Abstract
Most grape juice fermentation takes place when yeast cells are in a nondividing state called the stationary phase. Under such circumstances, we aimed to identify the genetic determinants controlling longevity, known as the chronological life span. We identified commercial strains with both short (EC1118) and long (CSM) life spans in laboratory growth medium and compared them under diverse conditions. Strain CSM shows better tolerance to stresses, including oxidative stress, in the stationary phase. This is reflected during winemaking, when this strain has an increased maximum life span. Compared to EC1118, CSM overexpresses a mitochondrial rhodanese gene-like gene, RDL2, whose deletion leads to increased reactive oxygen species production at the end of fermentation and a correlative loss of viability at this point. EC1118 shows faster growth and higher expression of glycolytic genes, and this is related to greater PKA activity due to the upregulation of the adenylate cyclase gene. This phenotype has been linked to the presence of a δ element in its promoter, whose removal increases the life span. Finally, EC1118 exhibits a higher level of protein degradation by autophagy, which might help achieve fast growth at the expense of cellular structures and may be relevant for long-term survival under winemaking conditions.
INTRODUCTION
Yeast growth during grape juice fermentation develops through the typical batch growth phases: lag phase, exponential growth, stationary phase, and death phase (6, 39). Most sugar fermentation takes place after cells enter the nondividing state, that is, the stationary phase. Therefore, the viability and vitality of Saccharomyces cerevisiae in the stationary phase are key factors in successful vinification. The yeast death phase during winemaking is still a poorly understood process that has been exclusively linked to the toxicity of the high ethanol concentration reached during fermentation (14). Some authors claim that cells start dying when sugar is still present (39), while others state that cells start dying only when all of the sugars have been consumed (6). In our experience, both situations can occur, depending on the environment. The same yeast strain grown in the same grape juice is highly viable at the end of fermentation at a low temperature, but the death phase starts in the presence of sugars when the temperature rises (9). Cell death leads to loss of cell integrity and to the release of cell contents, which could influence other microorganisms' growth; for instance, lactic acid bacteria, which carry out malolactic fermentation, can be inhibited (via medium-chain fatty acids) and stimulated (via nitrogenous components) by yeast lysis products (3). Spoilage microorganisms can likewise be affected. Besides, certain wines age in the presence of dead yeast (aging on lees), which confers wine chemical and color stability (25). Cell autolysis is central in these increasingly popular forms of winemaking. The impact of autolysis on sparkling wines has been studied in detail (12).
Survival in the stationary phase has been referred to as the chronological life span (CLS) and is one of the standard ways of studying aging in S. cerevisiae (22, 29). CLS is highly variable in natural isolates and tends to be shorter than in laboratory strains (37). Molecular causes of aging have been widely studied in several model organisms, including yeast (29). Traditionally, the cause of aging has been explained as an accumulation of molecules damaged by free radicals, mainly produced upon electron transport events in mitochondria (5). The impact of reactive oxygen species (ROS), such as superoxide, on CLS has been studied in yeast (19). Antioxidant proteins like superoxide dismutase Sod2p are essential for the life span. The high sugar concentration of grape juice is believed to cause repression of the oxidative phosphorylation machinery. However, in wine yeast grown on a synthetic medium with a high sugar concentration, ROS are produced and oxidative damage takes place (30). In the last few years, many authors have found that nutrient-sensing pathways, such as Sch9/TOR and Ras/cyclic AMP (cAMP)/protein kinase A (PKA), have a considerable impact on the life span (24). For instance, mutations in CYR1, the gene encoding adenylate cyclase, an enzyme involved in cAMP synthesis which activates PKA, extend the CLS of laboratory yeast strains (23). This result is related to the fact that a low-nutrient diet without malnutrition (dietary restriction) is an effective way of extending the life span. This situation would diminish the activity of these pathways, leading to less growth and lower metabolic rates, and develop mechanisms that extend the life span, such as stress resistance and autophagy. Autophagy consists of the sequestration of part of the cytoplasm by a double membrane that fuses to vacuoles to release nutrients (33). This phenomenon has also been described in sparkling wine production (12). Work done under laboratory conditions has linked aging to two relevant metabolites in winemaking: ethanol and acetic acid. The downregulation of alcohol dehydrogenase 2 by deacetylase Sir2p leads to higher ethanol levels, which shorten the CLS (20). Acetic acid has been identified as a proaging agent under laboratory conditions because of its ability to acidify media (8).
In this work, we aimed to identify the genetic determinants of stationary-phase viability and the CLS. To go about this, we compared the CLSs of a large number of industrial wine yeasts and we isolated model short-lived (EC1118) and long-lived (CSM) strains. We compared their stress resistance, vinification performance, autophagy, and transcriptomes to find differences in their behavior. Strain CSM's greater oxidative stress in stationary-phase tolerance correlates with its longer CLS, and we found that rhodanese-like proteins Rdl1/2p are players in this response. The higher metabolic rate of strain EC1118 also seems to be a determinant of its short life span. Finally, we also suggest that it is at least partially dependent on a particular allele of the adenylate cyclase gene CYR1.
MATERIALS AND METHODS
Yeast strains and growth media.
Table S1 in the supplemental material lists the industrial wine yeasts used in this work. Haploid strain C9 was a gift from M. Walker (47). Gene disruptions were performed by using the recyclable selection marker loxP-kanMX-loxP from plasmid pUG6 following the protocol of Güldener et al. (28). The marker was eliminated by transformation with the cre recombinase-containing plasmid YEp351-cre-cyh according to Delneri et al. (17). Table S2 in the supplemental material lists the oligonucleotides employed to amplify deletion cassettes and to check transformants.
For yeast growth, YPD medium (1% yeast extract, 2% Bacto peptone, 2% glucose) and synthetic complete (SC) medium (0.17% yeast nitrogen base, 0.5% ammonium sulfate, 2% glucose, 0.2% dropout mix with all the amino acids) were used (2). Selective plates contained 20 μg ml−1 Geneticin or 0.1 μg ml−1 cycloheximide. Red grape juice (Bobal variety) was a gift from Bodegas Murviedro and was sterilized overnight with 500 μg/liter dimethyl dicarbonate. MS300 synthetic grape juice was prepared as described by Riou et al. (40) but with an equimolar amount of glucose and fructose at 10%.
Yeast growth, CLS measurements, and stress conditions.
