (Carl Zeiss, Oberkochen, Germany) at 80 kV.
Phylogenetic analysis.
High-quality sequences of the 16S rRNA, groEL, and COI genes were aligned using the ClustalW software in BioEdit (10) and edited manually. A likelihood ratio test was performed using MrModeltest V.2.3 (38) to find the best-fitting models for the underlying molecular data. The Akaike criterion selected the GTR+I+G model for the 16S rRNA, groEL, and COI gene data. Under the evolutionary model, a Bayesian analysis with MrBayes (v.3.1.2) (19) was performed with four simultaneous Markov chains for each data set. For the 16S rRNA, groEL, and COI gene data, 10,000,000 generations were used; in total, 10,000 trees were obtained (samplefreq = 1,000), and the first 2,500 of these were considered the “burn in” and discarded. A maximum-parsimony analysis was performed with PAUP* v. 4.0b10 (47). Relative-rate tests were carried out using Kimura's two-parameter model in the program RRTree (43). For groEL gene sequences, translated amino acid sequences were analyzed.
Nucleotide sequence accession numbers.
The DNA sequences of the bacterial 16S rRNA gene and groEL gene as well as the mitochondrial cytochrome oxidase I (COI) host gene determined in this study were deposited in the DDBJ/EMBL/GenBank nucleotide sequence databases under accession numbers HE586112 to HE586117, HE586264 to HE586269, and HE586258 to HE586263, respectively.
RESULTS
General observation of the bacteriomes.
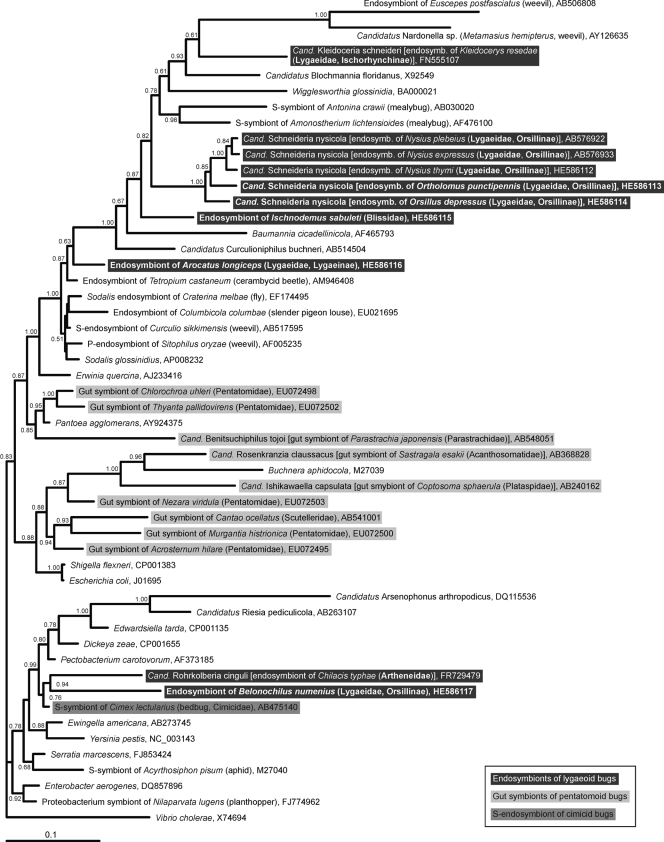
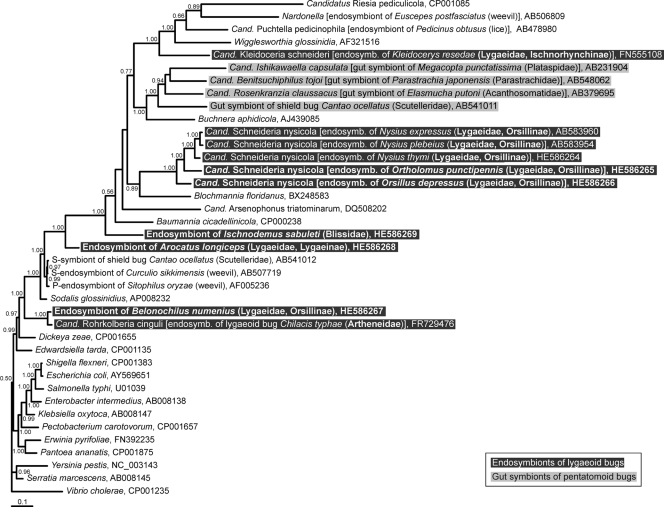
All dissected individuals of the five examined lygaeoid species possessed paired bacteriomes. The structures of the bacteriomes in the different species were different (for an overview, see Fig. 6). Characteristic appendages at the posterior part of the midgut, as have been observed in other lygaeoid and coreoid species, were not detected. All analyzed individuals were positive for the respective symbiotic bacteria, which consistently showed the same localization patterns and morphological characteristics in adults and nymphs (Table 1). All clones of each specific endosymbiont (only one RFLP type) were nearly identical to each other (99.6 to 99.9%) among the insect samples derived from geographically distant localities. The phylogenetic placements of the five lygaeoid endosymbionts were comparable in the 16S rRNA and groEL phylogenetic trees. The evolutionary substitution rates of the 16S rRNA and groEL gene sequences of the examined lygaeoid endosymbionts were higher than those of their free-living gammaproteobacterial relatives (Table 2). Cultivation experiments with standard microbiological media were unsuccessful (data not shown).
Fig 6.
