Abstract
The potential for sexual reproduction in Aspergillus oryzae was assessed by investigating the presence and functionality of MAT genes. Previous genome studies had identified a MAT1-1 gene in the reference strain RIB40. We now report the existence of a complementary MAT1-2 gene and the sequencing of an idiomorphic region from A. oryzae strain AO6. This allowed the development of a PCR diagnostic assay, which detected isolates of the MAT1-1 and MAT1-2 genotypes among 180 strains assayed, including industrial tane-koji isolates. Strains used for sake and miso production showed a near-1:1 ratio of the MAT1-1 and MAT1-2 mating types, whereas strains used for soy sauce production showed a significant bias toward the MAT1-2 mating type. MAT1-1 and MAT1-2 isogenic strains were then created by genetic manipulation of the resident idiomorph, and gene expression was compared by DNA microarray and quantitative real-time PCR (qRT-PCR) methodologies under conditions in which MAT genes were expressed. Thirty-three genes were found to be upregulated more than 10-fold in either the MAT1-1 host strain or the MAT1-2 gene replacement strain relative to each other, showing that both the MAT1-1 and MAT1-2 genes functionally regulate gene expression in A. oryzae in a mating type-dependent manner, the first such report for a supposedly asexual fungus. MAT1-1 expression specifically upregulated an α-pheromone precursor gene, but the functions of most of the genes affected were unknown. The results are consistent with a heterothallic breeding system in A. oryzae, and prospects for the discovery of a sexual cycle are discussed.
INTRODUCTION
The filamentous fungus Aspergillus oryzae is known as a koji mold and plays an important role in the Japanese food industry, where it has long been used for traditional fermentative food production, such as the manufacture of sake, soy sauce, and miso (33). More recently, the ability of the species to secrete large amounts of proteins has extended its industrial use to include heterologous protein production (28, 46, 63, 64). A. oryzae has no known sexual cycle and has traditionally been classified within the Fungi Imperfecti (the Deuteromycota), or “mitosporic” fungi (35). Therefore, it has not been possible to utilize the sexual cycle for strain improvement purposes, preventing, for example, the outcrossing of independent strains with industrially useful characteristics. However, sequencing of the A. oryzae genome has revealed the presence of a series of genes implicated in sexual reproduction, suggesting the possibility of cryptic sexuality in this supposedly asexual fungus (19, 39). Phylogenetic analysis also clearly demonstrates that A. oryzae, along with other asexual Aspergillus species, is a member of the fungal subphylum Pezizomycotina, which is defined by sexual reproduction involving the production of sexual ascospores within asci (15, 52).
Sexual reproduction in pezizomycete fungi typically occurs in two distinct manners. Homothallic fungi are self-fertile and can complete the sexual cycle without the need for a partner, whereas heterothallic fungi require a partner of a complementary mating type. Research over the past 20 years has revealed that the different forms of sexual breeding systems are determined principally by the presence within the genome of key “mating type” (MAT) genes (11). Heterothallic species contain a single MAT locus, and complementary mating types have different genes encoding members of the high-mobility-group (HMG) superfamily of proteins (41). MAT1-1 isolates characteristically contain a MAT1-1 gene encoding a protein with a MATα_HMG domain, whereas MAT1-2 isolates characteristically contain a MAT1-2 gene encoding a protein with a MATA_HMG domain (11, 41). In contrast, homothallic species typically contain MAT genes encoding both MATα_HMG and MATA_HMG domain proteins within the same genome (11, 50). These mating type proteins are thought to act as master regulatory transcription factors, controlling cell identity, for example, by regulating the expression of pheromone precursor and receptor genes (3, 4, 9, 11, 61). Mating type genes are also thought to regulate a wider series of genes together with later stages of sexual development, but only a limited number of investigations into such roles have been made for the Pezizomycotina, as reported by Pöggeler et al. (56), Klix et al. (34), and Bidard et al. (3).
With respect to the aspergilli, mating type genes have been identified from a series of both known sexual and asexual species of Aspergillus (19, 49, 50, 51, 58, 59). Within the aspergilli, MAT genes were first shown to be functional in the homothallic species Aspergillus nidulans, in which deletion of the resident MAT1 and MAT2 genes led to loss of sexual reproduction, while their overexpression suppressed vegetative growth and stimulated sexual differentiation under conditions normally unfavorable for sex (50). Subsequently, the MAT1-1 and MAT1-2 products of A. fumigatus were also shown to be functional by their ability, through heterologous complementation, to restore fertility to MAT deletant strains of A. nidulans, at a time when A. fumigatus was considered asexual (22, 57). Following the discovery of a sexual cycle in A. fumigatus (47), the MAT1-1 and MAT1-2 genes were shown to be required for sexual development in A. fumigatus, as evidenced by the sterility of MAT deletant strains (62). However, the genetic mechanisms by which mating type genes regulate sexual reproduction in the aspergilli still remain largely unknown, although a recent study has provided insight into the regulatory mechanisms controlling the expression of A. nidulans MAT2 (a synonym of matA) during the sexual cycle (10). In the aspergilli as a whole, more than 80 genes, including the MAT genes, have been shown to be involved in the regulation of sexual reproduction (12, 14). Significantly, the recovery of isolates of complementary MAT1-1 and MAT1-2 identities (containing MAT1-1 and MAT1-2 genes, respectively, in a heterothallic arrangement) for the species A. fumigatus, A. flavus, A. parasiticus, and A. nomius, previously considered “asexual,” has led to the discovery of functional sexual cycles in all of these species as a result of the setup of laboratory crosses between isolates of compatible mating types under appropriate conditions (15, 25, 26, 27, 47).
Regarding A. oryzae, the reference strain RIB40, used for genome sequencing, was found to contain a MAT1-1 α_HMG domain gene, i.e., to be a MAT1-1 strain (19). It was not determined, however, whether complementary MAT1-2 strains required for heterothallic mating might exist in nature. In this study, we first identified a MAT1-2 strain of A. oryzae containing the complementary MAT1-2 HMG gene at the MAT locus. Using a PCR diagnostic assay, we then demonstrated the existence of both MAT1-1 and MAT1-2 genotypes among a variety of reference and industrial strains used in the manufacture of sake, soy sauce, and miso. Finally, to explore the possible functionality of these mating type genes, we constructed a MAT replacement strain in which the original MAT1-1 gene was replaced by the MAT1-2 gene. This enabled us to use DNA microarray methodology to analyze possible differential expression by genes of opposite mating types within the same genetic background—the first such analysis of MAT gene expression in a supposedly asexual filamentous ascomycete fungus.
MATERIALS AND METHODS
Strains and media.
A. oryzae strains RIB40 (39) and AO6 (CBS108.24) (43) were used as DNA donors. Escherichia coli DH5α was used as the host for DNA manipulations. A. oryzae strain NSPlD1 (MAT1-1 niaD− sC− adeA− ΔargB::adeA− ΔligD::argB ΔpyrG::adeA) (42) was used as the host strain for replacement and disruption of MAT genes. For the initial mating type analysis of A. oryzae, the industrial strains AO1 (RIB128), RIB177, RIB430, RIB609, and RIB647 from the NRIB (National Research Institute of Brewing, Hiroshima, Japan) culture collection and strains IAM2609, IAM2735, IAM2959, and IAM2960 from the IAM culture collection (Japan Collection of Microorganisms, RIKEN BioResource Center, Wako, Japan) were used. In addition, previously described A. oryzae reference strains AO1 (ATCC 22788), AO2 (IAM 13883), AO3 (CBS 466.91), AO4 (ATCC 10196, ATCC 13791), AO5 (IFO 4278) (43), and AO8 from the University of Nottingham culture collection were used in initial MAT screening. In subsequent experiments, 164 further strains of tane-koji (koji mold starters; isolate codes starting with KBN) used in the manufacture of sake, soy sauce, and miso (Bio'c Co., Ltd., Toyohashi, Japan) were also analyzed. The strains studied are summarized in Tables S1 and S2 in the supplemental material. DPY medium (2% dextrin, 1% polypeptone, 0.5% yeast extract, 0.5% KH2PO4, and 0.05% MgSO4 · 7H2O), potato dextrose (PD) medium supplemented with 0.5% uridine, 0.2% uracil, and 1.5% agar, and Aspergillus complete medium (ACM) (49) were used to grow the A. oryzae strains. M medium [0.2% NH4Cl, 0.1% (NH4)2SO4, 0.05% KCl, 0.05% NaCl, 0.1% KH2PO4, 0.05% MgSO4 · 7H2O, 0.002% FeSO4 · 7H2O, and 2% glucose (pH 5.5)] supplemented with 0.5% uridine and 0.2% uracil was used as a selective medium in A. oryzae transformation (all percentages are weight per volume). DNA extraction from A. oryzae isolates and transformation of A. oryzae were achieved according to previously reported methods (33). The pyrG marker gene, used in the replacement and disruption of MAT genes, was excised using PD medium supplemented with 1 mg/ml 5-fluoroorotic acid (5-FOA), 0.5% uridine, 0.2% uracil, and 1.5% agar according to a method described previously (42).