Under laboratory conditions, the CLS experiments were adapted from Fabrizio et al. (22) and performed as follows. Precultures of the selected strains were grown overnight on YPD medium and inoculated into SC medium at an optical density at 600 nm (OD600) of 0.1. From day 3 of growth at 30°C, aliquots were taken, diluted, and plated. Colonies were counted, and percent survival was calculated by taking day 3 of growth as 100% survival.
For the microvinification experiments, cells from 2-day cultures in YPD medium were inoculated at a final concentration of 106/ml into filled-in, loosely closed, conical centrifuge tubes with 50 ml of grape juice. Incubation was performed with very low shaking at 22°C for natural juice and at 24°C for synthetic grape juice. The vinification process was monitored by determining cell viability and sugar consumption as previously described (49). Survival plots were drawn by taking the highest cell viability point (around 2 to 4 days) as 100% survival.
For the stress tests, cells were either grown in YPD medium for 2 days to test them under stationary-phase conditions or diluted in fresh YPD medium at an OD600 of 0.1 and grown to 0.5 when testing exponential-phase conditions. An aliquot of the culture was diluted and plated before application of stress, and its viability was considered 100%. The stress condition was applied to the remaining culture. Ethanol was added at 10% (vol/vol) for 1 h of incubation; for hyperosmotic stress, cells were spun and placed in YPD medium containing 20% glucose for 90 min, and heat shock was achieved by incubation for 1 h at 42°C. For oxidative stress, 5 mM H2O2 was added to the culture and cells were incubated for 30 min if they were growing exponentially or for 1 h in the case of stationary-phase cells.
Microscopy methods.
For propidium iodide staining, a 500-μl volume of cells was washed in phosphate-buffered saline (PBS). A 5-μl volume of a 1-mg/ml stock solution of the dye was added, and the mixture was incubated in darkness for 30 min. Cells were washed in PBS and visualized. Dihydrorhodamine 123 was added at 5 μg/ml from a 2.5-mg/ml stock solution in ethanol. Cells were viewed after a 2-h incubation at room temperature in the dark. Cells were visualized with a rhodamine filter under a Nikon eclipse 90i fluorescence microscope.
Metabolite and lipid peroxidation determinations.
Reduction of sugars during fermentation was measured by their reaction to dinitro-3,5-salycilic acid following a modified version of Miller's method (41). Ethanol and acetic acid were measured with the kits provided by r-Biopharm following the manufacturer's instructions.
Quantification of lipid peroxidation was carried out by the reaction of thiobarbituric acid with the malondialdehyde product of oxidized fatty acid breakage (18). Cells were collected and then extracted by vortexing with 1 volume of glass beads in 0.5 ml of 50 mM sodium phosphate buffer (pH 6.0)–10% trichloroacetic acid with FastPrep 24. After centrifugation, 300 μl of supernatants was mixed with 100 μl of 0.1 M EDTA and 600 μl of 1% thiobarbituric acid in 0.05 M NaOH and then incubated at 100°C for 15 min. Malondialdehyde was measured by absorbance at 535 nm.
Microarray analysis and Northern blot analysis.
RNA isolation was carried out as previously described (10). cDNA was synthesized by combining 20 μg of total RNA, 1 μg of oligo(dT) (Roche), dithiothreitol at 0.1 M, a deoxynucleoside triphosphate mixture (10 mM each; Invitrogen), aminoallyl-dUTP at 50 mM (Fermentas), RNase inhibitor, and Superscript III reverse transcriptase (Invitrogen). Aminoallyl-cDNA was purified by using the MinElute PCR purification kit (Qiagen) and following the manufacturer's instructions. A labeling reaction was produced by incubating 1.5 to 2 mg of aminoallyl-cDNA with 3 ml of the Cy3 and Cy5 fluorophores (Amersham) at a basic pH (0.2 M Na2CO3, pH 9) for 2 h at room temperature. Labeled cDNA was purified using the aforementioned kit. Yeast 6.4Kv7 microarray slides from the University Health Network Microarray Center (Toronto, Ontario, Canada) were used. Microarrays were prehybridized for 45 min at 42°C in a mixture of 3× SSC (0.15 M NaCl and 0.015 M sodium citrate), 0.1% (wt/vol) sodium dodecyl sulfate (SDS), and 0.1 mg/ml bovine serum albumin. Before hybridization, equal amounts of Cy5- and Cy3-labeled cDNA, 50% (vol/vol) formamide, 5× SSC, and 0.1% (wt/vol) SDS were mixed and denatured for 1 min at 95°C. The mixture was applied to the prehybridized microarrays and covered with a Hybri-slip (Sigma-Aldrich), and hybridization took place for 16 h in a humidified chamber in a water bath at 42°C. The hybridized microarrays were washed for 5 min at 42°C in 2× SSC–0.1% (wt/vol) SDS for 20 min at room temperature in 0.1× SSC–0.1% (wt/vol) SDS, for 6 min at room temperature in 0.1× SSC, and for 1 min at room temperature in 0.01× SSC. After washing, microarrays were immediately scanned with an Axon 4100A scanner at a resolution of 10 μm and the data were analyzed using the GENEPIX PRO 6.1 software package (Axon Instruments). Functional categories were identified using Funcassociate 2.0.
The array experiments were carried out in triplicate.
Total RNA was separated in formaldehyde agarose gels, blotted onto nylon membranes, and hybridized in accordance with reference 43.
Western blotting.
For Western blot analysis, cells were broken with a volume of glass beads in a buffer containing Tris-HCl at 0.1 M (pH 7.5), NaCl at 0.5 M, MgCl2 at 0.1 M, NP-40 at 1% (vol/vol), phenylmethylsulfonyl fluoride at 10 mM, and Complete Mini EDTA-free protease inhibitor cocktail (Roche). Protein concentration was measured by the Bradford method (7) by using the Bio-Rad protein assay and following the manufacturer's instructions. Extracts were diluted in loading buffer (Tris-HCl at 240 mM [pH 6.8], SDS at 8% [wt/vol], glycerol at 40%, β-mercaptoethanol at 10%).
To conduct the Western blot analysis, SDS-PAGE was done with an Invitrogen mini-gel device and the gel was blotted onto polyvinylidene difluoride membranes. The anti-aldehyde dehydrogenase (anti-ALDH) antibodies used were obtained from Rockland (Gilbertsville, PA). The ECL Western blotting detection system (Amersham) was used by following the manufacturer's instructions.
Microarray data accession number.
The microarray data obtained in this study were deposited in the Gene Expression Omnibus database under accession number GSE33068.
RESULTS
Study of the CLS of wine yeast.