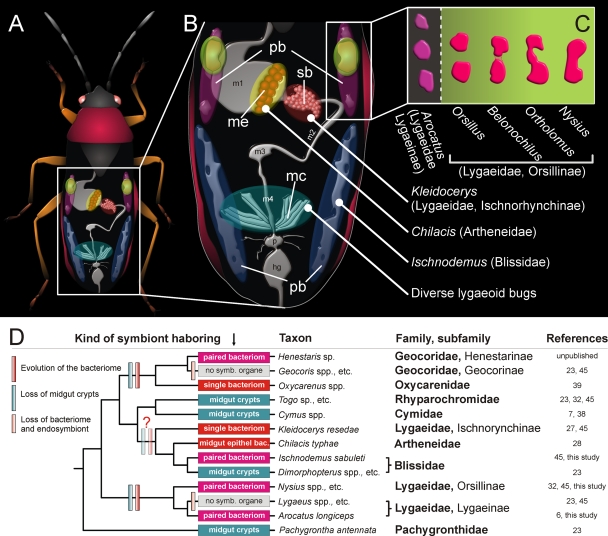
Schematic representation of the diverse symbiotic systems in lygaeoid bugs. (A) Overview. (B) Enlarged image of internal organs in the abdomen. (C) Bacteriomes types found in the subfamilies Lygaeinae and Orsillinae. Abbreviations: pb, paired bacteriomes; sb, single bacteriome; me, midgut epithelium; mc, midgut crypts; m1 to -4, midgut sections; p, pylorus; and hg, hindgut. (D) A hypothesis on the evolution of the symbiotic systems in lygaeoid bugs. The cladogram of the host insects is based on the molecular phylogeny (Fig. 5).
Table 2.
Relative-rate tests for the 16S rRNA and groEL gene sequences of the lineages of I. sabuleti, A. longiceps, B. numenius, O. depressus, and O. punctipennis endosymbionts and Escherichia coli and Pectobacterium carotovorum as free-living relatives, as well as Vibrio cholerae as an outgroup
| Host and gene | Organism(s) (accession no.) in: |
K1a | K2b | K1 − K2 (mean ± SD) | Rate ratio (K1/K2) | P valuec | ||
|---|---|---|---|---|---|---|---|---|
| Lineage 1 | Lineage 2 | Outgroup | ||||||
| Ischnodemus | ||||||||
| 16S rRNA | Endosymbiont of I. sabuleti (HE586115) | E. coli (J01695), P. carotovorum (AF373185) | V. cholerae (X74694) | 0.125 | 0.098 | 0.027 ± 0.0090 | 1.27 | 0.003 |
| groEL | Endosymbiont of I. sabuleti (HE586269) | E. coli (AY569651), P. carotovorum (CP001657) | V. cholerae (CP001235) | 0.174 | 0.139 | 0.036 ± 0.0212 | 1.26 | 0.093 |
| Arocatus | ||||||||
| 16S rRNA | Endosymbiont of A. longiceps (HE586116) | E. coli (J01695), P. carotovorum (AF373185) | V. cholerae (X74694) | 0.115 | 0.098 | 0.017 ± 0.0068 | 1.17 | 0.011 |
| groEL | Endosymbiont of A. longiceps (HE586268) | E. coli (AY569651), P. carotovorum (CP001657) | V. cholerae (CP001235) | 0.194 | 0.159 | 0.035 ± 0.0159 | 1.22 | 0.027 |
| Belonochilus | ||||||||
| 16S rRNA | Endosymbiont of B. numenius (HE586117) | E. coli (J01695), P. carotovorum (AF373185) | V. cholerae (X74694) | 0.167 | 0.096 | 0.071 ± 0.0088 | 1.74 | 1e−07 |
| groEL | Endosymbiont of B. numenius (HE586267) | E. coli (AY569651), P. carotovorum (CP001657) | V. cholerae (CP001235) | 0.178 | 0.159 | 0.019 ± 0.0149 | 1.12 | 0.197 |
| Orsillus | ||||||||
| 16S rRNA | Endosymbiont of O. depressus (HE586114) | E. coli (J01695), P. carotovorum (AF373185) | V. cholerae (X74694) | 0.143 | 0.099 | 0.044 ± 0.0095 | 1.44 | 5e−06 |
| groEL | Endosymbiont of O. depressus (HE586266) | E. coli (AY569651), P. carotovorum (CP001657) | V. cholerae (CP001235) | 0.210 | 0.159 | 0.051 ± 0.0194 | 1.32 | 0.008 |
| Ortholomus | ||||||||
| 16S rRNA | Endosymbiont of O. punctipennis (HE586113) | E. coli (J01695), P. carotovorum (AF373185) | V. cholerae (X74694) | 0.148 | 0.099 | 0.049 ± 0.0097 | 1.49 | 1e−06 |
| groEL | Endosymbiont of O. punctipennis (HE586265) | E. coli (AY569651), P. carotovorum (CP001657) | V. cholerae (CP001235) | 0.201 | 0.159 | 0.042 ± 0.0190 | 1.27 | 0.026 |
Estimated mean distance between lineage 1 and the last common ancestor of lineages 1 and 2.
Estimated mean distance between lineage 2 and the last common ancestor of lineages 1 and 2.
P values were generated using the program RRTree.
Endosymbiotic system of Ischnodemus sabuleti (Blissidae).