Identification of the MAT1-2 locus.
A PCR fragment was amplified from genomic DNA of A. oryzae strain AO6 using the degenerate primer pair MAT5-4 and MAT3-2 according to the method of Seymour et al. (60). These primers were designed to amplify conserved HMG domain-encoding sequences within pezizomycete MAT1-2 genes (60). A gene walk toward both ends of the idiomorph was then initiated by using the Gene Walker kit from Stratagene (La Jolla, CA) according to the manufacturer's instructions. Gene-specific primer sequences are provided in Table S3 in the supplemental material. The gene walk reached the end of the idiomorph upstream of the MAT1-2 gene but not downstream. A fragment containing the MAT1-2 gene and its flanking sequences was then amplified from strain AO6 genomic DNA by PCR using Expand Long Template Taq DNA polymerase (Roche, Rotkreuz, Switzerland) and primers AOmat-2R and AOmat-2F, based on the conserved region sequences of DNA lyase (gene identification [ID] AO080527000086) and Aoend4 (gene ID AO080527000085) (http://nribf21.nrib.go.jp/CFGD/search.cgi?dspQO=1&prj=02201), respectively. The amplified fragment was sequenced to confirm its identity.
Southern blot analysis.
After electrophoresis, the digested genomic DNAs were transferred to Hybond N+ membranes (GE Healthcare, Piscataway, NJ). ECL (enhanced chemiluminescence) direct nucleic acid labeling and detection (GE Healthcare) and a LAS-4000miniEPUV luminescent image analyzer (Fuji Photo Film, Tokyo, Japan) were used for detection. Probes were generated using the A. oryzae strain RIB40 genome as a template, and the primers are listed in Table S3 in the supplemental material.
PCR analysis of mating type genes.
The mating type of isolates was determined by a diagnostic PCR using primers MAT1-F and MAT1-R for the MAT1-1 gene and primers MAT2-F and MAT2-R for the MAT1-2 gene (see Table S3 in the supplemental material). PCR conditions involved the use of 20 ng genomic DNA in a 20-μl reaction volume with 5.0 U Ex Taq polymerase (TaKaRa, Otsu, Japan), 0.5 μM primers, 0.2 mM deoxynucleoside triphosphate (dNTP) mixture, and cycle parameters of initial denaturation at 94°C for 5 min, followed by 25 cycles of 94°C for 30 s, 53°C for 30 s, and 72°C for 1 min before storage at 4°C (all at a ramp rate of 1.0 kb/min). MAT diagnostic PCR was performed using genomic DNA from 164 tane-koji strains as templates (see Table S2 in the supplemental material). These strains were distinguished by several morphological characteristics, such as levels of conidiation, pigmentation and color, and enzyme productivity (data not shown). Also, the length of aerial hyphae was assessed, since this can be an important consideration in choosing tane-koji strains for particular commercial processes. Given the number of strains involved, a 5-point scale based on visual observation was used. A score of 1 indicated an aerial hypha length of ∼0.5 mm; 2, 0.5 mm to 1.5 mm; 3, 1.5 mm to 2.5 mm; 4, 2.5 mm to 3.5 mm; 5, >3.5 mm (see Table S2). The two-tailed Mann-Whitney U test, available from the VassarStats website (http://faculty.vassar.edu/lowry/VassarStats.html), was used to compare possible differences in the length of aerial hyphae between test groups.
Construction of plasmids for MAT gene replacement and disruption.
All plasmids used for A. oryzae transformation in this study were constructed by the MultiSite Gateway cloning system (Invitrogen, Carlsbad, CA). All primers used for constructing the plasmids and PCR products described below are listed in Table S3 in the supplemental material. The 1.5-kb 3′ end of the DNA lyase gene in RIB40 was amplified by PCR using primers aB4+lyase-2.5kF and MAT2+lyase-R, while the MAT1-2 locus in AO6 was amplified using primers lyase+MAT2dF and aB1+MAT2uR. Those two DNA fragments were united by fusion PCR according to previously described methods (29, 65) using primers aB4+lyase-2.5kF and aB1+MAT2uR. The 0.3-kb 3′ end of the MAT1-2 locus in AO6 was amplified by PCR using primers aB2+MAT2uF and fla+MAT2uR, while a 0.2-kb downstream region flanking the Aoend4 (SLA2) gene in RIB40 was amplified by PCR using primers MAT2+fla-F and aB3+SLA-0.1kR. Those two DNA fragments were united by fusion PCR using primers aB2+MAT2uF and aB3+SLA-0.1kR. The former and latter fusion PCR products were cloned by a BP Clonase reaction into the pDONR P4-P1R and pDONR P2R-P3 entry vectors, respectively. The resultant 5′ and 3′ entry clones and the center entry clone, pgEpG (containing the pyrG marker gene) (42), were subjected to an LR Clonase reaction in the presence of pDEST R4-R3 (the destination vector) to produce plasmid pgpM2. The replacement fragment for the MAT1-1 gene was amplified by PCR using plasmid pgpM2 as a template and primers aB4+lyase-2.5kF and aB3+SLA-0.1kR. The 1.5-kb 5′ region upstream of the MAT1-1 gene was amplified by PCR using primers aB4-5MAT1-1_F and aB1r-5MAT1-1_R. The 0.3-kb 5′ upstream region of the MAT1-1 gene was amplified by PCR using primers aB2r-MAT1-1_F and f-MAT1-1+p_R, while the 1.5-kb 3′ downstream region of the MAT1-1 gene was amplified by PCR using primers f-MAT1-1+p_F and aB3-MAT1-1_R. Those two DNA fragments were united by fusion PCR using primers aB2r-MAT1-1_F and aB3-MAT1-1_R. The 1.5-kb 5′ upstream region of the MAT1-1 gene and the fusion PCR product were cloned by a BP Clonase reaction into the pDONR P4-P1R and pDONR P2R-P3 entry vectors, respectively. The resultant 5′ and 3′ entry clones and the center entry clone pgEpG were subjected to the LR Clonase reaction in the presence of pDEST R4-R3 to produce plasmid pgdM1pG. The disruption fragment for the MAT1-1 gene was amplified by PCR using plasmid pgdM1pG as a template and primers aB4-5MAT1-1_F and aB3-MAT1-1_R.
RNA isolation and qRT-PCR analysis.
Approximately 2 × 106 conidia were inoculated into 100 ml DPY liquid medium supplemented with 0.5% uridine and 0.2% uracil. After 24 h of growth, 200 mg of mycelia was shifted onto DPY agar medium covered with a cellulose acetate membrane (Membrane Filter; Advantec Toyo, Tokyo, Japan) and was further cultivated for 48 h. Total RNA was extracted as described previously (32), followed by treatment with DNase I (TaKaRa, Otsu, Japan). First-strand cDNAs were synthesized with ReverTra Ace reverse transcriptase (Toyobo, Osaka, Japan) using oligo(dT)12–18 (Invitrogen) as the primer. Quantitative real-time PCRs (qRT-PCRs) were performed using a LightCycler FastStart DNA Master SYBR green I system (Roche Diagnostics, Indianapolis, IN) as instructed by the manufacturer. The primers used in this study are listed in Table S3 in the supplemental material. Transcript levels were quantified by qRT-PCR, and the means from three independent experiments were taken. Transcript levels were normalized against actin as the internal standard.