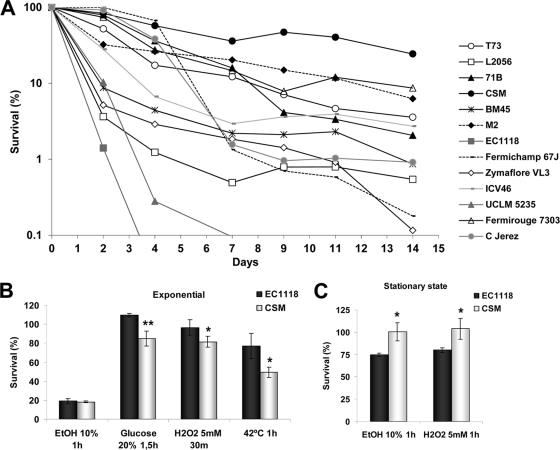
In order to gain a better understanding of the genetic determinants of longevity in industrial yeasts, we first compared a set of industrial strains from different origins (see Table S1 in the supplemental material) under standard laboratory conditions (22) to identify the long- and short-lived strains to be used as aging models. To go about this, a preculture was grown overnight in YPD medium and inoculated into SC medium to an OD600 of 0.1. After 3 days (time zero), aliquots were taken, diluted, and plated to determine cell viability. The time zero number of CFU/ml was taken as 100% survival (Fig. 1A), this being a commonly used time point in life span experiments (22). On day 3, all of the strains had stopped growing and their total cell numbers were similar, ranging from 76 ×106 to 156 ×106 CFU/ml (see Fig. S1a in the supplemental material). In general, we observed a wide variety of death profiles. The viability of a group of strains (EC1118, UCLM 5235, L2056, Zymaflore, and BM45) rapidly declined. EC1118 and UCLM5235 presented the shortest life span and a constant decrease in longevity. The other three strains displayed a different death profile with an extended long-term life span after an initial sudden loss of viability. Of the long-lived strains, Fermichamp exhibited high viability after the first 5 days, after which it fell sharply, while CSM presented the most remarkable long-term life span. We included a strain devoted to the biological aging of Sherry wines (C Jerez). Despite its highly different physiology, it exhibited an intermediate life span. Therefore, we chose a short-lived strain (EC1118) and a long-lived one (CSM) for comparison at several levels in the following experiments. Upon plotting of the day that cells reached a viability of 50% in relation to oxidative stress resistance (measured as the inhibition diameter caused by H2O2), we found a correlation between the two parameters (see Fig. S1b in the supplemental material), albeit a poor one. This indicates that oxidative stress tolerance is an important factor in survival but not the only one. To understand the differences between our model strains, we first tested their stress tolerance under standard laboratory conditions, i.e., applying stress conditions to those cells growing exponentially in rich YPD medium (Fig. 1B). Under these conditions, we found no difference in sensitivity to ethanol, but CSM was more sensitive to hyperosmotic stress (produced by a large amount of glucose), oxidative stress (caused by hydrogen peroxide), and heat shock stress (42°C). Unexpectedly, the long-lived strain had a lower basal stress response than the short-lived one. Next we tested the stresses that could prove relevant at the end of fermentation, such as high ethanol and oxidative stress, in stationary-phase cells grown in YPD medium for 2 days (Fig. 1C). We found that CSM was more tolerant and matched the generally accepted role of stress tolerance as a positive factor in the life span.
Fig 1.
Comparative CLSs and stress responses of strains. (A) CLS analysis of industrial yeast strains in SC medium. The assays were performed by making serial dilutions, plating an aliquot, and counting colonies. Counts of CFU/ml were done in relation to the values obtained from day 3 postinoculation and were considered 100% survival. The stress tolerance of strains EC1118 and CSM grown exponentially (B) or to the stationary phase (C) in YPD medium was also determined. Stress was applied, and serial dilutions were made and plated. Cell viability values were determined relative to viability before the stress conditions. Experiments were done in triplicate, and means and standard deviations are shown. *, P < 0.05; **, P < 0.01 (unpaired, two-tailed t test). EtOH, ethanol.
Differential behavior of strains EC1118 and CSM during winemaking.
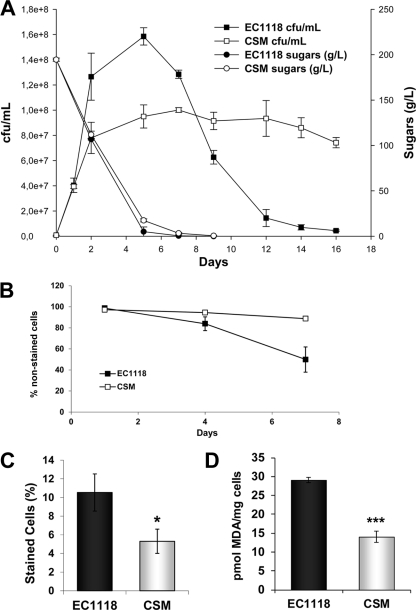
We compared the behavior of strains EC1118 and CSM under winemaking conditions during red grape juice fermentation (Fig. 2A). The two strains displayed very different cell viability patterns. EC1118 grew faster and reached a higher cell density but also went through a fast cell death phase, which started when the sugars were about to be depleted. CSM showed a totally different pattern: it reached a lower cell density, but cells remained viable for a longer time. Therefore, during winemaking, the CLS correlated with the results observed under laboratory conditions. Regarding glucose consumption, EC1118 underwent a faster pace of metabolism, although both strains finished vinification to dryness efficiently and produced similar final amounts of ethanol (data not shown). We repeated the experiment with synthetic grape juice and obtained similar results (see Fig. S2 in the supplemental material); in this case, however, the growth and glucose metabolism rates were lower, with greater differences between the two strains. The colony-counting method detected cells which were able to reenter a proliferative state. To determine whether there was also a difference in cell integrity, we stained cells with propidium iodide, which is taken up only by damaged cells (15). We compared EC1118 and CSM after glucose consumption in grape juice fermentation (taken as time zero). We found that the damaged cells of strain EC1118 also quickly increased compared to those of CSM (Fig. 2B), which correlates with the viable cell count. Hence, at least partial lysis took place during the aging process under our winemaking conditions. The amount of ethanol formed during this process did not change and was similar between the strains (data not shown); thus, death is dependent on neither different production nor ethanol consumption.
Fig 2.