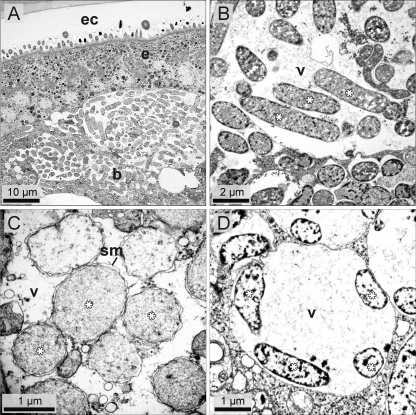
In I. sabuleti (Fig. 1A), a pair of white bacteriomes was found in the fat body, located close to the hypodermis of the posterior abdomen (Fig. 1B). The bacteriome was tubular in shape, with a length of up to 1 mm, and was pervaded by three muscle strands running in a dorso-ventral direction (Fig. 1C). A fine-branched net of tracheoles covered the whole organ. A direct connection to the female gonads was not observed. The top hits in DNA database searches with the symbiont sequences were 16S rRNA gene sequences of the mealybug endosymbiont Moranella endobia (CP002243), with 92% sequence identity, and groEL gene sequences of the sharpshooter endosymbiont Baumannia cicadellinicola (CP000238), with 82% sequence identity. The endosymbiont genes exhibited AT contents of 50% for the 16S rRNA gene and 62.3% for the groEL gene. Phylogenetic analysis revealed that the endosymbiont of I. sabuleti was distantly allied to Baumannia cicadellinicola (see Fig. 3 and 4), exhibiting no close phylogenetic affinity to other lygaeoid endosymbionts. In situ hybridization showed that the I. sabuleti endosymbiont was localized in the paired bacteriomes (Fig. 1C). In addition, the endosymbiont signals were also observed in ovarioles, forming an “infection zone” (Fig. 1D). Transmission electron microscopy showed that the bacteriome was filled with rod-shaped bacterial cells, located just beneath the cuticle-forming epidermis and encased in a thin epithelial layer (Fig. 2A). Most of the bacteriome was filled with vacuoles, which contained only the endosymbionts and no cell organelles (Fig. 2B).
Fig 1.
The endosymbiotic systems of lygaeoid bugs examined in this study. (A to D) Endosymbiotic system of Ischnodemus sabuleti. (A) An adult female. (B) A dissected bacteriome of tubular shape in the abdomen of I. sabuleti. (C) Localization of the endosymbiont of I. sabuleti visualized by fluorescence in situ hybridization (FISH). The bacteriome is pervaded by three muscle strands. (D) The “infection zone” at the tip of ovarioles of I. sabuleti where vertical transmission of the endosymbiont occurs. (E to G) Symbiotic system of Arocatus longiceps. (E) An adult female. (F) Three partial bacteriomes, reddish in color, on the left side of the dissected abdomen of A. longiceps. (G) FISH localization of the endosymbiont in the partial bacteriomes, ovarioles, and eggs. (H and I) Endosymbiotic system of Orsillus depressus. (H) An adult female. (I) Dissected internal organs of the fifth-instar nymph of O. depressus, including the digestive tract, ovaries, and bacteriomes. Each of the paired bacteriomes is clearly split into two parts. The endosymbiont may transferred from the bacteriomes to the ovary via the thin connection (inset, arrow). (K to M) Endosymbiotic system of Belonochilus numenius. (K) An adult female. (L) Two partial bacteriomes on each side of the abdomen of B. numenius. (M) Fused partial bacteriomes in B. numenius. (O and P) Endosymbiotic system of Ortholomus punctipennis. (O) An adult male. (P) Paired bacteriomes of O. punctipennis closely associated with ovaries. Each mature egg exhibits an orange “symbiont ball” at the anterior pole. (J, N, and Q) Fluorescence in situ hybridization of the bacteriomes of O. depressus, B. numenius, and O. punctipennis, respectively. Abbreviations: B, bacteriome; ms, muscle strand; ov, ovarioles; iz, infection zone; sb, “symbiont ball”; o, ovary; St, sternit; m1, midgut first section; m2, midgut second section; m3, midgut third section; hg, hindgut; p, pylorus; and t, tracheole. (Panels H and K are reprinted from reference 26.)
Fig 3.
Phylogenetic positions of the endosymbionts of the lygaeoid bugs on the basis of 16S rRNA gene sequences. A consensus tree inferred by the Bayesian method with 51 sequences (MrBayes; 1,372 bp, 603 variable sites, 396 parsimony-informative sites, 10,000,000 generations, 10,000 trees, samplefreq = 1,000, burn in = 2,500) is shown. The tree has been rooted with Vibrio cholerae as an outgroup. Support values of greater than 0.5 are indicated at the nodes.
Fig 4.
Phylogenetic positions of the endosymbionts of the lygaeoid bugs on the basis of groEL gene sequences. A consensus tree inferred by the Bayesian method with 39 sequences (MrBayes; 1,547 bp, 850 variable sites, 719 parsimony-informative sites, 10,000,000 generations, 10,000 trees, samplefreq = 1,000, burn in = 2,500) is shown. The tree has been rooted with Vibrio cholerae as an outgroup. Support values of greater than 0.5 are indicated at the nodes.
Fig 2.
Transmission electron microscopy of endosymbionts of Ischnodemus sabuleti, Arocatus longiceps, and Orsillus depressus. (A) The bacteriome of I. sabuleti is adjacent to the epidermis (e) of the posterior abdomen (ec, endocuticle). (B) Endosymbiont cells within the bacteriome of I. sabuleti embedded in vacuoles (v). (C) Endosymbiont cells of A. longiceps within the ovarial bacteriocyte, which are enclosed by a symbiosomal membrane (sm) in vacuoles (v). (D) Endosymbiont cells of O. depressus in a vacuole-like structure (v) within the bacteriocyte. Asterisks indicate endosymbiont cells.
Endosymbiotic system of Arocatus longiceps (Lygaeidae: Lygaeinae).