DNA microarray analysis.
Total RNA was extracted from MAT1-1 (NSPlD1) and MAT1-2 (NSPlD-M2) strains as described above using the RNeasy Plant minikit (Qiagen Sciences, Germantown, MD) and was dissolved in RNase-free water. DNA microarray experiments were conducted according to the Affymetrix GeneChip manual with an A. oryzae DNAchip (Affymetrix, Madison, WI) containing probe sets for 13,857 genes and 1,704 expressed sequence tags (ESTs) from A. oryzae strain RIB40 (48). The gene IDs used were those registered in the Comparative Fungal Genome Database (http://nribf21.nrib.go.jp/CFGD/gnm.cgi?prj=02201&gnm=aor01). The ESTs selected for the DNA microarray do not overlap with the genes registered in the genome database (Aspergillus oryzae EST DataBase [http://nribf21.nrib.go.jp/EST2/index.html]). Briefly, one cycle of cDNA synthesis was performed, and then biotin-labeled cRNA was amplified, followed by fragmentation. The fragmented cRNA was hybridized with the 25-mer oligonucleotide probe sets, each containing 16 probe pairs consisting of a perfect-match probe (PM) and a mismatch probe (MM). After washing and staining, the probes were scanned with a GeneChip Scanner 3000 (Affymetrix). Microarray data were normalized using intensity and detection P values. The trimmed mean signal of the array was scaled to the target signal of 500 with the “all probe sets” scaling option. Statistical analysis for signal intensities and “detection P values” was performed with the algorithm of GeneChip Operating Software (GCOS), version 1.4 (Affymetrix). Detection of the transcripts was assessed by the “detection call” (P, present; A, absent; M, marginal) on the basis of detection P values. The array data of the MAT1-1 and MAT1-2 strains were compared with each other. The comparative analysis was performed for each strain by GCOS, which calculates the “signal log ratio” and “change P values” to determine the “change call” (I, increasing; MI, marginally increasing; NC, no change; MD, marginally decreasing; D, decreasing). For each strain, genes with a detection call of “P” (expression detected) and a change call of “I” were selected as upregulated genes. A detection call P value of <0.001111 was regarded as indicating an increase (I); a P value of ≥0.001111 and <0.001333 or of >0.998667 and ≤0.998889 indicated marginal (M) results; a P value of ≥0.001333 and ≤0.998667 indicated no change (NC); and a P value of >0.998889 indicated a decrease (D). Fold changes were converted from the signal log ratio for each strain by using Microsoft Excel, and for the purposes of the present study, we investigated further all genes showing at least a 10-fold difference in expression between MAT1-1 and MAT1-2 strains as those subject to possible MAT regulation (see Tables 2 and 3). A detection P value of <0.04 was regarded as indicating the presence (P) of a gene; a P value of ≥0.04 and <0.06 was considered to indicate a marginal (M) result; and a P value of ≥0.06 was considered to indicate the absence (A) of a gene. Upregulated genes with detection calls of “A” in both strains were omitted.
Table 2.
Genes upregulated in the MAT1-1 mating type strain based on DNA microarray analysis
| Gene ID | Fold change (MAT1-1/MAT1-2) | Proteina |
|---|---|---|
| AO080503000328 | 55.7 | Acyl-CoA dehydrogenases |
| AO080508000390 | 55.7 | Predicted protein |
| AO080503000008 | 39.4 | AoPpgA, putative α-pheromone precursor |
| AO080508000389 | 27.9 | Predicted protein |
| AO080541000465 | 26.0 | Predicted protein |
| AO080523000564 | 24.3 | Predicted protein |
| AO080511000446 | 21.1 | Probable taurine catabolism dioxygenase |
| AO080521000309 | 19.7 | Predicted protein |
| AO080502000077 | 18.4 | Predicted protein |
| AO080521000174 | 18.4 | NptB, neutral protease NPII |
| AO080564000028 | 18.4 | NADP FAD-dependent oxidoreductase |
| AO080523000782 | 16.0 | Predicted protein |
| AoEST3991 | 16.0 | Predicted protein |
| AO080501000145 | 14.9 | Predicted protein |
| AO080513000200 | 14.9 | Predicted protein |
| AO080541000466 | 14.9 | Predicted protein |
| AO080531000014 | 13.9 | Cytochrome P450 |
| AoEST3853 | 13.9 | Predicted protein |
| AO080508000337 | 13.0 | Predicted protein |
| AO080523000711 | 13.0 | Cytochrome P450 |
| AO080541000488 | 13.0 | Predicted protein |
| AO080521000051 | 12.1 | Predicted protein |
| AO080523000026 | 11.3 | Chitosanase |
| AO070333000246 | 10.6 | Predicted protein |
Acyl-CoA, acyl coenzyme A; FAD, flavin adenine dinucleotide.
Table 3.
Genes upregulated in the MAT1-2 mating type strain based on DNA microarray analysis
| Gene ID | Fold change (MAT1-2/MAT1-1) | Protein |
|---|---|---|
| AO080505000070 | 84.4 | Predicted protein |
| AoEST5207 | 64.0 | Predicted protein |
| AO080515000084 | 18.4 | Predicted protein |
| AO080532000139 | 18.4 | Vesicle coat complex COPII, subunit Sec31 |
| AoEST6582 | 17.1 | Predicted protein |
| AO080523000595 | 14.9 | Predicted protein |
| AO080508000372 | 14.9 | Predicted protein |
| AO080511000543 | 11.3 | Predicted protein |
| AO080553000049 | 11.3 | Predicted protein |
Microarray data accession number.
Our DNA microarray data were submitted to the Gene Expression Omnibus public database (http://www.ncbi.nlm.nih.gov/geo/) with the accession number GSE33542.
RESULTS
Identification of a MAT1-2 gene in A. oryzae.
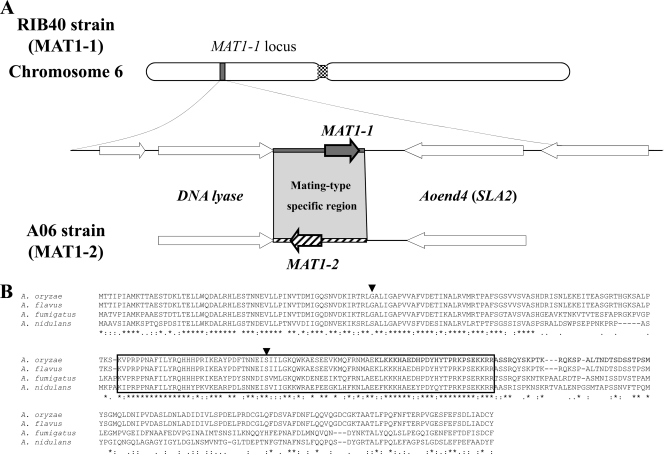
PCR using degenerate primers MAT5-4 and MAT3-2, designed to anneal to a sequence in the conserved HMG box of fungal MAT1-2 genes (60), led to the amplification of an approximately 300 bp fragment from A. oryzae strain AO6 genomic DNA but not from strain RIB40 genomic DNA harboring the MAT1-1 idiomorph. Sequencing of this 300-bp fragment confirmed homology to known MAT1-2 HMG genes (results not shown). A gene walk that reached the end of the idiomorph upstream of the MAT1-2 gene was therefore initiated, but the kit failed to yield any products downstream of the MAT1-2 gene. Given that gene synteny is fairly well conserved around the MAT locus in the aspergilli and many pezizomycete fungi (11, 12), primers AOmat-2R and AOmat-2F, located in the flanking DNA lyase region, predicted to be present downstream of the MAT1-2 gene (12), and in the upstream Aoend4 (SLA2) region, respectively, were designed. This primer pair successfully amplified the whole MAT1-2 idiomorph along with some conserved flanking sequences. A schematic diagram of the MAT loci of strains RIB40 and AO6 is shown in Fig. 1A. The locations of the two mating type genes at the MAT locus in A. oryzae are similar to those in other heterothallic aspergilli (12, 19, 49, 58). The predicted MAT1-2 gene sequence (1,069 bp, including two introns) was contained within a 3,276-bp MAT1-2 idiomorph, compared to a 3,130-bp MAT1-1 idiomorph (including the 1,168-bp MAT1-1 gene) located between the DNA lyase and Aoend4 (SLA2) genes (DDBJ accession number AB617942). An amino acid sequence alignment of MAT1-2 proteins from representative aspergilli is shown in Fig. 1B. The predicted A. oryzae MAT1-2 protein consists of 321 amino acids and contains a MATA_HMG domain, as conserved in other fungal MAT1-2 proteins. The amino acid sequence shows 100%, 63.0%, and 51.3% identity with the MAT1-2 mating type proteins of A. flavus, A. fumigatus, and A. nidulans, respectively. Sequencing of a limited number of 8 isolates, 4 from each mating type, suggested the presence of some fixed polymorphic differences within the flanking sequences (up to 1 kb away) between MAT1-1 and MAT1-2 isolates, which could indicate either a lack of sexual reproduction or the suppression of recombination at/around the MAT locus. However, sequencing of a higher number of isolates is required to confirm this observation.