Behavior of strains EC1118 and CSM during grape juice fermentation. (A) Natural grape juice fermentation. Cell viability, measured as CFU/ml, and total sugars over time are indicated. (B) Cell integrity measured by propidium iodide staining after grape juice fermentation had finished. Percentages of unstained cell counts under a fluorescence microscope are shown. (C) ROS production. Cells containing ROS were detected on day 13 of fermentation by the fluorescent probe dihydrorhodamine 123, and the percentage of stained cells is shown. (D) Lipid peroxidation measured as picomoles of malondialdehyde per milligram of cells 10 days after glucose consumption. Experiments were done in triplicate, and means and standard deviations are shown. *, P < 0.05; ***, P < 0.005 (unpaired, two-tailed t test).
Oxidative stress is a key factor in cell aging. To determine potential differences in oxidative damage in our model strains, we measured ROS at the end (13 days) of fermentation in MS300 synthetic must (Fig. 2C). We used this medium to avoid interference of florescence with red grape juice components. We observed a significantly higher incidence of ROS in EC1118. As a marker of oxidative damage, we measured lipid peroxidation 10 days after fermentation completion (Fig. 2D). Once again, EC1118 accumulated greater oxidative damage than the CSM strain. Therefore, EC1118 seemed less prepared to detoxify ROS under winemaking conditions, leading to increased damage, which may contribute to loss of viability. This correlated with a higher sensitivity to H2O2, as seen under laboratory conditions (Fig. 1C).
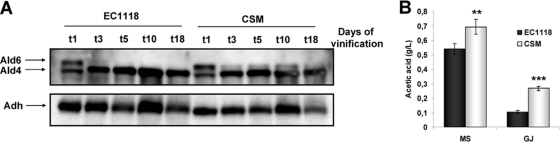
A well-known mechanism required for long-term survival is autophagy. A marker of nitrogen deficiency-induced autophagy is cytosolic ALDH Ald6p (36). Using an anti-ALDH antibody, we detected Ald6p and mitochondrial Ald4p (used as an internal loading control under laboratory conditions) during fermentation in synthetic grape juice (Fig. 3A). Ald4p remained relatively stable throughout fermentation and showed similar patterns between strains. Surprisingly, Ald6p disappeared quickly in strain EC1118, while CSM showed a lower degree of autophagy, as the Ald6p band was visible in later vinification stages. According to the yeast producer, EC1118 has a low nitrogen requirement while CSM has a high nitrogen requirement (http://www.lallemandwine.us/products/yeast_chart.php). EC1118 seemed to have better protein turnover, which may lead to enhanced nitrogen supply and therefore to growth at the expense of long-term survival. Despite its good autophagic mechanism, EC1118 reacted to the nitrogen levels in the medium, and nitrogen starvation extended the CLS (see Fig. S3 in the supplemental material), as previously observed with another commercial strain, L2056 (H. Orozco, E. Matallana, and A. Aranda, submitted for publication).
Fig 3.
EC1118 and CSM have different autophagy and acetic acid production levels. (A) Western blotting of ALDH (Ald) during synthetic grape juice fermentation by both strains. Alcohol dehydrogenase (Adh) was used as a loading control. (B) Acetic acid production after fermentation of synthetic grape juice (MS) and natural grape juice (GJ) by both strains. Experiments were done in triplicate, and the mean and standard deviation are shown. **, P < 0.01; ***, P < 0.005 (unpaired, two-tailed t test).
As Ald6p is not only the main cytosolic ALDH but also the main enzyme involved in acetate production during winemaking (38), we measured the amount of acetic acid present at the end of fermentation (when the level of sugars was below 2 g/liter) carried out with synthetic grape juice (the conditions described for Fig. 3A) and with natural grape juice (as described for Fig. 2A). In all cases, we found that acetic acid reached higher levels in strain CSM than in strain EC1118. Therefore, acetic acid production during fermentation may be modulated by the autophagic degradation of Ald6p. It is worth mentioning that acetic acid under winemaking conditions is not a key player in the life span (no correlation was found between higher acetic acid accumulation and a shorter CLS). This is possibly because of the highly buffered nature of grape juice due to the presence of a large amount of tartaric, malic, and citric acids, which allows the pH to remain stable throughout fermentation.
Comparative global analysis of strains EC1118 and CSM.
In order to find genetic determinants of the life span under winemaking conditions, we compared the transcriptomes of our reference strains in the stationary phase of fermentation on day 6 of growth in synthetic grape juice (see Fig. S2 in the supplemental material). Thirty-five genes were upregulated by a factor of 2 or more in CSM compared to those of EC1118. The functional categories of both aryl alcohol dehydrogenase (AAD genes) and the thiamine biosynthetic process (THI genes) were enriched (Table 1). Thiamine is a cofactor of several enzymes, like pyruvate dehydrogenase and decarboxylase, involved in carbon metabolism. This could potentially confer a selective advantage on CSM in late fermentation stages. THI and AAD are families of highly homologous and subtelomerically located genes (16). We measured the telomere lengths of both strains during fermentation (see Fig. S4 in the supplemental material) and found that CSM had slightly shorter telomeres than EC1118, which may cause a different chromatin structure, leading to higher expression of these genes. As previously shown with laboratory strains, telomere length did not correlate with the life span (4), as the strain with the longest telomeres (EC1118) showed a shorter life span. Genes with oxidoreductase activity were relatively abundant, including the genes in the ergosterol biosynthesis pathways (ERG5 and ERG11), which may confer better tolerance to oxidative stress on strain CSM (31). Two homologous genes, RDL1 and RDL2, were among the genes that do not belong to any functional category, which could be relevant in antioxidant protection, given their mitochondrial location (26); we studied them as described in the next section.
Table 1.
Comparative microarrays analysis of the gene expression between strains CSM and EC1118
| Categorya | P value | Genes upregulated in strain CSM (fold induction) |
|---|---|---|
| A | ||
| Thiamine biosynthetic process | 1.04e−6 | THI12 (10.88), THI11 (8.44), THI13 (6.71), THI5 (2.65) |
| Aryl alcohol dehydrogenase | 5.19e−13 | AAD15 (3.42), AAD10 (2.86), AAD6 (2.30), AAD14 (2.24), AAD4 (2.05), AAD3 (2.04) |
| Oxidoreductase activity | 4.82e−10 | ERG11 (2.70), ERG5 (2.66), FRE7 (2.28), MET12 (2.22), GTT1 (2.15), COX8 (2.05) |
| Others | HPA2 (4.16), RDL1 (3.92), SPG4 (3.55), RIM4 (2.97), YOR348c (2.94), RDL2 (2.82) | |
| B | ||
| Glucose metabolic process | 4.1e−12 | ENO2 (4.55), TDH3 (3.65), PFK1 (3.38), ENO1 (2.98), TDH2 (2.91), PGK1 (2.74), TPI1 (2.54) |
| Amino acid metabolic process | 8e−14 | ADH3 (4.50), ARG4 (4.09), ADH5 (4.06), MET16 (3.86), MET17 (3.71), SER33 (3.37), CYS3 (3.35), CPA1 (3.31), BAT1 (3.04), MET22 (2.92), ARO3 (2.59), MET14 (2.55) |
| Others | YIL060w (3.74), YCT1 (3.43), SPT4 (3.00), PMA1 (2.88), YCR023c (2.79), POL4 (2.63), HXT3 (2.62), JLP1 (2.60), ROG3 (2.56), PDR12 (2.54), CYR1 (2.49), HRK1 (2.37) |
A, functional categories overrepresented in the genes upregulated by a factor of 2 or more in strain CSM; B, functional categories overrepresented in the genes upregulated by a factor of 2 or more in strain EC1118. The induction level of each gene is shown in parentheses. In both categories, all of the genes upregulated more than 2.5-fold have been included.