The endosymbiont of A. longiceps (Fig. 1E) was localized in a paired region adjacent to the gonads, which consisted of three separate partial bacteriomes lying in a row at intervals of ca. 100 μm. In fresh specimens, the partial bacteriomes were distinguishable from fat body by their reddish coloration (Fig. 1F). This 3-fold occurrence of the bacteriomes was always found in females, while the number of partial bacteriomes was often reduced to two in males. The top BLAST hits against the DNA databases with the endosymbiont sequences were the 16S rRNA gene sequence of the tsetse endosymbiont Sodalis glossinidius strain “morsitans” (AP008232), with 97% sequence identity, and the groEL gene sequence of the endosymbiont of weevil Sitophilus oryzae (AF005236), with 86% sequence identity. AT contents were 45.8% for the 16S rRNA gene and 52.9% for the groEL gene, which were comparable to those of free-living gammaproteobacteria. Phylogenetic analysis placed the A. longiceps endosymbiont adjacent to Sodalis-allied insect endosymbionts, including the endosymbiont of cerambycid beetle Tetropium castaneum (Fig. 3). In situ hybridization detected the endosymbiont signals in all three partial bacteriomes on each side (Fig. 1G). The symbiotic organs were located close to ovarioles, which exhibited an infection zone in the germarium with tightly aggregated bacteria and a symbiont ball in the developing oocytes (Fig. 1G). Transmission electron microscopy of the ovarial bacteriocytes revealed that the endosymbionts were enclosed by a symbiosomal membrane (Fig. 2C) and embedded in vacuoles like in I. sabuleti.
Endosymbiotic system of Belonochilus numenius (Lygaeidae: Orsillinae).
The symbiotic configuration of B. numenius (Fig. 1K) was comparable to that of A. longiceps. A pair of deeply red-colored bacteriomes was developed at the proximal part of the abdomen, located close to the ovarioles. But only two, instead of three, separate partial bacteriomes resided on each site (Fig. 1L), which sometimes fused to each other in some individuals, resulting in a barbell-shaped appearance (Fig. 1 M).
Determination of the closest matches of the 16S rRNA sequences with those in the GenBank databases resulted in a 95% identity with the secondary endosymbiont of Cimex lectularius (AB475140). The comparison with the groEL gene exhibited the highest BLAST hit to “Candidatus Rohrkolberia cinguli” (FR729476), the endosymbiont of Chilacis typhae (Artheneidae), with a96% identity. The symbiont genes exhibited AT contents comparable to those of free-living gammaproteobacteria, with 47.7% for the 16S rRNA gene and 47.4% for the groEL gene. In phylogenetic trees, the B. numenius endosymbiont also clustered together with “Ca. R. cinguli,” as substantiated by 100% (16S rRNA gene) and 94% (groEL) support values (Fig. 3 and 4). FISH experiments illustrated the barbell-shaped bacteriome (Fig. 1N) and symbiont occurrence in female gonads (data not shown).
Endosymbiotic systems of Orsillus depressus and Ortholomus punctipennis (Lygaeidae: Orsillinae).
Two pairs of deeply red-colored bacteriomes, which accommodate endosymbiotic bacteria, were found in O. depressus (Fig. 1H) and O. punctipennis (Fig. 1O). In contrast to the fused bacteriomes of O. punctipennis (Fig. 1P and Q), whose shape is reminiscent of an hourglass or wasp waist, the symbiotic organ of O. depressus was always split into two separate partial bacteriomes (Fig. 1I and J). These bacteriomes were also associated with the female gonads (Fig. 1I and P). The dissection of an O. depressus larva just before adult molting revealed that the bacteriome and the gonads were conjoined by a thin, red connection, possibly indicating a symbiont transfer route from the bacteriome to the infection zone of the ovarioles (Fig. 1I).
The 16S rRNA gene sequences and the groEL sequences of both species showed the highest similarity (95 to 96% and 90%, respectively) to “Ca. Schneideria nysicola,” the endosymbiont of Nysius spp. (Lygaeidae: Orsillinae). AT contents of 51.0% and 51.6% for the 16S rRNA gene and 63.2% and 64.0% for groEL gene for Ortholomus and Orsillus, respectively, were equivalent to those of Kleidocerys and Nysius endosymbionts. Their close phylogenetic affinity to “Ca. Schneideria nysicola” was clearly seen in the phylogenetic trees (Fig. 3 and 4): the symbionts of O. depressus and O. punctipennis represented basal lineages in this highly supported gammaproteobacterial clade consisting only of endosymbionts of lygaeoid bugs of the subfamily Orsillinae. Electron microscopic examinations of the bacteriocytes of both species demonstrated that the rod-shaped bacteria were found in specific vacuole-like structures (Fig. 2D), which filled out the whole bacteriocyte.
Host-symbiont cospeciation between lygaeoid bugs and their endosymbionts.
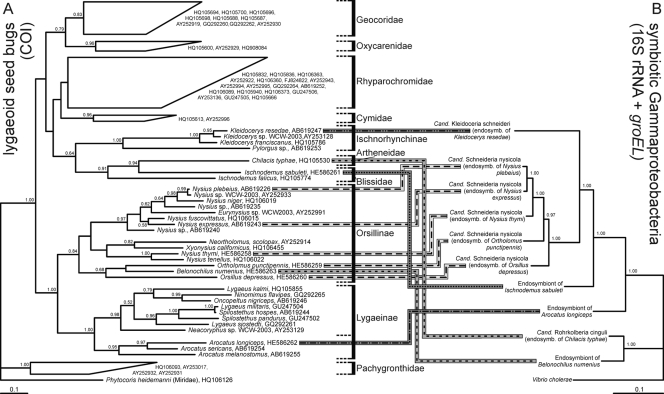
A 570-bp segment of the mitochondrial cytochrome oxidase I (COI) gene was amplified by PCR from the lygaeoid bugs, sequenced, and subjected to phylogenetic analysis together with other lygaeoid COI sequences deposited in the DNA databases. Altogether, the phylogenetic relationship (Fig. 5A) generally reflected the systematics of the lygaeoid bugs based on morphological characteristics (13). The only unexpected finding was the placement of the subfamily Ischnorhynchinae, which was not placed in the family Lygaeidae but clustered with the families Artheneidae and Blissidae.