Fig 1.
Mating type genes of Aspergillus oryzae. (A) Schematic arrangements of the MAT loci in A. oryzae strains RIB40 and AO6. Arrows represent putative ORFs in regions adjoining the MAT loci. (B) Amino acid sequence alignment of MAT1-2 proteins from A. oryzae, A. flavus, A. fumigatus, and A. nidulans. The conserved HMG domain is boxed. Arrowheads indicate the locations of conserved introns.
Distribution of mating types in industrial A. oryzae strains.
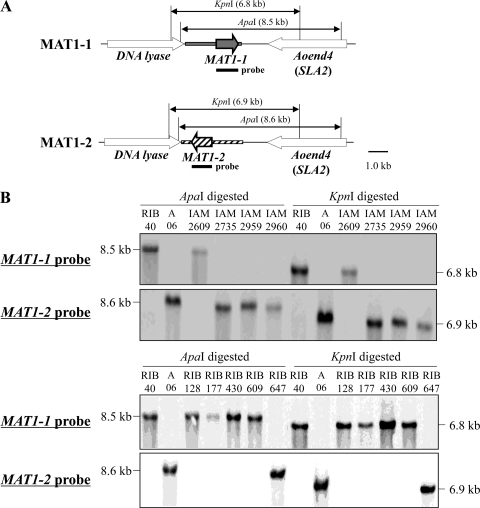
Using the DNA fragments of MAT1-1 and MAT1-2 genes from A. oryzae strains RIB40 and AO6, respectively, as probes, Southern blot analysis was carried out to identify the mating types of 11 industrial A. oryzae strains from the RIB and IAM culture collections (see Table S1 in the supplemental material). As shown in Fig. 2, a single MAT locus was detected, and four of five strains used for sake production (AO1 [RIB128], RIB177, RIB430, RIB609, and RIB647) were identified as MAT1-1 strains, while IAM2959, used for soy sauce production, was identified as a MAT1-2 strain. The nucleotide sequences of the MAT1-1 and MAT1-2 open reading frames (ORFs) from these strains were 100% identical to those of strains RIB40 and AO6, respectively (data not shown). Slight differences in the sizes of bands were observed (Fig. 2), presumably due to sequence divergence in the MAT locus region outside the MAT ORFs themselves. In parallel, initial screenings using the mating type diagnostic PCR (employing primers designed to amplify sequences from either MAT gene) determined that three of the reference strains belonged to the MAT1-1 genotype (AO1, AO2, and AO5) and three belonged to the MAT1-2 genotype (AO3, AO4, and AO8).
Fig 2.
Existence of both the MAT1-1 and MAT1-2 mating types in industrial A. oryzae strains from the RIB and IAM culture collections. (A) Construct for Southern blot analysis. (B) Mating type identification by Southern blot analysis. DNA fragments from the MAT1-1 and MAT1-2 genes were used as probes. MAT1-1 strains were RIB40, AO1 (RIB128), RIB177, RIB430, RIB609, and IAM2609. MAT1-2 strains were AO6, IAM2735, IAM2959, IAM2960, and RIB647.
In order to examine whether the mating type differs according to the use of the source strains for the production of sake, soy sauce, or miso, we further analyzed the distribution of mating types in 164 strains from tane-koji (koji mold starter) strains utilizing the MAT diagnostic PCR test. Isolates of the MAT1-1 and MAT1-2 genotypes were found to be present among strains used for the production of sake, soy sauce, and miso (Table 1). Strains used for sake and miso production showed a near-1:1 ratio of mating types, with no statistically significant bias (P, 0.20 and 0.43, respectively) from a balanced ratio. In contrast, tane-koji strains used for soy sauce showed a statistically significant dominance of the MAT1-2 mating type (P, 0.0019). Although the overall distribution of the two mating types in the tane-koji strains was close to a 1:1 ratio, there was a slight bias toward the MAT1-2 mating type (Table 1). When the height of aerial hyphae was scored on a 5-point scale (from a score of 1 for the shortest to 5 for the longest [see Materials and Methods]), the aerial hyphae of MAT1-2 mating type strains were significantly (P = 0.005) shorter than those of MAT1-1 mating type strains on average (2.38 ± 0.11 and 2.96 ± 0.13, respectively; see Table S2 in the supplemental material). These data may be correlated with the fact that strains with shorter aerial hyphae had been selected for manufacturing soy sauce.
Table 1.
Distribution of mating types in A. oryzae tane-koji strains as determined by mating type diagnostic PCR
| Strain group | % (no.) of strains of mating type: |
χ2 (df) | Contingency χ2 (df) | |
|---|---|---|---|---|
| MAT1-1 | MAT1-2 | |||
| All tane-koji strains | 40.6 (67) | 59.4 (97) | 5.49a (1) | |
| Sake | 43.2 (38) | 56.8 (50) | 1.64 (1) | |
| Soy sauce | 27.5 (14) | 72.5 (36) | 9.68b (1) | |
| Miso | 57.7 (15) | 42.3 (11) | 0.62 (1) | 7.05a (2) |
Significant (P = 0.05).
Significant (P = 0.01).
Generation of MAT1-2 and ΔMAT strains.
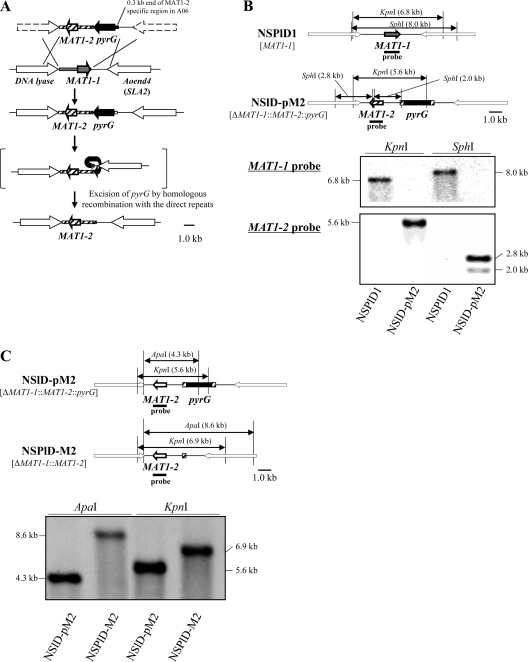
In order to examine the function of mating type genes, and especially for the detection of MAT-specific gene expression by use of DNA microarray and qRT-PCR analyses, it is very important to generate strains of opposite mating types with the same genetic background that differ only in their MAT loci. Hence, we generated a mating type replacement strain in which the original MAT1-1 gene was replaced with a MAT1-2 gene. The mating type genes were replaced by marker recycling techniques (42). A DNA fragment containing the MAT1-2 gene from A. oryzae strain AO6 and a pyrG marker gene was introduced into A. oryzae strain NSPlD1 (referred to below as the MAT1-1 strain), derived from strain RIB40 by homologous recombination (Fig. 3A). The pyrG marker gene was joined to direct repeats of a 0.3-kb region from strain AO6, which was located at the end of the MAT1-2-specific region. Integration of the MAT1-2 gene with the pyrG marker gene into the MAT locus of the resultant strain NSlD-pM2 was verified by Southern blot analysis (Fig. 3B). Subsequently, conidia of the NSlD-pM2 strain were spread onto PD agar medium containing 5-FOA, uridine, and uracil for positive selection of strains in which the pyrG marker gene was excised by homologous recombination with the direct repeats (Fig. 3A). Excision of the pyrG marker gene in the mating type replacement strain NSPlD-M2 (referred to below as the MAT1-2 strain) was verified by Southern blot analysis (Fig. 3C).