Regarding the relatively upregulated genes in strain EC1118, one remarkable finding was that the abundance of the functional categories related to metabolism in the 87 genes that were induced by 2-fold or more (Table 1, category B). Enrichment of glycolysis and amino acid (particularly sulfur amino acid) metabolism genes was noted. This stronger metabolism could explain why this strain had faster growth and a higher glucose consumption rate than CSM. The other genes found may also influence EC1118 metabolism; for instance, the plasma membrane proton pump gene PMA1 was induced, together with the two protein kinase genes HRK1 (2.37-fold) and PTK2 (1.65-fold), which stimulate activity (27). This may lead to the use of ATP, produced during enhanced glycolysis, to create a higher membrane potential, which would facilitate transport and therefore growth. The relative upregulation of the adenylate cyclase gene CYR1 (2.45-fold) would activate PKA, thus stimulating growth and reducing the life span (23), which will be further studied later.
Rhodanese-like protein Rdl2p and adenylate cyclase are relevant for the yeast life span under winemaking conditions.
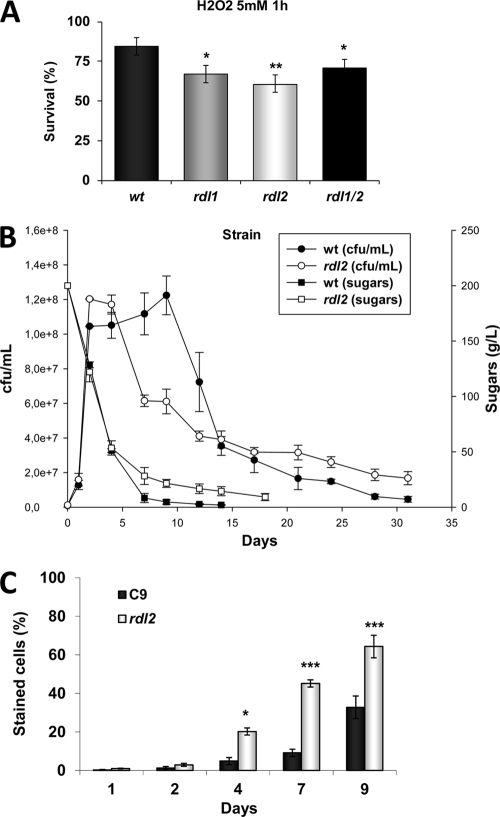
In strain CSM, two genes for rhodanese-like proteins (RDL1/2) were upregulated (Table 1, category A). Rhodanese enzymes are able to catalyze the transfer of sulfane sulfur from thiosulfate to cyanide (26). Rhodanese has been shown to play a role in antioxidant defense in bacteria (13) and mammals (34) and to be age related in humans (48). To test their role in oxidative stress in wine yeast, we deleted RDL1, RDL2, or both genes from haploid wine strain C9 derived from strain L2056 (47). We tested the viability of stationary-phase cultures in YPD medium after H2O2 shock (Fig. 4A). We saw increased sensitivity in both single mutants, although the rdl2Δ mutant was affected more. The double mutant was affected no more than the single mutants, suggesting that both proteins could act in the same protection pathway. To investigate the role of these proteins, we chose the rdl2Δ mutant, the most stress-sensitive one, to carry out fermentation of synthetic grape juice (which enabled us to measure ROS). We saw that the mutant strain grew slightly more than the parental one and started to metabolize sugars faster. However, its viability declined sharply in later fermentation stages, which is probably the cause of the lower sugar assimilation rate noted at this point (Fig. 4B). Note that viability dropped at around day 6 of fermentation in synthetic grape juice, which corresponds to the point at which the RDL2 gene was more highly expressed in strain CSM than in strain EC1118, suggesting that this protein is relevant for maintaining viability at this fermentation stage. However, it displayed an extended life span once fermentation finished, indicating a physiological change upon saturation, after which the Rdl2p function becomes important. Another change occurs once glucose is consumed, after which point Rdl2p seems to play a negative role. We measured ROS during fermentation and observed that oxidatively stressed cells progressively increased in the parental strain and vastly increased in the rdl2Δ mutant strain at the point where viability dropped (Fig. 4C). Therefore, Rdl2p plays a role in diminishing oxidative damage at the end of fermentation when mitochondria seem to play a role despite the still fermentative metabolism.
Fig 4.
Rhodanese Rdl2p plays a role in oxidative stress response, ROS levels, and life span. (A) Viability after oxidative stress caused by H2O2 of stationary-phase cultures of rdl1Δ, rdl2Δ and rdl1Δrdl2Δ mutants and their parental strain C9 in YPD medium. (B) Synthetic grape juice fermentation by the rdl2Δ mutant and C9. Cell viability, measured as CFU/ml, and total sugars are indicated over time. (C) ROS production measured during fermentation described in panel B. Experiments were done in triplicate, and means and standard deviations are provided. *, P < 0.05; **, P < 0.01; ***, P < 0.005 (unpaired, two-tailed t test). wt, wild type.