Fig 5.
Phylogenetic concordance between lygaeoid bugs and their endosymbionts. (A) A consensus tree inferred by the Bayesian method with 76 sequences of the mitochondrial cytochrome oxidase I (COI) gene (MrBayes; 562 bp, 10,000,000 generations, 10,000 trees, samplefreq = 1,000, burn in = 2,500). (B) A consensus tree inferred by the Bayesian method with 11 concatenated sequences of the 16S rRNA and groEL genes (MrBayes; 2909 bp, 100,000 generations, 1,000 trees; samplefreq = 100; burn in = 250). The insect and bacterial tree has been rooted with Phytocoris heidemanni (Heteroptera: Miroidea) and Vibrio cholerae as outgroups, respectively.
Comparison with the symbiont phylogeny inferred from concatenated sequences of the bacterial 16S rRNA and groEL genes (Fig. 5B; the topology presents only symbionts of lygaeoid bugs and no other symbionts or free-living relatives [for this purpose compare with Fig. 3]) hardly showed cospeciating patterns at the family level. The only cospeciating pattern was observed within the subfamily Orsillinae.
DISCUSSION
In the present study, we obtained new insights into endosymbioses of lygaeoid bugs by examining five different lygaeoid species: Ischnodemus sabuleti (Blissidae), Arocatus longiceps (Lygaeidae: Lygaeinae), and Belonochilus numenius, Orsillus depressus, and Ortholomus punctipennis (all Lygaeidae: Orsillinae). All these lygaeoid bugs consistently possess paired bacteriomes in the abdomen, but the structures and localizations of the symbiotic organs are remarkably different between these species (Fig. 6). In addition, the endosymbionts harbored by the bacteriomes are also phylogenetically diverse in these species (Fig. 5). These results highlight the complex evolutionary trajectories underlying the diverse endosymbiotic systems in this insect group.
In I. sabuleti, the bacteriomes are white and tubular, as described in an earlier study (45), and the endosymbiont is distantly related to Baumannia, the obligate endosymbiont of sharpshooters (34). In contrast, the paired and red bacteriomes of A. longiceps are subdivided into partial bacteriomes (6) and harbor a Sodalis-allied endosymbiont (2). The three analyzed species belonging to the Orsillinae (B. numenius, O. depressus, and O. punctipennis) possess paired red bacteriomes, as described for Nysius spp. (32). The endosymbionts of O. depressus and O. punctipennis are closely related to the Nysius endosymbiont, “Ca. Schneideria nysicola.” Unexpectedly, the endosymbiont of B. numenius is not related to “Ca. Schneideria nysicola” but is phylogenetically close to “Ca. Rohrkolberia cinguli,” the endosymbiont of Chilacis typhae (Artheneidae), which is harbored in enlarged epithelial cells of the midgut (28).
In regard to this diversity of the endosymbiotic systems, lygaeoid bugs offer an ideal study system for analysis of developmental and evolutionary traits of symbiotic organs and associated endosymbionts. Unfortunately, no high-resolution phylogeny of Lygaeoidea bugs based on multilocus gene sequences which would elucidate the status of different lygaeoid (sub)families has been available to date. Nevertheless, on the basis of the differently structured bacteriomes as well as their phylogenetically distinct endosymbionts described here and in previous studies (15, 27, 28, 32), we suggest that at least three major symbiotic systems have developed independently among these lygaeoid bugs: (i) the betaproteobacterial gut symbionts of the genus Burkholderia within crypts of the posterior midgut section in species of the Berytidae, Blissidae, Cymidae, Pachygronthidae, and Rhyparochromidae; (ii) the gammaproteobacterial endosymbionts “Ca. Kleidoceria schneideri” of Kleidocerys spp., “Ca. Schneideria nysicola” of Nysius spp., and the endosymbionts of I. sabuleti, A. longiceps, O. depressus, and O. punctipennis; and (iii) the gammaproteobacterial endosymbiont “Ca. Rohrkolberia cinguli” of C. typhae and the endosymbiont of B. numenius.
The symbiotic systems in the second group may be further subdivided into several subsystems of independent origins. A recent study showed that the endosymbiont of Kleidocerys spp., “Ca. Kleidoceria schneideri,” is phylogenetically distinct from the endosymbiont of Nysius spp., “Ca. Schneideria nysicola” (32). In this context, it may be notable that the subfamily Ischnorhynchinae does not cluster within the family Lygaeidae in our phylogenetic analysis based on COI gene sequences (Fig. 5), in contrast to the morphological phylogeny of Henry (12). On the other hand, the endosymbionts of O. depressus and O. punctipennis constitute a well-defined monophyletic group with the endosymbiont of Nysius spp. (“Ca. Schneideria nysicola”), forming an endosymbiont clade specific to members of the subfamily Orsillinae. All these species possess paired bacteriomes of similar color and structure, and our results favor the hypothesis of host-symbiont cospeciation among these species (Fig. 5), suggesting the origin of the endosymbiosis in their common ancestor.