Fig 3.
Replacement of the MAT1-1 gene. (A) Strategy for replacement of the MAT1-1 gene with MAT1-2 by pyrG marker recycling. pyrG was excised by homologous recombination with direct repeats of the 0.3-kb upstream region of the MAT1-2 gene. (B) Verification of MAT gene replacement by Southern blot analysis. DNA fragments from MAT1-1 and MAT1-2 were used as probes. (C) Southern blot analysis of the strain from which pyrG had been excised. A DNA fragment from the MAT1-2 gene was used as a probe.
In order to clearly define mating type-specific gene expression, we also generated a control strain in which the MAT1-1 gene had been disrupted (referred to below as the ΔMAT strain). A disruption fragment containing the 1.5-kb upstream and downstream regions of the MAT1-1 ORF and the pyrG marker gene was introduced into strain NSPlD1 (see Fig. S1A in the supplemental material), and its presence was verified in the ΔMAT1-1::pyrG strain (NSlD-pdM1) by Southern blot analysis (see Fig. S1B). By positive selection using a medium containing 5-FOA, the ΔMAT strain (NSPlD-dM1) was obtained, and excision of the pyrG marker gene was verified by Southern blot analysis (Fig. S1C).
No apparent phenotypic differences in growth and conidiation (on DPY, PD, and M agar media, each supplemented with 0.5% uridine and 0.2% uracil, at 30°C for 5 days) were observed between the MAT1-1 (NSPlD1), MAT1-2 (NSPlD-M2), and ΔMAT strains (data not shown). Measurement of the height of aerial hyphae showed no significant difference, suggesting that this phenotype was not linked to MAT1-2 expression (data not shown).
Analysis of the MAT1-1 and MAT1-2 strains for differential gene expression.
To analyze possible differential gene expression that is dependent on the mating type in A. oryzae, DNA microarray analysis was performed using the MAT1-1 (NSPlD1) and MAT1-2 (NSPlD-M2) strains. First, we determined the culture conditions under which the A. oryzae strains expressed mating type genes. When analyzed by semiquantitative RT-PCR, expression of the MAT1-1 and MAT1-2 genes could not be observed when strains were grown for 24 h or 48 h in DPY liquid medium (data not shown), but the “culture shift” method, in which the mycelia were transferred to an agar medium, allowed us to detect MAT gene expression by both semiquantitative RT-PCR and qRT-PCR (data not shown). DNA microarray analyses were therefore carried out using RNA extracted from cultures subjected to the same conditions.
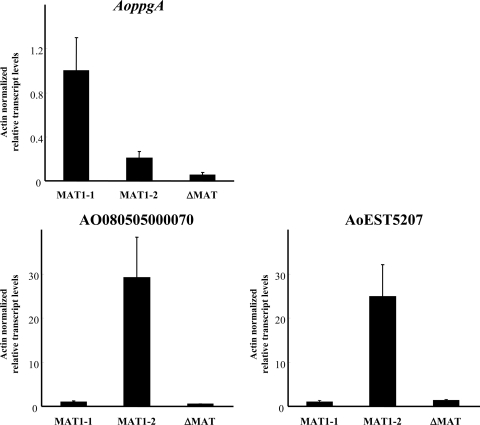
The resulting microarray analysis revealed a series of 1,155 genes that were upregulated >2-fold relative to each other in either the MAT1-1 (NSPlD1) or the MAT1-2 (NSPlD-M2) strain (596 genes were upregulated for MAT1-1 and 559 genes for MAT1-2). Full results are available from the Gene Expression Omnibus public database (http://www.ncbi.nlm.nih.gov/geo/), accession number GSE33542. For the purposes of this study, particular attention was given to those genes showing >10-fold upregulation, to reduce the risks of indirect changes in expression. Using these criteria, a total of 33 genes (listed in Tables 2 and 3) were identified. This marked difference in expression revealed that both the MAT1-1 and MAT1-2 genes are functional in regulating gene expression in A. oryzae. A. oryzae has genes encoding an α-pheromone precursor (AoppgA [AO080503000008]) and putative α- and a-pheromone receptors (AogprA [AO080550000138] and AogprB [AO080551000023], respectively). Among 24 genes upregulated in the MAT1-1 strain (Table 2), the mating type-specific gene AoppgA showed remarkable upregulation (39.4-fold) compared to the MAT1-2 strain, but most genes encoded predicted proteins of unknown function according to the annotation. The results from the microarray analysis of AoppgA expression were verified by qRT-PCR experiments. Transcripts of the putative α-pheromone precursor gene AoppgA were indeed more abundantly detected in the MAT1-1 strain than in the MAT1-2 and ΔMAT (NSPlD-dM1) strains (Fig. 4), although the differences in expression were not as marked as those detected by microarray analysis (by qRT-PCR analysis, AoppgA transcript levels were about 5 times greater in the MAT1-1 strain than in the MAT1-2 strain), possibly due to overlap in the probes used, resulting in higher microarray signals. In contrast, no significant differences in the transcript levels of putative pheromone receptor genes were observed between the MAT1-1 and MAT1-2 strains by DNA microarray analysis (data not shown). This is consistent with previous reports of analyses of pheromone receptor gene expression, which did not show differential expression in isolates of other heterothallic Pezizomycotina with different mating types (34, 49, 62).
Fig 4.
Analysis of gene expression in the MAT1-1, MAT1-2, and ΔMAT strains by qRT-PCR. qRT-PCR was performed for the AoppgA gene, putatively involved in the mating process, and for genes upregulated in the MAT1-2 strain (AO080505000070 and AoEST5207) to confirm the changes in gene expression detected by DNA microarray experiments. The actin transcript levels were used as internal standards, and the transcript levels for MAT1-1 were set at 100%. The data shown are means ± standard errors from three independent experiments.
Meanwhile, 9 genes were markedly (>10-fold) upregulated in the MAT1-2 (NSPlD-M2) strain (Table 3). These included two genes (AO080505000070 and AoEST5207) showing very high rises in expression (85- and 64-fold, respectively). However, these two genes were predicted to encode proteins of unknown function, as were all but one of the other genes (Table 3). It should be noted that no a-pheromone precursor gene has been identified yet in the aspergilli, most likely due to sequence divergence, because the a-factor pheromone sequence is poorly conserved in the Pezizomycotina (12, 14, 54). The results of microarray expression experiments were verified by qRT-PCR analysis for the AO080505000070 and AoEST5207 genes, which confirmed significant upregulation of these genes in the MAT1-2 strain compared to the MAT1-1 and ΔMAT (NSPlD-dM1) strains (Fig. 4).
DISCUSSION
Mating type (MAT) genes have been shown to be key regulators of sexual identity throughout the fungal kingdom and appear to be a prerequisite for sexual reproduction (36). Therefore, many studies assessing the asexual nature of particular fungal species have first investigated the presence of MAT genes as a possible indication of cryptic sexuality, and MAT loci have been detected in a series of supposedly asexual fungi (35). It should be noted, nonetheless, that the presence of mating type genes alone does not necessarily confer the ability to undergo sexual reproduction; many other genes are also required for mating, fruiting body development, and meiosis. Indeed, at least 75 genes have been shown to be involved in sexual reproduction in pezizomycete fungi such as the aspergilli (14).