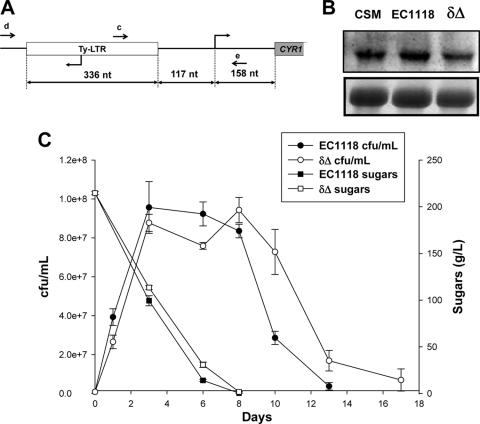
As mentioned in the previous section, the CYR1 gene, which codes for adenylate cyclase, was upregulated in strain EC1118. cAMP overproduction, which would cause PKA induction, may explain its shorter CLS. While the EC1118 genome was sequenced (35), we searched for distinct properties of this gene. In the promoter, 117 bp upstream from the transcription start site (32), we found a 336-bp δ element, a long terminal repeat (LTR) which marked an ancient transposition event by a Ty2 transposon (Fig. 5A). This element was not present in the S288c laboratory strain background, which was used in the initial genome sequencing project. By PCR, we tested the presence of this δ element in the industrial strains described in Fig. 1 using oligonucleotides c and e and by also checking the original allele by employing oligonucleotides d and e (Fig. 5A). Most of them presented a canonical sequence, but strains UCLM 5235 and ICV46 and sherry strain C (depicted in gray in Fig. 1) also had the δ element inserted into the CYR1 promoter. UCLM5235 had the second shortest CLS, so this element could be common in strains with an extremely short life span. This element contained a promoter, but its transcription ran in the direction opposite to that of CYR1. Thus, the higher CYR1 expression could be due to changes in chromatin structure or to the elimination or distancing of a repressor element. To gain a better understanding of its function in, and its influence on, the EC1118 life span, we deleted the two copies of the δ element using a recyclable kanMX cassette (28). This would replace the LTR with a shorter 71-bp loxP element. We measured the CYR1 mRNA of this mutant and its parental strain together with that of strain CSM (Fig. 5B). We found higher levels in strain EC1118 than in CSM, which was expected, given the array data. The δ element deletion lowered the levels of this transcript, suggesting that the shorter length of loxP at least partially restores the canonical arrangement or the chromatin structure of the CYR1 promoter. We carried out vinifications of grape juice with EC1118 and the EC1118 Δδ mutant to find that the mutant strain grew at a slightly lower rate, although it reached a similar cellular density to start a less pronounced death phase (Fig. 5C). Regarding glucose consumption, the mutant strain displayed a lower metabolic rate, although it finished fermentation efficiently. All of these aging and metabolism results fit with decreased PKA activity in the Δδ mutant, given its lower CYR1 expression.
Fig 5.
A δ element in the CYR1 promoter affects longevity. (A) Scheme of the LTR present in the CYR1 promoter of some industrial yeast strains. The oligonucleotides used to detect it by PCR are depicted. c, d, and e are oligonucleotides. nt, nucleotides. (B) Northern blot analysis of CYR1 gene expression in strains CSM and EC1118 and a mutant with both copies of the δ element replaced with a loxP site, EC1118 Δδ, at 6 days of fermentation in MS300 synthetic grape juice. rRNA was used as a loading control. (C) Natural grape juice fermentation by strain EC1118 and mutant strain EC1118 Δδ. Cell viability, measured as CFU/ml, and total sugars are indicated over time.
DISCUSSION
This study investigated determinants of wine yeast longevity in the stationary phase. Our approach involved the screening of a collection of industrial wine yeasts, most of which are commercial, with a view to identifying strains with extreme life spans under standard laboratory conditions of incubation in SC medium (Fig. 1A). We identified EC1118, a widely used commercial strain, as our short-lived strain model. On the other hand, CSM exhibited an extremely long life span. This behavior noted in the laboratory medium correlated with the behavior observed during natural grape juice fermentation. As expected, we observed an intermediate behavior of other strains with an intermediate CLS, such as L2056 or T73, during winemaking (data not shown). The life span was measured by the ability to reenter the cell cycle. However, we noticed that cell death during winemaking is linked to increased cell lysis, indicating that cellular content was leaked into the medium (Fig. 2B), and that may potentially affect the growth of malolactic bacteria or spoilage microorganisms. Therefore, the CLS studies done under laboratory conditions are a good approach to studying aging in a large number of strains. The comparison of these two strains indicates that oxidative stress resistance and metabolism control are factors which contribute to the CLS under winemaking conditions.
As in laboratory strains, stress tolerance is a key factor in aging (21). Strain CSM has increased tolerance to stress, but only under stationary-phase conditions (Fig. 1B), while the basal stress tolerance of EC1118 in the exponential phase is greater. The global stress response has been shown to change during the growth phase in a strain-dependent manner (42). CSM's greater tolerance to ethanol in the stationary phase may prove positive for survival after finalizing fermentation. It is worth mentioning that the two strains produce similar final amounts of alcohol. Under laboratory conditions, strain CSM shows enhanced tolerance to oxidative insult by hydrogen peroxide (Fig. 1B). During winemaking, we found that ROS production and the concomitant damage measured as lipid peroxidation correlate with EC1118's lower oxidative stress tolerance and shorter life span (Fig. 2). However, such greater ROS production may be caused by the larger number of dying cells. At this point, we are unable to state whether ROS production is the cause or the result of cell death. This differential stress tolerance does not seem to be based on transcriptional regulation, as no significant up- or downregulation of the oxidative stress genes was noted when the transcriptomes of the two strains were compared (Table 1). Nevertheless, the upregulation of the two genes (RDL1 and RDL2) related to the mitochondria in strain CSM was striking. Both proteins belong to a family of enzymes called rhodaneses that are able to transfer sulfur from thiosulfate into cyanide (26). There is no evidence that cyanide detoxification is relevant during vinification, but we believe that these proteins are probably involved in other processes. Deletion of both genes in a wine yeast strain, particularly RDL2, produces enhanced sensitivity to oxidative stress (Fig. 2). The short-term viability of mutant strain rdl2Δ diminishes during winemaking, although the long-term CLS is extended. In the last fermentation stages, we see increased ROS production in the mutant, indicating defective free radical metabolism or increased free radical production. This increase in oxidative damage may be behind the drop in the mutant strain's viability, but surviving cells may activate the antioxidant mechanism to cause elongation of their long-term life span. Rhodanese RhdA from Acetobacter is required for an oxidative stress response and has been linked to the repair of Fe-S clusters of certain proteins. An isoform of mitochondrial bovine rhodanese is able to oxidize thioredoxin by acting as a detoxification mechanism of the intramitochondrial reactive species (34).