The Arocatus lineage is the only group with a bacteriome-associated endosymbiosis in the subfamily Lygaeinae. No symbiotic organs have so far been reported to be present in the other members of this subfamily (32). Considering the similar structure and positioning of the bacteriomes, it is conceivable, although speculative, that the Arocatus endosymbiosis derives from the common ancestor of the Lygaeinae and the Orsillinae (Fig. 6D). On the other hand, the phylogenetic placement of the A. longiceps endosymbiont favors an alternative hypothesis that the Arocatus endosymbiosis has evolved independently of the endosymbiosis in the Orsillinae.
The endosymbiosis of I. sabuleti may also have developed independently, on the grounds that the bacteriome structure and the phylogenetic position of the endosymbiont are distinct from those in all the other described lygaeoid endosymbioses. However, current analyses of members of Henestarinae (Lygaeoidea: Geocoridae) also show morphological patterns of the bacteriome comparable to that in Ischnodemus (unpublished data). Notably, it was reported that some other Blissidae species, such as Dimorphopterus spinolae, possess midgut crypts with a Burkholderia gut symbiont instead of bacteriomes (23).
Whether possession of midgut crypts or a bacteriome is ancestral in the Ischnodemus lineage or generally all other described lygaeoid bugs cannot be answered adequately at present (Fig. 6D). A detailed phylogeny of Lygaeoidea is needed for a better understanding of development processes of symbiotic organs. At present, all analyzed bugs either lack conspicuous symbiotic structures or possess either midgut crypts or bacteriomes, but never both structures together. Consequently, more representatives of the lygaeoid bugs should be investigated for their symbiotic relationship, and more information about bacteriome development during embryogenesis is needed for conclusions about the evolution of bacteriomes in lygaeoid bugs. In addition, future studies have to clarify to what extent the lygaeoid symbionts can switch the host, as possibly happened in the cases of B. numenius and C. typhae. Although both species possess a phylogenetically closely related symbiont, they have anatomically diverse bacteriomes for their intracellular symbiont accommodation. Furthermore, these two lygaeoid bugs live monophagously on completely different host plants, and until recently they were geographically separated from each other (31, 50). The palearctic lygaeoid C. typhae spends most of its time on bulrush (Typha latifolia and T. angustifolia) and feeds on seeds at different stages of maturation, whereas the nearctic B. numenius is strictly associated with Platanus sp. as its host plant. Whether the similarity of the endosymbionts is a result of an occasional lateral transfer of “Ca. Rohrkolberia cinguli” from C. typhae to B. numenius or vice versa is still unknown. Nevertheless, a potential endosymbiont replacement in these lineages should be taken into account. However, for A. longiceps and B. numenius, it can be observed that vertically transmitted symbionts are not transferred horizontally among hosts easily. Although the two species coexist together on Platanus and feed on its seed balls, the endosymbiotic system of A. longiceps shows no morphological or phylogenetic relationship to B. numenius, even though they have similar feeding ecologies and overlap in the microhabitat. It should be mentioned that a third, heteropteran bug, called Corythucha ciliata (Miroidea: Tingidae), that strictly lives on Platanus does not have any endosymbiotic bacteria which are localized in specific bacteriomes (unpublished data).
As for all the lygaeoid endosymbionts, their biological roles in hosts are totally unknown. It seems likely that the endosymbionts aid in the supply of essential nutrients (amino acids, vitamins, etc.) for their hosts. Alternatively, the endosymbionts may be involved in degradation of toxic compounds derived from their food plants. Many plants protect themselves by the production and accumulation of defense compounds in leaves, roots, and seeds, such as hydrolyzable tannins and flavonoid glycosides in birches (11). Recently, it was shown that the obligate gut symbiont Ishikawaella capsulata of the plataspid bug Megacopta punctatissima is able to synthesize all essential amino acids and some vitamins and cofactors (37). Furthermore, it possesses a plasmid that carries genes for arginine metabolism and oxalate detoxification. Hence, the functional roles of Ishikawaella are comparable to those of the aphid endosymbiont Buchnera in aphids (4, 33), which is probably attributable to their common nutritional physiology as phloem sap feeders. Moreover, it is speculated that the gut symbiont of the shield bug Cantoa ocellatus (Scutelleridae), which feeds on plant sap as well, is involved in detoxification of plant defense substances (21). Whether such nutritional supplements (e.g., amino acids) are also necessary for the seed-sucking lygaeoid bugs will be examined in future studies. Likewise, further research is required to elucidate the physiological and morphological differences between lygaeoid species with bacteriomes and species with midgut crypts that apparently live on same host plants and also feed on seeds. In addition, the symbiotic systems of lygaeoid bugs should be compared with those of other insects that also feed on seeds, such as Curculio spp. (Coleoptera; Curculionidae) (48), regarding the structure and diversity of endosymbiosis as well as potential biological roles.
On the basis of the distinct genetic, phylogenetic, and histological traits described above, we propose the following names for the different endosymbionts. For the endosymbiont of I. sabuleti, we propose “Candidatus Ischnodemia utricula.” The generic name indicates the association with the host insect, whereas the specific name refers to the longitudinal structure of the paired bacteriomes. For the endosymbiont of A. longiceps, we propose the name “Candidatus Arocatia carayoni.” The generic name represents the association with the host insect, whereas the specific name honors Jacques Carayon, who first described the endosymbiosis of Arocatus in detail. Because of the close phylogenetic relationship between the C. typhae and B. numenius endosymbionts, we propose the designation “Candidatus Rohrkolberia belonochilicola” for the endosymbiont of B. numenius. The specific name indicates the association with Belonochilus bugs. For endosymbionts of Nysius spp., O. depressus, and O. punctipennis, we propose the strain names “Ca. Schneideria nysicola” strain nysicola, “Ca. Schneideria nysicola” strain orsillicola, and “Ca. Schneideria nysicola” strain ortholocola, respectively.