In the present study, we therefore investigated the possible presence and functionality of MAT genes in A. oryzae as part of long-term studies aiming to induce sexuality in this supposedly asexual species. The results were 3-fold. First, we were able to identify an isolate, AO6, containing a MAT1-2 gene encoding a MATA_HMG domain protein that was present at the same idiomorphic MAT locus as the MAT1-1 gene, previously identified by genome sequencing of A. oryzae strain RIB40, which encodes a MATα_HMG domain protein (19, 39, 41). Both the MAT1-1 and MAT1-2 genes appeared to encode putative functional proteins with no frameshift or deletion mutations, in contrast to, for example, the asexual species Candida parapsilosis, in which a defective MAT allele is present (38). The MAT genes had 100% sequence identity to those of the known sexual species Aspergillus (Petromyces) flavus (25), which is very closely related to A. oryzae according to phylogenetic analysis (43, 52). This first result is significant because it demonstrates that A. oryzae has the characteristic MAT locus organization of known sexual heterothallic pezizomycete species, an important indication of potential for sexual reproduction (15, 16).
Second, we developed a MAT diagnostic PCR assay and used it to determine the mating types of >180 reference and industrial strains of A. oryzae, including tane-koji strains used in the manufacture of sake, soy sauce, and miso. The overall results showed the presence of the MAT1-1 and MAT1-2 genotypes in a near-1:1 distribution, again consistent with either current potential for sexual reproduction or evidence of historic sexuality, or both (15, 16), although a slight bias toward the MAT1-2 genotype was observed in strains used for soy sauce production. MAT1-2 strains overall exhibited shorter aerial hyphae, although this is likely to be an artifact of the preponderance of the MAT1-2 mating type among soy sauce production strains, in which shorter aerial hyphae are favored. This morphological feature avoids the formation of highly entangled masses of hyphae, which can be inconvenient, since they may lower enzyme activity due to increases in temperature and moisture levels and may pose an increased risk of contamination by other microorganism, such as Bacillus species. We caution that some of the A. oryzae isolates might have been clonal; therefore, the total number of independent isolates assayed may in reality have been smaller. However, almost all isolates were obtained from different sources and exhibited morphological variation, and in addition, some were proven to be genetically distinct by amplified fragment length polymorphism (AFLP) fingerprinting (43). Thus, it is considered likely that the majority of isolates were nonclonal. It is noteworthy that the tane-koji starter cultures sold to brewing companies usually consist of strains that are mixed beforehand, and it is therefore possible that strains of opposite mating types might meet under industrial growth conditions. There have been similar reports of the presence of both MAT1-1 and MAT1-2 isolates in populations of other “asexual” pezizomycete fungi, including Alternaria species (2), Aspergillus species (49, 58), Rhynchosporium species (37, 66), Cercospora species (21), Coccidioides species (18, 40), and Penicillium chrysogenum (23, 24). The only exception has been the asexual species Acremonium chrysogenum, used to produce cephalosporin, for which only the MAT1-1 genotype could be found in a preliminary survey (55).
Third, and finally, we investigated whether the MAT genes identified in A. oryzae might prove to be functionally active in regulating gene expression. Using a MAT1-1 strain as the host, an isogenic strain was created in which the MAT1-1 idiomorph was replaced with the newly identified MAT1-2 idiomorph, including the MAT1-2 gene encoding a MATA_HMG domain protein. Comparisons in gene expression were then made between these two strains by use of DNA microarray and qRT-PCR methodologies under growth conditions under which the MAT genes were known to be expressed. It was found by microarray analysis that 1,155 genes were upregulated >2-fold in either the MAT1-1 host strain or the MAT1-2 gene replacement strain relative to each other. Of these, 33 genes were found to be upregulated more than 10-fold in either the MAT1-1 host strain or the MAT1-2 gene replacement strain, and upregulation was confirmed for certain genes by qRT-PCR. This showed that both the MAT1-1 and MAT1-2 genes are functional in regulating gene expression in A. oryzae, although our experimental design did not distinguish between direct and indirect transcriptional activation of these genes. Furthermore, the MAT genes acted in a mating type-dependent manner, suggesting that A. oryzae MAT genes potentially act as sexual determinants. Consistent with this hypothesis, MAT1-1 specifically upregulated the putative α-pheromone precursor gene AoppgA, in agreement with reports of MAT genes differentially regulating pheromone production in other heterothallic pezizomycete fungi (9, 11). However, most of the other genes upregulated in either the MAT1-1 or the MAT1-2 strain were of unknown function, so their role(s) in determining sexual identity and regulating sexual development remains to be investigated. Those showing major upregulation according to the microarray analysis (e.g., the 4 genes with >50-fold differences in expression [Tables 2 and 3]) are particularly interesting subjects for future study. Despite the anticipation that an a-pheromone precursor gene would be upregulated in the MAT1-2 strain, we failed to find any candidate genes encoding a C-terminal CAAX motif that might be part of an a-pheromone precursor (6, 7, 9). Thus, the search for the elusive putative a-pheromone precursor gene in the aspergilli continues (14).
Parallel studies using microarrays and RT-PCR to determine genes regulated by MAT expression have been reported from pezizomycete fungi with known sexual cycles. Pöggeler et al. (56) found that more than 100 genes were differentially regulated in a ΔSmta-1 mating type mutant strain of the homothallic species Sordaria macrospora. These included a pheromone precursor gene and elements of signal transduction pathways, with a cutoff value of >2-fold selected for significance. In subsequent experimental work by Klix et al. (34), additional ΔSmtA-1 and ΔSmtA-2 mating type mutants of S. macrospora were created, and microarray analyses showed altered expression of >900 genes, including pheromone precursor genes, with a >2-fold level again chosen for significance. Meanwhile, Bidard et al. (3) recently reported a genomewide analysis of the effects of MAT gene expression in the heterothallic species Podospora anserina. They found that 157 genes were differentially transcribed in isolates of different mating types (>2-fold), including known sex-related genes for production of pheromone precursors and receptors, and for enzymatic processing of propheromones. However, to our knowledge, the present study is the first to use DNA microarray analysis to examine differential gene expression that is dependent on mating type genes in a supposedly asexual filamentous ascomycete.
Our data, together with the discovery of a complement of genes that are likely to be required for sexual reproduction in the genome of A. oryzae (19), are good indicators of the potential for sexual reproduction in A. oryzae. We have also observed the expression of pheromone precursor and receptor genes, and elements of a pheromone response mitogen-activated protein (MAP) kinase signal transduction pathway, in two MAT1-1 (AO46 and AO10) and two MAT1-2 (AO6 and AO8) A. oryzae isolates of complementary mating types (M. Paoletti, E. U. Schwier, and P. S. Dyer, unpublished data). In addition, there is evidence of previous sexual reproduction by A. oryzae based on analysis of repeat-induced point mutations (RIP) in the genome (44), as also observed for the asexual species Aspergillus niger and Penicillium chrysogenum (5). Furthermore, sexual cycles have recently been detected in both A. flavus and A. parasiticus (25, 27), which are phylogenetically very closely related to A. oryzae (43, 52), and it has been suggested that A. flavus is an ancestor of A. oryzae (20). As a result, mating experiments are now being started using the congenic MAT1-1 and MAT1-2 strains constructed in the present study.
However, a major obstacle remains to be overcome before a sexual cycle might be induced in A. oryzae. Sexual reproduction in Aspergillus section Flavi (teleomorph genus, Petromyces), which includes A. oryzae, A. flavus, and A. parasiticus, requires the formation of sclerotia, spherical resistance structures formed for survival under adverse conditions (14). One of the key differences between A. flavus and A. oryzae is that most strains of A. oryzae have lost the ability to form sclerotia or have a much lower ability than A. flavus (45), perhaps as a result of domestication. Therefore, it will be necessary to reliably induce the formation of sclerotia in A. oryzae before sexuality may be realized. This might involve the manipulation of culture conditions, given that sclerotial formation is dependent on environmental conditions (14) and given the exacting demands of some aspergilli for sexual reproduction (14, 47), and/or the use of genetic manipulation techniques to induce sclerotial formation. At present, only one A. oryzae gene, encoding the bHLH transcription factor SclR (for sclerotium regulator), involved in the production of sclerotia has been identified (14, 30, 31). Deletion of sclR in certain strains observed to form sclerotia was found to result in sparse sclerotial production, while overexpression of sclR led to >5-fold production of sclerotia with increased formation of branched aerial hyphae (31). In related work, deletion of a zinc finger calcineurin response gene, crzA, was found to result in the production of mainly immature sclerotia in A. parasiticus (8), so a homolog might also be required for the maturation of sclerotia in A. oryzae. Genes linked to sclerotial formation have also been identified in the pezizomycete fungi Botrytis cinerea and Sclerotinia sclerotiorum; these might provide insight for studies with the aspergilli (1).