The life span is closely linked to metabolic activity. EC1118 is a more vigorous strain that grows and consumes glucose faster than CSM, which may be due to the relative upregulation of the genes involved in carbon (particularly glycolysis) and nitrogen metabolism (Table 1). This higher metabolism rate can cause imbalances, leading to greater ROS production by this strain. This difference in the metabolism rate may be related, at least partially, to the higher expression of the adenylate cyclase gene CYR1 (Table 1). Higher cAMP levels possibly lead to the activation of PKA, an inducer of a transcriptional program linked to growth and cell division which also stimulates glycolysis (44). High PKA activity has been linked to necrosis in mammals, and deletion of the PKA regulatory subunit BCY1 induces autolytic phenotypes in yeast under sparkling wine second fermentation conditions (46). We found that this difference may be caused by the presence of an ancient retrotransposition event in some wine yeasts which leaves a δ element in the promoter of CYR1 (Fig. 5). Replacing it with a shorter loxP sequence extends the life span but lowers the glucose metabolism rate. Therefore, this allele might be a good marker to identify those strains displaying rapid growth and sugar consumption with a potentially shorter life span and increased cell lysis.
This work also identifies another molecular difference between our model strains. EC1118 displays a higher degradation of autophagy marker Ald6p (Fig. 3). Cytosolic ALDH is degraded when nitrogen is low, but only by autophagy (36). This higher protein turnover rate may be why its nitrogen requirements are lower than those of CSM. Ald6p degradation is also a way of controlling acetic acid production, whose impact on the organoleptic quality of wine is negative. Studying autophagy with simpler markers, such as alkaline phosphatase, could be a good way to select yeast with low nitrogen requirements and potentially low acetic acid production. Autophagy usually tends to be required to achieve a full life span (29) but could prove detrimental to cells under certain conditions. For instance, the csc1-1 allele, which causes a constitutive autophagy phenotype, displays accelerated autolysis (11). Therefore, balanced autophagy is vital for an optimal life span. Autophagy may degrade a protein or an organelle, which may prove useful for survival under winemaking conditions. Autophagy is repressed by the PKA and TOR nutrient-sensing pathways (45, 46). In this case, EC1118 shows increased autophagy and Cyr1p; thus, PKA does not seem to be a determinant in wine strains under grape juice fermentation conditions. This may relate to the facts that PKA responds to carbon sources and that grape juice fermentation takes place in an environment where glucose is not limiting. Nitrogen is the limiting nutrient during vinification, and TOR responds to nitrogen levels (24); hence, this pathway is likely the key factor in controlling autophagy under such conditions. Therefore, finding inhibitors of TOR could be a good way of increasing autophagy and of obtaining better adaptation to low-nitrogen fermentation with potentially lower acetate production due to Al6p degradation. Finally, the food additive caffeine, which is generally regarded as safe, is able to inhibit TOR and has been proposed to be a good candidate for this very purpose (1).
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by grants from the Spanish Ministry of Science (AGL2008-00060) and CSIC (PIE 200970I028). H.O. is supported by an F.P.I. fellowship.
Footnotes
Published ahead of print 10 February 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Abeliovich H, Gonzalez R. 2009. Autophagy in food biotechnology. Autophagy 5:925–929 [DOI] [PubMed] [Google Scholar]
- 2. Adams A, Gottschling DE, Kaiser C, Stearns T. 1998. Methods in yeast genetics: a laboratory course manual (1997 edition). Cold Spring Harbor Laboratory Press, Plainview, NY [Google Scholar]
- 3. Alexandre H, Costello PJ, Remize F, Guzzo J, Guilloux-Benatier M. 2004. Saccharomyces cerevisiae-Oenococcus oeni interactions in wine: current knowledge and perspectives. Int. J. Food Microbiol. 93:141–154 [DOI] [PubMed] [Google Scholar]
- 4. Ashrafi K, Sinclair D, Gordon JI, Guarente L. 1999. Passage through stationary phase advances replicative aging in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 96:9100–9105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Balaban RS, Nemoto S, Finkel T. 2005. Mitochondria, oxidants, and aging. Cell 120:483–495 [DOI] [PubMed] [Google Scholar]
- 6. Boulton RB. 1996. Principles and practices of winemaking. Chapman & Hall, New York, NY [Google Scholar]
- 7. Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 8. Burtner CR, Murakami CJ, Kennedy BK, Kaeberlein M. 2009. A molecular mechanism of chronological aging in yeast. Cell Cycle 8:1256–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cardona F, Carrasco P, Perez-Ortin JE, del Olmo M, Aranda A. 2007. A novel approach for the improvement of stress resistance in wine yeasts. Int. J. Food Microbiol. 114:83–91 [DOI] [PubMed] [Google Scholar]
- 10. Carrasco P, Querol A, del Olmo M. 2001. Analysis of the stress resistance of commercial wine yeast strains. Arch. Microbiol. 175:450–457 [DOI] [PubMed] [Google Scholar]
- 11. Cebollero E, Martinez-Rodriguez A, Carrascosa AV, Gonzalez R. 2005. Overexpression of csc1-1. A plausible strategy to obtain wine yeast strains undergoing accelerated autolysis. FEMS Microbiol. Lett. 246:1–9 [DOI] [PubMed] [Google Scholar]
- 12. Cebollero E, Rejas MT, Gonzalez R. 2008. Autophagy in wine making. Methods Enzymol. 451:163–175 [DOI] [PubMed] [Google Scholar]
- 13. Cereda A, Carpen A, Picariello G, Tedeschi G, Pagani S. 2009. The lack of rhodanese RhdA affects the sensitivity of Azotobacter vinelandii to oxidative events. Biochem. J. 418:135–143 [DOI] [PubMed] [Google Scholar]
- 14. D'Amore T, Panchal CJ, Russell I, Stewart GG. 1990. A study of ethanol tolerance in yeast. Crit. Rev. Biotechnol. 9:287–304 [DOI] [PubMed] [Google Scholar]
- 15. Deere D, et al. 1998. Flow cytometry and cell sorting for yeast viability assessment and cell selection. Yeast 14:147–160 [DOI] [PubMed] [Google Scholar]
- 16. Delneri D, Gardner DC, Oliver SG. 1999. Analysis of the seven-member AAD gene set demonstrates that genetic redundancy in yeast may be more apparent than real. Genetics 153:1591–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Delneri D, et al. 2000. Exploring redundancy in the yeast genome: an improved strategy for use of the cre-loxP system. Gene 252:127–135 [DOI] [PubMed] [Google Scholar]