ACKNOWLEDGMENTS
We thank S. Geimer and R. Grotjahn for assistance with electron microscopy analysis, as well as D. Scholz and B. Westermann for the opportunity to use the fluorescence microscope and for providing help. We also thank A. Kirpal for technical assistance and W. Rabitsch for providing B. numenius. Special thanks go to Gerhard Strauss for taking pictures of O. depressus and B. numenius. We gratefully acknowledge H. Feldhaar for reading and correcting the manuscript. Finally, we thank the two reviewers for their valuable comments.
Footnotes
Published ahead of print 3 February 2012
REFERENCES
- 1. Abe Y, Mishiro K, Takanashi M. 1995. Symbiont of brown winged green bug, Plautia stali Scott. Jpn. J. Appl. Entomol. Z. 39:109–115 [Google Scholar]
- 2. Aksoy S, Rio RV. 2005. Interactions among multiple genomes: tsetse, its symbionts and trypanosomes. Insect Biochem. Mol. Biol. 35:691–698 [DOI] [PubMed] [Google Scholar]
- 3. Baumann P. 2005. Biology bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu. Rev. Microbiol. 59:155–189 [DOI] [PubMed] [Google Scholar]
- 4. Bourtzis K, Miller TA. 2003. Insect symbiosis. CRC Press, Boca Raton, FL [Google Scholar]
- 5. Buchner P. 1965. Endosymbiosis of animals with plant microorganisms. Interscience Publishers, New York, NY [Google Scholar]
- 6. Carayon J. 1974. Formes nouvelles d'endosymbiose chez les Hémiptères. C. R. Hebd. Seances Acad. Sci. D 278:1495–1498 [Google Scholar]
- 7. Cobben RH. 1968. Evolutionary trends in Heteroptera, part I. Eggs, architecture of the shell, gross embryology and eclosion. Centre for Agricultural Publication and Documentation, Wageningen, Netherlands [Google Scholar]
- 8. Fukatsu T, Hosokawa T. 2002. Capsule-transmitted gut symbiotic bacterium of the Japanese common plataspid stinkbug, Megacopta punctatissima. Appl. Environ. Microbiol. 68:389–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Glasgow H. 1914. The gastric caeca and the caecal bacteria of the Heteroptera. Biol. Bull. 3:101–171 [Google Scholar]
- 10. Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95–98 [Google Scholar]
- 11. Haukioja E. 2003. Putting the insect into the birch-insect interaction. Oecologia 136:161–168 [DOI] [PubMed] [Google Scholar]
- 12. Henry TJ. 1997. Phylogentic analysis of family groups within the infraorder Pentatomorpha (Hemiptera: Heteroptera), with emphasis on the Lygaeoidea. Ann. Entomol. Soc. Am. 90:275–301 [Google Scholar]
- 13. Henry TJ. 2009. Biodiversity of Heteroptera, p 223–263 In Foottit RG, Adler PH. (ed), Insect biodiversity, 1st ed Wiley-Blackwell, Chichester, United Kingdom [Google Scholar]
- 14. Hirose E, Panizzi AR, De Souza JT, Cattelan AJ, Aldrich JR. 2006. Bacteria in the gut of southern green stink bug (Heteroptera: Pentatomidae). Ann. Entomol. Soc. Am. 99:91–95 [Google Scholar]
- 15. Hosokawa T, Koga R, Kikuchi Y, Meng XY, Fukatsu T. 2010. Wolbachia as a bacteriocyte-associated nutritional mutualist. Proc. Natl. Acad. Sci. U. S. A. 107:769–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hosokawa T, Kikuchi Y, Shimada M, Fukatsu T. 2007. Obligate symbiont involved in pest status of host insect. Proc. Biol. Sci. 274:1979–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hosokawa T, Kikuchi Y, Nikoh N, Shimada M, Fukatsu T. 2006. Strict host-symbiont cospeciation and reductive genome evolution in insect gut bacteria. PLoS Biol. 4:e337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hosokawa T, et al. 2010. Phylogenetic position and peculiar genetic traits of a midgut bacterial symbiont of the stinkbug Parastrachia japonensis. Appl. Environ. Microbiol. 76:4130–4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huelsenbeck JP, Ronquist F. 2001. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755 [DOI] [PubMed] [Google Scholar]
- 20. Kaiwa N, et al. 2011. Bacterial symbionts of the giant jewel stinkbug Eucorysses grandis (Hemiptera: Scutelleridae). Zoolog. Sci. 28:169–174 [DOI] [PubMed] [Google Scholar]
- 21. Kaiwa N, et al. 2010. Primary gut symbiont and secondary, Sodalis-allied symbiont of the scutellerid stinkbug Cantao ocellatus. Appl. Environ. Microbiol. 76:3486–3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kikuchi Y, Hosokawa T, Fukatsu T. 2007. Insect-microbe mutualism without vertical transmission: a stinkbug acquires a beneficial gut symbiont from the environment every generation. Appl. Environ. Microbiol. 73:4308–4316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kikuchi Y, Hosokawa T, Fukatsu T. 2011. An ancient but promiscuous host-symbiont association between Burkholderia gut symbionts and their heteropteran hosts. ISME J. 5:446–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kikuchi Y, et al. 2009. Host-symbiont co-speciation and reductive genome evolution in gut symbiotic bacteria of acanthosomatid stinkbugs. BMC Biol. 7:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kikuchi Y, Meng XY, Fukatsu T. 2005. Gut symbiotic bacteria of the genus Burkholderia in the broad-headed bugs Riptortus clavatus and Leptocorisa chinensis (Heteroptera: Alydidae). Appl. Environ. Microbiol. 71:4035–4043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kuechler SM, Strauss G. 2010. Belonochilus numenius (Say, 1832) (Heteroptera: Lygaeidae)—bald auch in Mitteleuropa? Beitraeg. Entomofaun. 11:27–33 [Google Scholar]