Within an industrial setting, sexual reproduction has been used to breed strains with desirable characteristics, to generate novel variation allowing selection for significantly increased metabolite production, to restore the vigor of impaired strains, and to investigate the genetic basis of traits of interest. Specific examples include selection for flocculation in Saccharomyces cerevisiae, increased alkaloid production in Claviceps purpurea, improved thermotolerance in Pleurotus species, and investigation of resistance to fungicides in Tapesia yallundae (13, 17, 53). It is hoped that future research with A. oryzae might lead to a “sexual revolution” in this species, as seen for other aspergilli previously assumed to be asexual (15, 47), thus providing an important new method for strain improvement in this widely utilized industrial fungus.
Supplementary Material
ACKNOWLEDGMENTS
M.P., D.B.A., and P.S.D. thank the Biotechnology and Biological Sciences Research Council (United Kingdom) for providing funding to support this research. J.M. thanks the Japan Society for the Promotion of Science for providing funding for Grant-in-Aid Challenging Exploratory Research.
M.P., D.B.A., and P.S.D. thank Elke Schwier for experimental assistance with preliminary screening work.
Footnotes
Published ahead of print 10 February 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Amselem J, et al. 2011. Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet. 7:e1002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Berbee ML, Payne BP, Zhang G, Roberts RG, Turgeon BG. 2003. Shared ITS DNA substitutions in isolates of opposite mating type reveal a recombining history for three presumed asexual species in the filamentous ascomycete genus Alternaria. Mycol. Res. 107:169–182 [DOI] [PubMed] [Google Scholar]
- 3. Bidard F, et al. 2011. Genome-wide gene expression profiling of fertilization competent mycelium in opposite mating types in the heterothallic fungus Podospora anserina. PLoS One 6:e21476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bobrowicz P, Pawlak R, Correa A, Bell-Pedersen D, Ebbole DJ. 2002. The Neurospora crassa pheromone precursor genes are regulated by the mating type locus and the circadian clock. Mol. Microbiol. 45:795–804 [DOI] [PubMed] [Google Scholar]
- 5. Braumann I, Van Den Berg M, Kempken F. 2008. Repeat induced point mutation in two asexual fungi, Aspergillus niger and Penicillium chrysogenum. Curr. Genet. 53:287–297 [DOI] [PubMed] [Google Scholar]
- 6. Caldwell GA, Wang SH, Naider F, Becker JM. 1994. Consequences of altered isoprenylation targets on a-factor export and bioactivity. Proc. Natl. Acad. Sci. U. S. A. 91:1275–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Caldwell GA, Naider F, Becker JM. 1995. Fungal lipopeptide mating pheromones: a model system for the study of protein prenylation. Microbiol. Rev. 59:406–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chang PK. 2008. Aspergillus parasiticus crzA, which encodes calcineurin response zinc-finger protein, is required for aflatoxin production under calcium stress. Int. J. Mol. Sci. 9:2027–2043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Coppin E, de Renty C, Debuchy R. 2005. The function of the coding sequences for the putative pheromone precursors in Podospora anserina is restricted to fertilization. Eukaryot. Cell 4:407–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Czaja W, Miller KY, Miller BL. 2011. Complex mechanisms regulate developmental expression of the matA (HMG) mating type gene in homothallic Aspergillus nidulans. Genetics 189:795–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Debuchy R, Berteaux-Lecellier V, Silar P. 2010. Mating systems and sexual morphogenesis in ascomycetes, p 201–253 In Borkovich KA, Ebbole DJ. (ed), Cellular and molecular biology of filamentous fungi. American Society for Microbiology, Washington, DC [Google Scholar]
- 12. Dyer PS. 2007. Sexual reproduction and significance of MAT in the aspergilli, p 123–142 In Heitman J, Kronstad JW, Taylor JW, Casselton LA. (ed), Sex in fungi: molecular determination and evolutionary principles. American Society for Microbiology, Washington, DC [Google Scholar]
- 13. Dyer PS, Hansen J, Delaney A, Lucas JA. 2000. Genetic control of resistance to the sterol 14α-demethylase inhibitor fungicide prochloraz in the cereal eyespot pathogen Tapesia yallundae. Appl. Environ. Microbiol. 66:4599–4604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dyer PS, O'Gorman CM. 2012. Sexual development and cryptic sexuality in fungi: insights from Aspergillus species. FEMS Microbiol. Rev. 36:165–192 [DOI] [PubMed] [Google Scholar]
- 15. Dyer PS, O'Gorman CM. 2011. A fungal sexual revolution: Aspergillus and Penicillium show the way. Curr. Opin. Microbiol. 14:649–654 [DOI] [PubMed] [Google Scholar]
- 16. Dyer PS, Paoletti M. 2005. Reproduction in Aspergillus fumigatus: sexuality in a supposedly asexual species? Med. Mycol. 43:S7–S14 [DOI] [PubMed] [Google Scholar]
- 17. Esser K. 1986. Genetics of strain improvement in filamentous fungi with respect to biotechnology, p 143–153 In Vanek Z, Hostalek Z. (ed), Overproduction of microbial metabolites. Butterworths, Stonetown, MA [Google Scholar]
- 18. Fraser JA, et al. 2007. Evolution of the mating type locus: insights gained from the dimorphic primary fungal pathogens Histoplasma capsulatum, Coccidioides immitis, and Coccidioides posadasii. Eukaryot. Cell 6:622–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Galagan JE, et al. 2005. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438:1105–1115 [DOI] [PubMed] [Google Scholar]
- 20. Geiser DM, Pitt JL, Taylor JW. 1998. Cryptic speciation and recombination in the aflatoxin-producing fungus Aspergillus flavus. Proc. Natl. Acad. Sci. U. S. A. 95:388–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Groenewald M, Groenewald JZ, Harrington TC, Abeln ECA, Crous PW. 2006. Mating type gene analysis in apparently asexual Cercospora species is suggestive of cryptic sex. Fungal Genet. Biol. 43:813–825 [DOI] [PubMed] [Google Scholar]
- 22. Große V, Krappmann S. 2008. The asexual pathogen Aspergillus fumigatus expresses functional determinants of Aspergillus nidulans sexual development. Eukaryot. Cell 7:1724–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Henk DA, et al. 23 September 2011. Speciation despite globally overlapping distributions in Penicillium chrysogenum: the population genetics of Alexander Fleming's lucky fungus. Mol. Ecol. [Epub ahead of print.] doi:10.1111/j.1365-294X.2011.05244.x [DOI] [PubMed] [Google Scholar]
- 24. Hoff B, Pöggeler S, Kück U. 2008. Eighty years after its discovery, Fleming's Penicillium strain discloses the secret of its sex. Eukaryot. Cell 7:465–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Horn BW, Moore GG, Carbone I. 2009. Sexual reproduction in Aspergillus flavus. Mycologia 101:423–429 [DOI] [PubMed] [Google Scholar]
- 26. Horn BW, Moore GG, Carbone I. 2011. Sexual reproduction in aflatoxin-producing Aspergillus nomius. Mycologia 103:174–183 [DOI] [PubMed] [Google Scholar]
- 27. Horn BW, Ramirez-Prado JH, Carbone I. 2009. Sexual reproduction and recombination in the aflatoxin-producing fungus Aspergillus parasiticus. Fungal Genet. Biol. 46:169–175 [DOI] [PubMed] [Google Scholar]
- 28. Ito K, et al. 2007. Microbial production of sensory-active miraculin. Biochem. Biophys. Res. Commun. 360:407–411 [DOI] [PubMed] [Google Scholar]
- 29. Jin FJ, Maruyama J, Juvvadi PR, Arioka M, Kitamoto K. 2004. Development of a novel quadruple auxotrophic host transformation system by argB gene disruption using adeA gene and exploiting adenine auxotrophy in Aspergillus oryzae. FEMS Microbiol. Lett. 239:79–85 [DOI] [PubMed] [Google Scholar]