- 18. Espindola S, Gomes DS, Panek AD, Eleutherio EC. 2003. The role of glutathione in yeast dehydration tolerance. Cryobiology 47:236–241 [DOI] [PubMed] [Google Scholar]
- 19. Fabrizio P, et al. 2004. Superoxide is a mediator of an altruistic aging program in Saccharomyces cerevisiae. J. Cell Biol. 166:1055–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fabrizio P, et al. 2005. Sir2 blocks extreme life-span extension. Cell 123:655–667 [DOI] [PubMed] [Google Scholar]
- 21. Fabrizio P, et al. 2003. SOD2 functions downstream of Sch9 to extend longevity in yeast. Genetics 163:35–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fabrizio P, Longo VD. 2003. The chronological life span of Saccharomyces cerevisiae. Aging Cell 2:73–81 [DOI] [PubMed] [Google Scholar]
- 23. Fabrizio P, Pozza F, Pletcher SD, Gendron CM, Longo VD. 2001. Regulation of longevity and stress resistance by Sch9 in yeast. Science 292:288–290 [DOI] [PubMed] [Google Scholar]
- 24. Fontana L, Partridge L, Longo VD. 2010. Extending healthy life span—from yeast to humans. Science 328:321–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fornairon-Bonnefond C, Salmon JM. 2003. Impact of oxygen consumption by yeast lees on the autolysis phenomenon during simulation of wine aging on lees. J. Agric. Food Chem. 51:2584–2590 [DOI] [PubMed] [Google Scholar]
- 26. Foster MW, Forrester MT, Stamler JS. 2009. A protein microarray-based analysis of S-nitrosylation. Proc. Natl. Acad. Sci. U. S. A. 106:18948–18953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Goossens A, de La Fuente N, Forment J, Serrano R, Portillo F. 2000. Regulation of yeast H(+)-ATPase by protein kinases belonging to a family dedicated to activation of plasma membrane transporters. Mol. Cell. Biol. 20:7654–7661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Güldener U, Heck S, Fielder T, Beinhauer J, Hegemann JH. 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24:2519–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kaeberlein M. 2010. Lessons on longevity from budding yeast. Nature 464:513–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Landolfo S, Politi H, Angelozzi D, Mannazzu I. 2008. ROS accumulation and oxidative damage to cell structures in Saccharomyces cerevisiae wine strains during fermentation of high-sugar-containing medium. Biochim. Biophys. Acta 1780:892–898 [DOI] [PubMed] [Google Scholar]
- 31. Landolfo S, et al. 2010. Oleic acid and ergosterol supplementation mitigates oxidative stress in wine strains of Saccharomyces cerevisiae. Int. J. Food Microbiol. 141:229–235 [DOI] [PubMed] [Google Scholar]
- 32. Nagalakshmi U, et al. 2008. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320:1344–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. 2009. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat. Rev. Mol. Cell Biol. 10:458–467 [DOI] [PubMed] [Google Scholar]
- 34. Nandi DL, Horowitz PM, Westley J. 2000. Rhodanese as a thioredoxin oxidase. Int. J. Biochem. Cell Biol. 32:465–473 [DOI] [PubMed] [Google Scholar]
- 35. Novo M, et al. 2009. Eukaryote-to-eukaryote gene transfer events revealed by the genome sequence of the wine yeast Saccharomyces cerevisiae EC1118. Proc. Natl. Acad. Sci. U. S. A. 106:16333–16338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Onodera J, Ohsumi Y. 2004. Ald6p is a preferred target for autophagy in yeast, Saccharomyces cerevisiae. J. Biol. Chem. 279:16071–16076 [DOI] [PubMed] [Google Scholar]
- 37. Qin H, Lu M. 2006. Natural variation in replicative and chronological life spans of Saccharomyces cerevisiae. Exp. Gerontol. 41:448–456 [DOI] [PubMed] [Google Scholar]
- 38. Remize F, Andrieu E, Dequin S. 2000. Engineering of the pyruvate dehydrogenase bypass in Saccharomyces cerevisiae: role of the cytosolic Mg(2+) and mitochondrial K(+) acetaldehyde dehydrogenases Ald6p and Ald4p in acetate formation during alcoholic fermentation. Appl. Environ. Microbiol. 66:3151–3159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ribe'reau-Gayon P, Dubourdieu D, Done'che B. 2006. Handbook of enology, 2nd ed John Wiley, Hoboken, NJ [Google Scholar]
- 40. Riou C, Nicaud JM, Barre P, Gaillardin C. 1997. Stationary-phase gene expression in Saccharomyces cerevisiae during wine fermentation. Yeast 13:903–915 [DOI] [PubMed] [Google Scholar]
- 41. Robyt JF, Whelan WJ. 1972. Reducing value methods for maltodextrins. I. Chain-length dependence of alkaline 3,5-dinitrosalicylate and chain-length independence of alkaline copper. Anal. Biochem. 45:510–516 [DOI] [PubMed] [Google Scholar]
- 42. Rossouw D, Olivares-Hernandes R, Nielsen J, Bauer FF. 2009. Comparative transcriptomic approach to investigate differences in wine yeast physiology and metabolism during fermentation. Appl. Environ. Microbiol. 75:6600–6612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 44. Smets B, et al. 2010. Life in the midst of scarcity: adaptations to nutrient availability in Saccharomyces cerevisiae. Curr. Genet. 56:1–32 [DOI] [PubMed] [Google Scholar]
- 45. Stephan JS, Yeh YY, Ramachandran V, Deminoff SJ, Herman PK. 2009. The Tor and PKA signaling pathways independently target the Atg1/Atg13 protein kinase complex to control autophagy. Proc. Natl. Acad. Sci. U. S. A. 106:17049–17054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tabera L, Munoz R, Gonzalez R. 2006. Deletion of BCY1 from the Saccharomyces cerevisiae genome is semidominant and induces autolytic phenotypes suitable for improvement of sparkling wines. Appl. Environ. Microbiol. 72:2351–2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Walker ME, et al. 2003. Application of the reuseable, KanMX selectable marker to industrial yeast: construction and evaluation of heterothallic wine strains of Saccharomyces cerevisiae, possessing minimal foreign DNA sequences. FEMS Yeast Res. 4:339–347 [DOI] [PubMed] [Google Scholar]
- 48. Yi H, et al. 2010. Identification of Rack1, EF-Tu and Rhodanese as aging-related proteins in human colonic epithelium by proteomic analysis. J. Proteome Res. 9:1416–1423 [DOI] [PubMed] [Google Scholar]
- 49. Zuzuarregui A, del Olmo M. 2004. Expression of stress response genes in wine strains with different fermentative behavior. FEMS Yeast Res. 4:699–710 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.