- 27. Kuechler SM, Dettner K, Kehl S. 2010. Molecular characterization and localization of the obligate endosymbiotic bacterium in the birch catkin bug Kleidocerys resedae (Heteroptera: Lygaeidae, Ischnorhynchinae). FEMS Microbiol. Ecol. 73:408–418 [DOI] [PubMed] [Google Scholar]
- 28. Kuechler SM, Dettner K, Kehl S. 2011. Characterization of an obligate intracellular bacterium in the midgut epithelium of the bulrush bug Chilacis typhae (Heteroptera, Lygaeidae, Artheneinae). Appl. Environ. Microbiol. 77:2869–2876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kuskop M. 1924. Bakteriensymbiosen bei Wanzen (Hemiptera: Heteroptera). Arch. Protistenkde. 47:1–35 [Google Scholar]
- 30. Lane DJ. 1991. 16S and 23S rRNA sequencing, p 115–148 In Stackenbrandt E, Goodfellow M. (ed), Nucleic acid techniques in bacterial systematics. John Wiley and Sons, New York, NY [Google Scholar]
- 31. Matocq A. 2008. Présence en France et en Corse d'un Hétéroptère néarctique, Belonochilus numenius (Say, 1831) (Hemiptera, Lygaeidae, Orsillinae). Bull. Soc. Entomol. Fr. 113:533–534 [Google Scholar]
- 32. Matsuura Y, et al. 4 August 2011. Evolution of symbiotic organs and endosymbionts in lygaeid stinkbugs. ISME J. doi:10.1038/ismej.2011.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McCutcheon JP, McDonald BR, Moran NA. 2009. Convergent evolution of metabolic roles in bacterial co-symbionts of insects. Proc. Natl. Acad. Sci. U. S. A. 106:15394–15399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Moran NA, Dale C, Dunbar H, Smith WA, Ochman H. 2003. Intracellular symbionts of sharpshooters (Insecta: Hemiptera: Cicadellinae) form a distinct clade with a small genome. Environ. Microbiol. 5:116–126 [DOI] [PubMed] [Google Scholar]
- 35. Moran NA, McCutcheon JP, Nakabachi A. 2008. Genomics and evolution of heritable bacterial symbionts. Annu. Rev. Genet. 42:165–190 [DOI] [PubMed] [Google Scholar]
- 36. Müller HJ. 1956. Experimentelle Studien an der Symbiose von Coptosoma scutellatum Geoffr. (Hem. Heteropt.). Z. Morphol. Ökol. Tiere. 44:459–482 [Google Scholar]
- 37. Nikoh N, Hosokawa T, Oshima K, Hattori M, Fukatsu T. 2011. Reductive evolution of bacterial genome in insect gut environment. Genome Biol. Evol. 3:702–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nylander JAA. 2004. MrModeltest, version 2. Evolutionary Biology Centre, Uppsala University, Uppsala, Sweden [Google Scholar]
- 39. Pericart J. 1998. Hémiptères Lygaeidae euro-méditerranéens. Faune de France, vol 84 Fédération Française des Sociétés de Sciences Naturelles, Paris, France [Google Scholar]
- 40. Prado SS, Almeida RP. 2009. Phylogenetic placement of pentatomid stink bug gut symbionts. Curr. Microbiol. 58:64–69 [DOI] [PubMed] [Google Scholar]
- 41. Prado SS, Hung KY, Daugherty MP, Almeida RP. 2010. Indirect effects of temperature on stink bug fitness, via maintenance of gut-associated symbionts. Appl. Environ. Microbiol. 76:1261–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rabitsch W. 2010. True bugs (Hemiptera, Heteroptera), p 407–433 In Roques A, et al. (ed), Alien terrestrial arthropods of Europe. BioRisk 4(1). Pensoft Publishers, Sofia, Bulgaria [Google Scholar]
- 43. Robinson-Rechavi M, Huchon D. 2000. RRTree: relative-rate tests between groups of sequences on a phylogenetic tree. Bioinformatics 16:296–297 [DOI] [PubMed] [Google Scholar]
- 44. Rosenkranz W. 1939. Die Symbiose der Pentatomiden. Z. Morphol. Ökol. Tiere. 36:279–309 [Google Scholar]
- 45. Schneider G. 1940. Beiträge zur Kenntnis der symbiontischen Einrichtungen der Heteropteren. Z. Morphol. Ökol. Tiere. 36:565–644 [Google Scholar]
- 46. Simon C, et al. 1994. Evolution, weighting and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann. Entomol. Soc. Am. 87:651–701 [Google Scholar]
- 47. Swofford DL. 2000. PAUP*: phylogenetic analysis using parsimony (*and other methods). Sinauer Associates, Sunderland, MA [Google Scholar]
- 48. Toju H, et al. 2010. “Candidatus Curculioniphilus buchneri,” a novel clade of bacterial endocellular symbionts from weevils of the genus Curculio. Appl. Environ. Microbiol. 76:275–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wachmann E, Melber A, Deckert J. 2007. Wanzen, band 3. Die Tierwelt Deutschlands, vol 78 Goecke and Evers, Keltern, Germany [Google Scholar]
- 50. Wheeler AGJ, Fetter JE. 1987. Chilacis typhae (Heteroptera: Lygaeidae) and the subfamily Artheneinae new to North America. Proc. Entomol. Soc. Wash. 89:244–249 [Google Scholar]