- 30. Jin FJ, Takahashi T, Machida M, Koyama Y. 2009. Identification of a basic helix-loop-helix type transcription regulator gene in Aspergillus oryzae. Appl. Environ. Microbiol. 75:5943–5951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jin FJ, et al. 2011. SclR, a basic helix-loop-helix transcription factor, regulates hyphal morphology and promotes sclerotial formation in Aspergillus oryzae. Eukaryot. Cell 10:945–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kimura S, Maruyama J, Takeuchi M, Kitamoto K. 2008. Monitoring global gene expression of proteases and improvement of human lysozyme production in the nptB gene disruptant of Aspergillus oryzae. Biosci. Biotechnol. Biochem. 72:499–505 [DOI] [PubMed] [Google Scholar]
- 33. Kitamoto K. 2002. Molecular biology of the Koji molds. Adv. Appl. Microbiol. 51:129–153 [DOI] [PubMed] [Google Scholar]
- 34. Klix V, et al. 2010. Functional characterization of MAT1-1-specific mating-type genes in the homothallic ascomycete Sordaria macrospora provides new insights into essential and non-essential sexual regulators. Eukaryot. Cell 9:894–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kück U, Pöggeler S. 2009. Cryptic sex in fungi. Fungal Biol. Rev. 23:86–90 [Google Scholar]
- 36. Lee SC, Ni M, Li W, Shertz C, Heitman J. 2010. The evolution of sex: a perspective from the fungal kingdom. Microbiol. Mol. Biol. Rev. 74:298–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Linde CC, Zala M, Ceccarelli S, McDonald BA. 2003. Further evidence for sexual reproduction in Rhynchosporium secalis based on distribution and frequency of mating-type alleles. Fungal Genet. Biol. 40:115–125 [DOI] [PubMed] [Google Scholar]
- 38. Logue ME, Wong S, Wolfe KH, Butler G. 2005. A genome sequence survey shows that the pathogenic yeast Candida parapsilosis has a defective MTLa1 allele at its mating type locus. Eukaryot. Cell 4:1009–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Machida M, et al. 2005. Genome sequencing and analysis of Aspergillus oryzae. Nature 438:1157–1161 [DOI] [PubMed] [Google Scholar]
- 40. Mandel MA, Barker BH, Kroken S, Rounsley SD, Orbach MJ. 2007. Genomic and population analyses of the mating type loci in Coccidioides species reveal evidence for sexual reproduction and gene acquisition. Eukaryot. Cell 6:1189–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Martin T, et al. 2010. Tracing the origin of the fungal α1 domain places its ancestor in the HMG-box superfamily: implication for fungal mating-type evolution. PLoS One 5:e15199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Maruyama J, Kitamoto K. 2008. Multiple gene disruptions by marker recycling with highly efficient gene-targeting background (ΔligD) in Aspergillus oryzae. Biotechnol. Lett. 30:1811–1817 [DOI] [PubMed] [Google Scholar]
- 43. Montiel D, et al. 2003. Genetic differentiation of the Aspergillus section Flavi complex using AFLP fingerprints. Mycol. Res. 107:1427–1434 [DOI] [PubMed] [Google Scholar]
- 44. Montiel MD, Lee HA, Archer DB. 2006. Evidence of RIP (repeat-induced point mutation) in transposase sequences of Aspergillus oryzae. Fungal Genet. Biol. 43:439–445 [DOI] [PubMed] [Google Scholar]
- 45. Murakami H. 1971. Classification of the Koji mold. J. Gen. Appl. Microbiol. 17:281–309 [Google Scholar]
- 46. Nakajima K, et al. 2006. Extracellular production of neoculin, a sweet-tasting heterodimeric protein with taste-modifying activity, by Aspergillus oryzae. Appl. Environ. Microbiol. 72:3716–3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. O'Gorman CM, Fuller HT, Dyer PS. 2009. Discovery of a sexual cycle in the opportunistic fungal pathogen Aspergillus fumigatus. Nature 457:471–474 [DOI] [PubMed] [Google Scholar]
- 48. Ohno A, Maruyama J, Nemoto T, Arioka M, Kitamoto K. 2011. A carrier fusion significantly induces unfolded protein response in heterologous protein production by Aspergillus oryzae. Appl. Microbiol. Biotechnol. 92:1197–1206 [DOI] [PubMed] [Google Scholar]
- 49. Paoletti M, et al. 2005. Evidence for sexuality in the opportunistic fungal pathogen Aspergillus fumigatus. Curr. Biol. 12:1242–1248 [DOI] [PubMed] [Google Scholar]
- 50. Paoletti M, et al. 2007. Mating type and the genetic basis of self-fertility in the model fungus Aspergillus nidulans. Curr. Biol. 17:1384–1389 [DOI] [PubMed] [Google Scholar]
- 51. Pel HJ, et al. 2007. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat. Biotechnol. 25:221–231 [DOI] [PubMed] [Google Scholar]
- 52. Peterson SW. 2008. Phylogenetic analysis of Aspergillus species using DNA sequences from four loci. Mycologia 100:205–226 [DOI] [PubMed] [Google Scholar]
- 53. Pöggeler S. 2001. Mating-type genes for classical strain improvement of ascomycetes. Appl. Microbiol. Biotechnol. 56:589–601 [DOI] [PubMed] [Google Scholar]
- 54. Pöggeler S. 2011. Function and evolution of pheromones and pheromone receptors in filamentous ascomycetes, p 73–96 In Pöggeler S, Wöstermeyer J. (ed), The Mycota. XIV. Evolution of fungi and fungal-like organisms. Springer-Verlag, Heidelberg, Germany [Google Scholar]
- 55. Pöggeler S, Hoff B, Kück U. 2008. Asexual cephalosporin C producer Acremonium chrysogenum carries a functional mating type locus. Appl. Environ. Microbiol. 74:6006–6016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pöggeler S, et al. 2006. Microarray and real-time PCR analyses reveal mating-type dependent gene expression in a homothallic fungus. Mol. Genet. Genomics 275:492–503 [DOI] [PubMed] [Google Scholar]
- 57. Pyrzak W, Miller KY, Miller BL. 2008. Mating type protein Mat1-2 from asexual Aspergillus fumigatus drives sexual reproduction in fertile Aspergillus nidulans. Eukaryot. Cell 7:1029–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ramirez-Prado JH, Moore GG, Horn BW, Carbone I. 2008. Characterization and population analysis of the mating-type genes in Aspergillus flavus and Aspergillus parasiticus. Fungal Genet. Biol. 45:1292–1299 [DOI] [PubMed] [Google Scholar]
- 59. Rydholm C, Dyer PS, Lutzoni F. 2007. DNA sequence characterization and molecular evolution of MAT1 and MAT2 mating-type loci of the self-compatible ascomycete mold Neosartorya fischeri. Eukaryot. Cell 6:868–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Seymour FA, et al. 2005. Breeding systems in the lichen-forming fungal genus Cladonia. Fungal Genet. Biol. 42:554–563 [DOI] [PubMed] [Google Scholar]
- 61. Shen WC, Bobrowicz P, Ebbole DJ. 1999. Isolation of pheromone precursor genes of Magnaporthe grisea. Fungal Genet. Biol. 27:253–263 [DOI] [PubMed] [Google Scholar]
- 62. Szewczyk E, Krappmann S. 2010. Conserved regulators of mating are essential for Aspergillus fumigatus cleistothecium formation. Eukaryot. Cell 9:774–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tsuchiya K, et al. 1992. High level expression of the synthetic human lysozyme gene in Aspergillus oryzae. Appl. Microbiol. Biotechnol. 38:109–114 [DOI] [PubMed] [Google Scholar]
- 64. Tsuchiya K, et al. 1994. High level secretion of calf chymosin using a glucoamylase-prochymosin fusion gene in Aspergillus oryzae. Biosci. Biotechnol. Biochem. 58:895–899 [DOI] [PubMed] [Google Scholar]
- 65. Yu JH, et al. 2004. Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 41:973–981 [DOI] [PubMed] [Google Scholar]
- 66. Zaffarano PL, McDonald BA, Zala M, Linde CC. 2006. Global hierarchical gene diversity analysis suggests the Fertile Crescent is not the center of origin of the barley scald pathogen Rhynchosporium secalis. Phytopathology 96:941–950 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.